SUMMARY
Tissue-specific differentiation programs become dysregulated during cancer evolution. The transcription factor Nkx2-1 is a master regulator of pulmonary differentiation that is downregulated in poorly differentiated lung adenocarcinoma. Here we use conditional murine genetics to determine how the identity of lung epithelial cells changes upon loss of their master cell fate regulator. Nkx2-1 deletion in normal and neoplastic lung causes not only loss of pulmonary identity but also conversion to a gastric lineage. Nkx2-1 is likely to maintain pulmonary identity by recruiting transcription factors Foxa1 and Foxa2 to lung-specific loci thus preventing them from binding gastrointestinal targets. Nkx2-1-negative murine lung tumors mimic mucinous human lung adenocarcinomas, which express gastric markers. Loss of the gastrointestinal transcription factor Hnf4α leads to de-repression of the embryonal protoncogene Hmga2 in Nkx2-1-negative tumors. These observations suggest that loss of both active and latent differentiation programs is required for tumors to reach a primitive, poorly differentiated state.
INTRODUCTION
When a normal cell sustains an oncogenic mutation, its differentiation state begins to change. The ultimate differentiation state acquired by a cancer over the course of its evolution often predicts prognosis and therapeutic response. Lung adenocarcinomas exhibit a diverse array of differentiation states (Travis et al., 2011), and tumors which have diverged the most dramatically from normal lung confer the worst prognosis (Russell et al., 2011; Yoshizawa et al., 2011). Lung adenocarcinomas treated with targeted therapies undergo radical differentiation state changes that affect their sensitivity to standard drug regimens (Sequist et al., 2011). Nevertheless, the molecular regulators of lung adenocarcinoma differentiation remain poorly understood.
The transcription factor Nkx2-1/TTF1 has emerged as a candidate regulator of lung adenocarcinoma differentiation. Nkx2-1 is a highly conserved homeodomain-containing transcription factor that is expressed at the onset of lung and thyroid development (Boggaram, 2009). The primordial lung buds arise from the ventral wall of the anterior foregut at day E9.5, invade into the surrounding splanchnic mesoderm, and undergo branching morphogenesis to form the mature lung (Costa et al., 2001). In mice harboring a targeted deletion of Nkx2-1, lung buds are initiated but there is a complete failure of branching morphogenesis, resulting in the formation of dilated sacs lined by epithelial cells lacking markers of pulmonary differentiation (Kimura et al., 1996; Minoo et al., 1999).
NKX2-1 is expressed in 75-85% of human lung adenocarcinomas (Kunii et al., 2011; Stenhouse et al., 2004). NKX2-1-negative tumors confer a worse prognosis and have an altered differentiation state compared to NKX2-1-positive tumors (Barletta et al., 2008; Berghmans et al., 2006; Travis et al., 2011). These correlations suggest that NKX2-1 may enforce a lineage-specific differentiation program on lung adenocarcinomas that restrains their malignant potential. The NKX2-1 gene is genomically amplified in 10-15% of human lung adenocarcinomas, indicating that it can also act as a lineage-survival oncogene in a subset of tumors (Kendall et al., 2007; Kwei et al., 2008; Tanaka et al., 2007; Weir et al., 2007), likely by activating targets such as LMO3 (Watanabe et al., 2013) and ROR1 (Yamaguchi et al., 2012). NKX2-1 was also identified as an oncogene in T-cell acute lymphoblastic leukemia (Homminga et al., 2011), and other NKX family members regulate tumorigenesis in a variety of tissues (Abate-Shen et al., 2008; Yu et al., 2012).
We have previously shown that Nkx2-1 restrains the progression of a mouse model of lung adenocarcinoma. In this model, Nkx2-1 positive tumors are initiated by expression of the KrasG12D oncogene, and simultaneous loss of the p53 tumor suppressor enables progression to a metastatic state over time (Winslow et al., 2011). Stochastic loss of Nkx2-1 expression is observed in poorly differentiated, metastatic tumors that upregulate the proto-oncogene Hmga2, whose expression is normally restricted to embryonic tissues (Fusco and Fedele, 2007). Re-expression of Nkx2-1 in lung adenocarcinoma cell lines inhibited Hmga2 expression and reduced tumorigenesis after transplantation into mice. These results indicated that Nkx2-1 restrained the ability of Kras-driven lung tumors to evolve to a poorly differentiated, Hmga2-positive state.
