Summary
To mine possibly hidden causal single nucleotide polymorphisms (SNPs) in the etiology of melanoma, we investigated the association of SNPs in 76 M/G1 transition genes with melanoma risk using our published genome-wide association study (GWAS) dataset with 1804 melanoma cases and 1,026 cancer-free controls. We found multiple SNPs with P < 0.01 and performed validation studies for 18 putative functional SNPs in PSMB9 in other two GWAS datasets. Two SNPs (rs1351383 and rs2127675) were associated with melanoma risk in the GenoMEL dataset (P = 0.013 and 0.004, respectively), but failed validation in the Australia dataset. Genotype-phenotype analysis revealed these two SNPs were significantly correlated with mRNA expression levels of PSMB9. Further experiments revealed that the promoter SNP rs2071480, which is in high LD with rs1351383 and rs2127675, involved in influencing transcription factor binding and gene expression. Taken together, our data suggested that functional variants in PSMB9 may contribute to melanoma susceptibility.
Keywords: GWAS, Cell cycle, PSMB9, Polymorphism, melanoma
Introduction
Although the number of nevi and sun exposure (sunburn sensitivity) as well as other pigmentation-related characteristics are the known risk factors for melanoma (Titus-Ernstoff et al., 2005), numerous studies have suggested that genetic variants in genes involved in several biologic pathways also have an impact on melanoma susceptibility (Curtin et al., 2005; Figl et al., 2010; Zhang et al., 2010). Recently, genome-wide association studies (GWASs) have identified several new promising causal genetic variants or loci for melanoma risk and verified a few genes that were identified in earlier studies of high-risk melanoma kindreds (Barrett et al., 2011; Bishop et al., 2009; Brown et al., 2008; Falchi et al., 2009; Macgregor et al., 2011). However, these GWAS-level significant SNPs explain only a small proportion of heritable melanoma risk. To identify additional novel but low-penetrance causal variants, the pathway-based analysis of GWAS data has been widely applied. Such analyses appear to be effective in detecting SNPs that confer relatively small but measurable and potentially biologically significant risk (Donnelly, 2008; Elbers et al., 2009; Hong et al., 2009; Luo et al., 2010), thereby helping us to better understand the mechanisms underlying melanoma etiology.
The cell cycle consists of a series of events that occur during cell division and duplication. It can be briefly divided in two periods: interphase and mitosis (including G1, S, G2 and M phases, linked by cell-cycle checkpoints, G1/S transition, and M/G1 transition). The dysfunction of cell cycle often leads to disordered cell growth and ultimately carcinogenesis (Malumbres and Barbacid, 2007). The mutation or abnormal expression of cell-cycle related genes have been reported in studies of many cancers, such as lung cancer, breast cancer and melanoma (Malumbres and Barbacid, 2007). The M/G1 transition is one of the important cell-cycle processes, which includes exiting from mitosis and onset of the G1 phase. The genes function of this process have been involved in the development of melanoma and other cancers (Anantha and Borowiec, 2009; Clarke et al., 2009; Du et al., 2004; Khan et al., 2008). Currently, genetic variants in these genes have also been studied in several cancers (Cunningham et al., 2009; Frank et al., 2008; Ma et al., 2011; Xiong et al., 2009). However, most of these early published candidate-gene studies have investigated only a few genes or variants in this cell cycle process. Recent data from melanoma GWASs provide a unique opportunity for us to elucidate the impact of other unknown low-penetrance variants of the genes involved in the cell-cycle control pathway on melanoma development.
In the present study, we first performed a pathway-based analysis using our published GWAS dataset to evaluate the association between genetic variants in 76 M/G1 transition genes and melanoma risk. Then, we selected significant SNPs with putative functions for validation in other two GWAS datasets. Additional validations of the most promising functional variants were further performed through bioinformatics and laboratory approaches to provide biological supports for our findings.
Results
The distribution of the major melanoma risk factors between cases and controls is presented in Supplementary Table S1, which shows that cases were more likely to have light skin / hair color, more moles, more frequent severe sunburns and dysplastic nevi than were controls (P < 0.001 for all significant variables).
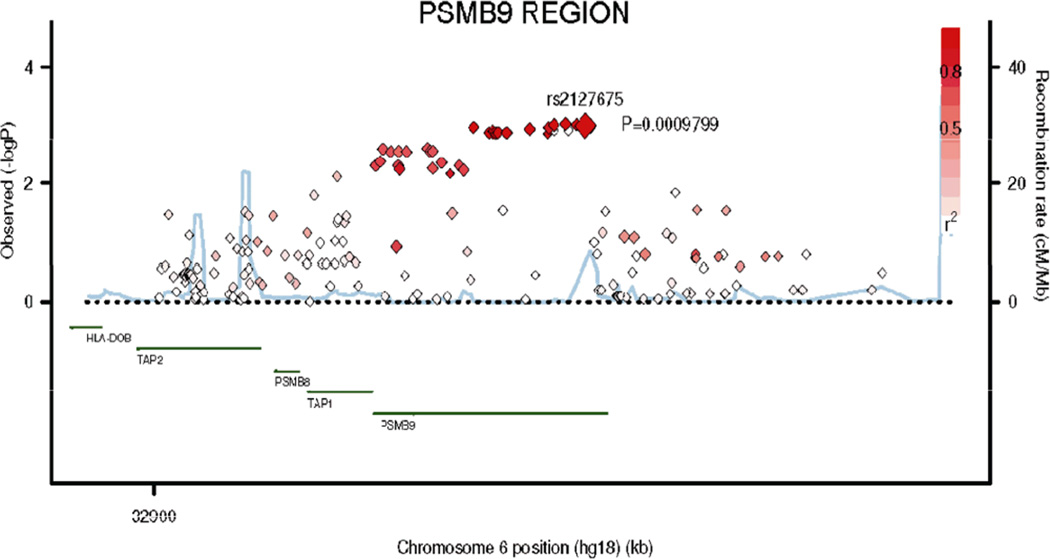
In the present study, a total of 1,149 SNPs in 76 M/G1 transition-related genes were extracted from our GWAS dataset (Supplementary Table S2). The gene-based test had been performed with the VEGAS method (Liu et al., 2010), which revealed seven genes with P-value < 0.05. A list of the SNPs with P value < 0.01 in the discovery set and their assigned genes are shown in Supplemental Table S3, including 68 SNPs in eight genes. There were 34 SNPs with P value < 0.05 after corrections for multiple testing by Benjamin and Hochberg FDR method (Benjamini and Hochberg, 1995). Most of the 68 SNPs (57/68 = 83.8%) were mapped within the PSMB9 gene region on chromosome 6, and the gene-based P value of PSMB9 was 0.003 according to the VEGAS method. Therefore, we focused on this region by selecting 18 SNPs with some putative functions for the in silico replication (Supplementary Figure S1). Validation results are shown in Table 1 that used actual genotyping data for all of the 18 SNPs in the three datasets. Two significant SNPs in the discovery dataset were replicated in the GenoMEL (UK) dataset: rs1351383 in the first intron of PSMB9 (Pdiscovery = 0.005, Preplication in UK = 0.013, Pjoint = 2.29×10−4), rs2127675 in the 3’ flanking of PSMB9 (Pdiscovery = 0.001, Preplication in UK = 0.004, Pjoint = 1.29×10−5) but replication failed in the Australian GWAS dataset. In the meta-analysis of the three datasets, we found that the Pfix value for rs1351383 in the fixed effect model was 0.052 and Pfix value for rs2127675 was 0.006 (Table 1). However, no significance remained in the random model (P(R) = 0.255 and P(R) = 0.163, respectively), likely due to large heterogeneity after combining with the Australian dataset). The regional association plot for the PSMB9 region in the discovery set is presented in Figure 1 with additional 163 imputed SNPs.
