Abstract
Cellular membrane deformation and the associated redistribution of membrane-bound proteins are important aspects of membrane function. Current model membrane approaches for studying curvature sensing are limited to positive curvatures, and often require complex and delicate experimental setups. To overcome these challenges, we fabricated a wavy substrate imposing a range of curvatures onto an adhering lipid bilayer membrane. We examined the curvature sorting of several peripheral proteins binding to the wavy membrane and observed them to partition into distinct regions of curvature. Furthermore, single molecule imaging experiments suggested that curvature sensing of proteins on low-curvature substrates requires cooperative interactions.
Keywords: supported lipid bilayers, curvature sorting, peripheral proteins, N-BAR, ENTH, CTB
INTRODUCTION
Active deformation of cell membranes is an important aspect of their functions. Transitions between different shapes have been postulated to be accompanied by and controlled through the sorting of phospholipids and membrane proteins1. Growing support for this notion has been contributed by research on the membrane curvature sensing and generation2, 3 of a myriad of peripheral membrane proteins4–7. The malfunctions of curvature-sensing proteins have been implicated in defective cellular functions and diseases8–11.
For example, the protein Epsin is believed to be involved in clathrin-mediated endocytosis12, and the distribution of ENTH (Epsin N-terminal homology) domains on membranes has been found to be sensitive to membrane curvature13. Furthermore, N-terminal helix containing BINAmphiphysin- Rvs (N-BAR) domains, forming crescent-shaped dimers4, 5 consisting of α-helical bundles, have been shown to preferentially localize on positively curved membranes7, 14. Electron microscopy (EM) studies have furthermore demonstrated that these proteins can tubulate liposomes4, 5, 15, 16.
In addition to positive curvature sensitivity, recent studies have demonstrated the localization of cell divisions protein DivIVA in the negatively curved intracellular leaflets of the poles of Bacillus subtilis cells17, 18. Furthermore, bacterial toxins, including cholera toxin and Shiga toxin, were reported to induce inward membrane invaginations19, 20 and to segregate away from positive curvature regions in vitro21, 22.
To investigate the mechanisms of membrane curvature sensing and generation, a variety of experimental approaches have been established, including a recently developed biochemical vesiculation assay23, the single liposome curvature (SLiC) assay14, and a membrane tether system where cylindrically shaped membranes are pulled from pipette-aspirated giant unilamellar vesicles (GUVs)21, 24. Shortcomings of all of these systems are that only positively curved outer leaflets can be accessed with ease, these techniques tend to be time consuming and technically challenging, and some of them suffer from low signal-to-noise ratios.
In order to overcome these challenges, we engineered a solid-supported wavy membrane to quantitatively investigate the distribution of several peripheral membrane proteins in a spatially varying membrane curvature field. Two-dimensional solid-supported curved membranes have previously been fabricated on polydimethylsiloxane25, 26 and quartz surface27 to examine the spatial distributions of liquid ordered and disordered lipid phases. Here we use a related approach to demonstrate that an engineered wavy platform with continuous curvatures bearing positive and negative regions allows evaluation of the curvature sensitivities of a variety of proteins. We compare findings for this wavy surface to the more established tether/ GUV protein sorting system.
