Abstract
Background:
LINE-1 methylation level is a surrogate marker of global DNA methylation. LINE-1 methylation in primary colorectal cancers (CRCs) is highly variable and strongly associated with a poor prognosis. However, no study has examined LINE-1 methylation levels of metastatic CRCs in relation to prognosis or assessed the heterogeneity of LINE-1 methylation level within the primary CRCs.
Methods:
Pyrosequencing was used to quantify LINE-1 methylation level in 42 liver metastases, 26 matched primary tumours, and 6 matched lymph node (LN) metastases. KRAS, BRAF, and PIK3CA mutation status and microsatellite instability (MSI) status were also examined.
Results:
The distribution of LINE-1 methylation level in liver metastases was as follows: mean, 67.3; range, 37.1–90.1. Primary tumours showed LINE-1 methylation levels similar to those of matched liver and LN metastases. The difference in LINE-1 methylation level between superficial areas and invasive front areas was within 7.0 in all six cases evaluated. Prognostic impact of LINE-1 hypomethylation in liver metastases on overall survival was not observed. The concordance rate was 94% for KRAS, 100% for BRAF, 88% for PIK3CA, and 97% for MSI.
Conclusion:
Alteration of LINE-1 methylation level may occur in early CRC tumorigenesis, and the LINE-1 methylation level is relatively stable during CRC progression.
Keywords: LINE-1, colorectal cancer, epigenetics
Colorectal cancer (CRC), the third and fourth most common cancer in women and men, respectively, causes 610 000 deaths per year worldwide (Ferlay et al, 2010). Nearly 50% of patients with CRC develop tumour recurrence (Van Cutsem et al, 2006; Yoo et al, 2006; Lee et al, 2007), and the primary cause of death is liver metastasis (Foster, 1984). Genetic testing for KRAS somatic mutations in patients with advanced CRC has become a routine clinical practice (Benvenuti et al, 2007; Amado et al, 2008; Freeman et al, 2008; Karapetis et al, 2008; Lievre et al, 2008), and epidermal growth factor receptor (EGFR) inhibitors are now recommended only for use in patients with KRAS wild-type CRC tumours (Allegra et al, 2009). However, even among patients with KRAS wild-type tumours, some patients do not respond to EGFR inhibitors. One reason for this may be discordance of the KRAS mutation status between primary tumours and corresponding metastases (Albanese et al, 2004; Oliveira et al, 2007; Richman et al, 2011). In addition, there is emerging evidence that mutations in BRAF and PIK3CA are associated with resistance to EGFR-targeted agents (Di Nicolantonio et al, 2008; Sartore-Bianchi et al, 2009; De Roock et al, 2010; Maughan et al, 2011). Thus, a thorough understanding of molecular alterations in primary tumours and corresponding metastases of CRCs may have considerable clinical implications.
The development of CRC is a multistep process arising from the accumulation of genetic and epigenetic alterations (Worthley et al, 2007). The chromosomal instability pathway and microsatellite instability (MSI) pathway are the two recognised pathways of CRC carcinogenesis (Markowitz and Bertagnolli, 2009). Along with genetic mutations, epigenetic changes such as DNA methylation and histone modification are important for carcinogenesis and tumour development (Issa, 2004; Ushijima, 2005; Esteller, 2006). Cancer cells exhibit two types of DNA methylation alterations: global DNA hypomethylation and site-specific CpG island promoter hypermethylation (Jones and Baylin, 2007; Taby and Issa, 2010; Rodriguez-Paredes and Esteller, 2011). Global DNA hypomethylation appears to have a crucial role in genomic instability (Bestor, 2005), leading to cancer development and progression (Gaudet et al, 2003; Holm et al, 2005; Suzuki et al, 2006). Because the LINE-1 retrotransposon constitutes a substantial portion (approximately 17%) of the human genome, the methylation status of LINE-1 reflects the global DNA methylation level (Cordaux and Batzer, 2009).
The tumour LINE-1 methylation level is attracting interest as a useful marker for predicting CRC prognosis. Ogino et al (2008b) previously found that the degree of LINE-1 hypomethylation is linearly associated with aggressive tumour behaviour and observed an approximately 5-fold increase in cancer-specific mortality associated with tumours at the low end of the methylation spectrum compared with those at the high end. In addition, LINE-1 hypomethylated CRC has been associated with young age at onset (Baba et al, 2010) and CRC familial clustering (Goel et al, 2010). However, no study has yet examined the LINE-1 methylation status in metastatic tumours, including both metachronous tumours and synchronous tumours, or assessed the heterogeneity of the LINE-1 methylation level within a CRC tumour.
The aims of this study were to: (1) evaluate the LINE-1 methylation status in metastatic lesions from CRCs; the mutational status of the KRAS, BRAF, and PIK3CA genes; and the MSI status; (2) compare the LINE-1 methylation level of the superficial area with that of the invasive front area within the primary tumour; and (3) examine the prognostic impact of LINE-1 methylation levels in metastatic liver tumours.