Based on these observations, we have employed the lung as a model system to characterize how epithelial cells react to the loss of their master fate regulator. We have used a conditional allele of Nkx2-1 (Kusakabe et al., 2006) to determine the consequences of Nkx2-1 deletion in the normal lung and in autochthonous murine lung adenocarcinomas. We have found that normal and neoplastic epithelial cells adopt a gastric differentiation state after Nkx2-1 deletion perhaps reflecting the embryologic origins of the lung. We have implicated the relocalization of the transcription factors Foxa1 and Foxa2 from pulmonary to gastrointestinal genes as a mechanism for this change in differentiation. Finally, we show that the loss of two master regulators of differentiation, Nkx2-1 and Hnf4α, can have a profound effect on tumor burden, demonstrating a direct connection between transcriptionally controlled differentiation programs and tumor growth.
RESULTS
Nkx2-1 controls differentiation state in lung adenocarcinoma
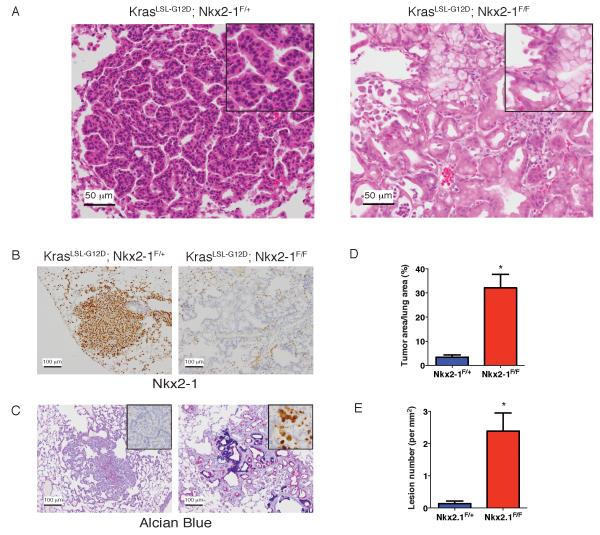
We generated mice in which Cre recombinase can activate a conditional allele of oncogenic Kras (KrasLSL-G12D/+, hereafter KrasLSL-G12D) (Jackson et al., 2001) and simultaneously delete Nkx2-1 (Kusakabe et al., 2006). We infected lung epithelial cells of KrasLSL-G12D; Nkx2-1F/F mice and KrasLSL-G12D; Nkx2-1F/+ controls with adenovirus expressing Cre (Ad-Cre). Simultaneous KrasG12D activation and Nkx2-1 deletion yielded invasive adenocarcinomas in the peripheral lung within 2-4 weeks of initiation that exhibited a dramatically altered differentiation state compared to Nkx2-1 positive tumors (Figure 1A). Control tumors express Nkx2-1 and its target pro-surfactant protein C (proSPC), whereas tumors in KrasLSL-G12D; Nkx2-1F/F mice do not (Figure 1B and Figure S1A). Nkx2-1-positive tumors were organized into predominantly papillary structures (Figure 1A, left), but Nkx2-1-deleted tumors exhibited a distinct glandular growth pattern (Figure 1A, right). Nkx2-1-negative tumor cells produced abundant mucin, including Muc5AC (Figure 1C, right), whereas control tumors were non-mucinous (Figure 1C, left). Transcript levels of Spdef, which encodes a transcription factor that promotes mucinous differentiation in the lung (Chen et al., 2009; Maeda et al., 2011) were elevated in Nkx2-1-negative lung tumors relative to controls (Figure S1B).
Figure 1. Nkx2-1 regulates differentiation state and inhibits tumor initiation in KrasG12D-driven lung adenocarcinoma.
(A) Hematoxylin and eosin (H&E) stain of lung tumors in KrasLSL-G12D; Nkx2-1F/+ (left) and KrasLSL-G12D; Nkx2-1F/F mice (right). Scale bar, 50 μm.
(B) – (C) Analysis of lung tumors in KrasLSL-G12D; Nkx2-1F/+ (left) and KrasLSL-G12D;Nkx2-1F/F mice (right). Scale bar, 100 μm.
(B) Immunohistochemistry (IHC) for Nkx2-1.
(C) Alcian blue/PAS stain for mucin. Inset: IHC for Muc5AC.
(D) Quantitation of tumor burden six weeks after Ad-Cre infection in KrasLSL-G12D;Nkx2-1F/F (n=6) mice and KrasLSL-G12D; Nkx2-1F/+ (n=7) controls. *p=0.001.