Table 1.
Top hit (P < 0.01) SNPs of PSMB9 with putative functions in the M/G1 transition of cell cycle pathway in the discovery dataset and replication datasets
| SNP information | Discovery dataset from MD Anderson |
Replication dataset from GenoMEL |
Replication dataset from Australia |
Meta-analysis of the three datasets |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNPa | Minor allele |
Major allele |
MAF | Beta | SE | P | Beta | SE | P | Beta | SE | P | Beta | SE | P |
| PSMB9 | rs2071480 | A | C | 0.38 | 0.16 | 0.06 | 0.005 | NA | NA | NA | −0.08 | 0.08 | 0.351 | NA | NA | NA |
| PSMB9 | rs1351383 | C | A | 0.38 | 0.16 | 0.06 | 0.005 | 0.11 | 0.05 | 0.013 | −0.04 | 0.04 | 0.28 | 0.05 | 0.03 | 0.051 |
| PSMB9 | rs1351382 | G | C | 0.38 | 0.17 | 0.06 | 0.004 | NA | NA | NA | −0.04 | 0.04 | 0.281 | 0.08 | 0.03 | 0.012 |
| PSMB9 | rs4713600 | A | C | 0.38 | 0.16 | 0.06 | 0.004 | 0.03 | 0.03 | 0.391 | −0.04 | 0.04 | 0.249 | 0.06 | 0.02 | 0.029 |
| PSMB9 | rs3763348 | A | G | 0.40 | 0.17 | 0.06 | 0.002 | NA | NA | NA | −0.08 | 0.08 | 0.323 | 0.09 | 0.05 | 0.053 |
| PSMB9 | rs3763346 | G | A | 0.40 | 0.17 | 0.06 | 0.003 | NA | NA | NA | −0.08 | 0.08 | 0.335 | 0.09 | 0.05 | 0.056 |
| PSMB9 | rs2071534 | A | G | 0.40 | 0.17 | 0.06 | 0.003 | NA | NA | NA | −0.08 | 0.08 | 0.335 | 0.09 | 0.05 | 0.06 |
| PSMB9 | rs2071477 | G | A | 0.38 | 0.16 | 0.06 | 0.005 | 0.03 | 0.03 | 0.37 | −0.04 | 0.04 | 0.248 | 0.06 | 0.02 | 0.029 |
| PSMB9 | rs2071476 | G | A | 0.40 | 0.17 | 0.06 | 0.003 | NA | NA | NA | −0.08 | 0.08 | 0.335 | NA | NA | NA |
| PSMB9 | rs9276815 | A | G | 0.30 | 0.17 | 0.06 | 0.006 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| PSMB9 | rs6930981 | G | A | 0.40 | 0.17 | 0.06 | 0.003 | NA | NA | NA | −0.08 | 0.08 | 0.335 | NA | NA | NA |
| PSMB9 | rs9276899 | A | G | 0.32 | 0.20 | 0.06 | 0.001 | NA | NA | NA | −0.09 | 0.08 | 0.259 | NA | NA | NA |
| PSMB9 | rs2170185 | G | A | 0.32 | 0.19 | 0.06 | 0.001 | NA | NA | NA | −0.09 | 0.08 | 0.259 | NA | NA | NA |
| PSMB9 | rs2127675 | G | A | 0.32 | 0.19 | 0.06 | 0.001 | 0.17 | 0.06 | 0.004 | −0.03 | 0.04 | 0.494 | 0.08 | 0.03 | 0.006 |
| PSMB9 | rs9276911 | A | C | 0.32 | 0.19 | 0.06 | 0.001 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| PSMB9 | rs9276912 | A | G | 0.32 | 0.19 | 0.06 | 0.001 | NA | NA | NA | −0.09 | 0.08 | 0.268 | 0.10 | 0.05 | 0.046 |
| PSMB9 | rs4947259 | A | G | 0.32 | 0.19 | 0.06 | 0.001 | NA | NA | NA | −0.09 | 0.08 | 0.279 | NA | NA | NA |
| PSMB9 | rs1124799 | T | A | 0.40 | 0.16 | 0.06 | 0.004 | NA | NA | NA | −0.07 | 0.08 | 0.366 | 0.13 | 0.05 | 0.004 |
Genotyping data was used for all of the 18 SNPs in the three GWAS datasets.
Figure 1.
Regional association plot in the 20-kb neighborhood of PSMB9 in MD Anderson discovery dataset. The left-hand Y-axis shows the association P-value of individual SNPs in the discovery set, which is plotted as −log10 (P) against chromosomal base-pair position. The righth- and Y-axis shows the recombination rate estimated from the HapMap CEU population. Genotyped SNPs were plotted as diamonds, and imputed SNPs were colored grey. Red diamonds indicates the SNP of rs2127675; red: moderate LD (r2 ≥ 0.5 but <0.8) and white: no LD (r2 < 0.2). The genomic coordinate is in NCBI35/hg18.
We then applied four genetic models to these two SNPs in our discovery GWAS dataset. It should be noted that this might overestimate the genetic effect when just using the discovery dataset due to the “Winner’s course” (Zollner and Pritchard, 2007). For PSMB9 rs2127675, subjects carrying the AG or GG genotype had an increased risk of melanoma (P = 8.00 × 10−4, OR = 1.37, 95% CI: 1.12–1.68; P = 1.93 × 10−3, OR = 1.65, 95% CI: 1.19–2.27, respectively), when compared with those with the AA genotype. The association was more significant under the dominant model (P = 1.65 × 10 −4, OR = 1.42, 95% CI: 1.17–1.73). When stratified by skin color, nevi, and moles status, significant associations were found mainly in subgroups of light skin color or with moles (P = 0.005 and 5.37×10−4, respectively). Similar results were found for SNP rs1351383, which may be due to the fact that these two SNPs are in the same block with a strong LD (r2 = 0.79) (Table 2).
Table 2.
