Abstract
Background:
Paired related homoeobox 1 (PRRX1) has been identified as a new epithelial-mesenchymal transition (EMT) inducer in breast cancer. However, the function of PRRX1 in colorectal cancer (CRC) has not been elucidated.
Methods:
We utilised ectopic PRRX1-expressing cell lines to analyse the function of PRRX1 in CRC. The clinical significance of PRRX1 was also examined on three independent CRC case sets.
Results:
PRRX1 induced EMT and the stem-like phenotype in CRC cells. In contrast to studies of breast cancer, abundant expression of PRRX1 was significantly associated with metastasis and poor prognosis in CRC.
Conclusion:
PRRX1 is an indicator of metastasis and poor prognosis in CRC cases. Further investigation is required to uncover the signalling network regulating PRRX1.
Keywords: colorectal cancer, PRRX1, epithelial-to-mesenchymal transition, EMT, metastasis
The first event in cancer metastasis is the movement of cancer cells away from the primary tumour tissue. Thereafter, cancer cells invade surrounding tissues and intravasate into vessels. Several reports have shown that the invasive capability of cancer cells is acquired by transformation to the mesenchymal phenotype (the epithelial-mesenchymal transition (EMT; Nieto, 2011). Moreover, EMT may produce cancer stemness properties (Alison et al, 2011). Inducers such as TWIST1, (Kang and Massague, 2004) SNAI1, (Pena et al, 2009) ZEB1, and ZEB2 (Chua et al, 2007) are reported to be associated with EMT in cancer cells.
Most recently, Ocana et al (2012) reported that a newly identified EMT inducer, paired related homoeobox 1 (PRRX1), promoted full EMT in cancer cells. However, they focused on the mesenchymal-to-epithelial transition (MET; Thiery et al, 2009) They showed that the loss of PRRX1 was required for cancer metastasis. Downregulation of PRRX1 caused the acquisition of MET. Low PRRX1 expression levels were significantly associated with metastasis and poor prognosis in the analysis of clinical samples of breast cancer and lung squamous cell carcinoma. Furthermore, the stemness phenotype was suppressed by PRRX1-induced EMT, and PRRX1 had to be downregulated to activate stem cell properties and allow colonisation. These findings were clearly distinct from previous analyses of EMT and the stemness phenotype. We noted that the function of PRRX1 in colorectal cancer (CRC) cells had not been elucidated. Thus, the purpose of the present study was to verify that PRRX1 induced EMT in CRC cells. We also examined the clinical significance of PRRX1 in CRC cases.
Materials and methods
Patients and sample collection
A total of 173 CRC samples were obtained during surgery. All patients underwent resection of the primary tumour at Kyushu University Beppu Hospital and affiliated hospitals between 1992 and 2007. Full informed consent was obtained from all individuals according to the local ethical review board approval. Detailed information is described in the Supplementary Material.
Cell lines
The human CRC cell lines DLD-1 and COLO-205 were obtained from the Japanese Cancer Research Bank (Tokyo, Japan) and maintained in RPMI 1640 medium containing 10% foetal bovine serum, 100 units per ml penicillin, and 100 μg ml−1 streptomycin sulphate. All cells were cultured in a humidified 5% CO2 incubator at 37 °C.
PRRX1 expression lentiviral vector
To generate PRRX1 expression lentiviral vectors, we amplified the insert (full-length human PRRX1; NM_022716.2) by PCR from human reference cDNA. Lentiviruses were produced by transient transfection of HEK293T cells with pCMV-VSV-G-RSV-Rev, pCAG-HIVgp, and either CSII-CMV-HOTAIR or CSII-CMV-MCS (empty) plasmid DNAs (5′-XhoI and 3′-EcoRI sites) plus Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol.
Quantitative real-time reverse-transcription (qRT)-PCR
qRT-PCR was performed in a LightCycler 480 instrument (Roche Applied Science, Basel, Switzerland) using a LightCycler 480 Probes Master kit or LightCycler 480 SYBR Green I Master kit (Roche Applied Science), following the manufacturer's protocol. The detailed protocol and the primer sequences used here are described in the Supplementary Material and Supplementary Table 1.
