Abstract
Background:
Gemcitabine-based chemotherapy is the standard treatment for pancreatic cancer. However, the issue of resistance remains unresolved. The aim of this study was to identify microRNAs (miRNAs) that govern the resistance to gemcitabine in pancreatic cancer.
Methods:
miRNA microarray analysis using gemcitabine-resistant clones of MiaPaCa2 (MiaPaCa2-RGs), PSN1 (PSN1-RGs), and their parental cells (MiaPaCa2-P, PSN1-P) was conducted. Changes in the anti-cancer effects of gemcitabine were studied after gain/loss-of-function analysis of the candidate miRNA. Further assessment of the putative target gene was performed in vitro and in 66 pancreatic cancer clinical samples.
Results:
miR-320c expression was significantly higher in MiaPaCa2-RGs and PSN1-RGs than in their parental cells. miR-320c induced resistance to gemcitabine in MiaPaCa2. Further experiments showed that miR-320c-related resistance to gemcitabine was mediated through SMARCC1, a core subunit of the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex. In addition, clinical examination revealed that only SMARCC1-positive patients benefited from gemcitabine therapy with regard to survival after recurrence (P=0.0463).
Conclusion:
The results indicate that miR-320c regulates the resistance of pancreatic cancer cells to gemcitabine through SMARCC1, suggesting that miR-320c/SMARCC1 could be suitable for prediction of the clinical response and potential therapeutic target in pancreatic cancer patients on gemcitabine-based therapy.
Keywords: microRNA, miR-320c, pancreatic cancer, gemcitabine resistance, SMARCC1
Pancreatic cancer is one of the leading causes of tumour-related mortalities. The prognosis of patients after complete resection is poor, and >50% of patients develop tumour recurrence at distant or locoregional sites, with an estimated 5-year survival of only 20% (Li et al, 2004). Treatment modalities for pancreatic cancer include surgery, chemotherapy, radiation therapy, and combination therapy. Gemcitabine-based chemotherapy has formed the core of the multimodal therapy and improved the prognosis of patients with pancreatic cancer (Oettle et al, 2007), but its effect is modest because of high drug resistance. The selection of patients who derive a true benefit from gemcitabine could be an important stepping stone toward improvement of outcome of pancreatic cancer.
Several molecular mechanisms in drug resistance in pancreatic cancer have been elucidated. We have previously described that RRM1, the gene that encodes the regulatory subunit of ribonucleotide reductase, is strongly associated with gemcitabine resistance in pancreatic cancer (Nakahira et al, 2007; Akita et al, 2009). However, the development of drug resistance appears to be a multifactorial process, so our understanding is still fragmentary.
Recently, several studies have indicated that microRNAs (miRNAs) regulate this drug resistance (Tomimaru et al, 2010; Tomokuni et al, 2011). miRNAs are endogenous, single-stranded, non-coding RNAs and modulators of gene expression in the post-transcriptional phase, composed of 18–25 nucleotides. Currently, 1600 human miRNAs have been identified (miRBase 19, http://www.mirbase.org/). miRNAs are predicted to control the activity of approximately 30% of all protein-coding genes in mammals, and each miRNA can regulate up to 100 different messenger RNAs. Currently, the most promising miRNAs in association with pancreatic cancer drug resistance against gemcitabine are miR-15a (Zhang et al, 2010), miR-21 (Park et al, 2009; Ali et al, 2010; Giovannetti et al, 2010; Hwang et al, 2010), miR-34 (Ji et al, 2009), miR-200b and miR-200c (Li et al, 2009; Ali et al, 2010), miR-214 (Zhang et al, 2010), miR-221 (Park et al, 2009), and members of the let7 family (Li et al, 2009). However, only selected miRNAs have been investigated for their role in drug resistance in pancreatic cancer.
In this study, we developed gemcitabine-resistant cell clones from human pancreatic cancer cell lines and performed comprehensive expression profiling of miRNAs. The results indicate that miR-320c confers resistance to gemcitabine in pancreatic cancer cells through SMARCC1.
Materials and methods
Cell lines
The human pancreatic cell lines MiaPaCa2 and PSN1 were obtained from the Japan Cancer Research Resources Bank (Tokyo, Japan). These cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U ml−1 penicillin, and 100 mg ml−1 streptomycin at 37 °C in a humidified incubator with 5% CO2 in air.
Establishment of gemcitabine-resistant cell clones
Gemcitabine-resistant cells were generated by exposure to gradually increasing concentrations of the drug for 2 months as described previously (Goan et al, 1999; Davidson et al, 2004). Parental MiaPaCa2 cells (MiaPaCa2-P) and parental PSN1 cells (PSN1-P) were exposed to gemcitabine at an initial concentration of 1 ng ml−1. When cells adapted to the drug, the gemcitabine concentration was increased. The final concentrations were 20 ng ml−1 gemcitabine for MiaPaCa2 and 10 ng ml−1 gemcitabine for PSN1. Through this process, we successfully established gemcitabine-resistant cells (Nakahira et al, 2007). Limiting the dilution of the established cells allowed the production of MiaPaCa2 cell clones and PSN1 cell clones that were resistant to gemcitabine. Four clones (MiaPaCa2-RGs: MiaPaCa2-RG1, MiaPaCa2-RG2, MiaPaCa2-RG3, and MiaPaCa2-RG4) and three clones (PSN1-RGs: PSN1-RG1, PSN1-RG2, and PSN1-RG3) from each line were used in these experiments.
Drugs and reagents
Gemcitabine was kindly supplied by Eli Lilly Pharmaceuticals (Indianapolis, IN, USA). In reference to previous studies, polyclonal rabbit anti-human SMARCC1 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for western blot analysis (DelBove et al, 2011) and monoclonal mouse anti-human SMARCC1 antibody (Santa Cruz Biotechnology Inc.) for immunohistochemistry (Andersen et al, 2009) were used as primary antibodies. Monoclonal rabbit anti-human actin for western blot analysis was purchased from Sigma-Aldrich Co. (St. Louis, MO, USA).
Transfection
The precursor oligonucleotide of hsa-miR-320c (pre-miR-320c), antisense oligonucleotide inhibitor of hsa-miR-320c (anti-miR-320c), SMARCC1 small interfering (si) RNA oligonucleotide (siSMARCC1), and their scrambled oligonucleotides were obtained from Ambion Inc. (Austin, TX, USA). Pre/anti-miR-320c and siSMARCC1 were transfected using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the protocol provided by the manufacturer. Each scrambled oligonucleotide was transfected in the same way as a matched negative control.
RNA extraction
Total RNA, including the small RNA fraction, was isolated from cell lines with Trizol reagent (Invitrogen) as previously described (Yang et al, 2009). The quality of the RNA was assessed with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) at 260 and 280 nm (A260/280) wavelengths.