Associations of PSMB9 SNPs rs1351383 and rs2127675 (validated in the GenoMEL study samples) with risk of melanoma in the MD Anderson Melanoma GWAS (1957 subjects [931 cases and 1024 controls] who had questionnaire data)
|
SNPs Genotype |
rs1351383 Case/Control |
OR (95% CI) | Pa | Genotype |
rs2127675 Case/Control |
OR (95% CI) | Pa | ||
|---|---|---|---|---|---|---|---|---|---|
| AA | 290/405 | 1.00 | AA | 358/476 | 1.00 | ||||
| AC | 486/467 | 1.44 (1.16–1.78) | 8.00×10−4 | AG | 458/444 | 1.37 (1.12–1.68) | 2.69×10−3 | ||
| CC | 155/154 | 1.59 (1.19–2.12) | 1.93×10−3 | GG | 115/106 | 1.65 (1.19–2.27) | 2.38×10−3 | ||
| Additive | -- | 1.29 (1.12–1.49) | 3.23×10−4 | Additive | -- | 1.31 (1.13–1.52) | 2.63×10−4 | ||
| Dominant | -- | 1.48 (1.21–1.81) | 1.65×10−4 | Dominant | -- | 1.42 (1.17–1.73) | 4.33×10−4 | ||
| Recessive | -- | 1.28 (0.99–1.67) | 0.063 | Recessive | -- | 1.40 (1.03–1.90) | 0.030 | ||
|
Genotype Known risk factor |
AA | AC+CC | AA | AG+GG | |||||
| Skin color | |||||||||
| ≤ 5 (fair) | 264/337 | 571/510 | 1.45 (1.16–1.80) | 1.08×10−3 | 326/393 | 509/454 | 1.68 (1.15–2.45) | 5.19×10−3 | |
| > 5 (dark) | 26/68 | 70/111 | 1.83 (0.98–3.42) | 0.057 | 32/83 | 64/96 | 2.16 (1.19–3.92) | 0.011 | |
| Dysplastic nevi | |||||||||
| > 1 (yes) | 255/402 | 587/611 | 1.56 (1.26–1.92) | 3.49×10−5 | 317/472 | 525/541 | 1.50 (1.22–1.83) | 9.49×10−5 | |
| ≤ 1 (no) | 35/3 | 54/10 | 0.46 (0.08–2.48) | 0.362 | 41/4 | 48/9 | 0.56 (0.10–3.07) | 0.508 | |
| Moles (counts) | |||||||||
| > 1 | 215/206 | 503/287 | 1.75 (1.35–2.26) | 1.93×10−5 | 273/239 | 445/254 | 1.55 (1.21–1.98) | 5.37×10−4 | |
| ≤ 1 | 75/199 | 138/334 | 1.15 (0.80–1.65) | 0.441 | 85/237 | 128/296 | 1.25 (0.88–1.78) | 0.212 | |
Adjusted for age, sex, moles, hair/eye/skin color, freckle, untanned, dysplastic and tendency to sunburn and family history.
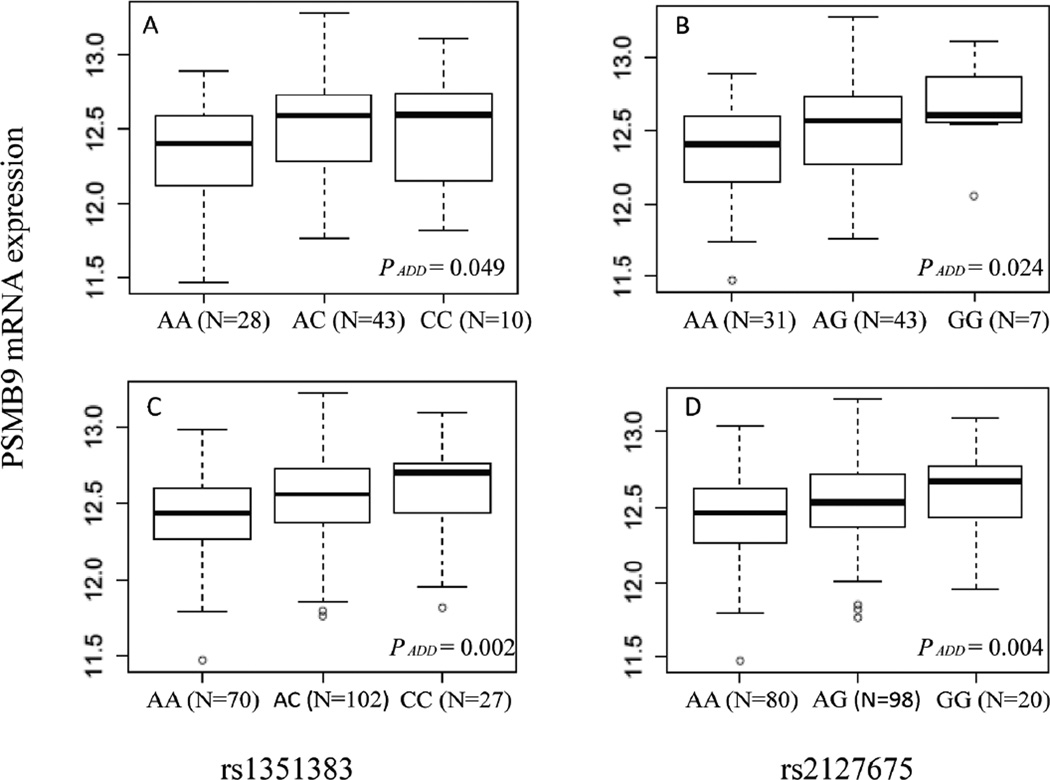
In addition, we evaluated the mRNA expression of PSMB9 by the genotypes of rs2127675 and rs1351383 in 270 lymphoblastoid cell lines derived from the HapMap populations (Figure 2). The risk genotypes of rs2127675 AG/GG were shown to be associated with higher expression levels of PSMB9 (Ptrend = 0.024 in the CEU population and Ptrend = 0.004 in all unrelated populations, respectively) than the common AA genotype. Similar results were found for rs1352383: risk AC/CC genotypes were associated with higher expression levels, compared with the common genotype AA (Ptrend = 0.049 in the CEU population and Ptrend = 0.002 in all unrelated populations, respectively).
Figure 2.
Box-plots of PSMB9 mRNA expression by the genotypes of rs1351383 (A, for 81 CEU cell lines; C, for 199 HapMap unrelated cell lines, after excluding missing data) and rs2127675 (B, for 81 CEU cell lines; D, for 198 unrelated cell lines, after excluding missing data). (CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; other ethnic groups included CHB: Han Chinese in Beijing, China; JPT: Japanese in Tokyo, Japan; YRI: Yoruban in Ibadan, Nigeria).