RESULTS AND DISCUSSION
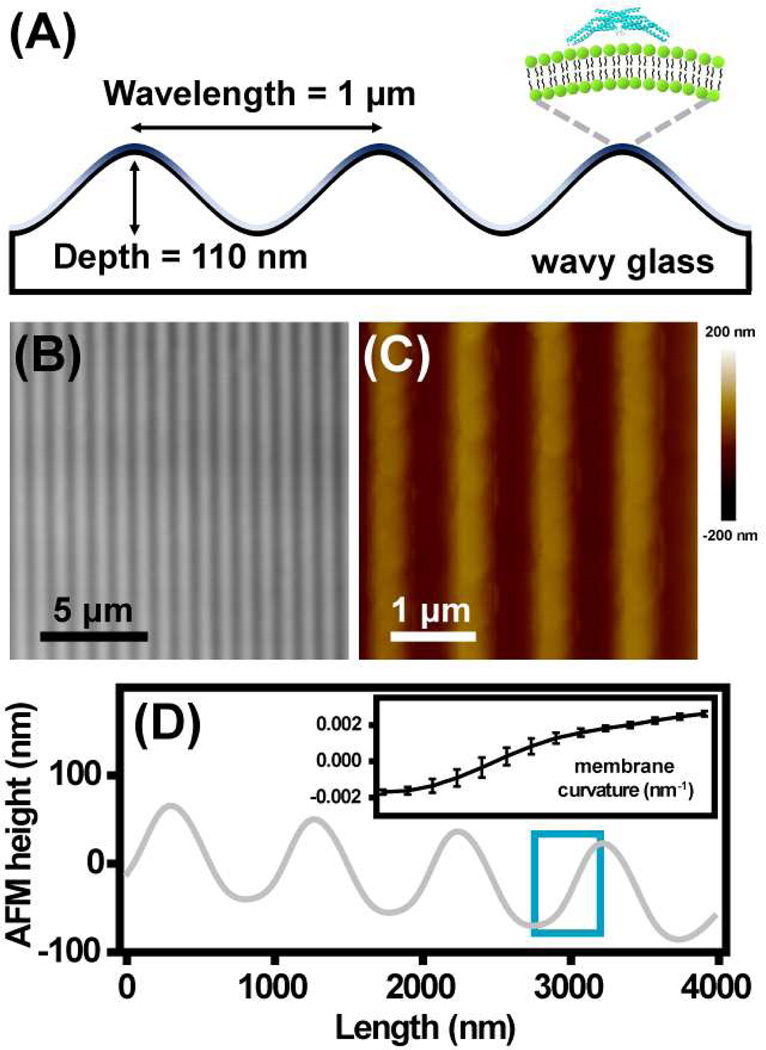
In order to study curvature sorting of peripheral proteins among regions of both positive and negative membrane curvature, a wavy glass substrate was fabricated via a combination of photolithography and wet etching to generate periodic topographic features with wavelength of about 1 µm and depth of 110 nm (Fig. 1A-1B). The surface topography was characterized using atomic force microscopy (AFM) (Fig. 1C-1D) which allowed computation of local surface curvatures (inset in Fig. 1D). The variation in surface area density caused by the projection of the wavy surface into the imaging plane26 was estimated to be 0.3%. As becomes more obvious below, this effect is small compared to fluorescence patterns resulting from curvature partitioning. Wavy fluid membranes composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) were obtained by the fusion of small unilamellar vesicles (SUVs) onto the glass surfaces.
Figure 1. Surface topography of wavy glass surface.
(A) Schematic cross-section view of the surface with topography pattern of a wavelength of 1 µm and depth of 110 nm supporting a fluid lipid bilayer membrane. (B) Transmitted light and (C) AFM images of a wavy glass surface with average depth of 110 nm. (D) AFM height profile quantified from (C). Note that the aspect ratio of the surface profile is adjusted for display purposes. The average curvature range within a half wavelength highlighted in the cyan box is shown in the inset. The error bars represent standard deviations from five regions with identical curvatures on a single substrate.
We examined the lateral distributions of ENTH, N-BAR domains, and cholera toxin subunit B (CTB) on wavy membranes to evaluate the effect of signs of curvatures. Proteins bearing fluorescent tags were incubated with supported wavy membranes, and the spatial distribution was visualized via confocal fluorescence microscopy. ENTH domains exhibited preferential partitioning into positively curved membrane regions (Fig. 2A). For ENTH experiments, membranes consisted of 1% L-α-phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) in a background of DOPC lipids. This curvature-dependent distribution was not observed for the fluorescent PtdIns(4,5)P2 analog BODIPY-TMR PtdIns(4,5)P2 embedded in the curved membranes (in the absence of ENTH), indicating that protein sorting was not influenced by the distribution of PtdIns(4,5)P2 (data not shown, but confer Ref.13). The N-BAR domains of both Endophilin and BIN1 (Bridging INtegrator 128, also called amphiphysin 2) preferentially distributed into positively curved membrane regions (Fig. 2B-2C). For Endophilin and BIN1 experiments, membranes were composed of 5% 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) in a background of DOPC. The low content of negatively charged lipids was chosen in order to maintain a large fraction of mobile proteins and lipids (see below). In striking contrast, CTB was found to partition into negatively curved membrane regions (Fig. 2D). For CTB membranes consisted of a mixture of 1% GM1 and 99% DOPC.
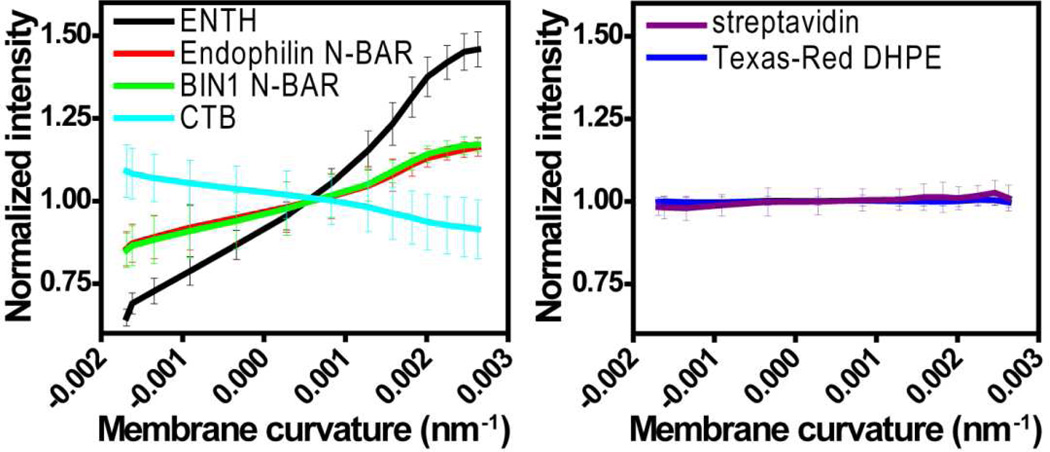
Figure 2. Curvature sensing proteins exhibit preferential partitioning on wavy membranes.