Real-time quantitative reverse-transcription-PCR for miRNA expression
The reverse transcription (RT) reaction was performed with the TaqMan MicroRNA RT Kit (Applied Biosystems, Foster City, CA, USA), and real-time quantitative (q) PCR was performed with TaqMan MicroRNA Assays (Applied Biosystems) using the ABI7900HT system (Applied Biosystems). The expression of the target miRNA was normalised relative to that of the endogenous control, RNU48. Data were analysed according to the comparative Ct method (Schmittgen et al, 2004).
Real-time qRT-PCR for messenger RNA expression
Complementary DNA was synthesised from 8.0 μg total RNA using the SuperScript first-strand synthesis system (Invitrogen), according to the instructions supplied by the manufacturer. Real-time quantitative PCR was performed using designed oligonucleotide primers and the LightCycler 480 Real-Time PCR system (Roche Diagnostics, Mannheim, Germany). For detection of the amplification products, the LightCycler-DNA master SYBR green I (Roche Diagnostics) was used as described previously (Yamamoto et al, 2004), and the amount of target gene expression was calculated. The expression of the target gene was normalised relative to the expression of GAPDH, which was used as an endogenous control. The designed PCR primers were as follows: GAPDH forward primer 5′-GTCGGAGTCAACGGATTTGGT-3′ and GAPDH reverse primer 5′-GCCATGGGTGGAATCATATTGG-3′ and SMARCC1 forward primer 5′-TCATGCGGATGCTCCTACCA-3′ and SMARCC1 reverse primer 5′-AAACCTCCGCCATCCCTGTT-3′.
MiRNA microarray experiments
The purified RNAs obtained from MiaPaCa2-P, MiaPaCa2-RGs (MiaPaCa2-RG1, MiaPaCa2-RG2, MiaPaCa2-RG3, and MiaPaCa2-RG4), PSN1-P, and PSN1-RGs (PSN1-RG1, PSN1-RG2, and PSN1-RG3) were used as samples and assessed as being of high quality by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and NanoDrop (NanoDrop Technologies). Next, 500 ng of extracted total RNA was labeled with Hy5 using the miRCURY LNA Array microRNA Power Labeling kit (Exiqon, Vedbaek, Denmark). The labeled RNAs were hybridised onto 3D-Gene Human miRNA Oligo chips containing 1011 anti-sense probes printed in duplicate spots (Toray, Kamakura, Japan). The annotation and oligonucleotide sequences of the probes conformed to the miRBase miRNA database (http://microrna.sanger.ac.uk/sequences/). After stringent washes, the fluorescent signals were scanned with the ScanArray Express Scanner (Perkin Elmer, Waltham, MA, USA) and analysed using GenePix Pro version 5.0 (Molecular Devices, Sunnyvale, CA, USA). The raw data for each spot were normalised by substitution with the mean intensity of the background signal determined by the signal intensities of all blank spots with 95% confidence intervals. Measurements of both duplicate spots with signal intensities >2 s.ds. of the background signal intensity were considered to be valid. The relative expression level of a given miRNA was calculated by comparing the signal intensities of the averaged valid spots with their mean value throughout the microarray experiments after normalisation by their median values adjusted equivalently.
Construction of reporter plasmids and evaluation of luciferase reporter activity
To construct a luciferase reporter plasmid, a SMARCC1-3′UTR fragment containing the miR-320c target site was subcloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) located 3′ to the firefly luciferase gene. Nucleotide sequences of the constructed plasmids were confirmed by DNA sequencing analysis. For luciferase reporter assays, MiaPaCa2-P were seeded in 96-well plates and then cotransfected with the pmirGLO-SMARCC1-3′UTR construct and either pre-miR-320c or scrambled oligonucleotide for negative control (Ambion) using Lipofectamine 2000 (Invitrogen). Assays were conducted 48 h after transfection using Dual-Glo Luciferase Assay System (Promega). Firefly luciferase signals were normalised to renilla luciferase signals. All transfection experiments were conducted in triplicate.
Western blot analysis
Western blot analysis was performed as described previously (Yamamoto et al, 2003). Briefly, total protein was extracted from cells grown to semiconfluence in radio-immunoprecipitation assay buffer (Thermo Fisher Scientific, Inc., Rockford, IL, USA). Aliquots of total protein (12 μg) were electrophoresed on sodium dodecyl sulfate polyacrylamide, 10% Tris-HCl gels (Bio-Rad Laboratories Inc., Hercules, CA, USA). The separated proteins were transferred to polyvinylidene difluoride membranes (Millipore Co., Billerica, MA, USA) and incubated with primary antibodies for 1 h.
Growth inhibitory assay
The growth inhibitory assay was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma-Aldrich Co.) assay as described previously (Eguchi et al, 2000). In brief, cells were incubated for 72 h under several concentrations of gemcitabine. After reincubation for 4 h with MTT solution, an acid–isopropanol mixture was added to dissolve the resulting formazan crystals. The absorbance of the plate was measured in a microplate reader at a wavelength of 550 nm with a 650-nm reference, and the results were expressed as a percentage of absorbance relative to that of untreated controls.
Patients and specimens
The study subjects were 66 patients with pancreatic cancer, recruited as described previously (Akita et al, 2009). Between January 1992 and March 2008, 166 patients underwent surgery for pancreatic cancer at Osaka University Hospital, Osaka, Japan. We excluded 100 patients for the following reasons: tumours were not resectable in 26 patients because of liver metastases or peritoneal carcinomatosis; surgery resulted in R1 (residual microscopic cancer) or R2 (residual macroscopic cancer) resections in 21 patients; chemotherapy or chemoradiotherapy was provided preoperatively to 37 patients; lack of neutral-buffered formalin-fixed and paraffin-embedded tumour blocks or/and clinical follow-up information for study purposes in 14 cases; and radiotherapy or immunotherapy was provided postoperatively to 2 patients. Because the natural history of variant pancreatic neoplasms differs from that of the usual pancreatic ductal adenocarcinoma, patients with intraductal papillary mucinous neoplasms, mucinous cystic adenocarcinomas, and medullary adenocarcinomas were excluded from this study. Supplementary Table S1 summarises the characteristics of the 66 patients who were enrolled. They included 31 men and 35 women with a mean age of 65.3±7.6 years (±s.d.). All patients had R0 (no residual cancer) resections by pancreaticoduodenectomy in 53 patients, distal pancreatectomy in 11 patients, and other resections in 2 patients. The histopathological grading showed well, moderately, and poorly differentiated adenocarcinoma in 26, 30, and 10 patients, respectively. The UICC-TNM classification was 2, 1, and 63 patients with pT1, pT2, and pT3; 28, 32, and 6 patients with pN0, pN1, and pM1lym; and 1, 1, 26, 32, and 6 patients with stage IA, IB, IIA, IIB, and IV, respectively. None of the patients had received neoadjuvant therapy preoperatively. All 66 patients were followed until disease recurrence and/or death. The median follow-up period was 17.0 months (3.5–147.7), the 5-year survival rate was 25.0%, and recurrence of disease was observed in 51 patients. Treatment with gemcitabine was carried out in 26 patients; 3 patients received it as adjuvant chemotherapy, and 23 patients received it after disease recurrence. Radiation therapy was not carried out during all the follow-up period.