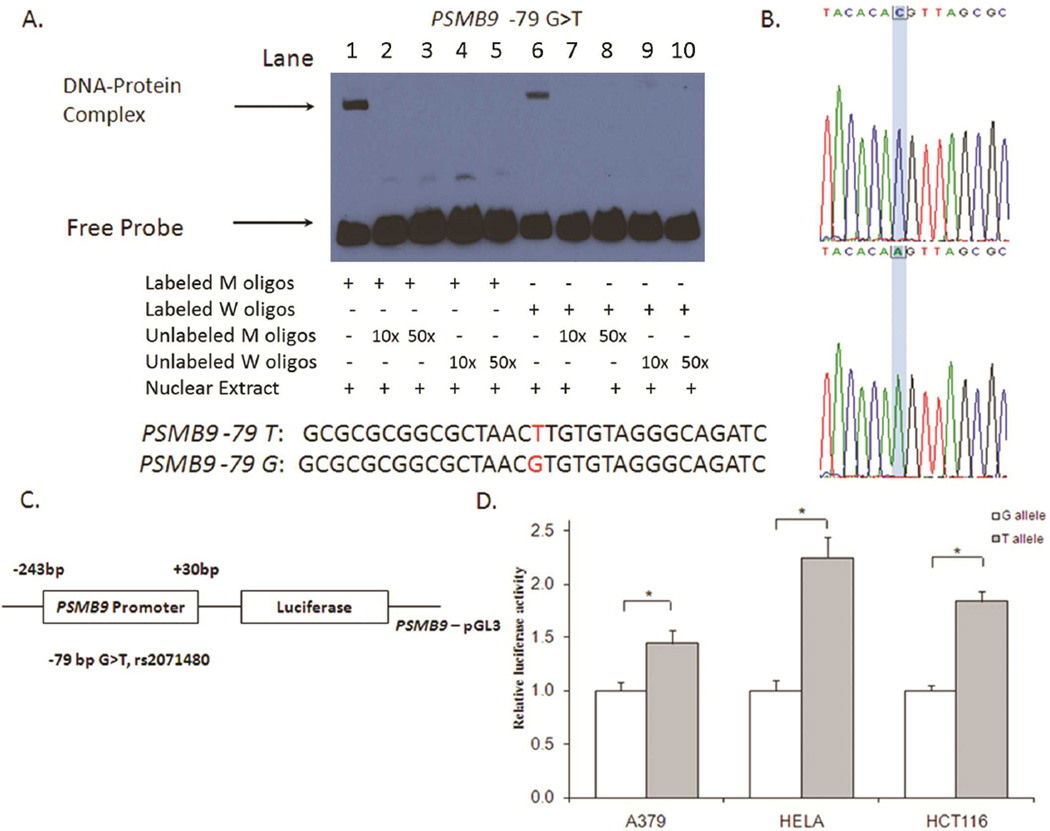
Although rs2071480 was not included in the GenoMEL (UK) GWAS dataset, this SNP was in high LD with rs1351383 and rs2127675 (r2= 0.99 and r2=0.79, respectively) and was at −79bp upstream of the transcription start site of PSMB9. This SNP was also predicted to be located at the putative transcription factor binding sites by SNPinfo. Therefore, we further examined whether rs2071480 could change the binding affinity of transcriptional factors by the electrophoresis migration shift assay (EMSA). As shown in Figure 3A, the nuclear proteins prepared from A375 melanoma cells were able to bind to both oligo probes containing either rare or common or alleles of this SNP (Figure 3A, lanes 2 and 6). Compared with the common G allele-specific shifted band (DNA-Protein complex) in lane 6, the relative intensity of shifted band for the variant T allele (lane 1) was slightly increased (the intensity ratios of shifted/upshifted bands are 0.16 and 0.30, respectively), indicating that the nuclear protein binding activity with T-allele oligo was stronger than that with the G allele oligo. However no shifted band was observed when 10× or 50× unlabeled probes with either the T or G allele were added to compete with the labeled probes, which indicated there was not a large difference between the binding activities of probes with different alleles (Figure 3A, lanes 2, 3, 4, 5 and lanes 7, 8, 9, 10).
Figure 3.
Function assays for SNP rs2071480 in the promoter of PSMB9. (A) Electrophoretic mobility shift assay with biotin-labeled oligo contained the wild type (W) allele, variant (M) allele for the putative functional SNP rs2071480 and nuclear extracts from A375 cells. (B) PCR sequence of rs2071480 (C > A). (C) Schematic of the position of rs2071480 SNP relative to the transcription start site. (D) Luciferase expression of constructs in A375, Hela and HCT116 cell lines. Values represent fold change of luciferase activity relative to the wild-type constructs as 1. Each bar represents the mean of triplicate transfected plates plus standard deviation. * P < 0.001, compared with the wildtype construct.
To further test for the effect of rs2071480 on the promoter activity of the PSMB9 gene, the promoter sequence containing either G or T allele was inserted into the pGL3 vector (Figure 3B and 3C). We selected two clones that were identical to each other except for the SNP site and compared their promoter activities among A375, Hela and HCT116 cancer cell lines. As shown in Figure 3D, the luciferase activities driven by the construct containing T allele increased 1.4–2.3 folds compared with those driven by G allele construct (P < 0.01). The expression analysis also showed that GT/TT genotypes were associated with higher mRNA expression levels of PSMB9 in HapMap lymphoblastoid cell lines (Ptrend = 0.047 for 84 CEU samples and Ptrend = 0.004 for 201 unrelated samples, respectively; Supplementary Figure S2). These results were consistent with that of the EMSA assay (Figure 3.A).
To provide further evidence for the association between PSMB9 expression levels and melanoma risk, we mined the microarray data at NCBI's Gene Expression Omnibus (GEO: http://www.ncbi.nlm.nih.gov/geo/). There was one previous study that had detected the differences in global gene expression profiles among seven normal skin, 18 benign nevi and 45 primary melanoma tissues (Talantov et al., 2005). By using that expression dataset (GEO accession number: GSE3189), we compared the difference in PSMB9 mRNA expression levels between melanoma and benign tissues. As shown in Supplementary Figure S3, the PSMB9 expression levels in primary melanoma were significantly higher than those in normal skin (P = 0.013) and benign nevi (P = 0.005), supporting a role of PSMB9 over expression in skin carcinogenesis.
Discussion
In this GWAS-based pathway study, we investigated the association between genetic variants in the M/G1 transition-related genes and melanoma risk, using three published GWAS datasets. In the discovery phase, we first found that multiple SNPs with P-value < 0.01 located in the PSMB9 region, and we then selected 18 putative functional SNPs in this region to perform validation in other two GWAS datasets. We found that two SNPs, rs1351383 and rs2127675, also showed significant associations with melanoma risk in the GenomMEL GWAS dataset, but the validation failed in the Australian GWAS dataset. Further mRNA expression analyses and functional assays provided evidence that these SNPs might be associated with melanoma risk by increasing PSMB9 mRNA expression. Stratified analysis indicated the risk associated with these SNPs was more evident in subgroups with moles and fair skin, suggesting these PSMB9 SNPs may play a role in susceptibility to melanoma associated with pigmentogenesis and mole development. To our knowledge, this report provides the first evidence from large GWAS datasets for associations between PSMB9 SNPs and melanoma risk in a non-Hispanic white population.