(E) Quantification of the number of neoplastic lesions per mm2 of lung. Lungs were analyzed 2 weeks after Ad-Cre infection of KrasLSL-G12D; Nkx2-1F/F (n=6) mice and KrasLSL-G12D; Nkx2-1F/+ (n=4) controls. * p<0.0007.
Data are represented as mean +/− SEM.
See also Figure S1
The changes observed upon engineered Nkx2-1 deletion recapitulate some, but not all of the changes that take place in tumors from KrasLSL-G12D; p53F//F mice that stochastically downregulate Nkx2-1 (Winslow et al., 2011). Like the engineered Nkx2-1-deficient tumors generated here, Nkx2-1-negative tumors from that study often had a glandular architecture and produced more mucin than Nkx2-1-positive tumors (Figure S1C). However, some tumors with stochastic Nkx2-1 loss also progressed to a poorly differentiated state in which cells did not form glands and produced little if any mucin ((Winslow et al., 2011) and data not shown). Furthermore, Nkx2-1-negative tumors from KrasLSL-G12D; p53F//F mice typically expressed the embryonal proto-oncogene Hmga2, which was not detectable in most Nkx2-1-deleted tumors (Figure S1D). This suggests that multiple genetic or epigenetic changes, in addition to Nkx2-1 loss, may be required for the emergence of the poorly differentiated Hmga2-positive tumors from Nkx2-1-positive tumors.
Tumors arising in KrasLSL-G12D; Nkx2-1F/F mice bear a striking morphologic resemblance to mucinous lung adenocarcinoma in humans (Figure S1E), a subtype that has not been previously modeled in the mouse. This subtype comprises ~5-10% of human lung adenocarcinomas (Hata et al., 2010; Kunii et al., 2011), typically lacks NKX2-1 expression (Figure S1E), and harbors activating mutations in KRAS in 60-80% of cases (Finberg et al., 2007). Mucinous adenocarcinomas have been hypothesized to arise from Kras-mutant, Nkx2-1-positive precursor lesions (Garfield, 2008). Our data raise the possibility that loss of Nkx2-1 expression may be sufficient for the transition from precursor lesions such as atypical alveolar hyperplasia (AAH) to mucinous adenocarcinoma. Consistent with these results, KrasG12D overexpression in Nkx2-1 heterozygous mice has been reported to induce lung adenocarcinomas with areas of mucinous differentiation (Maeda et al., 2012).
Nkx2-1 limits tumor initiation by KrasG12D
Nkx2-1 appears to function as either an oncogene or an inhibitor of tumor progression in different contexts (Kendall et al., 2007; Kwei et al., 2008; Tanaka et al., 2007; Weir et al., 2007; Winslow et al., 2011). We therefore quantitated the effect of Nkx2-1 loss on overall lung tumor burden in vivo. At 6 weeks after initiation, tumor burden was nine-fold higher in KrasLSL-G12D; Nkx2-1F/F mice than KrasLSL-G12D; Nkx2-1F/+ controls (Figure 1D). We obtained similar results in p53- deficient tumors (Figure S1F). KrasLSL-G12D; Nkx2-1F/F mice exhibited a significantly greater number of neoplastic lesions at 2 weeks post-initiation than control mice (Figure 1E). In contrast, the difference in proliferation between the two groups was modest (Figure S1G,), and apoptosis was virtually undetectable (Figure S1H). Thus, Nkx2-1 loss augments tumor burden predominantly by increasing the number of lesions initiated by KrasG12D. Of note, tissue-specific Nkx2-1 deletion also enhances chemically-induced thyroid tumorigenesis (Hoshi et al., 2009).
Since we have previously implicated stochastic Nkx2-1 loss in the progression of KrasG12D; p53-deficient lung adenocarcinomas to a metastatic state (Winslow et al., 2011), we determined whether engineered Nkx2-1 loss promotes metastasis in KrasG12D; p53-proficient lung tumors, which rarely, if ever, metastasize (Jackson et al., 2001). However, no metastases were observed up to 33 weeks after tumor initiation. We also asked whether Nkx2-1 deletion could enhance metastasis in KrasG12D;p53-deficient lung adenocarcinomas (Jackson et al., 2005), but we did not detect a significant difference in the proportion of mice with metastatic disease (Figure S1I). Tumors arising in KrasLSL-G12D; p53F/F; Nkx2-1F/F mice tumors were initially similar to those that arose in KrasLSL-G12D;Nkx2-1F/F mice, but progressed to a poorly differentiated (Hmga2-positive) state over time (Figure S1J). All metastases identified in these mice were high grade and Hmga2-positive (Figure S1J). Thus, Nkx2-1 deletion is not sufficient to induce the full complement of differentiation-state changes required for metastasis. Additional genetic and epigenetic changes, including de-repression of Hmga2, are likely required for tumors reach a highly metastatic state.