Proteins were incubated on fluid wavy membranes (containing a variety of negatively charged phospholipids or GM1 in a background of DOPC lipids) and visualized via confocal fluorescence microscopy. (A) Partitioning of ENTH-GFP into positive curvature regions identified by transmitted light imaging (see upper edge). Fluorescence distributions of (B) Endophilin N-BAR-Alexa Fluor 488 and (C) BIN1 N-BAR-Alexa Fluor 488 showing enrichment in positive curvature regions. (D) Preferential partitioning of CTB-Alexa Fluor 555 into negative curvature regions identified via transmitted light imaging (see upper edge). No significant curvature preference was observed for (E) streptavidin-FITC bound to membranes containing 1% cap-biotin PE, and (F) 0.1% of the lipid fluorophore Texas-Red DHPE in a wavy DOPC membrane.
In line with published results21, 29, two molecules used as controls, streptavidin (protein) and Texas-Red DHPE (lipid), were observed to be insensitive to membrane curvatures (Fig. 2E-2F).
In order to quantify protein distributions with respect to membrane curvature, confocal images were normalized with respect to the average fluorescence intensity of the image frame. Averaging the normalized intensities in corresponding curvature regions substantially improved signal-to-noise ratios. It was found that protein density varied monotonically in the curvature range under investigation, yielding an increase of the normalized (relative to the average) fluorescence intensity from 0.79 (at the most negative curvature) to 1.46 (at the most positive curvature) for ENTH, 0.92 to 1.16 for Endophilin N-BAR, 0.91 to 1.17 for BIN1 N-BAR, and a decrease from 1.05 (at the most negative curvature) to 0.91 (at the most positive curvature) for CTB (left panel of Fig. 3). In contrast, streptavidin and Texas-Red DHPE showed little correlation with membrane curvature (right panel of Fig. 3).
Figure 3. Analysis of protein and lipid distributions with respect to membrane curvatures.
Fluorescence images from confocal microscopy were normalized to the mean intensity of the image and analyzed as a function of membrane curvature. Left panel: ENTH (black), Endophilin N-BAR (red), BIN1 N-BAR (green), and CTB (cyan). Right panel: streptavidin (purple), and Texas-Red DHPE (blue). Error bars represent standard deviations from at least six different regions on the substrate for three different bilayer preparations.
Interestingly, quantitative analysis of protein fluorescence intensities revealed higher curvature sensitivity of ENTH domains compared to Endophilin N-BAR and BIN1 N-BAR (left panel of Fig. 3) despite the fact that for Endophilin two curvature sorting mechanisms (scaffolding and amphipathic helix insertion) might be simultaneously at work, whereas for ENTH likely only one sorting mechanism (amphipathic helix insertion) applies23.
The negative curvature partitioning of CTB is in accordance with its crystal structure21, 30, which suggests negative (convex) curvature of the membrane binding interface of the protein. Patchy features observed in CTB fluorescence distributions (Fig. 2D) were attributed to aggregation of the CTB-GM1 complex, consistent with previously reported well-defined clusters observed in AFM studies31.
The interpretation of fluorescence intensity distributions as resulting from thermodynamic equilibration requires the experimental demonstration that protein locations equilibrate on accessible time scales. To so do, we examined the lateral mobility of proteins on wavy membranes. The lateral diffusion of ENTH domains was investigated via fluorescence recovery after photobleaching (FRAP) measurements using our solid-supported wavy membrane and was compared to measurements in a tether-GUV system (Fig. 4A-4B). ENTH domains showed high (> 0.97) recovery fractions in both systems, with diffusion coefficients of 1.4 and 2.9 µm2/s in wavy membrane and tether-GUV system, respectively. Similar diffusion coefficients and recovery fractions were found for N-BAR domain proteins on wavy membranes (not shown).
Figure 4. ENTH domain exhibits lateral mobility in the wavy membrane and tether-GUV systems.
(A) Time-lapse recovery of ENTH was examined from 2D photobleaching experiments. Error bars represent standard deviation from three bilayer preparations and five measurements in total. (B) Time-lapse recovery fraction of ENTH bound on the membrane tether generated from single GUV containing Texas-Red DHPE (red) and ENTH-GFP (green). Error bars represent standard deviations from three tethers. Gray lines in (A) and (B) represent the best fit curves. (C) Two representative trajectories of single ENTH domains on the wavy membrane. ENTH domain was observed to diffuse across both the positive-curvature (red dashed lines) and negative-curvature regions (blue dashed lines) of the wavy membrane. (D) The spatial distribution of single moving ENTH domains on the wavy membrane with positive and negative curvature regions. More than 400 steps and three different wavy membranes were analyzed.