Immunohistochemical staining
Immunohistochemical staining for SMARCC1 in 66 pancreatic cancer samples was performed using the method described previously (Kondo et al, 1999). Briefly, formalin-fixed, paraffin-embedded 4-μm-thick sections were deparaffinised in xylene, then treated with an antigen-retrieval procedure, and incubated in methanol containing 0.3% hydrogen peroxide to block endogenous peroxidase. After incubation with normal protein block serum, the sections were incubated overnight at 4 °C with an anti-SMARCC1 antibody as the primary antibody. Thereafter, the sections were detected with avidin–biotin complex reagents (Vector Laboratory Inc., Burlingame, CA, USA) and diaminobenzidine. All sections were counterstained with haematoxylin. The positivity for SMARCC1 staining was defined as detectable nuclear staining of >10% of cancer cells.
Statistical analysis
Data are expressed as means±s.d. Clinicopathological parameters were compared using the χ2 test, and continuous variables were compared using the Student's t-test. Survival curves were computed using the Kaplan–Meier method, and differences between survival curves were compared using the log-rank test. A P value <0.05 denoted the presence of a statistically significant difference. Statistical analysis was performed using JMP software version 8.0.2 (SAS Institute Inc., Cary, NC, USA).
Results
Characteristics of established gemcitabine-resistant cells
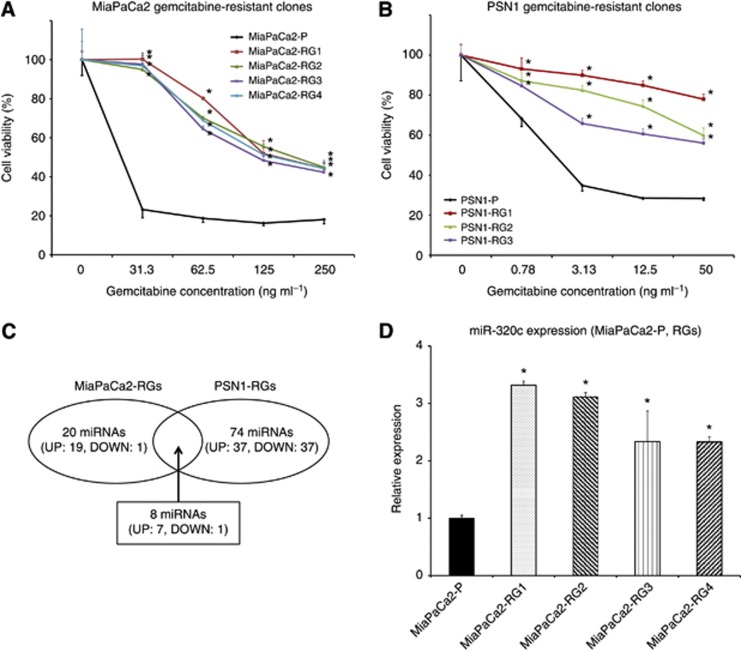
The morphology of MiaPaCa2-RGs resembled that of MiaPaCa2-P. Although MiaPaCa2-RGs showed similar growth curves compared with MiaPaCa2-P in the absence of gemcitabine (data not shown), MiaPaCa2-RGs were significantly resistant to gemcitabine compared with MiaPaCa2-P, which MTT assays confirmed (Figure 1A). On the other hand, the morphology of PSN1-RGs was slightly spindle-like in comparison with that of PSN1-P, and the growth rate of PSN1-RGs was slightly slower than that of PSN1-P in the absence of gemcitabine (data not shown). MTT assays showed significant resistance to gemcitabine in PSN1-RGs compared with PSN1-P (Figure 1B).
Figure 1.
Characteristics of gemcitabine-resistant MiaPaCa2 cell clones (MiaPaCa2-RGs) and PSN1 cell clones (PSN1-RGs). (A, B) MTT assay showed significantly lower antitumour effect of gemcitabine in MiaPaCa2-RGs than in parental MiaPaCa2 cells (MiaPaCa2-P) and in PSN1-RGs than in parental PSN1 cells (PSN1-P). Data are mean±s.d. of triplicate independent experiments. *P<0.05 compared with parental cells. (C) Schematic diagram of the results of microarray analysis. The protocol identified eight miRNAs in common with >1.5 average fold relative to parental, keeping adequate expression quantities and excluding miRNA*s both in MiaPaCa2-RGs and PSN1-RGs. (D) Real-time qRT–PCR demonstrated significantly higher miR-320c expression in MiaPaCa2-RGs than in MiaPaca2-P. Data are mean±s.d. of triplicate independent experiments. *P<0.05.
miR-320c expression was significantly higher in gemcitabine-resistant clones than in parental cells
To identify the candidate miRNAs related to resistance to gemcitabine, miRNA microarray analysis was performed using a MiaPaCa2 combination (MiaPaCa2-P and MiaPaCa2-RGs) and PSN1 combination (PSN1-P and PSN1-RGs). The analysis showed that, among the 1011 miRNAs, the miRNA expression levels of 20 (2.0%) in MiaPaCa2-RGs (Supplementary Table S2) and 74 (7.3%) in PSN1-RGs (Supplementary Table S3), respectively, were altered by >1.5 average fold relative to parental, keeping adequate expression quantities and excluding miRNA*s. Furthermore, eight miRNAs were identified in common both in MiaPaCa2-RGs and PSN1-RGs (Figure 1C). These miRNAs are listed with average fold change and P values of MiaPaCa2-RGs relative to MiaPaCa2-P and PSN1-RGs relative to PSN1-P in Table 1. Among them, miR-320c showed the highest alteration (1.97 average fold change; 2.20-fold increase in MiaPaCa2-RGs, and 1.73-fold increase in PSN1-RGs) and was statistically significant (P=0.0033 in MiaPaCa2-RGs, P=0.0366 in PSN1-RGs). The results of real-time qRT-PCR for miR-320c confirmed the upregulation in MiaPaCa2-RGs (Figure 1D). Therefore, miR-320c was selected for further analysis.