Nkx2-1 deletion induces mucinous alveolar hyperplasia in the adult lung
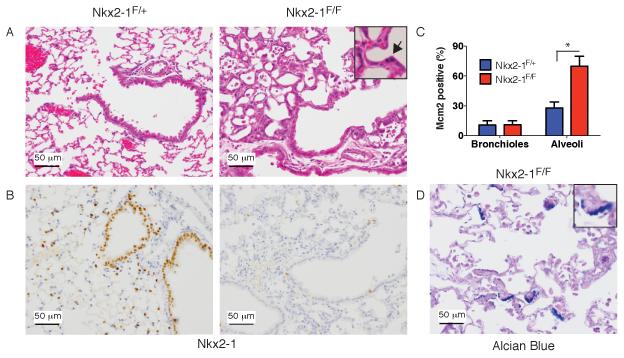
To evaluate the role of Nkx2-1 in regulating the differentiation of normal adult lung epithelium, we used a tamoxifen-inducible CreERT2 fusion expressed from the ubiquitous Rosa26 locus (Ventura et al., 2007) to delete Nkx2-1 throughout the adult lung epithelium. Nkx2-1 deletion induced diffuse epithelial (cytokeratin 8-positive) hyperplasia in the alveolar space (Figure 2A-B and S2A). Hyperplastic cells lacked expression of canonical Nkx2-1 targets such as proSPC and Clara Cell Secretory Protein (CCSP) (Figure S2A-B). At the highest dose of tamoxifen tested, mice became dyspneic and moribund within a week of deletion, possibly as a result of decreased surfactant production and diffuse epithelial hyperplasia throughout the alveoli (Figure S2C).
Figure 2. Nkx2-1 deletion induces alveolar epithelial hyperplasia in the adult lung.
(A-B) Lungs from RosaCreERT2; Nkx2-1F/+ (left) or Nkx2-1F/F (right) mice 11 days after tamoxifen. Scale bar, 50 μm.
(A) H&E stain. Inset: arrow marks mitosis in hyperplastic epithelium.
(B) IHC for Nkx2-1.
(C) Quantitation of proliferation marker Mcm2 in RosaCreERT2; Nkx2-1F/+ (n=4) and RosaCreERT2; Nkx2-1F/F mice (n=7) 1-2 weeks after tamoxifen administration. Type 2 pneumocytes were scored in the alveoli of Nkx2-1F/+ mice and hyperplastic (recombined) cells were scored in Nkx2-1F/F mice. *p<0.0001 Data are plotted as mean +/− SD.
(D) Alcian blue/PAS stain of alveolar hyperplasia 4 months after Nkx2-1 deletion. Scale bar, 50 μm.
See also Figure S2
In contrast to the alveoli, there was no morphologic evidence of proliferation in the airways. At 1-2 weeks after tamoxifen administration, hyperplastic alveolar cells had a far higher Mcm2-positive rate than normal type 2 pneumocytes, whereas Nkx2-1-deleted airway cells showed no increase in Mcm2 positivity (Figure 2C). Longer term studies show that the proliferation rate of hyperplastic alveolar cells declines over the course of 8 weeks (Figure S2D-E), and Nkx2-1 deleted cells never give rise to macroscopic tumors, even after several months of observation.
Nkx2-1 deletion also altered the differentiation state of epithelial cells in the adult lung. Although the hyperplastic alveolar cells were predominantly non-mucinous at 1-2 weeks after recombination, we began to observe mucin accumulation in these cells at 4-8 weeks (Figure 2D). We also observed an increase in the number of airways (including distal bronchioles) with mucin-positive cells in mice that survived for >1 month after tamoxifen-mediated Nkx2-1 deletion (Figure S2F-G).
Taken together, these data show that Nkx2-1 inhibits proliferation in a cell type-specific manner in the adult lung, but that its loss is not sufficient for frank tumor formation. In addition, Nkx2-1 represses mucinous differentiation throughout the pulmonary epithelium.