Motivated by recent reports on concentration-dependent curvature sorting mechanisms6, 32, we next evaluated sorting behavior through single molecule tracking experiments. In order to enable single molecule imaging, a low protein solution concentration of 0.05 nM was used. Single ENTH domains were observed to diffuse across both positive and negative curvature regions (Fig. 4C). The spatial probability distribution (Fig. 4D) shows that single ENTH protein domains were not preferentially localized in certain curvature regions, contrary to our observations from ensemble imaging experiments (left panel of Fig. 3). In order to be able to compare membrane coverage fractions comparing single molecule and ensemble imaging experiments, membrane binding isotherms were measured, resulting in a dissociation constant of 290 nM for ENTH (Fig. S1), comparable to a value measured via surface plasmon resonance33. Considering our protein solution concentration range for ensemble experiments, it follows that between 14 and 25% of available binding sites were covered (similar coverage fractions were determined for N-BAR domain proteins; not shown). This coverage range can be compared to a protein coverage fraction of ~0.01% for single molecule imaging experiments; i.e., the protein density in single molecule and ensemble imaging experiments differed by three orders of magnitude. Since single protein molecules in the present work were not observed to sense membrane curvature on low curvature surfaces contrary to our ensemble observations, there appears to be a threshold of protein surface coverage beyond which proteins start sensing membrane curvatures cooperatively on low curvature surfaces.
Solid-supported lipid bilayer membranes bear the potential for defect formation and subsequent protein immobilization in those defects. If defects were distributed preferentially in particular curvature regions, this could falsely suggest membrane mediated curvature sensing of proteins. Single molecule imaging also allowed us to examine the locations of immobile molecules. No preferential binding of immobile proteins to particular geometries suggested negligible possibility of permanent trapping of molecules in specific curvature ranges (Fig. S2). This observation further supports the notion that the fluorescence distributions reported here reflect protein distributions at thermodynamic equilibrium.
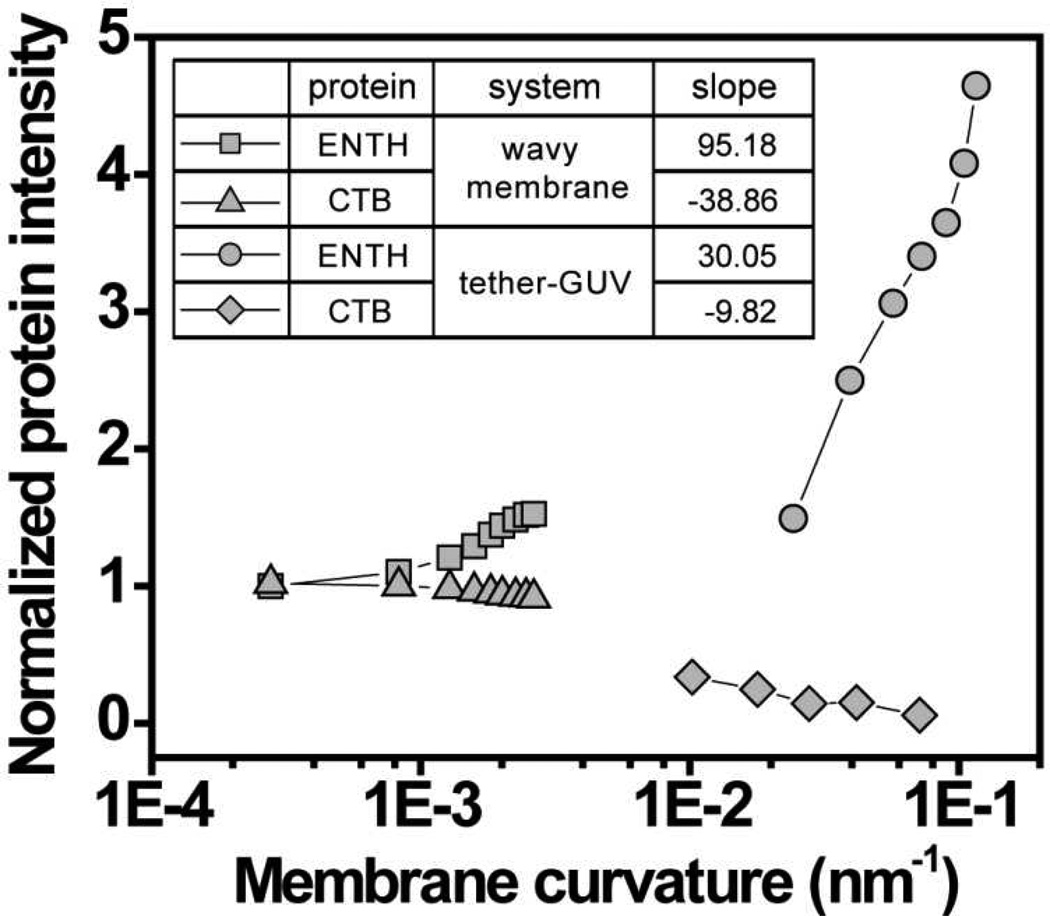
Next, we compared curvature sensing between free floating and solid-supported membrane systems. The sensing ability of solid-supported wavy membranes was compared to a tether-GUV system, where a high curvature membrane was generated by pulling a tubule from a single GUV13, 21. Note that membrane curvatures accessible in the tether-GUV system range from roughly 0.01 to 0.11 nm−1, compared to −0.002 to 0.003 nm−1 on wavy membranes. For both ENTH and CTB, solid-supported wavy membranes were observed to possess higher sorting ability compared to tether-GUV system, resulting in higher slopes of normalized fluorescence intensities as a function of membrane curvature (Fig. 5). One important difference between these two systems is that out-of-plane undulations that occur in freely suspended membranes are significantly suppressed in bilayers strongly adhering to a solid substrate. It can be hypothesized that the membrane curvature stored in undulations leads to a renormalization of curvature sorting coefficients that would be found for a non-undulating membrane. However, future research will have to further explain differences in curvature sensitivity of proteins on solid supported versus freely suspended membranes.