PSMB9, also known as LMP2, is located at 6q21, the high-risk region of major histocompatibility complex. It encodes one subunit of immunoproteasome, which involves in the antigen processing and presentation. Down-regulation of antigen-processing molecules, found in a variety of cancers, may change the spectrum of peptides presented by MHC molecules and may be associated with immune escape of tumors (Igney and Krammer, 2002). However, the association between PSMB9 expression levels and melanoma risk has been inconclusive. One previous study had reported down-expression of antigen processing molecules in malignant melanoma using the immunohistochemical method, but it did not find significant difference in PSMB9 expression levels between nevi and primary melanoma lesions (Kageshita et al., 1999). Another study also reported that the down-regulation of LMP7 and TAP2, but not PSMB9, was correlated with levels of the HLA class I surface expression in melanoma cell lines (Mendez et al., 2008). However, Dannull and colleagues found that down-regulation of immunoproteasome by siRNA transfection of LMP2, LMP7 and MECL-1 could stimulate the enhanced antimelanoma CTL activity (Dannull et al., 2007). By mining the GEO database, we found that mRNA expression levels of PSMB9 increased significantly in melanoma tissues, compared with that of the normal skin and benign nevi. This may provide some additional support for our finding of an association between genotypes and an elevated expression of PSMB9, which would increase melanoma risk. Further functional validations are warranted.
The roles of genetic variants of PSMB9 have been investigated in several non-melanoma cancers and autoimmune diseases. Up to now, most of these studies focused on limited number of exonic variants. One PSMB9 variant of Arg60His (rs17587), which may influence the gene’s functions (Mishto et al., 2006), had been reported to be associated with hypertension in adolescents (Honcharov et al., 2009), multiple sclerosis (Mishto et al., 2010), acute anterior uveitis (Maksymowych et al., 1997), Mycobacterium tuberculosis infection (Lv et al., 2011), insulin-dependent diabetes mellitus (Deng et al., 1995), and cervical carcinoma (Deshpande et al., 2008). Another exonic variant of Ile32Val (rs241419) was found to be weakly associated with colorectal cancer risk in a UK study (Webb et al., 2006), but it was not replicated in a study of German families (Frank et al., 2008). We have investigated the association of these variants with melanoma risk in the present study but no significant association was observed (data was not shown). In addition, other cell cycle-related genes and SNPs reported before (Choudhury et al., 2004; Darieva et al., 2010; Lan et al., 2006) were not replicated in the present study.
It should be noted that we did not find association that could reach genome-wide significance level and none SNPs were replicated in the Australian GWAS dataset. Although there are several SNPs in PSMB9 that could pass the multiple comparison correction (FDR = 0.045), we also cannot exclude possibility that these SNPs are not be true causal SNPs of melanoma. Considering the confounding influence of sun exposure on melanoma risk, one possible reason for the replication failure might be due to the difference in sun-exposure between the three study populations living at different latitudes (Chang et al., 2009). Further stratification analysis by sun-exposure may help to estimate genetic effects of these variants on melanoma risk in Australia population. Heterogeneity of age at onset of the disease may be another reason for the failure of replication. The median onset age for the patients included in MD Anderson study was 51 years (Amos et al., 2011), while for the Australia study, there were nearly half of cases (1064 cases) with onset age less than 40 (Macgregor et al., 2011). Previous studies have discussed the influence of birth cohort effect on melanoma incidence (Jemal et al., 2001). In the present study, although we did not have enough information to compare the birth year of patients in both the discovery study and the Australia study, considering the different median age, we cannot exclude the possible influence of cohort effects on the validation results. We also compared the MAF of the two SNPs between the three studies. The MAF of rs1351383 in the controls of MD Anderson, GenoMEL and Australia melanoma studies was 0.38, 0.42 and 0.42 respectively, while the MAF of rs2127675 in the controls of the three studies was 0.32, 0.36 and 0.37 respectively. The increased allele frequency in the Australian population might have decreased the power of the replication studies, leading to the replication failure (Greene et al., 2009). As an alternative to population replication, supportive evidence from functional assays may add additional biological plausibility to the weak associations and support identification of a true causal variant (Khoury et al., 2009). Therefore, further studies on the mechanisms underlying the observed associations are warranted to assess whether these or other untyped variants within this region may directly contribute to melanoma risk.
In conclusion, our findings suggest that functional genetic variants in PSMB9 may influence the development of melanoma by increasing gene expression. Further functional assays and replication in additional population are warranted to verify these results.
Methods and Materials
Study populations of the discovery set
For the discovery phase, we used the melanoma GWAS dataset from the MD Anderson Cancer Center that has been recently described (Amos et al., 2011). Briefly, the study participants consisted of 1804 non-Hispanic White patients and 1026 controls, as a part of an ongoing melanoma investigation, who were recruited at M.D. Anderson between March 1998 and August 2008. In addition, both GWAS data and risk-factor questionnaire data were available for 931 melanoma patients and 1,026 cancer-free controls (friends and relatives of other cancer patients visiting the clinics), who were genetically unrelated and frequency-matched on age and sex. The study protocol was approved by the Institutional Review Board at MD Anderson, and a written informed consent was obtained from all participants.
GWAS genotyping data
One-time whole blood samples were used for DNA extraction by various methods (including Gentra, Qiagen, and phenol/chloroform). DNA samples were genotyped using the Illumina Omnil-Quad array and the genotypes were called using the BeadStudio algorithm at the John Hopkins University Center for Inherited Disease Research (CIDR). Standard quality control (QC) procedures were applied to both samples and SNPs which had been described in the published paper (Amos et al., 2011). Briefly, SNPs were included, if they had a minor allele frequency (MAF) > 0.01, call rate ≥ 95%, and Hardy-Weinberg equilibrium in controls with P ≥ 1×10−5. We excluded the duplicated samples, related (IBD) samples or outliers identified by principle component analysis (PCA).