Nkx2-1 deletion in established tumors alters differentiation and induces proliferation
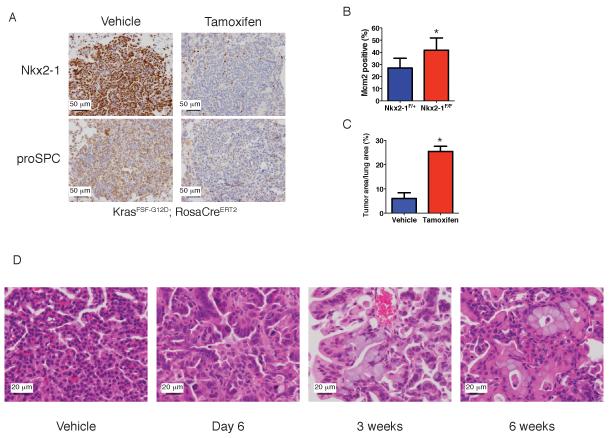
Deletion of Nkx2-1 at the time of tumor initiation might impact the differentiation state of the cell of origin and thereby alter the phenotype of the subsequent tumor. It was therefore critical to develop a system to study the consequences of Nkx2-1 deletion in established tumors. In order to temporally separate KrasG12D activation from Nkx2-1 deletion, we used a conditional allele of KrasG12D that is activated by Flp recombinase (KrasFSF-G12D) (Young et al., 2011). We delivered adenovirus expressing Flp (Ad-Flp) to the lungs of KrasFSF-G12D;Nkx2-1F/F;RosaCreERT2 mice, allowed tumors to grow for 2-7 months, and then treated the mice with tamoxifen to delete Nkx2-1. Expression of Nkx2-1 and its target proSPC was lost in most tumor cells within a week after tamoxifen administration (Figure 3A and S3A). Six days after Nkx2-1 deletion, we observed a significant increase in tumor cell proliferation compared to controls (Figure 3B), with no increase in apoptosis (Figure S3B). Thus, Nkx2-1 deletion in established tumors leads to rapid changes in the cell cycle state of tumor cells.
Figure 3. Nkx2-1 deletion in established tumors induces proliferation and differentiation state changes.
(A) IHC for Nkx2-1 (top) and proSPC (bottom) in tumors from KrasFSF-G12D;RosaCreERT2 mice six days after injection of vehicle (Nkx2-1F/+ mice) or tamoxifen (Nkx2-1F/F mice). Scale bar, 50 μm.
(B) Quantitation of proliferation marker Mcm2 in tumors of KrasFSF-G12D;RosaCreERT2 mice six days after injection of vehicle (Nkx2-1F/+ mice, n=12 tumors) or tamoxifen (Nkx2-1F/F mice, n=8 tumors). Data are represented as mean +/− SD. *p=0.002
(C) Quantitation of tumor burden in KrasFSF-G12D; RosaCreERT2 mice 6 weeks after injection of vehicle (Nkx2-1F/+, n=6 mice) or tamoxifen (Nkx2-1F/F mice, n=4 mice). Mice were treated three months after tumor initiation. Low dose tamoxifen was administered to avoid lethal alveolar hyperplasia, yielding ~50% tumor cell recombination. Data are represented as mean +/− SEM. *p<0.001
(D) Lung tumors (H&E) from KrasFSF-G12D; RosaCreERT2; Nkx2-1F/F mice at six days to six weeks after tamoxifen administration. Vehicle: tumor from Nkx2-1F/+ mouse. Scale bar, 20 μm.
See also Figure S3
Nkx2-1 deletion also promoted long term tumor growth. Six weeks after Nkx2-1 deletion in KrasFSF-G12D mice, the total burden of neoplastic cells was about four fold higher than controls (Figure 3C). Nkx2-1 deletion had similar consequences in the KrasLA2 model of lung adenocarcinoma (Figure S3C), in which tumors arise via spontaneous recombination of a latent KrasG12D allele (Johnson et al., 2001). These data show that Kras-driven tumorigenesis is enhanced by loss of Nkx2-1 during tumor progression as well as at the time of initiation.
Finally, we tracked sequential changes in tumor differentiation state after Nkx2-1 loss. At 6-10 days after Nkx2-1 deletion in tumors from KrasFSF-G12D mice, tumor cells became elongated, nuclear size increased, and chromatin appeared less compact (Figure 3D). By 3 weeks after deletion, many tumor cells had begun to produce mucin (Figure 3D). Within six weeks of deletion, many Nkx2-1-deleted cells had reorganized themselves into glandular structures within otherwise papillary Nkx2-1-positive tumors (Figure 3D). Nkx2-1 deletion also induced mucin production and glandular rearrangements in tumors of KrasLA2 mice (Figure S3E). Thus, tumor cells undergo dramatic and progressive changes in their differentiation state upon Nkx2-1 loss. These changes occurred in essentially all tumors, suggesting that they are the direct consequence of Nkx2-1 deletion and not secondary mutations.