Figure 5. Solid-supported wavy membrane shows higher capacity for curvature sorting compared to tether-GUV system.
Normalized ENTH (squares) and CTB (triangles) intensities as a function of membrane curvature in solid-supported wavy membrane were compared with tether-GUV system (circles and diamonds; data reproduced from refs. 13 and 21, respectively). Experimental data points were normalized to the value found for zero curvature, and only the positive curvature regime is represented, in logarithmic form. The inset compares the slopes of linear sorting versus curvature plots.
CONCLUSIONS
In this work, we have fabricated a wavy membrane system which can be utilized as a platform for investigating the sorting of curvature-sensing molecules. The ability to establish continuous curvatures including negative and positive regimes on a single substrate allowed us to study molecules with a variety of curvature preferences. We demonstrated that ENTH and N-BAR domains prefer to partition into positive curvature areas, while CTB partitions into negative ones. Single molecule tracking results suggested that curvature sorting on substrates with low membrane curvature requires protein cooperativity. Due to its solid-substrate based nature, our system may also serve as curvature-differentiated patterned substrates to study the redistribution of cell surface receptors in live cells. Moreover, these wavy membranes could provide insights for studying cellular mechanical responses in different curvature regions.
MATERIALS AND METHODS
Protein purification and fluorophore labeling
ENTH-GFP was purified as previously described13. BIN1 and Endophilin N-BAR domains were expressed as GST fusion proteins. In Endophilin N-BAR, the mutation A247C using site-directed mutagenesis allowed fluorophore labeling (in a protein where the natural cysteine was eliminated through a C108S mutation). For protein expression, BL21-Codon Plus (DE3)-RIL cells (Stratagene, La Jolla, CA) were transformed with the plasmid of interest, grown in LB media, induced with isopropyl-β-D-thiogalactoside (IPTG), and harvested via centrifugation. Pellets were resuspended in a pH 7.4 buffer containing 300 mM NaCl, 20 mM Tris, 1 mM dithiothreitol (Fisher, Pittsburgh, PA), and 0.2 mM phenylmethylsulfonyl fluoride (Thermo Scientific, Logan, UT). The cells were lysed by tip-sonication and centrifuged at 4 °C. The supernatant was applied to a GST affinity column (GE, Piscataway, NJ), followed by protease digestion to cleave off the GST moiety. The product was further purified by ion-exchange chromatography. Alexa Fluor 488 fluorophore was conjugated to cysteine residue 247 in Endophilin N-BAR and endogenous cysteine residues at positions 57 and 95 in BIN1 N-BAR via Alexa Fluor 488-C5-maleimide (Invitrogen, Carlsbad, CA).