Materials and methods
Study subjects
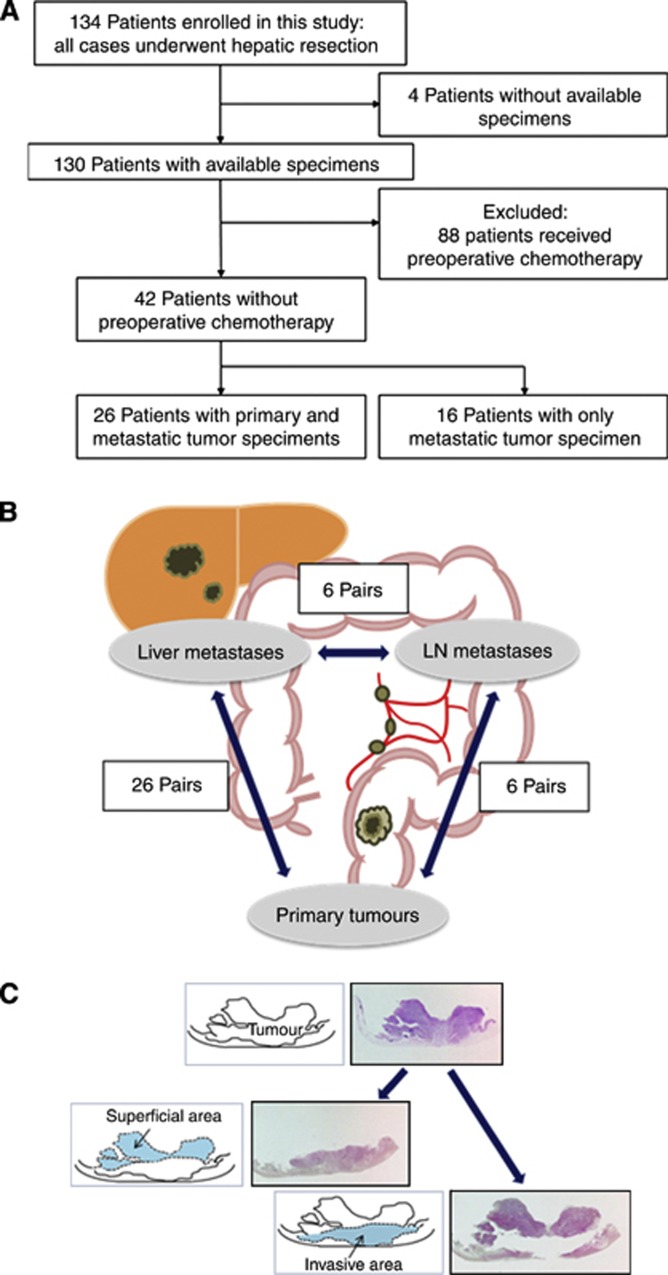
A total of 134 consecutive patients with liver metastases from CRC who underwent hepatic resection at Kumamoto University Hospital between 2000 and 2012 were initially enrolled in this study. Four patients were excluded owing to the unavailability of adequate tissue specimens. Eighty-eight patients were excluded because they had undergone preoperative chemotherapy (Figure 1A). We analysed 42 liver metastases from patients who had not undergone preoperative chemotherapy, 26 matched primary tumours, and 6 matched lymph node (LN) metastases (Figure 1B). Patients were observed at 1–6-month intervals until death or 31 December 2012, whichever came first. Tumour staging was performed according to the International Union Against Cancer TNM system. Written informed consent was obtained from each subject, and the study procedures were approved by the institutional review board.
Figure 1.
(A) Flow chart of the study population. (B) In this study, we examined the LINE-1 methylation level in 26 primary tumours, 26 matched liver metastases, and 6 matched LN metastases. (C) Macrodissection to separate the superficial area from the invasive front area.
DNA extraction and sodium bisulfite treatment
Hematoxylin- and eosin-stained slides of the tumours were reviewed by one pathologist, who marked the areas of the tumour and normal tissue. In addition, superficial areas and invasive front areas were marked separately in six cases (Figure 1C). Genomic DNA was extracted from tumour lesions enriched with neoplastic cells without adjacent normal tissue using a formalin-fixed, paraffin-embedded tissue kit (Qiagen, Valencia, CA, USA). Genomic DNA was extracted from the tumour and modified with sodium bisulfite using an EpiTect Bisulfite Kit (Qiagen).
Whole-genome amplification
Whole-genome amplification is a useful technique for preserving original study material for many different assays and future studies. In whole-genome amplification, genomic DNA is amplified by PCR using primers with a random sequence of 15 nucleotides. Each PCR mix contained 40 pmol of the random primers, 1.0 nmol of each dNTP, 2.0 mmol l−1 of MgCl2, 1 × PCR buffer (Applied Biosystems, Foster City, CA, USA), 0.25 U of AmpliTaq Gold 360 (Applied Biosystems), and 5 μl of template DNA solution in a total volume of 50 μl. PCR conditions were as follows: initial denaturation at 95 °C for 10 min; 50 cycles of 95 °C for 60 s, annealing (37 °C for 2 min), ramping from 37–55 °C (0.1 °C s−1), 55 °C for 2 min, and 68 °C for 30 s; and a final extension at 72 °C for 7 min.
Pyrosequencing to measure LINE-1 methylation
PCR and subsequent pyrosequencing for LINE-1 were performed as previously described by Ogino et al (2008a, 2008b) using the PyroMark Kit (Qiagen) (Baba et al, 2010; Irahara et al, 2010). This assay amplifies a region of the LINE-1 element (position 305–331 in accession no. X58075), which includes four CpG sites. The PCR conditions were 45 cycles of 95 °C for 20 s, 50 °C for 20 s, and 72 °C for 20 s, followed by 72 °C for 5 min. The biotinylated PCR product was purified and made single stranded to act as a template in a pyrosequencing reaction using the Pyrosequencing Vacuum Prep Tool (Qiagen). Pyrosequencing reactions were performed in the PyroMark Q24 System (Qiagen). The nucleotide dispensation order was 5′-ACTCAGTGTGTCAGTCAGTTAGTCTG-3′. The non-CpG cytosine in LINE-1 repetitive sequences has been documented to be rarely methylated. Thus, complete conversion of cytosine at a non-CpG site ensured successful bisulfite conversion. The amount of C relative to the sum of the amounts of C and T at each CpG site was calculated as a percentage (i.e., 0–100%). The average of the relative amounts of C in the four CpG sites was used as the overall LINE-1 methylation level in a given tumour. We have validated our LINE-1 methylation pyrosequencing assay in the published literature (Iwagami et al, 2012).