Immunoblotting
Total protein was extracted from PRRX1-expressing cell lines and mock cell lines. Aliquots of total protein (40 μg) were electrophoresed in 10% concentrated polyacrylamide gel, and then electrophoresed and electroblotted as previously described (Ieta et al, 2007). Detailed information can be found in the Supplementary Material.
Cell proliferation and cancer cell invasion assays
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (Roche Applied Science) was used to evaluate cell proliferation. The BD BioCoat Tumor Invasion System, 8 μm pores (BD Bioscience, San Jose, CA, USA), was used to evaluate invasive capacity, following the manufacturer's protocol. Detailed information is provided in the Supplementary Material.
Soft agar and sphere formation assay
Detailed information is provided in the Supplementary Material.
Meta-analysis and gene set enrichment analysis
We obtained CRC expression profiles from the National Center for Biotechnology Information Gene Expression Omnibus database (accession codes GSE17536 and GSE14333) and analysed these expression profiles using gene set enrichment analysis (GSEA; Subramanian et al, 2005). Detailed information is available in the Supplementary Material.
Statistical analysis
For continuous variables, data were expressed as means±s.d. The relationship between PRRX1 mRNA expression and clinicopathological factors was analysed using a χ2-test and a Student's t-test. Findings were considered significant when the P-value was <0.05. All tests were performed using JMP software (SAS Institute Inc., Cary, NC, USA).
Results
Ectopic PRRX1 promoted the EMT in CRC cells
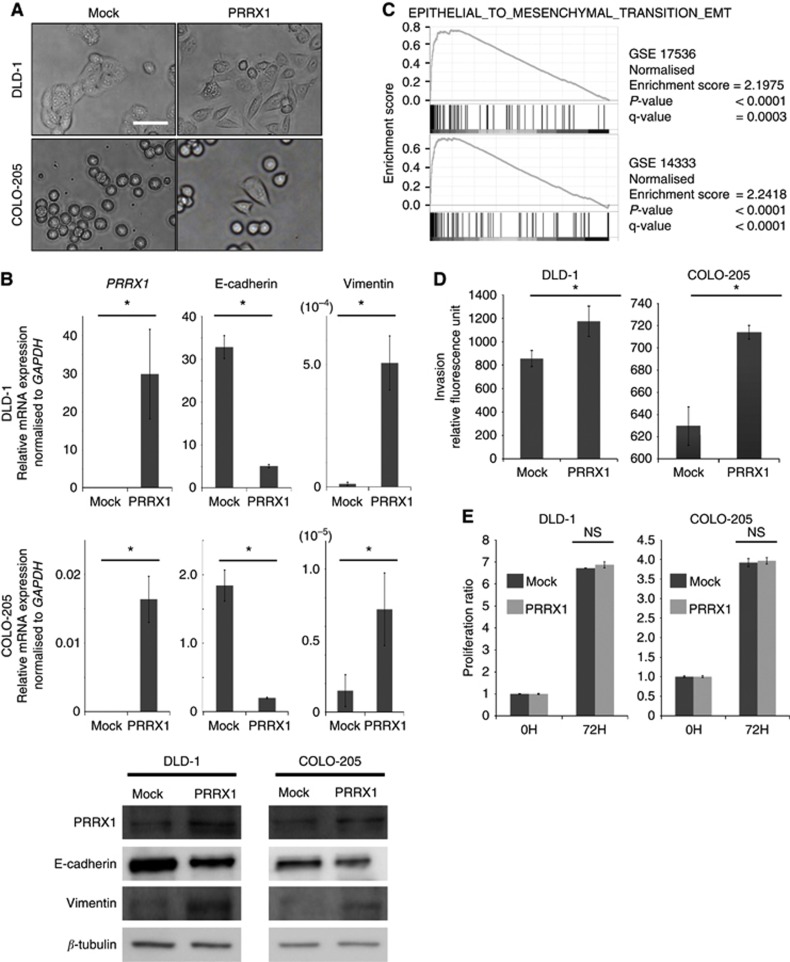