Wavy glass surface
An SU8 2 (MicroChem, Newton, MA) diluted to 50 wt% by gamma-Butyrolactone thin film on a clean glass substrate was fabricated via spin coating at 3000 rpm for 30 s. The cover slip was then baked at 65 °C for 1 min and at 95 °C for another 1 min (soft bake). Subsequently, a PDMS mold with the desired spatial features34 was imprinted onto the cover slip using capillary force lithography34, 35. The substrate was illuminated with UV (97435 Oriel flood exposure source with 6285 Newport 500W mercury lamp) at 100 mJ/cm2, followed by post exposure bake (PEB) at 65 °C for 1 min and 95 °C for another 1 min. Next, the substrate was heated at 200 °C for 30 min (hard bake). It was then etched with a CF4 and O2 gas plasma (Trion Technology, Clearwater, FL) until the glass surface was partially exposed (the remaining SU8 film creates pattern features during wet etch). The substrate was immersed in a buffer oxide etch (BOE, consisting of six parts of 40% NH4F and one part of 49% HF solutions) for 2–3 min (wet etch). The remaining SU8 2 film on the substrate was then removed by plasma etching for 20 min. To smooth any rough edges produced by the anisotropic etch, the cover slip was immersed in BOE for 30 s36. Transmitted light images were registered using fluorescent microspheres to assign regions of valleys and hills on the wavy surface26.
Supported lipid bilayer
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), L-α- phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) (Brain, ammonium salt), 1,2-dioleoyl-sn-glycero- 3-phospho-L-serine (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (Biotinyl Cap PE), brain ganglioside GM1, and extruder accessories were purchased from Avanti Polar Lipids (Alabaster, AL). Texas Red-1,2-dihexadecanoyl-sn-glycero-3- phosphoethanolamine triethylammonium salt (Texas-Red DHPE), streptavidin-FITC, and Alexa Fluor 555-conjugated CTB (CTB-A555) were purchased from Invitrogen (Carlsbad, CA). Calcium- and magnesium-free 150 mM NaCl and 7.5 mM phosphate pH 7.4 buffer was used in the preparation of vesicle solution.
Phospholipids were dissolved in chloroform and spread on the walls of a round bottom flask and evacuated in a desiccator to produce an even lipid film. The lipid film was rehydrated, sonicated for 30 min, extruded 21 times through a polycarbonate filter with 50 nm pores at room temperature. A supported lipid bilayer was formed by small unilamellar vesicle (SUV) fusion onto the hydrophilic wavy glass substrate. Before vesicle fusion, the wavy glass substrate was cleaned by sonicating sequentially in 2% Hellmanex solution (Hellma, Mullheim, Germany) for one hour, and 30 min each for water and ethanol. For the curvatures in our wavy surface (minimum curvature of radius ~500 nm), the adhesion energy between glass substrate and lipid bilayer is large enough to overcome the bilayer’s bending energy37. Thus, together with the recent finding showing the conformity of membranes to 100 nm silica beads38, we could infer that lipid bilayers follow the topography of the underlying wavy substrates.
Atomic force microscopy (AFM) and membrane curvature
AFM was performed using an Agilent 5420 microscope (Santa Clara, CA), and all accessories were purchased from Veeco (Plainview, NY). A silicon nitride probe with a spring constant of 3 N/m was used in tapping mode with scanning frequencies from 0.5–1.5 Hz. Surface curvature (C) was determined from the height profile (z) by using the definition of curvature in Eq. 1, where z′ and z″ are first and second derivatives with respect to plane coordinates, respectively.
| (Eq. 1) |
To evaluate the contribution from varying protein (or lipid) density due to the imaging of the surface topography, the area projection of the surface into a plane was calculated with the help of MATLAB (MathWorks, Natick, MA). The resulting spatially varying area density was then convolved with the point spread function of the confocal microscope.
Confocal microscopy
Protein or lipid partitioning on the wavy membrane was visualized by IX81 inverted confocal microscope (Olympus, Center Valley, PA) with Kalman imaging filter. The imaging chambers were produced from MatTek dishes (MatTek, Ashland, MA) by removing the original cover slip of the dish and replacing it with the fabricated wavy glass substrate attached to the dish through a thin layer of grease (Dow Corning, Midland, MI). The liquid volume covering the wavy surface was 200 µL. Fluid wavy membranes were incubated with 50–100 nM ENTH-GFP, 200–300 nM Endophilin N-BAR-Alexa Fluor 488, 200–300 nM BIN1 N-BAR-Alexa Fluor 488, or 150–500 nM CTB-Alexa Fluor 555 for 20 min. Images were normalized to the mean intensity of the frame and analyzed as a function of membrane curvature within a half wavelength.
Protein adsorption isotherm
To determine the coverage fraction of proteins on supported lipid bilayers, adsorption isotherms were determined. Supported lipid bilayers formed by vesicle fusion as mentioned above were incubated with a series of protein concentrations at room temperature. Fluorescence intensity of proteins on the bilayer surface was quantified by confocal microscopy imaging. The data was fitted with the Langmuir isotherm to obtain dissociation constant (Kd) and maximum surface coverage.
Single-molecule tracking
Dishes and buffer used for single-molecule imaging were illuminated via a 100 W UV lamp (Ultra Violet Products, Upland, CA) for at least 2 hours before fluorescence imaging experiments to reduce background fluorescence levels. Single molecules were imaged via a 60x 1.45NA TIRF lens (Olympus, Center Valley, PA) on an inverted microscope system (IX71, Olympus, Center Valley, PA) equipped with an EM-CCD camera (Hamamatsu, Bridgewater, NJ) using a 488 nm laser (50 mW, Coherent, Santa Clara, CA). Particle localization and tracking were performed with the help of MATLAB to generate single-molecule trajectories.