Pyrosequencing for KRAS, BRAF, and PIK3CA mutations
Pyrosequencing for KRAS, BRAF, and PIK3CA mutations was performed as previously described using the PyroMark kit (Qiagen) (Shigaki et al, 2012, 2013). We used PCR amplification and the pyrosequencing primers shown in Supplementary Table 1.
Microsatellite instability analysis
Microsatellite instability analysis was performed using the BAT25 primer set (sense, 5′-TCGCCTCCAAGAATGTAAGT-3′ and antisense, 5′-TCTGCATTTTAACTATGGCTC-3′) and the BAT26 primer set (sense, 5′-TGACTACTTTTGACTTCAGCC-3′ and antisense, 5′-AACCATTCAACATTTTTAACCC-3′). The sense primer was labelled with FAM. PCR was performed in a final volume of 25 μl containing 1 × PCR buffer, 2.0 mmol l−1 of MgCl2, 0.2 μmol l−1 of dNTPs, 0.2 μmol l−1 of each primer, 1.25 U of AmpliTaq Gold 360, and 1 μl of DNA. The thermal conditions were 95 °C/15 min followed by 45 cycles (95 °C/30 s, 60 °C/30 s, and 72 °C/30 s) and a final extension at 72 °C/7 min. The dye-labelled PCR products were analysed with an ABI PRISM 3100 Genetic Analyzer using Genescan 3·7 software (Applied Biosystems). A total of 0.5 μl of the PCR product was mixed with 9.25 μl of highly deionized formamide and 0.25 μl of DNA Size Standard LIZ 500/(−250) (both obtained from Applied Biosystems, Darmstadt, Germany). This mixture was denatured for 3 min at 95 °C, immediately put on ice, and separated using an ABI 3130 Genetic Analyzer. Results were analysed using GeneMapper software (Applied Biosystems).
Both tumoural and normal samples were analysed. Negative (HT29 cells) and positive (HCT116 and HCT15 cells) controls for MSI were included in all analyses. Samples were divided into two groups: those with one or more of the two markers displaying MSI and those with no instability (microsatellite-stable).
Statistical methods
For the statistical analyses, we used JMP software (version 9; SAS Institute, Cary, NC, USA). All P-values were two-sided. To compare the means, we performed the t-test assuming unequal variances. For the survival analysis, the Kaplan–Meier method was used to assess the survival time distribution, and the log-rank test was used.
Results
Pyrosequencing assay for LINE-1 methylation status in liver metastases
We first quantified the LINE-1 methylation level in 42 liver metastases and obtained valid results in all cases. Representative pyrograms for LINE-1 are shown in Supplementary Figure 1. The distribution of the LINE-1 methylation level in the 42 liver metastases on a 0–100 scale was as follows: mean, 67.3; median, 68.5; s.d., 10.9; range, 37.1–90.1; and interquartile range, 59.5–74.3. The LINE-1 methylation level was not associated with any clinical, epidemiological, or pathological characteristics including sex, age, year of operation, tumour size, detection of metastases, postoperative chemotherapy, and stage (all P>0.079) (Table 1). Importantly, the LINE-1 methylation level did not differ between synchronous tumours and metachronous tumours (P=0.217).
Table 1. LINE-1 methylation level and clinical or epidemiological features of liver metastases from colorectal cancer.
| Clinical or epidemiological features | N (%)a | LINE-1 methylation levelmean (%) | P-value |
|---|---|---|---|
| All cases |
42 |
67.3 |
|
|
Age | |||
| <65 | 25 (60.0%) | 67.2 | 0.798 |
| ⩽65 |
17(40.0%) |
67.5 |
|
|
Sex | |||
| Male | 26 (61.9%) | 67.7 | 0.786 |
| Female |
16 (38.1%) |
66.6 |
|
|
Year of operation | |||
| 2001–2006 | 24 (57.1%) | 67.8 | 0.657 |
| 2007–2012 |
18 (42.9%) |
66.6 |
|
|
Tumour size (mm) | |||
| 35< | 21 (50.0%) | 67.0 | 0.841 |
| ⩾35 |
21 (50.0%) |
67.6 |
|
|
Detection of metastases | |||
| Synchronous | 15 (35.7%) | 64.7 | 0.217 |
| Metachronous |
27 (64.3%) |
68.8 |
|
|
Postoperative chemotherapy | |||
| Yes | 32 (76.2%) | 68.9 | 0.079 |
| No |
10 (23.8%) |
62.4 |
|
|
Stage | |||
| I | 4 (9.5%) | 70.7 | 0.161 |
| II | 11 (26.2%) | 72.7 | |
| III | 11 (26.2%) | 65.0 | |
| IV | 16 (38.1%) | 64.4 | |
There is no significant relation between LINE-1 and any of the clinical or epidemiological features examined.
% indicates the proportion of cases with a specific clinical or epidemiological feature.