To examine whether PRRX1 induced EMT in CRC cells, we utilised ectopic PRRX1-expressing CRC cell lines. PRRX1-expressing cells showed a more spindle-like shape than did mock cells (Figure 1A). Both PRRX1-expressing cell lines had a significantly lower level of E-cadherin gene expression and a higher level of vimentin gene expression than did mock cells (Figure 1B). We also analysed the expression of well-known EMT transcription factors (Supplementary Figure 1). The data revealed that ectopic expression of PRRX1 significantly affected the expression of EMT transcription factors TWIST1 and ZEB1, but the directionality of the changes was not consistent between the cell lines. However, GSEA on two published clinical data sets of CRC (GSE17536 (Smith et al, 2010) and GSE14333 (Jorissen et al, 2009)) suggested that PRRX1 expression was significantly correlated with the EMT signature (Figure 1C). To further investigate the participation of PRRX1 in cancer invasiveness, invasion assays were carried out. PRRX1-expressing cells were more invasive than were mock cells in both cell lines (Figure 1D). On the other hand, cell proliferation was not affected by ectopic PRRX1 expression (Figure 1E). Our data indicated that PRRX1 enhanced the invasive CRC phenotype via EMT induction in vitro.
Figure 1.
PRRX1 expression supports an invasive and mesenchymal phenotype in CRC cells. (A) Phase-contrast images showing the phenotype of mock-transfected cells of DLD-1 and COLO-205 cells or of those in which PRRX1 has been ectopically expressed. Scale bars: 100 μm. (B) Real-time RT–PCR (upper panel, DLD-1; middle panel, COLO-205) and western blot (lower panel: left, DLD-1; right, COLO-205) analyses of mock and PRRX1-expressing cells show the repression of E-cadherin and the activation of vimentin following PRRX1 ectopic expression. (C) Enrichment plots of gene expression signatures for ‘EPITHELIAL_TO_MESENCHYMAL_TRANSITION_EMT' according to PRRX1 expression levels with data sets, GSE17536, and GSE14333. (D) Invasive properties of DLD-1 cells (left panel) and COLO-205 cells (right panel), with or without PRRX1 expression. (E) Cell proliferation ratio of DLD-1 cells (left panel) and COLO-205 cells (right panel) with or without PRRX1 expression. Histograms represent the means±s.d. of three independent experiments. *statistically significant (P⩽0.05).