Study populations of the replication set from GenoMEL
One replication was performed in silico utilizing GWAS from the GenoMEL Consortium (Barrett et al., 2011; Bishop et al., 2009). The GenoMEL GWAS utilizes samples collected from multiple centers across Europe and Israel in two phases. Phase 1 of the original GenoMEL GWAS consisted of samples collected from 8 centres across 6 different European countries. These were supplemented with controls from the Wellcome Trust Case-Control Consortium. Standard QC measures were applied to both samples and SNPs, giving a total of 1,353 cases and 3,571 controls. Phase 2 of the GenoMEL GWAS was collected across 10 centers (4 not in Phase 1) in 8 different European countries and Israel, supplemented again by samples from the Wellcome Trust Case Control Consortium. After QC, 1,450 cases and 4,047 controls remained. Most GenoMEL Phase 1 samples were genotyped on the llumina HumanHap300 BeadChip version 2 duo array (with 317k tagging SNPs), with the exception of the French cases, which were genotyped on the Illumina Humancnv370k array. The GenoMEL Phase 2 samples were genotyped on the Illumina 610k array. The Australian data used in Phase 1 were dropped, as these samples were included in the Australian GWAS as another replication set. SNP quality control were applied to each genotyping platform separately. SNPs were excluded, if they had a call rate < 97%, or Hardy-Weinberg equilibrium P < 1×10−20 or recommendation for exclusion by Wellcome Trust Case-Control Consortium (WTCC). The imputed p-values from the Phase 1 data were available for replication.
Study populations of the replication set from Australia
The second in silico replication included 2,168 melanoma cases selected from the Queensland, Australia study of Melanoma: Environment and Genetic Associations (Q-MEGA) and the Australian Melanoma Family Study (AMFS), for which 1242 patient samples were typed on the Omni1-Quad and 926 typed on the Hap610 arrays (Macgregor et al., 2011). Three Australian Caucasian sample populations were used as controls (n=4,387), for which 431 were typed on the Omni1-Quad and 3956 were typed on the Hap610 arrays. Cases and controls were combined into a single dataset for quality control analysis (including principal component analysis for outlier removal) and imputation. SNPs were excluded for MAF < 0.01, call rate < 95%, or Hardy-Weinberg equilibrium in controls with P < 1×10−6. Imputation was based on the 1000 Genomes Project data, which helped recover the full sample size for SNPs that were only typed on a subset of the arrays (Macgregor et al., 2011).
Gene and SNP selection
In the re-analysis of M.D. Anderson Melanoma GWAS, genes involved in the M/G1 transition process of the cell cycle were selected based on the following criteria: genes that have been reported to be involved in the M/G1 transition process; genes that have been included in the M/G1 transition of cell cycle pathway in the Web of Amigo (http://amigo.geneontology.org/, as of 12/03/2010) (Carbon et al., 2009); and genes that have been covered by the Illumina Omni1-Quad BeadChips (Illumina, San Diego, CA). As a result, there were a total of 1149 SNPs in 76 genes in the M/G1 transition of cell cycle pathway available from our GWAS database. The assignment of a SNP to a gene was defined by Illumina annotation file “Human 1M Quad_gene_annotation.txt”, which annotated all SNPs to their closest gene regardless how far a SNP is away from the gene.
To search for putative functional SNPs for these with P < 0.01 obtained for the initial association analyses, we used SNPinfo (http://snpinfo.niehs.nih.gov/snpfunc.htm) (Xu and Taylor, 2009) to identify any putative functional SNPs based on the HapMap phase II data. To obtain more confident prediction, we validated the functional findings of SNPinfo with other bioinformatics softwares. For example, for SNPs that were predicted to affect transcription factor binding sites in the promoter region, we further evaluated their effects on the transcription factor binding in TFSEARCH (Heinemeyer et al., 1998) (http://molsun1.cbrc.aist.go.jp/research/db/TFSEAR CH.html). Only SNPs that were identified to be functional in two or more bioinformatics software were further validated by the laboratory functional assays.
For replication, we selected the 18 most significantly associated SNPs (P < 0.01) in the PSMB9 gene region with putative function and evaluated their associations with melanoma using the genotyping data of two additional GWASs from UK and Australia.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts from melanoma cell line A375 were prepared according to the method of Andrews and Faller (Andrews and Faller, 1991). Complementary single-stranded oligonucleotides for rs2071480 of PMSB9 (5’-GCGCGCGGCGCTAACTTGTGTAGGGCAGATC −3’ for the T allele and 5’-GCGCGCGGCGCTAACGTGTGTAGGGCAGATC −3’ for the G allele) were biotin-labeled using the 3’-end biotin labeling kit (Thermo Scientific, Rockford, IL) and re-annealed before performing the DNA binding. The binding of DNA and protein was performed by using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL). The DNA-protein complexes were separated on 6% polyacrylamide gel, and the products were detected by Stabilized Streptavidin-Horseradish Peroxidase Conjugate (Thermo Scientific). The competition assays were performed 50-fold excess of unlabeled wild-type and mutation oligonucleotides, respectively. The intensity of shifted bands in scanned films was determined by using TotalLab™ program (Nonlinear Dynamics Ltd., UK).
Transient Transfection and Luciferase Reporter assay
PCR fragments containing T allele of SNP rs2071480 were amplified from genomic DNA isolated from homozygous T carriers using the following primers: forward primer 5'-AAGCTAGCATCTGAGAATCTCGGGAGCA −3', reverse primer 5'-TTAAGCTTGGTTTCCAACCTGGGACAG −3'. The PCR products were then cloned into the pGL3-Basic vector (Promega, Madison, WI, USA) between NheI and HindIII sites and verified by directly sequencing. The common allele G of rs2071480 was introduced into the recombination vector by QuickChange site-directed mutagenesis kit (Cat # 200518; Stratagene, La Jolla, CA) using the forward mutagenic primer 5’-TTTGCGCGCGGCGCTAACGTGTGTAGGGCAGATCT-3’ and reverse mutagenic primer 5’-AGATCTGGCCTACACACGTTAGCGCCGCCCGCAAA-3’ according to the manufacturer’s protocol. The clones containing the expected G allele of rs2071480 were verified by direct sequencing.
Three cancer cell lines (A375 from melanoma, HeLa from cervical cancer and HCT116 from colon cancer) were placed on 24-well plates at 1.0×105 cells per well with DMEM or 1x RPMI 1640 culture medium containing 10% fetal bovine serum and allowed to grow for one day prior to transfection (50–70% confluence). Transfection experiments were performed using FuGENE HD (Invitrogen, Carlsbad, CA, USA). Each transfection was performed in triplicates. Dual Luciferase Kit (Promega) was used to detect the activity of firefly luciferase and Renilla luciferase.
Genotype-phenotype correlation analysis of PSMB9 expression
We also analyzed PSMB9 mRNA expression by genotypes of rs1351383, rs2127675, and rs2071480 based on the transcript expression profiling data of 270 lymphoblastoid cell lines from CEU and other HapMap samples (including 90 CEU samples, 90 CHB/JPT samples and 90 YRI samples) (NCBI GEO accession ID: GSE7792) (Stranger et al., 2007). The expression data and genotyping data were available for rs1351383 in 81 CEU and 199 unrelated lymphoblastoid cell lines, for rs2127675 in 81 CEU and 198 unrelated lymphoblastoid cell lines, and for rs2071480 in 84 CEU and 201 unrelated lymphoblastoid cell lines.