Table 1. Common up- or downregulated miRNAs both in MiaPaCa2-RGs and PSN1-RGs.
| |
|
Fold change (relative to MiaPaCa2-P) |
Fold change (relative to PSN1-P) |
|
||
|---|---|---|---|---|---|---|
| miR no. | Fold change (average) | MiaPaCa2-RGs (average) | P-value | PSN1-RGs (average) | P-value | Ref sequence ID |
|
Common upregulated miRNAs both in MiaPaCa2-RGs and PSN1-RGs | ||||||
| hsa-miR-320c | 1.97 | 2.20 | 0.0033 | 1.73 | 0.0366 | MIMAT0005793 |
| hsa-miR-29a | 1.85 | 2.13 | 0.0097 | 1.57 | 0.2751 | MIMAT0000086 |
| hsa-miR-10a | 1.69 | 1.64 | 0.0202 | 1.73 | 0.0606 | MIMAT0000253 |
| hsa-miR-30c | 1.68 | 1.54 | 0.0222 | 1.81 | 0.0384 | MIMAT0000244 |
| hsa-miR-30a | 1.65 | 1.51 | 0.0243 | 1.79 | 0.2634 | MIMAT0000087 |
| hsa-miR-29b | 1.58 | 1.53 | 0.0026 | 1.63 | 0.3311 | MIMAT0000100 |
| hsa-miR-320a |
1.56 |
1.51 |
0.0142 |
1.61 |
0.0556 |
MIMAT0000510 |
|
Common downregulated miRNAs both in MiaPaCa2-RGs and PSN1-RGs | ||||||
| hsa-miR-1246 | 3.65 | 1.97 | 0.0085 | 5.32 | 0.1209 | MIMAT0005898 |
Abbreviations: MiaPaCa-P=parental MiaPaCa2 cells; MiaPaCa2-RGs=gemcitabine-resistant clones of MiaPaCa2; miR and miRNA=microRNA; PSN1-P=parental PSN1 cells; PSN1-RGs=gemcitabine-resistant clones of PSN1.
Gain-of-function and loss-of-function of miR-320c alters the resistance of MiaPaCa2 cells to gemcitabine
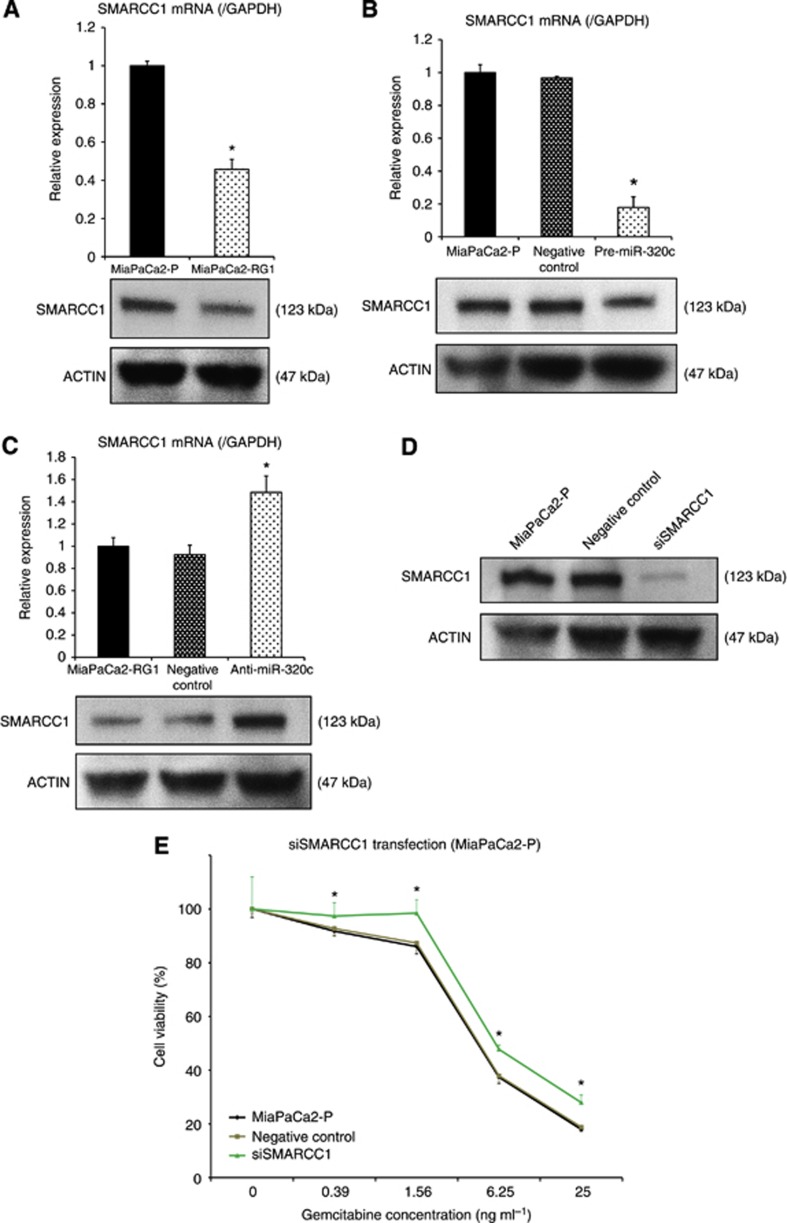
To evaluate the effect of miR-320c on the response to gemcitabine in MiaPaCa2 cells, pre-miR-320c was first transfected into MiaPaCa2-P. Real-time qRT-PCR showed that transfection of pre-miR-320c markedly increased the miR-320c expression level for over 72 h (Figure 2A). The MTT assay demonstrated that transfection of pre-miR-320c into MiaPaCa2-P induced resistance to gemcitabine treatment (Figure 2C). To further assess the effect of miR-320c on the gemcitabine resistance, anti-miR-320c was transfected into MiaPaCa2-RG1. Real-time qRT-PCR showed sufficient inhibition of miR-320c expression for over 72 h (Figure 2B), and the MTT assay demonstrated significant reduction of viability of anti-miR-320c-transfected cells compared with the control cells (Figure 2D). These results indicate that, at least partially, miR-320c induces gemcitabine resistance in MiaPaCa2 cells.