Ectopic PRRX1 promoted the stemness phenotype and anchorage-independent growth of CRC cells
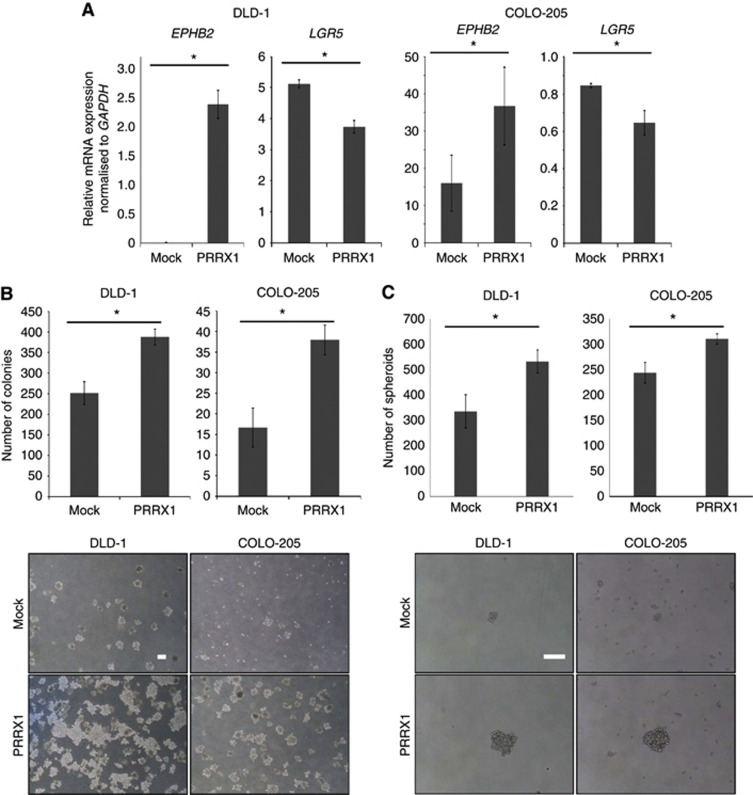
With regard to the previously reported stemness properties induced by loss of PRRX1 (Ocana et al, 2012), we verified the association between expression of PRRX1 and representative CRC stem cell markers (Barker et al, 2009; Merlos-Suarez et al, 2011). Ectopic PRRX1 expression upregulated EPHB2, an intestinal stem cell marker, but LGR5 was adversely decreased (Figure 2A). Next, a sphere formation assay was carried out to investigate the impact of PRRX1 on the stem-like phenotype of CRC cells. PRRX1-expressing cell lines showed a significant increase in colony formation compared with mock cells (Figure 2B). Further, soft agar colony-formation assays revealed the higher capability of anchorage-independent growth of PRRX1-expressing cells compared with mock cells (Figure 2C). Therefore, the current findings indicated that PRRX1 induced the stemness phenotype in CRC in at least one pathway.
Figure 2.
PRRX1 led CRC cells to express stem-like properties. (A) Real-time RT–PCR (upper panel, DLD-1; lower panel, COLO-205) analyses of mock and PRRX1-expressing cells on EPHB2 and LGR5, representative CRC stem cell markers. (B) Upper panel, quantitation of the soft agar colony-formation assay. Lower panel, representative images depicting colonies formed by mock and PRRX1-expressing cells. Scale bars: 100 μm. (C) Upper panel, sphere formation assay. Lower panel, representative images depicting spheroids formed by mock and PRRX1-expressing cells. Scale bars: 100 μm. Histograms represent the means±s.d. of three independent experiments. *statistically significant (P⩽0.05).
Abundant PRRX1 expression was correlated with poor prognosis and metastasis in CRC cases
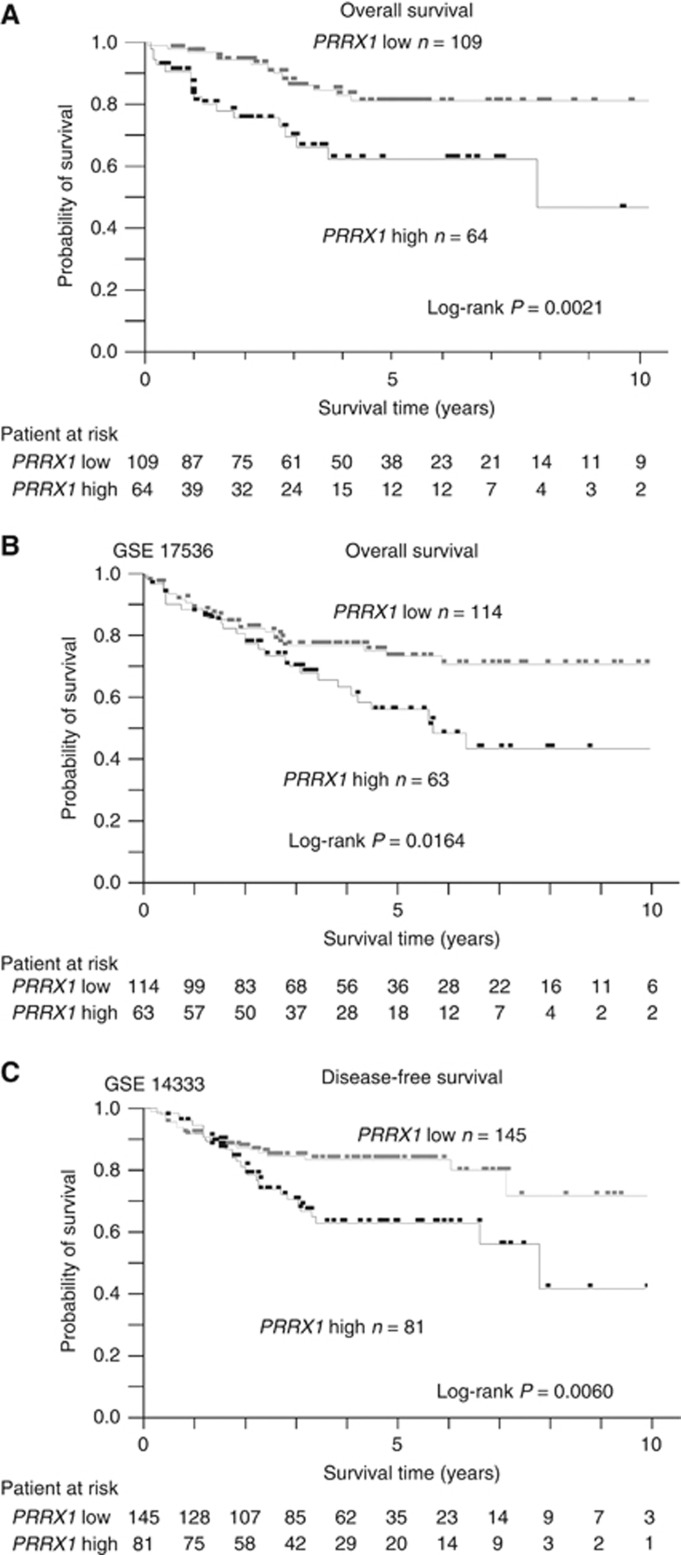