LINE-1 methylation level between primary and metastatic lesions of CRCs
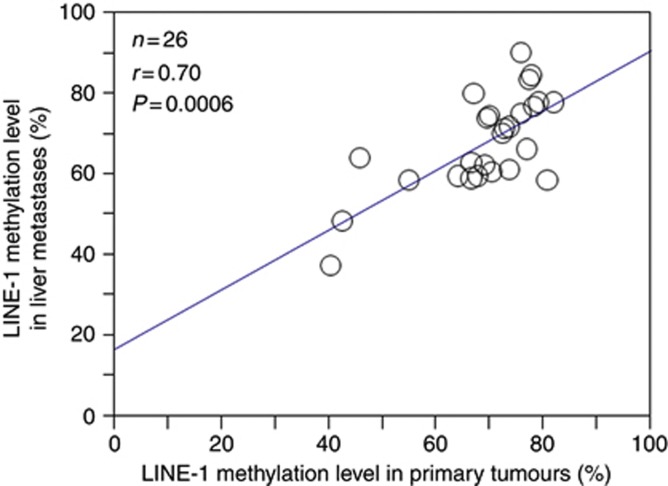
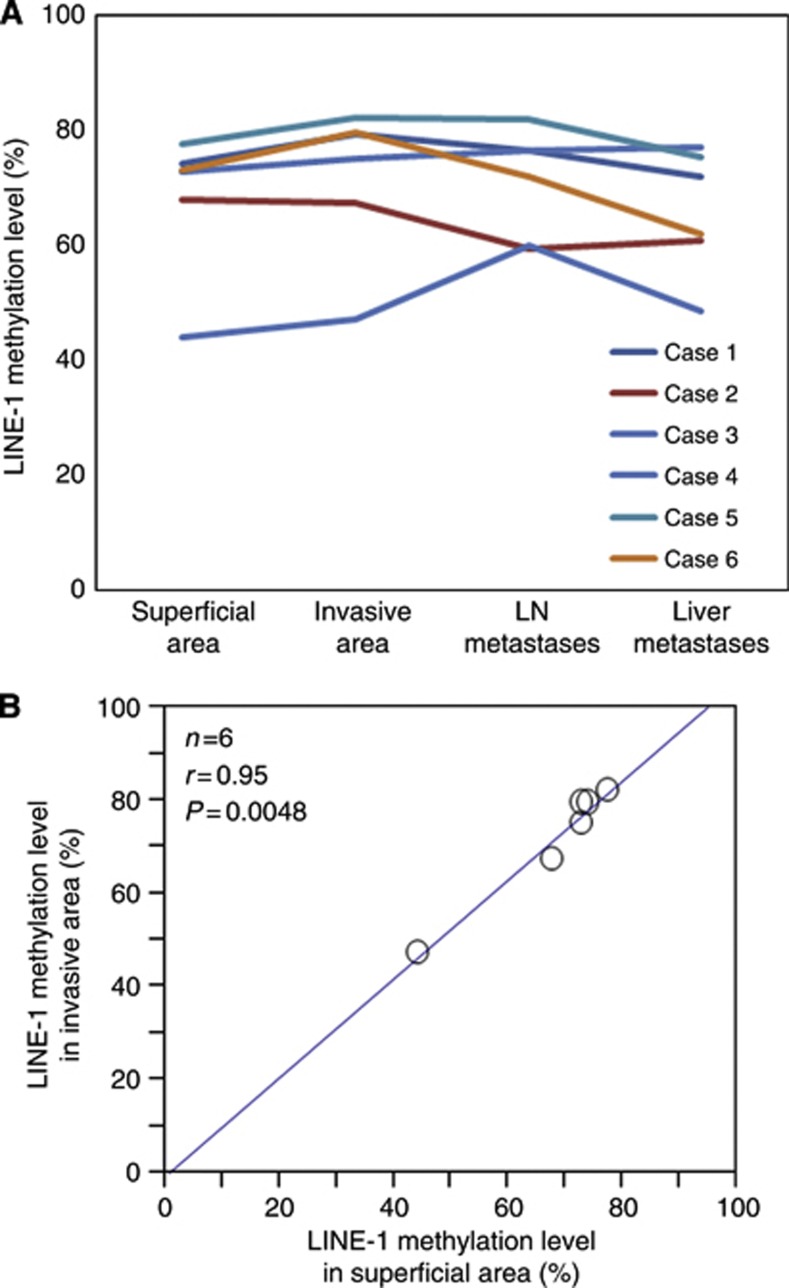
Next, we examined the LINE-1 methylation level in 26 matched primary tumours. The distribution of the LINE-1 methylation level in the 26 matched primary tumours was as follows: mean, 69.0; median, 70.5; s.d., 11.3; and range, 40.4–82.2. The LINE-1 methylation level of liver metastases was significantly associated with that of matched primary tumours (n=26; Pearson's correlation coefficient r=0.70; P=0.0006) (Figure 2). The distribution of the LINE-1 methylation level in the six matched LN metastases was as follows: mean, 71.0; median, 71.9; s.d., 9.37; and range, 56.3–81.9. In six cases, the difference in the LINE-1 methylation level between primary tumours and LN metastases ranged from 0.3 to 12.8 (Figure 3A). Given that the LINE-1 methylation level in CRC primary tumours was highly variable (23.1–90.3 in 869 tumours) (Ogino et al, 2008a), this difference may be so small as to be insignificant.
Figure 2.
Correlation between the LINE-1 methylation level in primary tumours and that in liver metastases. The LINE-1 methylation level in liver metastases was significantly associated with that in matched primary tumours (n=26; Pearson's correlation coefficient r=0.70; P=0.0006).
Figure 3.
LINE-1 methylation level in superficial and invasive areas of primary tumours, matched LN metastases, and matched liver metastases (n=6). (A) In each case, the LINE-1 methylation level of the LN and liver metastases was similar to that in the matched primary tumours. (B)The LINE-1 methylation level in superficial areas was significantly associated with that in the matched invasive areas within primary tumours (n=6; Pearson's correlation coefficient, r=0.95; P=0.0048).