Figure 2.
miR-320c induced gemcitabine resistance in MiaPaCa2 cells. (A, B) Real-time qRT-PCR confirmed overexpression (MiaPaCa2-P transfected with pre-miR-320c) and suppression (MiaPaCa2-RG1 transfected with anti-miR-320c) of miR-320c for >72 h. (C, D) MTT assay showed significant changes in resistance to gemcitabine by gain-of-function of miR-320c in MiaPaCa2-P and loss-of-function in MiaPaCa2-RG1. Data are mean±s.d. of triplicate independent experiments. *P<0.05.
miR-320c inhibits the response to gemcitabine by targeting SMARCC1
Few studies have reported miR-320 expression in gastrointestinal cancers, and little is known about the function of this miRNA. As putative targets of miR-320c, 539 genes were predicted by TargetScan. Among them, SMARCC1, a component of the chromatin remodeling complex, also known as a tumour suppressor, was selected for further analysis. SMARCC1 expression was lower in MiaPaCa2-RG1 than in MiaPaCa2-P (Figure 3A). We investigated direct binding of miR-320c and the SMARCC1 gene by luciferase assay in MiaPaCa2-P and observed reduction of the luciferase activity in the pre-miR-320c-treated cells in comparison with negative control (Supplementary Figure S1). Pre-miR-320c transfection decreased SMARCC1 expression, and anti-miR-320c transfection increased it (Figures 3B and C), suggesting that SMARCC1 is one of the target genes of miR-320c in MiaPaCa2 cells. Next, we used siRNA for SMARCC1 to validate its involvement in the resistance to gemcitabine. Knockdown of SMARCC1 was confirmed by western blot analysis (Figure 3D). The MTT assay demonstrated that transfection of siSMARCC1 enhanced the resistance of MiaPaCa2-P to gemcitabine (Figure 3E). These results suggest that SMARCC1 mediates, at least in part, the miR-320c-related resistance to gemcitabine.
Figure 3.
The miR-320c–related resistance to gemcitabine treatment is mediated through SMARCC1. (A) Real-time qRT-PCR and western blot analysis confirmed the significantly low SMARCC1 expression in MiaPaCa2-RG1 compared with MiaPaCa2-P. (B) SMARCC1 gene and protein expression was confirmed to be significantly suppressed by pre-miR-320c transfection in real-time qRT-PCR and western blot analysis. (C) real-time qRT-PCR and western blot analysis showed enhancement of SMARCC1 gene and protein expression levels by anti-miR-320c transfection. (D) Knockdown of SMARCC1 in MiaPaCa2-P was confirmed in western blot analysis. (E) MTT assay showed that knockdown of SMARCC1 induced resistance to gemcitabine treatment. Data are mean±s.d. of triplicate independent experiments. *P<0.05.
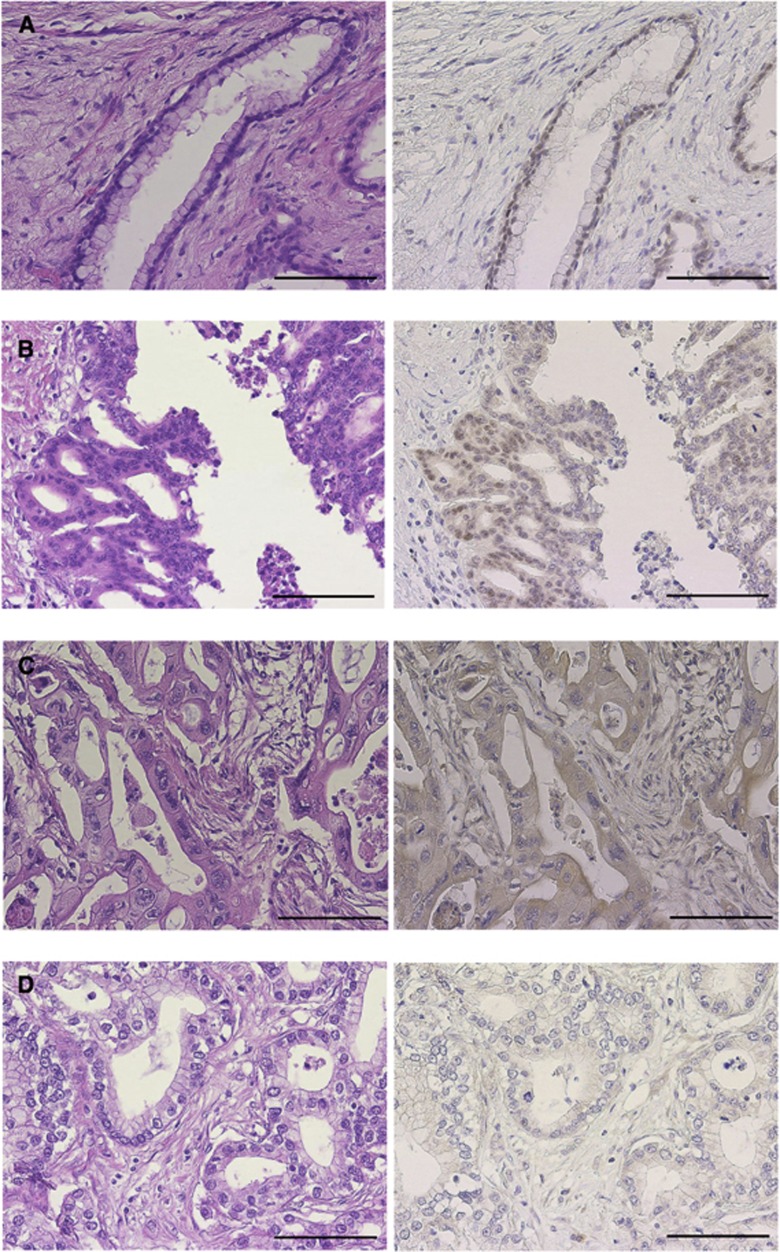
SMARCC1 expression in clinical pancreatic cancer samples
Pancreatic cancer samples of 66 patients who underwent R0 resection were immunohistochemically stained for SMARCC1 expression. Whereas the expression of SMARCC1 in pancreatic cancer lesions varied among the patients, a homogeneous staining for SMARCC1 was observed in the nucleus in normal pancreatic duct cells (Figure 4A). Although SMARCC1 has shown to appear not only in the nucleus but also in the cytoplasm in the previous study (Andersen et al, 2009), functional SMARCC1 protein is considered to localise in the nucleus, therefore we defined SMARCC1-positive samples as having the spotted granular nuclear pattern (Figure 4B) and SMARCC1-negative samples as having the cytoplasmic pattern (not stained in the nucleus) (Figure 4C) or the negative pattern (not stained in the nucleus or the cytoplasm) (Figure 4D) in pancreatic cancer lesions. Among the 66 patients examined, 31 (47.0%) showed positive staining whereas 35 (53.0%) patients were negative for SMARCC1.
Figure 4.
Immunohistochemistry for SMARCC1 in clinical samples. (A–D) Haematoxylin and eosin staining on the left side and SMARCC1 staining on the right side. (A) A normal pancreatic duct sample. SMARCC1 expression was identified in the nucleus homogeneously in normal pancreatic duct cells. (B) A representative SMARCC1-positive sample. SMARCC1 staining was in the spotted granular nuclear pattern in pancreatic carcinoma cells. (C, D) Representative SMARCC1-negative samples. SMARCC1 staining was in the cytoplasmic pattern (not stained in the nucleus) or in the negative pattern (not stained in the nucleus and the cytoplasm) in pancreatic carcinoma cells. Bar=100 μm.