To determine how PRRX1 expression influenced cancer metastasis and disease prognosis in CRC, we analysed the relationship between the PRRX1 expression level, clinicopathological factors, and prognostic data. PPRX1 mRNA expression levels of 173 CRC tissues were assessed by qRT-PCR. Cases were subdivided into two groups according to PRRX1 mRNA expression level by the minimum P-value approach, which is a comprehensive method to find the optimal risk separation cut-off point in continuous measurements (Mizuno et al, 2009). The PRRX1 high expression group (N=64) had a significantly poorer overall survival rate than did the PRRX1 low expression group (N=109) (Figure 3A). Clinicopathological factors were compared between the high and low PRRX1 mRNA expression groups (Supplementary Table 2). The high PRRX1 expression group showed a higher risk for invasion. In addition, univariate and multivariate analyses of lymph node metastasis revealed that the PRRX1 expression level was an independent indicator for lymph node metastasis (Supplementary Table 3). However, there was a discrepancy between our data on CRC and those of Ocana et al (2012) who showed that the low expression group had a significantly poorer prognosis in breast cancer and lung squamous cell carcinoma. We also analysed two other independent data sets described above (GSE17536 and GSE14333). In accordance with our results, the high PRRX1 expression group showed poorer prognosis than did the low PRRX1 expression group in both data sets (Figures 3B and 2C).
Figure 3.
Abundant PRRX1 expression results in higher metastasis rate and poor prognosis in CRC cases. (A) Kaplan–Meier overall survival curves for 173 patients with CRC according to PRRX1 mRNA expression. (B) Kaplan–Meier overall survival curves for 177 patients with CRC (GSE17536) according to PRRX1 expression. (C) Kaplan–Meier disease-free survival curves for 226 patients with CRC (GSE 14333) according to PRRX1 expression. *statistically significant (P⩽0.05).