Heterogeneity of LINE-1 methylation level
To evaluate the heterogeneity of the LINE-1 methylation level within primary tumours, we investigated the LINE-1 methylation levels in both superficial areas and invasive front areas (Figure 1C). We found that the LINE-1 methylation level in superficial areas was significantly associated with that in invasive front areas (n=6; P=0.0048) (Figure 3B). The difference between the methylation level in the superficial area and that in the invasive front area was within 7.0 in all six cases, suggesting the absence of heterogeneity of the LINE-1 methylation level within a given primary tumour.
KRAS, BRAF, and PIK3CA mutations and MSI status in primary and metastatic lesions of CRCs
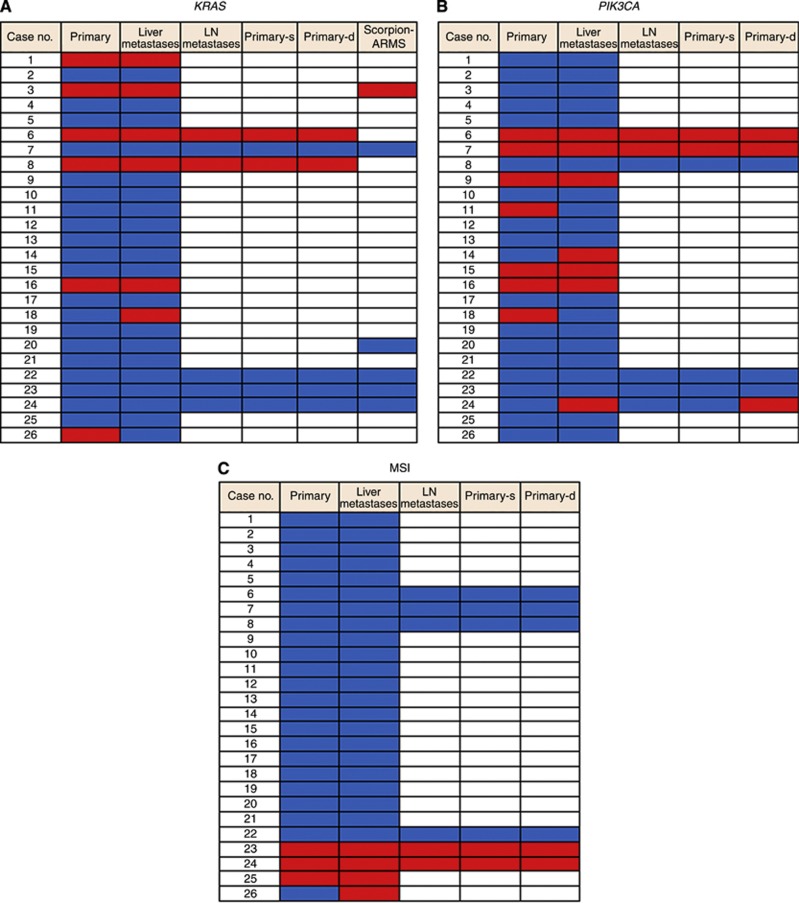
For a better understanding of the differences in molecular alterations between primary and metastatic tumours, we examined the KRAS, BRAF, and PIK3CA mutational status and the MSI status in 26 primary tumours, 26 matched liver metastases, and 6 LN metastases. KRAS mutation was detected in 6 out of 26 primary lesions: 5 in c.35G>A (codon 12 GAT) and 1 in c.35G>T (codon 12 GTT). In 6 out of 26 cases, the KRAS mutational status in primary tumours was confirmed by using a TheraScreen K-RAS Mutation Kit (Qiagen), the Scorpion Amplified Refractory Mutation System technology, which incorporates a unique bifunctional fluorescent primer/probe molecule (Scorpion probe). The results supported the reliability of our pyrosequencing assay for KRAS-mutation detection (Figure 4A). KRAS mutation was detected in 8 out of 32 metastatic lesions: 7 in c.35G>A (codon 12 GAT) and 1 in c.35G>T (codon 12 GTT). Twenty-four cases showed a concordant KRAS mutation status in the primary tumour and metastatic tumour, whereas two cases (6%) showed a discordant mutation status. In addition, we evaluated the KRAS mutation status in superficial areas and invasive front areas within six primary tumours, and found that all six cases showed a concordant mutation status between the two lesions.
Figure 4.
Gene mutation analysis in 26 paired primary CRCs and liver metastases and 6 matched LN metastases. The wild type is shown in blue, and the mutational type is shown in red. The white square denotes the lack of a specimen. (A) KRAS mutation status. Six primary tumours showed the KRAS mutation type. Thirty liver and LN metastases showed a KRAS status identical to that of the matched primary tumours. The KRAS status was confirmed by Scorpion Amplified Refractory Mutation System (ARMS) technology and supported the reliability of the pyrosequencing assay for KRAS mutation detection in six cases. (B) PIK3CA mutation status. Seven primary tumours showed the PIK3CA mutation type. Twenty-eight liver and LN metastases showed a PIK3CA status identical to that of the matched primary tumours. (C) Microsatellite instability status. Three primary tumours showed MSI. Thirty-one liver and LN metastases showed an MSI status identical to that of the matched primary tumours.
The mutational status in BRAF exon 15 (V600E) was examined in 58 tumour lesions (26 primary tumours, 26 liver metastases, and 6 LN metastases) of CRC by pyrosequencing technology. Valid results were obtained in all 58 tissues. All 58 lesions harboured the wild type at codon 600 in BRAF exon 15.