SMARCC1 expression was not associated with overall and disease-free survival
Of all the 66 patients, the median overall survival was 17.0 months (3.5–147.7), and the median disease-free survival was 11.1 months (2.0–147.7). There were no significant differences between the groups who were SMARCC1 expression positive and negative with respect to age, sex, histopathological type (well/mod/poor), tumour size, tumour location (head/body/tail), pathological depth of tumour (pT1/T2/T3), and whether or not gemcitabine was used as chemotherapy. However, pathological lymph node metastasis and pathological stage were significantly different in the two groups (P=0.0383, P=0.0383, respectively) (Supplementary Table S4). The Kaplan–Meier overall survival estimates were not significantly different for patients who were SMARCC1 positive compared with those with SMARCC1-negative expression (median overall survival: 1.693 vs 2.189 years, P=0.5585; Supplementary Figure S2A). With regard to disease-free survival, there was no significant difference between the SMARCC1-positive and -negative groups (median disease-free survival, 0.956 vs 1.334 years, P=0.5633; Supplementary Figure S2B).
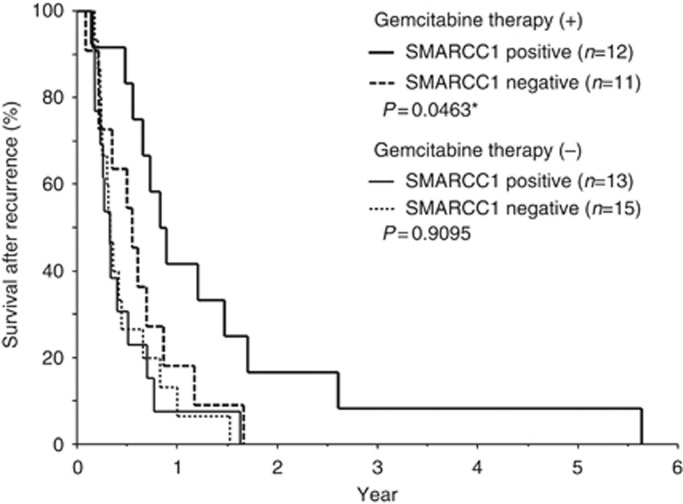
SMARCC1 was a useful predictor of clinical response to gemcitabine therapy
Of the 66 patients, 26 received therapy with single-agent gemcitabine. In 23 patients, this treatment was initiated at the time of tumour recurrence. To elucidate the relationship between SMARCC1 expression and gemcitabine therapy, we used survival after recurrence, which represented the period from starting gemcitabine therapy or other therapies in 51 patients with relapse, until death. There were no significant differences between patients with and without gemcitabine therapy in clinicopathological factors (Table 2). First, we examined the survival benefit of gemcitabine. The 23 patients who were treated with gemcitabine had a significantly better survival than those who did not (P=0.0046; Supplementary Figure S3). After dividing patients who were treated with gemcitabine into SMARCC1-positive and -negative groups, only patients who were SMARCC1 positive benefited from gemcitabine therapy (P=0.0463). The relationship between SMARCC1 and survival after recurrence was not significant in patients treated without gemcitabine therapy (P=0.9095; Figure 5).
Table 2. Relationship between gemcitabine therapy and clinicopathological factors.
| |
Gemcitabine therapy |
|
|
|---|---|---|---|
| Treated (n=23) | Not treated (n=28) | P-value | |
| Age (<65 : ⩾ 65 years) |
13 : 10 |
12 : 16 |
0.3314 |
| Sex (male:female) |
11 : 12 |
14 : 14 |
0.8772 |
| Histopathological type (well or mod:poor) |
21 : 2 |
22 : 6 |
0.2134 |
| Tumour size (<27 : ⩾27 mm) |
12 : 11 |
12 : 16 |
0.5071 |
| Tumour location (head:body or tail) |
18 : 5 |
22 : 6 |
0.9786 |
| Pathological depth of invasion pT (T1 or T2:T3) |
2 : 21 |
1 : 27 |
0.4390 |
| Pathological lymph node metastasis pN (negative:positive) |
5 : 18 |
11 : 17 |
0.1790 |
| Pathological stage (IA or IB or IIA:IIB or IV) |
5 : 18 |
11 : 17 |
0.1790 |
| SMARCC1 expression (−:+) | 11 : 12 | 15 : 13 | 0.6830 |
Abbreviations: mod=moderately differentiated; poor=poorly differentiated; well=well differentiated.
Figure 5.
Relationship between SMARCC1 expression and survival after recurrence. Survival after recurrence curves showed a significantly better survival rate for SMARCC1-positive patients than for SMARCC1-negative patients treated with gemcitabine therapy (*P=0.0463), but survival was not significantly different in patients treated without gemcitabine therapy (P=0.9095).
Discussion
Several studies have examined the involvement of miR-320c in various types of cancer. It has been reported that miR-320c is upregulated in breast cancer (Yan et al, 2008), retinoblastoma (Zhao et al, 2009), and malignant transformed bronchial epithelial cells (Shen et al, 2009; Duan et al, 2010), whereas it is downregulated in lung cancer (Gao et al, 2011) and in cholangiocarcinoma (Chen et al, 2009). It has also been reported that miR-320 is regulated by PTEN in mammary stromal fibroblasts (Bronisz et al, 2012), correlates with recurrence-free survival in colon cancer (Schepeler et al, 2008), and inhibits proliferation in leukaemia (Schaar et al, 2009). Regarding the association of miR-320 and drug resistance, it has recently been reported that miR-320 facilitates chemotherapeutic drug-triggered apoptosis in cholangiocarcinoma (Chen et al, 2009). The present study identified miR-320c as one of the common upregulated miRNAs in gemcitabine-resistant pancreatic cancer cells compared with their parental cells, and we showed that miR-320c induced the resistance to gemcitabine. Among the putative targets of miR-320c by TargetScan, we focussed on SMARCC1, a switch/sucrose nonfermentable (SWI/SNF)-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1, also known as BAF155 (BRG1-associated factor 155), as a candidate molecule responsible for miR-320-mediated drug resistance because of recent studies suggesting a role of chromatin remodeling in some cancers. Drug resistance is the major cause of treatment failure in cancer, yet the multifactorial mechanisms responsible for resistance remain largely unknown. Recently, several studies reported the contribution of chromatin remodeling in drug resistance in various types of cancer, such as the DEK oncogene in melanoma (Khodadoust et al, 2009), remodeling and spacing factor 1 in ovarian cancer (Choi et al, 2009), enhancer of zeste homolog 2 in pancreatic cancer (Ougolkov et al, 2008), and chromatin remodeling at the topoisomerase II-beta promoter in neuroblastoma (Das et al, 2010). Unlike DNA mutations, which are essentially irreversible in cancer, chromatin alterations, including both histone modifications and nucleosome remodeling, are potentially reversible and thus might constitute attractive therapeutic targets (Wilson and Roberts, 2011). The SWI/SNF chromatin remodeling complex is a 2-Mda multisubunit complex first identified in yeast and highly conserved among eukaryotes (Peterson, 1996). Transcriptional activation and efficient transcription of genes require dynamic structural changes in chromatin, and the ATP-dependent SWI/SNF complex is involved in chromatin restructuring (Percipalle and Farrants, 2006).