Fluorescence recovery after photobleaching (FRAP)
FRAP measurements were performed on an inverted IX71 fluorescence microscope (Olympus) equipped with an EM-CCD camera (Hamamatsu) for solid-supported membrane or on IX81 inverted confocal microscope (Olympus) for the tether-GUV system. Pre- and post- bleach images were acquired with attenuated illumination using an OD1.0 neutral density filter (Thorlabs, Newton, NJ). On the solid-supported membrane, a circular area with diameter of 21.6 µm was bleached for 30–60 sec. The recovery curve was fitted to a two-dimensional diffusion model by Soumpasis39 to obtain the diffusion coefficient (D). For the tether-GUV system, GUVs were prepared as described13, except that 50% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (Avanti) was included. The membrane tether was photobleached, and the recovery curve was fitted to a one-dimensional diffusion model32.
Supplementary Material
Acknowledgements
We thank Drs. W. Cho, P. De Camilli, and R. Langen for supplying expression plasmids, C. Zhu for assistance with data fitting, and Dr. Z. Fakhraai for AFM usage. Wavy surfaces were prepared at the Penn Wolf Nanofabrication Facility.
Funding Sources
NSF grant MCB 0718569 and PENN MRSEC No. DMR11-20901 (T.B.), and NIH Training Grant GM08275 (B.R.C.)
Footnotes
Supporting Information. The adsorption isotherm of ENTH domain; the distribution of immobile ENTH or N-BAR molecules via single-molecule tracking. This material is available free of charge via the Internet at http://pubs.acs.org.
REFENRENCES
- 1.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart T, Capraro BR, Zhu C, Das SL. Thermodynamics and Mechanics of Membrane Curvature Generation and Sensing by Proteins and Lipids. Annual Review of Physical Chemistry, Vol 62. 2011;Vol. 62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonny B. Mechanisms of Membrane Curvature Sensing. Annual Review of Biochemistry, Vol 80. 2011;Vol. 80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 4.Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, Langen R, T McMahon H. Mechanism of endophilin N-BAR domain-mediated membrane curvature. Embo Journal. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. Embo Journal. 2006;25:2889–2897. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorre B, Callan-Jones A, Manzi J, Goud B, Prost J, Bassereau P, Roux A. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:173–178. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinrich MC, Capraro BR, Tian AW, Isas JM, Langen R, Baumgart T. Quantifying Membrane Curvature Generation of Drosophila Amphiphysin N-BAR Domains. Journal of Physical Chemistry Letters. 2011;1:3401–3406. doi: 10.1021/jz101403q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verstreken P, Kjaerulff O, Lloyd TE, Atkinson R, Zhou Y, Meinertzhagen IA, Bellen HJ. Endophilin mutations block clathrin-mediated endocytosis but not neurotransmitter release. Cell. 2002;109:101–112. doi: 10.1016/s0092-8674(02)00688-8. [DOI] [PubMed] [Google Scholar]
- 9.Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, Kokunai Y, Tsuburaya R, de la Grange P, Dembele D, Francois V, Precigout G, Boulade-Ladame C, Hummel MC, de Munain AL, Sergeant N, Laquerriere A, Thibault C, Deryckere F, Auboeuf D, Garcia L, Zimmermann P, Udd B, Schoser B, Takahashi MP, Nishino I, Bassez G, Laporte J, Furling D, Charlet-Berguerand N. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nature Medicine. 2011;17:720–U112. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 10.Toussaint A, Cowling BS, Hnia K, Mohr M, Oldfors A, Schwab Y, Yis U, Maisonobe T, Stojkovic T, Wallgren-Pettersson C, Laugel V, Echaniz-Laguna A, Mandel JL, Nishino I, Laporte J. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta Neuropathologica. 2011;121:253–266. doi: 10.1007/s00401-010-0754-2. [DOI] [PubMed] [Google Scholar]
- 11.Rikhy R, Kumar V, Mittal R, Krishnan KS. Endophilin is critically required for synapse formation and function in Drosophila melanogaster. Journal of Neuroscience. 2002;22:7478–7484. doi: 10.1523/JNEUROSCI.22-17-07478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nature Reviews Molecular Cell Biology. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 13.Capraro BR, Yoon Y, Cho W, Baumgart T. Curvature Sensing by the Epsin N-Terminal Homology Domain Measured on Cylindrical Lipid Membrane Tethers. Journal of the American Chemical Society. 2010;132:1200–1201. doi: 10.1021/ja907936c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegard P, Gether U, Stamou D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. Embo Journal. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 16.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 17.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro L, McAdams HH, Losick R. Why and How Bacteria Localize Proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, Aly MRE, Fraisier V, Florent J-C, Perrais D, Lamaze C, Raposo G, Steinem C, Sens P, Bassereau P, Johannes L. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- 20.Ewers H, Romer W, Smith AE, Bacia K, Dmitrieff S, Chai WG, Mancini R, Kartenbeck J, Chambon V, Berland L, Oppenheim A, Schwarzmann G, Feizi T, Schwille P, Sens P, Helenius A, Johannes L. GM1 structure determines SV40-induced membrane invagination and infection. Nature Cell Biology. 2010;12:11–U36. doi: 10.1038/ncb1999. [DOI] [PubMed] [Google Scholar]