PIK3CA mutation was detected in 14 out of 26 primary lesions and 13 out of 32 metastatic lesions (Figure 4B). Twenty-eight liver and LN metastases showed a PIK3CA mutational status identical to that of the matched primary tumours. A discordant status of PIK3CA mutation between the superficial area and invasive front area was observed in only one case.
The MSI pathway is a recognised pathway of CRC development. Thus, we examined the MSI status in 26 primary tumours and 32 metastatic tumours. Microsatellite instability was detected in 3 out of 26 primary tumours and 4 out of 32 metastatic tumours (Figure 4C). Thirty-one liver and LN metastases showed an MSI status identical to that of the matched primary tumours. In addition, heterogeneity of the MSI status was not observed between superficial areas and invasive front areas within six primary tumours.
LINE-1 methylation level was not associated with KRAS, PIK3CA, and MSI status in metastatic tumours (P=0.068 for KRAS, P=0.54 for PIK3CA, and P=0.57 for MSI; Table 2).
Table 2. LINE-1 methylation level and molecular features of liver metastases from colorectal cancer.
| Molecular features | N | LINE-1 methylation level mean (%) | P-value |
|---|---|---|---|
| All cases |
26 |
67.8 |
|
|
KRAS mutation | |||
| Negative | 20 | 65.7 | 0.068 |
| Positive |
6 |
75.1 |
|
|
PIK3CA mutation | |||
| Negative | 19 | 66.5 | 0.544 |
| Positive |
7 |
71.4 |
|
|
MSI status | |||
| Negative | 22 | 67.1 | 0.570 |
| Positive | 4 | 71.9 | |
Abbreviation: MSI=microsatellite instability.
LINE-1 methylation level in liver metastases and patient survival
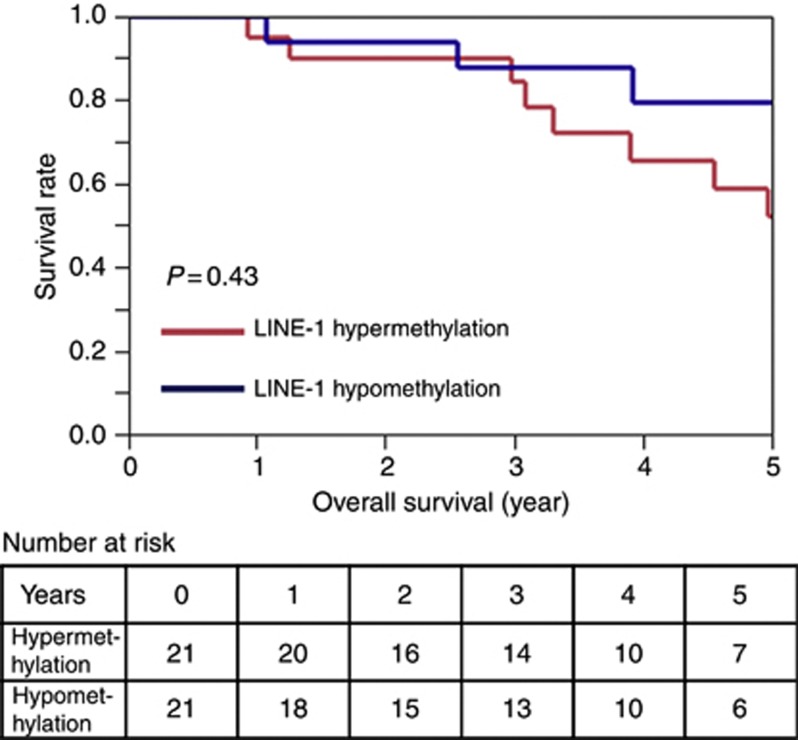
Overall survival was defined as the time between the date of the hepatic resection and the date of death. During an adequate follow-up period among the 42 patients, there were 12 deaths, including 10 deaths that were confirmed to be attributable to CRC. The median follow-up time for censored patients was 3·9 years. The LINE-1 methylation level in liver metastases was divided into two groups: the hypermethylation group (⩾69.0, n=21) and the hypomethylation group (37.1–68.5, n=21). Kaplan–Meier analysis showed a similar overall mortality rate between the two groups (P=0.432) (Figure 5).
Figure 5.
Kaplan–Meier curves for overall survival in the two groups of LINE-1 methylation in metastatic CRC tumours. The hypomethylation (37.1–68.5%, n=21) group experienced an overall mortality similar to that of the hypermethylation group (⩾69.0%, n=21).
Discussion
This is the first study to compare LINE-1 methylation levels (i.e., global DNA methylation levels) in primary CRC tumours with those in liver metastases including both metachronous tumours and synchronous tumours. Liver metastases showed LINE-1 methylation levels similar to those of primary tumours. In addition, we compared the LINE-1 methylation level of superficial areas with that of invasive front areas within primary tumours and found the absence of heterogeneity of the LINE-1 methylation level. These results may suggest that CRC cells acquire the tumoural characteristic of global DNA hypomethylation during the early stage of tumour development and that this characteristic is preserved throughout the process of cancer invasion and metastasis.
The tumour LINE-1 methylation level is attracting interest as a useful marker for predicting CRC prognosis (Ogino et al, 2008b). In addition, the LINE-1 methylation status may be a predictive marker for the response to fluoropyrimidines in a certain subgroup of CRC (Kawakami et al, 2011). In clinical practice, molecular analyses of tumour samples are generally performed using either biopsy specimens obtained before surgery or surgically resected tissue. Because tissue from metastatic and/or recurrent tumours is not easily accessible, results obtained from analysis of the primary tumour are always used in clinical decision-making regarding treatment of metastatic/recurrent CRCs. In this study, we have shown that LINE-1 methylation levels in metastatic liver tumours are similar to those in primary tumours, allowing for the potential clinical application of the LINE-1 methylation level in CRCs. Further studies are required to establish whether LINE-1 methylation can be used in personalised medicine for CRC.