Statistical analysis
The distribution differences of demographic variables and known risk factors between melanoma patients and controls were assessed by the χ2 test. The associations between alleles or genotypes of each tagSNP and melanoma risk were primarily evaluated using the allelic test in PLINK1.07 (Purcell et al., 2007). Benjamini and Hochberg FDR method was used for the multiple testing corrections (Benjamini and Hochberg, 1995). LD patterns among tagSNPs were evaluated by Haploview (Barrett et al., 2005). We adjusted for the five largest principal components of genetic variation to control potential effects of population structure. For the luciferase assay, significant differences between groups were determined by Student’s t test. For the in silico meta-analysis, betas from each study were combined using the inverse variance method. T-test and Wilcoxon-Mann-Whitney test were applied to compare the difference in mRNA expression levels of PSMB9 between different genotypes or different tissues. Unless specified otherwise, all other statistical analyses were performed using SAS 9.1.3 (SAS Institute Inc., Cary, NC), and all statistical tests were two sided, with a P < 0.05 set as the level of statistical significance.
Supplementary Material
Significance.
Cell cycle transitions are normally tightly regulated, and aberrant cell cycle regulation often leads to cancer development. In this genome-wide association study (GWAS)-based pathway study, we comprehensively investigated the associations between genetic variants in M/G1 transition-related genes and melanoma risk, using our published GWAS dataset. We then performed validation studies for identified potential functional SNPs in PSMB9 using other two GWAS datasets and functional experiments. Our results indicated that genetic variants of PSMB9 contribute to melanoma risk possibly by modulating gene expression. Further functional assays and replication in additional population are warranted.
Acknowledgments
We thank Yawei Qiao, Min Zhao, Jianzhong He, Kejing Xu and Hongxia Ma for their laboratory assistance, Hongping Yu, Yujing Huang, Ming Yin and Ziyuan Zhou for their help in data analysis, and Dakai Zhu for his technical support. This study was supported in part by National Institutes of Health grants R01 CA100264, R01ES011740 and R01CA131274 (Q. Wei), UT MD Anderson Cancer Center SPORE in melanoma P50 CA093459 (E. Grimm) and P30 CA016672 (M.D. Anderson Cancer Center). HHSN268200782096C (Christopher Amos), Cancer Center Core Grant P30 CA016672 (to M. D. Anderson Cancer Center), GenoMEL has received substantial funding from the European Commission under the 6th Framework Programme, Contract nr: LSHC-CT-2006-018702, from the National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115) and from Cancer Research UK (C588/A4994, C588/A10589 and CC8216/A6129). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
This work was also supported by the Melanoma Research Alliance, the National Institutes of Health/National Cancer Institute (CA88363, CA83115, CA122838, CA87969, CA055075, CA100264, CA133996 and CA49449), the National Health and Medical Research Council of Australia (NHMRC) (107359, 200071, 241944, 339462, 380385, 389927,389875, 389891, 389892,389938, 402761, 443036, 442915, 442981, 496610, 496675, 496739, 552485, 552498), the Cancer Councils NSW, Victoria and Queensland, the Cancer Institute New South Wales, the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), Cerylid Biosciences (Melbourne), the Australian Cancer Research Foundation, The Wellcome Trust (WT084766/Z/08/Z), donations from Neville and Shirley Hawkins, and the Marit Peterson Fund for Melanoma Research. NKH and GWM are supported by the NHMRC Fellowships scheme. SM is the recipient of a Career Development Award from the NHMRC (496674, 613705). MHL is supported by Cancer Australia grant 1011143. The AMFS, QMEGA, QIMR and endometriosis studies gratefully acknowledge its participants and the hard work of all its research interviewers, research assistants, technicians, project managers, data as well as sample managers and examiners. We thank Endometriosis Associations for supporting study recruitment and Sullivan Nicolaides and Queensland Medical Laboratory for pro bono collection and delivery of blood samples and other pathology services for assistance with blood collection.
Footnotes
Conflict of Interest Statement
None declared.
Reference
- Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, Kosoy R, Zhang M, Qureshi AA, Vattathil S, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20:5012–5023. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantha RW, Borowiec JA. Mitotic crisis: the unmasking of a novel role for RPA. Cell Cycle. 2009;8:357–361. doi: 10.4161/cc.8.3.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, Akslen LA, Armstrong BK, Avril MF, Azizi E, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YM, Barrett JH, Bishop DT, Armstrong BK, Bataille V, Bergman W, Berwick M, Bracci PM, Elwood JM, Ernstoff MS, et al. Sun exposure and melanoma risk at different latitudes: a pooled analysis of 5700 cases and 7216 controls. International journal of epidemiology. 2009;38:814–830. doi: 10.1093/ije/dyp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury AD, Xu H, Baer R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J Biol Chem. 2004;279:33909–33918. doi: 10.1074/jbc.M403646200. [DOI] [PubMed] [Google Scholar]
- Clarke LE, Fountaine TJ, Hennessy J, Bruggeman RD, Clarke JT, Mauger DT, Helm KF. Cdc7 expression in melanomas, Spitz tumors and melanocytic nevi. Journal of cutaneous pathology. 2009;36:433–438. doi: 10.1111/j.1600-0560.2008.01077.x. [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Vierkant RA, Sellers TA, Phelan C, Rider DN, Liebow M, Schildkraut J, Berchuck A, Couch FJ, Wang X, et al. Cell cycle genes and ovarian cancer susceptibility: a tagSNP analysis. British journal of cancer. 2009;101:1461–1468. doi: 10.1038/sj.bjc.6605284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, Leboit PE, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Dannull J, Lesher DT, Holzknecht R, Qi W, Hanna G, Seigler H, Tyler DS, Pruitt SK. Immunoproteasome down-modulation enhances the ability of dendritic cells to stimulate antitumor immunity. Blood. 2007;110:4341–4350. doi: 10.1182/blood-2007-04-083188. [DOI] [PubMed] [Google Scholar]
- Darieva Z, Clancy A, Bulmer R, Williams E, Pic-Taylor A, Morgan BA, Sharrocks AD. A competitive transcription factor binding mechanism determines the timing of late cell cycle-dependent gene expression. Mol Cell. 2010;38:29–40. doi: 10.1016/j.molcel.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng GY, Muir A, Maclaren NK, She JX. Association of LMP2 and LMP7 genes within the major histocompatibility complex with insulin-dependent diabetes mellitus: population and family studies. Am J Hum Genet. 1995;56:528–534. [PMC free article] [PubMed] [Google Scholar]