- 21.Tian A, Baumgart T. Sorting of Lipids and Proteins in Membrane Curvature Gradients. Biophysical Journal. 2009;96:2676–2688. doi: 10.1016/j.bpj.2008.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safouane M, Berland L, Callan-Jones A, Sorre B, Romer W, Johannes L, Toombes GES, Bassereau P. Lipid Cosorting Mediated by Shiga Toxin Induced Tubulation. Traffic. 2010;11:1519–1529. doi: 10.1111/j.1600-0854.2010.01116.x. [DOI] [PubMed] [Google Scholar]
- 23.Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon H, Kozlov M. Membrane Fission Is Promoted by Insertion of Amphipathic Helices and Is Restricted by Crescent BAR Domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorre B, Callan-Jones A, Manneville JB, Nassoy P, Joanny JF, Prost J, Goud B, Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramaniam AB, Lecuyer S, Ramamurthi KS, Losick R, Stone HA. Particle/Fluid Interface Replication as a Means of Producing Topographically Patterned Polydimethylsiloxane Surfaces for Deposition of Lipid Bilayers. Advanced Materials. 2010;22:2142–2147. doi: 10.1002/adma.200903625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanii B, Smith AM, Butti R, Brozell AM, Parikh AN. Bending membranes on demand: Fluid phospholipid bilayers on topographically deformable substrates. Nano Letters. 2008;8:866–871. doi: 10.1021/nl073085b. [DOI] [PubMed] [Google Scholar]
- 27.Parthasarathy R, Yu CH, Groves JT. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir. 2006;22:5095–5099. doi: 10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- 28.Ge K, Minhas F, Duhadaway J, Mao NC, Wilson D, Buccafusca R, Sakamuro D, Nelson P, Malkowicz SB, Tomaszewski J, Prendergast GC. Loss of heterozygosity and tumor suppressor activity of BIN1 in prostate carcinoma. International Journal of Cancer. 2000;86:155–161. doi: 10.1002/(sici)1097-0215(20000415)86:2<155::aid-ijc2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 29.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegard P, Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nature Chemical Biology. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 30.Chinnapen DJF, Chinnapen H, Saslowsky D, Lencer WI. Rafting with cholera toxin: endocytosis and trafficking from plasma membrane to ER. Fems Microbiology Letters. 2007;266:129–137. doi: 10.1111/j.1574-6968.2006.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Shi J, Parikh AN, Shreve AP, Chen LH, Swanson BI. Evidence for cholera aggregation on GM1-decorated lipid bilayers. Colloids and Surfaces B-Biointerfaces. 2004;33:45–51. [Google Scholar]
- 32.Zhu C, Das SL, T B. Nonlinear sorting, curvature generation, and crowding of Endophilin N-BAR on tubular membranes. Biophysical Journal. 2012;102:1837–1845. doi: 10.1016/j.bpj.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. Journal of Biological Chemistry. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- 34.Suh KY, Kim YS, Lee HH. Capillary Force Lithography. Advanced Materials. 2001;13:1386–1389. [Google Scholar]
- 35.Zhang Y, Lin C-T, Yang S. Fabrication of Hierarchical Pillar Arrays from Thermoplastic and Photosensitive SU-8. Small. 2010;6:768–775. doi: 10.1002/smll.200901843. [DOI] [PubMed] [Google Scholar]
- 36.Saragih AS, Ko TJ. Fabrication of passive glass micromixer with third-dimensional feature by employing SU8 mask on micro-abrasive jet machining. International Journal of Advanced Manufacturing Technology. 2009;42:474–481. [Google Scholar]
- 37.Cremer PS, Boxer SG. Formation and spreading of lipid bilayers on planar glass supports. Journal of Physical Chemistry B. 1999;103:2554–2559. [Google Scholar]
- 38.Mornet S, Lambert O, Duguet E, Brisson A. The formation of supported lipid bilayers on silica nanoparticles revealed by cryoelectron microscopy. Nano Letters. 2005;5:281–285. doi: 10.1021/nl048153y. [DOI] [PubMed] [Google Scholar]
- 39.Soumpasis DM. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophysical Journal. 1983;41:95–97. doi: 10.1016/S0006-3495(83)84410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.