Whether the LINE-1 methylation level is associated with the CRC tumour stage remains controversial. Sunami et al (2011) showed a linear correlation between LINE-1 demethylation progression and TNM stage progression, thus suggesting that the genomic methylation level continuously decreases during CRC development and progression. In contrast, Ogino et al (2008a) demonstrated that LINE-1 methylation levels were not associated with tumour stage (stage I–IV) in 869 population-based CRC tumours. In the current study, we found identical LINE-1 methylation levels between primary tumours and liver/LN metastases, as well as the absence of heterogeneity of the LINE-1 methylation level within primary tumours; these results are in agreement with those of the previous study by Matsunoki et al (2012). Interestingly, we also found that LINE-1 methylation levels did not differ between synchronous tumours and metachronous tumours, supporting the view that the LINE-1 methylation level is continuously preserved even during the long-term natural history of CRC development. These findings by Ogino et al (2008a), Matsunoki et al (2012), and us may suggest that the onset of LINE-1 demethylation occurs at an early stage of CRC development and that the LINE-1 methylation level is relatively stable during CRC progression.
To the best of our knowledge, this is the first study to examine the prognostic significance of the LINE-1 methylation level in metastatic tumours. Given the strong relationship between LINE-1 hypomethylation in primary CRC tumours and a poor prognosis (Ogino et al, 2008b), we first speculated that LINE-1 hypomethylation in metastatic CRC tumours might be associated with a shorter overall survival. However, we found no association between the LINE-1 methylation level and clinical outcome. One possible reason for this result may be the effect of postoperative chemotherapy. In this study, 30 patients received various types of postoperative chemotherapy, whereas 12 patients received no postoperative chemotherapy. Kawakami et al (2011) showed that patients with low LINE-1 methylation levels who were treated with adjuvant chemotherapy survived for a longer period of time than did patients treated by surgery alone, whereas a survival benefit from chemotherapy was not observed for patients with high LINE-1 methylation levels. Thus, the prognostic impact of the LINE-1 methylation level in metastatic tumours on clinical outcome may be influenced by the presence or absence of postoperative chemotherapy or the type of chemotherapy. In addition, we acknowledge that this study is limited by its small sample size. We are currently planning a further large-scale study on LINE-1 methylation levels, chemotherapeutic effects, and patient survival in CRC.
With the development of cancer therapies that specifically target molecular alterations that mediate cancer progression, genotyping of patients with advanced CRC has become a component of routine clinical practice. Specifically, KRAS mutational testing has been incorporated into several clinical practice guidelines for the treatment of patients with metastatic CRC. The National Comprehensive Cancer Network recommends testing either the primary tumour or a site of metastasis. This recommendation is based on several studies that found a high KRAS mutational concordance (>95%) between primary CRCs and metastases. Vakiani et al (2012) reported that primary tumours and matched metastases showed a high concordance rate (>90%) for five genes, including the KRAS, BRAF, and PIK3CA genes. Yim (2012) reported that primary tumours and matched metastases showed a high concordance rate (86%) for MSI. In the current study, the concordance rates of the KRAS, BRAF, and PIK3CA mutations and the MSI status were almost identical to those of previous reports. We also examined the heterogeneity within primary tumours and found that the KRAS and BRAF status and the MSI status were concordant within the primary tumour in all cases. Interestingly, the PIK3CA mutation status in primary tumours showed discordance between the superficial area and invasive front area in only one case. A model in which alterations in KRAS and BRAF occur early in CRC pathogenesis is proposed. Considering that the time at which PIK3CA mutation occurs is unknown, our finding on the discordance of the PIK3CA mutation should be confirmed by independent cohorts with larger sample sizes.
The relationship between LINE-1 methylation level and molecular alterations in CRCs is important. Ogino et al (2008a) have shown that LINE-1 methylation level is higher in MSI-positive tumours than in MSI-negative tumours, and higher in BRAF mutation tumours than in BRAF wild-type tumours in primary CRCs. Baba et al (2010) have demonstrated no significant relationship between KRAS and PIK3CA mutations and LINE-1 methylation level in primary CRCs. In this study of metastatic CRC tumours, LINE-1 methylation level was not associated with MSI status and KRAS and PIK3CA mutations. However, this study was limited by small sample size. In addition, BRAF mutation was not detected in 26 metastatic CRC tumours. Our findings need to be confirmed by independent studies in the future.