The SWI/SNF chromatin remodeling complex consists of a catalytic ATPase subunit, core subunits, and variant subunits. SMARCC1 is contained in the core subunits. The SWI/SNF complexes have a widespread role in tumour suppression (Wilson and Roberts, 2011). Inactivating deletion and mutations in SWI/SNF subunits have been reported at high frequency in various cancers, such as SNF5 in rhabdoid tumours (Versteege et al, 1998), BAF180 and ARID1A in renal cell carcinoma (Varela et al, 2011), ARID1A (Jones et al, 2010; Wiegand et al, 2010) and BAF155 (DelBove et al, 2011) in ovarian carcinoma, and BRG1, BRM, ARID1A, ARID1B, and BAF180 in pancreatic cancer (Shain et al, 2012). In addition, some SWI/SNF subunit deficiencies correlate with malignant potential, including drug resistance and shorter survival in melanoma (Lin et al, 2009) and in ovarian carcinoma (Katagiri et al, 2012) and glucocorticoid resistance in acute lymphoblastic leukaemia (Pottier et al, 2008), and as a severe risk factor for histologically malignant gastric cancer (Yamamichi et al, 2007). Some studies have reported that SMARCC1 deficiency prevents DNA damage–induced cell death (Ahn et al, 2011) and predicts short-term survival of colorectal cancer (Andersen et al, 2009). In addition, knockdown of SMARCC1 promotes self-renewal gene expression in embryonic stem cells (Schaniel et al, 2009). In the present study, we used MiaPaCa2-P and MiaPaCa2-RG1 and showed that knockdown of SMARCC1 induced gemcitabine resistance, and both gain-of-function and loss-of-function of miR-320c inversely altered the expression level of SMARCC1 protein. Although SMARCC1 may be only one of the responsible molecules, the molecule is shown to be involved, at least partly, in the miR-320c-related drug-resistance.
Evaluating the expression of miR-320c in clinical specimens may be crucial in predicting the drug-resistance, yet SMARCC1 may be practically easier and more useful than miR-320c. Thus, in the present study, we evaluated the clinical importance of SMARCC1 rather than miR-320c. We have previously reported RRM1 expression as the beneficial predictor of the clinical response to gemcitabine in pancreatic cancer patients after a complete resection (Akita et al, 2009). The present study revealed a significant association between SMARCC1 expression and the clinical response to gemcitabine therapy in completely resected pancreatic cancer patients. Therefore, RRM1, the key enzyme involved in DNA synthesis, and SMARCC1, the core subunit of the SWI/SNF chromatin remodeling complex, appear to make a contribution to drug-resistance mechanisms in separate processes and not to depend on each other. SMARCC1 expression could be a newly independent predictor of the clinical response to gemcitabine in pancreatic cancer patients.
In conclusion, we demonstrated in the present study that miR-320c inhibited the anti-cancer effect of gemcitabine in pancreatic cells and that SMARCC1 mediated this effect. The response to gemcitabine in MiaPaCa2 cells was controlled by genetic manipulation of miR-320c and SMARCC1. In addition, clinical examination revealed that only patients who were SMARCC1 positive benefited from gemcitabine therapy with regard to survival after recurrence. Considered together, the results suggest that miR-320c/SMARCC1-mediated gemcitabine resistance is a potential legitimate target for the treatment of pancreatic cancer.
Acknowledgments
We thank Nariaki Matsuura (Department of Molecular Pathology, Graduate School of Medicine, Osaka University, Osaka, Japan) for advice on immunohistochemical analysis.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Ahn J, Ko M, Lee C, Kim J, Yoon H, Seong RH. Srg3, a mouse homolog of BAF155, is a novel p53 target and acts as a tumor suppressor by modulating p21(WAF1/CIP1) expression. Oncogene. 2011;30 (4:445–456. doi: 10.1038/onc.2010.424. [DOI] [PubMed] [Google Scholar]
- Akita H, Zheng Z, Takeda Y, Kim C, Kittaka N, Kobayashi S, Marubashi S, Takemasa I, Nagano H, Dono K, Nakamori S, Monden M, Mori M, Doki Y, Bepler G. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. 2009;28 (32:2903–2909. doi: 10.1038/onc.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70 (9:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sorensen FB, Verspaget HW, Simon R, Kruhoffer M, Aaltonen LA, Laurberg S, Orntoft TF. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100 (3:511–523. doi: 10.1038/sj.bjc.6604884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ, Martin CK, Li F, Yu L, Fernandez SA, Pecot T, Rosol TJ, Cory S, Hallett M, Park M, Piper MG, Marsh CB, Yee LD, Jimenez RE, Nuovo G, Lawler SE, Chiocca EA, Leone G, Ostrowski MC. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol. 2012;14 (2:159–167. doi: 10.1038/ncb2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, Li L, Huang DD, Ding J, Shen F, Zhou WP, Wu MC, Wang HY. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50 (2:358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Choi JH, Sheu JJ, Guan B, Jinawath N, Markowski P, Wang TL, Shih IeM. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69 (4:1407–1415. doi: 10.1158/0008-5472.CAN-08-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das CM, Zage PE, Taylor P, Aguilera D, Wolff JE, Lee D, Gopalakrishnan V. Chromatin remodelling at the topoisomerase II-beta promoter is associated with enhanced sensitivity to etoposide in human neuroblastoma cell lines. Eur J Cancer. 2010;46 (15:2771–2780. doi: 10.1016/j.ejca.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64 (11:3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- DelBove J, Rosson G, Strobeck M, Chen J, Archer TK, Wang W, Knudsen ES, Weissman BE. Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics. 2011;6 (12:1444–1453. doi: 10.4161/epi.6.12.18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Jiang Y, Zhang H, Wu Y. MiR-320 and miR-494 affect cell cycles of primary murine bronchial epithelial cells exposed to benzo[a]pyrene. Toxicol In Vitro. 2010;24 (3:928–935. doi: 10.1016/j.tiv.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Eguchi H, Nagano H, Yamamoto H, Miyamoto A, Kondo M, Dono K, Nakamori S, Umeshita K, Sakon M, Monden M. Augmentation of antitumor activity of 5-fluorouracil by interferon alpha is associated with up-regulation of p27Kip1 in human hepatocellular carcinoma cells. Clin Cancer Res. 2000;6 (7:2881–2890. [PubMed] [Google Scholar]
- Gao W, Shen H, Liu L, Xu J, Shu Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2011;137 (4:557–566. doi: 10.1007/s00432-010-0918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, Danesi R, Campani D, Verheul HM, Boggi U. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70 (11:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- Goan YG, Zhou B, Hu E, Mi S, Yen Y. Overexpression of ribonucleotide reductase as a mechanism of resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell line. Cancer Res. 1999;59 (17:4204–4207. [PubMed] [Google Scholar]
- Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, Kim SW, Del Chiaro M, Peters GJ, Giaccone G. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PloS One. 2010;5 (5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, Xu L. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PloS One. 2009;4 (8:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih IeM, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr., Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330 (6001:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri A, Nakayama K, Rahman MT, Rahman M, Katagiri H, Nakayama N, Ishikawa M, Ishibashi T, Iida K, Kobayashi H, Otsuki Y, Nakayama S, Miyazaki K. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol. 2012;25 (2:282–288. doi: 10.1038/modpathol.2011.161. [DOI] [PubMed] [Google Scholar]
- Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz DM, Soengas MS. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69 (16:6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Yamamoto H, Nagano H, Okami J, Ito Y, Shimizu J, Eguchi H, Miyamoto A, Dono K, Umeshita K, Matsuura N, Wakasa K, Nakamori S, Sakon M, Monden M. Increased expression of COX-2 in nontumor liver tissue is associated with shorter disease-free survival in patients with hepatocellular carcinoma. Clin Cancer Res. 1999;5 (12:4005–4012. [PubMed] [Google Scholar]
- Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363 (9414:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69 (16:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wong RP, Martinka M, Li G. Loss of SNF5 expression correlates with poor patient survival in melanoma. Clin Cancer Res. 2009;15 (20:6404–6411. doi: 10.1158/1078-0432.CCR-09-1135. [DOI] [PubMed] [Google Scholar]
- Nakahira S, Nakamori S, Tsujie M, Takahashi Y, Okami J, Yoshioka S, Yamasaki M, Marubashi S, Takemasa I, Miyamoto A, Takeda Y, Nagano H, Dono K, Umeshita K, Sakon M, Monden M. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer. 2007;120 (6:1355–1363. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- Oettle H, Post S, Neuhaus P, Gellert K, Langrehr J, Ridwelski K, Schramm H, Fahlke J, Zuelke C, Burkart C, Gutberlet K, Kettner E, Schmalenberg H, Weigang-Koehler K, Bechstein WO, Niedergethmann M, Schmidt-Wolf I, Roll L, Doerken B, Riess H. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297 (3:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- Ougolkov AV, Bilim VN, Billadeau DD. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin Cancer Res. 2008;14 (21:6790–6796. doi: 10.1158/1078-0432.CCR-08-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38 (7:e190–e199. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Farrants AK. Chromatin remodelling and transcription: be-WICHed by nuclear myosin 1. Curr Opin Cell Biol. 2006;18 (3:267–274. doi: 10.1016/j.ceb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Peterson CL. Multiple SWItches to turn on chromatin. Curr Opin Genet Dev. 1996;6 (2:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- Pottier N, Yang W, Assem M, Panetta JC, Pei D, Paugh SW, Cheng C, Den Boer ML, Relling MV, Pieters R, Evans WE, Cheok MH. The SWI/SNF chromatin-remodeling complex and glucocorticoid resistance in acute lymphoblastic leukemia. J Natl Cancer Inst. 2008;100 (24:1792–1803. doi: 10.1093/jnci/djn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar DG, Medina DJ, Moore DF, Strair RK, Ting Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp Hematol. 2009;37 (2:245–255. doi: 10.1016/j.exphem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27 (12:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sorensen FJ, Kruhoffer M, Laurberg S, Kauppinen S, Orntoft TF, Andersen CL. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68 (15:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32 (4:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, Maitra A, Pollack JR. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci USA. 2012;109 (5:E252–E259. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YL, Jiang YG, Greenlee AR, Zhou LL, Liu LH. MicroRNA expression profiles and miR-10a target in anti-benzo[a] pyrene-7, 8-diol-9, 10-epoxide-transformed human 16HBE cells. Biomed Environ Sci. 2009;22 (1:14–21. doi: 10.1016/S0895-3988(09)60016-7. [DOI] [PubMed] [Google Scholar]
- Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103 (10:1617–1626. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomokuni A, Eguchi H, Tomimaru Y, Wada H, Kawamoto K, Kobayashi S, Marubashi S, Tanemura M, Nagano H, Mori M, Doki Y. miR-146a suppresses the sensitivity to interferon-alpha in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2011;414 (4:675–680. doi: 10.1016/j.bbrc.2011.09.124. [DOI] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469 (7331:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394 (6689:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih IeM, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363 (16:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11 (7:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Yamamichi N, Inada K, Ichinose M, Yamamichi-Nishina M, Mizutani T, Watanabe H, Shiogama K, Fujishiro M, Okazaki T, Yahagi N, Haraguchi T, Fujita S, Tsutsumi Y, Omata M, Iba H. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 2007;67 (22:10727–10735. doi: 10.1158/0008-5472.CAN-07-2601. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kondo M, Nakamori S, Nagano H, Wakasa K, Sugita Y, Chang-De J, Kobayashi S, Damdinsuren B, Dono K, Umeshita K, Sekimoto M, Sakon M, Matsuura N, Monden M. JTE-522, a cyclooxygenase-2 inhibitor, is an effective chemopreventive agent against rat experimental liver fibrosis1. Gastroenterology. 2003;125 (2:556–571. doi: 10.1016/s0016-5085(03)00904-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nagano H, Sakon M, Wada H, Eguchi H, Kondo M, Damdinsuren B, Ota H, Nakamura M, Marubashi S, Miyamoto A, Dono K, Umeshita K, Nakamori S, Yagita H, Monden M. Partial contribution of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)/TRAIL receptor pathway to antitumor effects of interferon-alpha/5-fluorouracil against hepatocellular carcinoma. Clin Cancer Res. 2004;10 (23:7884–7895. doi: 10.1158/1078-0432.CCR-04-0794. [DOI] [PubMed] [Google Scholar]
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14 (11:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Chen CL, Chau GY, Chiou SH, Su CW, Chou TY, Peng WL, Wu JC. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50 (5:1464–1474. doi: 10.1002/hep.23221. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J, Xu J, Cheng JQ, Lin JY, Ma X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25 (1:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.