Discussion
We have shown that PRRX1, a newly identified EMT inducer, enhances the mesenchymal and stem-like phenotype of CRC cells. Moreover, high expression of PRRX1 was significantly correlated to metastasis and poor prognosis. Our results on the relationship between PRRX1 expression and cancer prognosis differ substantially from those of Ocana et al (2012). Thus, our results suggest that PRRX1 expression is associated with different phenotypes in different types of cancer cells. For example, breast cancer cells acquire MET and stem-like phenotypes by loss of PRRX1(Ocana et al, 2012). In contrast, CRC cells show EMT and stem-like phenotypes when they express PRRX1. Our data on CRC agree with a very recent report that showed that PRRX1 has a central role in pancreatic regeneration and carcinogenesis (Reichert et al, 2013). In the current study, two PRRX1-expressing cell lines differed in their expression profiles of EMT transcription factors (Supplementary Figure 1). We speculate that this is because PRRX1 induces EMT independently of classical EMT pathways as shown by Ocana et al (2012). The pathways activating EMT and stemness properties in intestinal tissue (Yan et al, 2012; Zhu et al, 2013) might be diverse, and further studies are required.
In the current study, we focused on expression of PRRX1, a gene that regulates EMT and stem cell properties. Our results demonstrated different roles for PRRX1 in CRC from those in breast cancer. It has been suggested that transforming growth factor-β (Ocana et al, 2012) and microRNA (Yang et al, 2013) regulate the expression of PRRX1. However, details are lacking and further analysis is required to uncover the signalling network regulating PRRX1. Such data will be important for the development of new therapeutic approaches to control cancer metastases.
Acknowledgments
This work was supported in part by the following grants and foundations: CREST, Japan Science and Technology Agency (JST) and the Funding Program for Next Generation World-Leading Researchers (LS094). We also thank T Shimooka, M Kasagi, T Kawano, and S Kono for their technical assistance and H Miyoshi (RIKEN BioResource Center) for providing lentivirus vector plasmid DNA.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Alison MR, Lim SM, Nicholson LJ. Cancer stem cells: problems for therapy. J Pathol. 2011;223 (2:147–161. doi: 10.1002/path.2793. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457 (7229:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26 (5:711–724. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- Ieta K, Ojima E, Tanaka F, Nakamura Y, Haraguchi N, Mimori K, Inoue H, Kuwano H, Mori M. Identification of overexpressed genes in hepatocellular carcinoma, with special reference to ubiquitin-conjugating enzyme E2C gene expression. Int J Cancer. 2007;121 (1:33–38. doi: 10.1002/ijc.22605. [DOI] [PubMed] [Google Scholar]
- Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhoffer M, Orntoft TF, Andersen CL, Gruidl M, Kamath VP, Eschrich S, Yeatman TJ, Sieber OM. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15 (24:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118 (3:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Munoz P, Clevers H, Sancho E, Mangues R, Batlle E. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell stem cell. 2011;8 (5:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Ann Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer cell. 2012;22 (6:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Pena C, Garcia JM, Larriba MJ, Barderas R, Gomez I, Herrera M, Garcia V, Silva J, Dominguez G, Rodriguez R, Cuevas J, de Herreros AG, Casal JI, Munoz A, Bonilla F. SNAI1 expression in colon cancer related with CDH1 and VDR downregulation in normal adjacent tissue. Oncogene. 2009;28 (49:4375–4385. doi: 10.1038/onc.2009.285. [DOI] [PubMed] [Google Scholar]
- Reichert M, Takano S, von Burstin J, Kim SB, Lee JS, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim AD, Stanger BZ, Rustgi AK. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 2013;27 (3:288–300. doi: 10.1101/gad.204453.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, Eschrich S, Kis C, Levy S, Washington MK, Heslin MJ, Coffey RJ, Yeatman TJ, Shyr Y, Beauchamp RD. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138 (3:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102 (43:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139 (5:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, Kuo CJ. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109 (2:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, Cogdell D, Nykter M, Broaddus R, Rodriguez-Aguayo C, Lopez-Berestein G, Liu J, Shmulevich I, Sood AK, Chen K, Zhang W. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer cell. 2013;23 (2:186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang YF, Kek C, Bulavin DV. Apoptosis differently affects lineage tracing of lgr5 and bmi1 intestinal stem cell populations. Cell stem cell. 2013;12 (3:298–303. doi: 10.1016/j.stem.2013.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.