In summary, we found that liver and LN metastases showed LINE-1 methylation levels similar to those of the primary tumours. In addition, heterogeneity of the LINE-1 methylation level within primary tumours was not observed. Taken together, these results show that the onset of LINE-1 demethylation occurs at an early stage of CRC development and that the LINE-1 methylation level is relatively stable during CRC progression.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Albanese I, Scibetta AG, Migliavacca M, Russo A, Bazan V, Tomasino RM, Colomba P, Tagliavia M, La Farina M. Heterogeneity within and between primary colorectal carcinomas and matched metastases as revealed by analysis of Ki-ras and p53 mutations. Biochem Biophys Res Commun. 2004;325:784–791. doi: 10.1016/j.bbrc.2004.10.111. [DOI] [PubMed] [Google Scholar]
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, Schernhammer ES, Hunter DJ, Giovannucci EL, Fuchs CS, Ogino S. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer. 2010;9:125. doi: 10.1186/1476-4598-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- Bestor TH. Transposons reanimated in mice. Cell. 2005;122:322–325. doi: 10.1016/j.cell.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Esteller M. The necessity of a human epigenome project. Carcinogenesis. 2006;27:1121–1125. doi: 10.1093/carcin/bgl033. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Foster JH. Treatment of metastatic disease of the liver: a skeptic's view. Semin Liver Dis. 1984;4:170–179. doi: 10.1055/s-2008-1040656. [DOI] [PubMed] [Google Scholar]
- Freeman DJ, Juan T, Reiner M, Hecht JR, Meropol NJ, Berlin J, Mitchell E, Sarosi I, Radinsky R, Amado RG. Association of K-ras mutational status and clinical outcomes in patients with metastatic colorectal cancer receiving panitumumab alone. Clin Colorectal Cancer. 2008;7:184–190. doi: 10.3816/CCC.2008.n.024. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Goel A, Xicola RM, Nguyen TP, Doyle BJ, Sohn VR, Bandipalliam P, Rozek LS, Reyes J, Cordero C, Balaguer F, Castells A, Jover R, Andreu M, Syngal S, Boland CR, Llor X. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology. 2010;138:1854–1862. doi: 10.1053/j.gastro.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, 3rd, Jaenisch R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell. 2005;8:275–285. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Irahara N, Nosho K, Baba Y, Shima K, Lindeman NI, Hazra A, Schernhammer ES, Hunter DJ, Fuchs CS, Ogino S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ida S, Nagai Y, Ishimoto T, Iwatsuki M, Sakamoto Y, Miyamoto Y, Baba H. Pyrosequencing assay to measure LINE-1 methylation level in esophageal squamous cell carcinoma. Ann Surg Oncol. 2012;19:2726–2732. doi: 10.1245/s10434-011-2176-3. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Matsunoki A, Kaneko M, Saito K, Watanabe G, Minamoto T. Long interspersed nuclear element-1 hypomethylation is a potential biomarker for the prediction of response to oral fluoropyrimidines in microsatellite stable and CpG island methylator phenotype-negative colorectal cancer. Cancer Sci. 2011;102:166–174. doi: 10.1111/j.1349-7006.2010.01776.x. [DOI] [PubMed] [Google Scholar]
- Lee WS, Yun SH, Chun HK, Lee WY, Yun HR, Kim J, Kim K, Shim YM. Pulmonary resection for metastases from colorectal cancer: prognostic factors and survival. Int J Colorectal Dis. 2007;22:699–704. doi: 10.1007/s00384-006-0218-2. [DOI] [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunoki A, Kawakami K, Kotake M, Kaneko M, Kitamura H, Ooi A, Watanabe G, Minamoto T. LINE-1 methylation shows little intra-patient heterogeneity in primary and synchronous metastatic colorectal cancer. BMC Cancer. 2012;12:574. doi: 10.1186/1471-2407-12-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL, Kay E, Mitchell JK, Madi A, Jasani B, James MD, Bridgewater J, Kennedy MJ, Claes B, Lambrechts D, Kaplan R, Cheadle JP. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–2114. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008a;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008b;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, Capelinha AF, Duval A, Hamelin R, Machado JC, Schwartz S, Jr., Carneiro F, Seruca R. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26:158–163. doi: 10.1038/sj.onc.1209758. [DOI] [PubMed] [Google Scholar]
- Richman SD, Chambers P, Seymour MT, Daly C, Grant S, Hemmings G, Quirke P. Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost-effectiveness of multiple sample testing. Anal Cell Pathol (Amst) 2011;34:61–66. doi: 10.3233/ACP-2011-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- Shigaki H, Baba Y, Watanabe M, Miyake K, Murata A, Iwagami S, Ishimoto T, Iwatsuki M, Yoshida N, Baba H.2012KRAS and BRAF mutations in 203 esophageal squamous cell carcinomas: pyrosequencing technology and literature review Ann Surg Oncoldoi: 10.1245/s10434-012-2819-z [DOI] [PMC free article] [PubMed]
- Shigaki H, Baba Y, Watanabe M, Murata A, Ishimoto T, Iwatsuki M, Iwagami S, Nosho K, Baba H. PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res. 2013;19 (9:2451–2459. doi: 10.1158/1078-0432.CCR-12-3559. [DOI] [PubMed] [Google Scholar]
- Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- Vakiani E, Janakiraman M, Shen R, Sinha R, Zeng Z, Shia J, Cercek A, Kemeny N, D'Angelica M, Viale A, Heguy A, Paty P, Chan TA, Saltz LB, Weiser M, Solit DB. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Nordlinger B, Adam R, Kohne CH, Pozzo C, Poston G, Ychou M, Rougier P. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42:2212–2221. doi: 10.1016/j.ejca.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Worthley DL, Whitehall VL, Spring KJ, Leggett BA. Colorectal carcinogenesis: road maps to cancer. World J Gastroenterol. 2007;13:3784–3791. doi: 10.3748/wjg.v13.i28.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim KL. Microsatellite instability in metastatic colorectal cancer: a review of pathology, response to chemotherapy and clinical outcome. Med Oncol. 2012;29:1796–1801. doi: 10.1007/s12032-011-0050-6. [DOI] [PubMed] [Google Scholar]
- Yoo PS, Lopez-Soler RI, Longo WE, Cha CH. Liver resection for metastatic colorectal cancer in the age of neoadjuvant chemotherapy and bevacizumab. Clin Colorectal Cancer. 2006;6:202–207. doi: 10.3816/CCC.2006.n.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.