- Deshpande A, Wheeler CM, Hunt WC, Peyton CL, White PS, Valdez YE, Nolan JP. Variation in HLA class I antigen-processing genes and susceptibility to human papillomavirus type 16-associated cervical cancer. J Infect Dis. 2008;197:371–381. doi: 10.1086/524300. [DOI] [PubMed] [Google Scholar]
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Elbers CC, Van Eijk KR, Franke L, Mulder F, Van Der Schouw YT, Wijmenga C, Onland-Moret NC. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol. 2009;33:419–431. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- Falchi M, Bataille V, Hayward NK, Duffy DL, Bishop JA, Pastinen T, Cervino A, Zhao ZZ, Deloukas P, Soranzo N, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figl A, Scherer D, Nagore E, Bermejo JL, Botella-Estrada R, Gast A, Thirumaran RK, Planelles D, Hemminki K, Schadendorf D, et al. Single-nucleotide polymorphisms in DNA-repair genes and cutaneous melanoma. Mutat Res. 2010;702:8–16. doi: 10.1016/j.mrgentox.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Frank B, Burwinkel B, Bermejo JL, Forsti A, Hemminki K, Houlston R, Mangold E, Rahner N, Friedl W, Friedrichs N, et al. Ten recently identified associations between nsSNPs and colorectal cancer could not be replicated in German families. Cancer Lett. 2008;271:153–157. doi: 10.1016/j.canlet.2008.05.043. [DOI] [PubMed] [Google Scholar]
- Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4:e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honcharov SV, Dosenko V, Khaitovych MV, Moibenko OO. [Allele polymorphism of genes coding proteasome subunits is associated with an enhanced risk for arterial hypertension in adolescents] Fiziol Zh. 2009;55:3–10. [PubMed] [Google Scholar]
- Hong MG, Pawitan Y, Magnusson PK, Prince JA. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum Genet. 2009;126:289–301. doi: 10.1007/s00439-009-0676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igney FH, Krammer PH. Immune escape of tumors: apoptosis resistance and tumor counterattack. Journal of leukocyte biology. 2002;71:907–920. [PubMed] [Google Scholar]
- Jemal A, Devesa SS, Hartge P, Tucker MA. Recent trends in cutaneous melanoma incidence among whites in the United States. J Natl Cancer Inst. 2001;93:678–683. doi: 10.1093/jnci/93.9.678. [DOI] [PubMed] [Google Scholar]
- Kageshita T, Hirai S, Ono T, Hicklin DJ, Ferrone S. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: association with disease progression. The American journal of pathology. 1999;154:745–754. doi: 10.1016/S0002-9440(10)65321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AN, Gregorie CJ, Tomasi TB. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol Immunother. 2008;57:647–654. doi: 10.1007/s00262-007-0402-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, Bertram L, Boffetta P, Butterworth AS, Chanock SJ, Dolan SM, Fortier I, Garcia-Closas M, Gwinn M, Higgins JP, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170:269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan B, Liu BY, Cheng XH, Zhang J, Wang KK, Zhu ZG. Analysis of gene expression profiles in gastric cancer cell cycle. Zhonghua Zhong Liu Za Zhi. 2006;28:568–571. [PubMed] [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Peng G, Zhu Y, Dong H, Amos CI, Xiong M. Genome-wide gene and pathway analysis. Eur J Hum Genet. 2010;18:1045–1053. doi: 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Yan B, Yang H, Liu J, Zhong W, Li K, Chen Z, Xu C. LMP2/LMP7 gene variant: a risk factor for intestinal Mycobacterium tuberculosis infection in the Chinese population. Journal of gastroenterology and hepatology. 2011;26:1145–1150. doi: 10.1111/j.1440-1746.2011.06693.x. [DOI] [PubMed] [Google Scholar]
- Ma H, Chen J, Pan S, Dai J, Jin G, Hu Z, Shen H, Shu Y. Potentially functional polymorphisms in cell cycle genes and the survival of non-small cell lung cancer in a Chinese population. Lung cancer. 2011;73:32–37. doi: 10.1016/j.lungcan.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Montgomery GW, Liu JZ, Zhao ZZ, Henders AK, Stark M, Schmid H, Holland EA, Duffy DL, Zhang M, et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet. 2011;43:1114–1118. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksymowych WP, Jhangri GS, Gorodezky C, Luong M, Wong C, Burgos-Vargas R, Morenot M, Sanchez-Corona J, Ramos-Remus C, Russell AS. The LMP2 polymorphism is associated with susceptibility to acute anterior uveitis in HLA-B27 positive juvenile and adult Mexican subjects with ankylosing spondylitis. Ann Rheum Dis. 1997;56:488–492. doi: 10.1136/ard.56.8.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle kinases in cancer. Current opinion in genetics & development. 2007;17:60–65. doi: 10.1016/j.gde.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Mendez R, Rodriguez T, Del Campo A, Monge E, Maleno I, Aptsiauri N, Jimenez P, Pedrinaci S, Pawelec G, Ruiz-Cabello F, et al. Characterization of HLA class I altered phenotypes in a panel of human melanoma cell lines. Cancer Immunol Immunother. 2008;57:719–729. doi: 10.1007/s00262-007-0411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishto M, Bellavista E, Ligorio C, Textoris-Taube K, Santoro A, Giordano M, D’alfonso S, Listi F, Nacmias B, Cellini E, et al. Immunoproteasome LMP2 60HH variant alters MBP epitope generation and reduces the risk to develop multiple sclerosis in Italian female population. PLoS One. 2010;5:e9287. doi: 10.1371/journal.pone.0009287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishto M, Santoro A, Bellavista E, Sessions R, Textoris-Taube K, Dal Piaz F, Carrard G, Forti K, Salvioli S, Friguet B, et al. A structural model of 20S immunoproteasomes: effect of LMP2 codon 60 polymorphism on expression, activity, intracellular localisation and insight into the regulatory mechanisms. Biological chemistry. 2006;387:417–429. doi: 10.1515/BC.2006.056. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, De Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- Titus-Ernstoff L, Perry AE, Spencer SK, Gibson JJ, Cole BF, Ernstoff MS. Pigmentary characteristics and moles in relation to melanoma risk. Int J Cancer. 2005;116:144–149. doi: 10.1002/ijc.21001. [DOI] [PubMed] [Google Scholar]
- Webb EL, Rudd MF, Sellick GS, El Galta R, Bethke L, Wood W, Fletcher O, Penegar S, Withey L, Qureshi M, et al. Search for low penetrance alleles for colorectal cancer through a scan of 1467 non-synonymous SNPs in 2575 cases and 2707 controls with validation by kin-cohort analysis of 14 704 first-degree relatives. Hum Mol Genet. 2006;15:3263–3271. doi: 10.1093/hmg/ddl401. [DOI] [PubMed] [Google Scholar]
- Xiong XD, Qiu FE, Fang JH, Shen Y, Liang C, Jiang W, Zhuang SM. Association analysis between the Cdc6 G1321A polymorphism and the risk for non-Hodgkin lymphoma and hepatocellular carcinoma. Mutat Res. 2009;662:10–15. doi: 10.1016/j.mrfmmm.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Qureshi AA, Guo Q, Han J. Genetic variation in DNA repair pathway genes and melanoma risk. DNA Repair (Amst) 2010;10:111–116. doi: 10.1016/j.dnarep.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner S, Pritchard JK. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.