Nkx2-1 prevents activation of gastric differentiation in lung adenocarcinoma
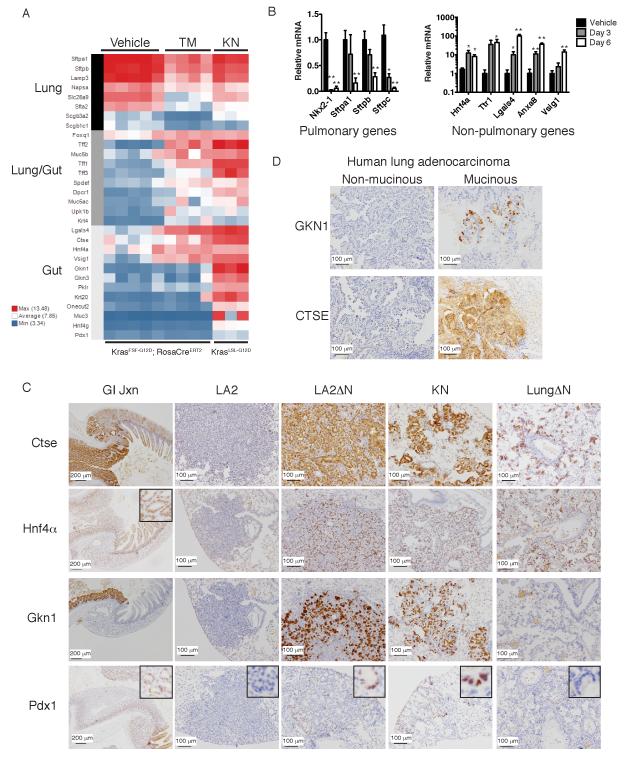
To identify genes regulated by Nkx2-1 in lung tumors, we harvested RNA from 6-7 month old KrasFSF-G12D lung tumors at 6 days after tamoxifen-induced Nkx2-1 deletion (designated “TM”) and compared their mRNA expression profile with vehicle-treated control tumors by Affymetrix exon array. 669 genes exhibited a significant change (at least 2 fold, p<0.05) in expression levels 6 days after Nkx2-1 deletion (Table S1), including 363 upregulated and 306 downregulated genes. Many canonical Nkx2-1 targets, as well as other genes expressed in the distal lung epithelium, declined after Nkx2-1 deletion (Figure 4A). Metacore pathway analysis (Table S2) showed that proliferation-associated genes were enriched in Nkx2-1 deleted tumors.
Figure 4. Nkx2-1 represses gastric differentiation in lung adenocarcinoma.
(A) Heat map illustrating differences in expression of tissue-restricted genes between Nkx2-1-positive and negative lung adenocarcinomas. Vehicle: tumors from KrasFSF-G12D; RosaCreERT2; Nkx2-1F/+ mice six days after treatment with vehicle (n=5). TM: tumors from KrasFSF-G12D; RosaCreERT2; Nkx2-1F/F mice six days after treatment with tamoxifen (n=4). KN: tumors from KrasLSL-G12D; Nkx2-1F/F mice 3-4 months after initiation by Ad-Cre (n=3).
(B) qRT-PCR for lung- or GI-restricted genes. Vehicle: KrasG12D-driven tumors 6-7 months after initiation by Ad-Flp or Ad-Cre (n=9 tumors). Day 3-6: Tumors from KrasFSF-G12D; RosaCreERT2; Nkx2-1F/F mice 3 or 6 days after tamoxifen (n=7 or 9 tumors, respectively). Data are represented as mean +/− SEM. *p<0.05 versus Vehicle (Nkx2-1-positive tumors), **p<0.001.
(C) IHC for GI-restricted proteins Cathepsin E (Ctse), Hnf4α, Gastrokine 1 (Gkn1), and Pdx1. GI Jxn: junction between stomach (left side of image) and duodenum (right side of image). LA2: tumor from KrasLA2; RosaCreERT2; Nkx2-1F/F mouse 10 weeks post vehicle. LA2ΔN: tumor from KrasLA2; RosaCreERT2; Nkx2-1F/F mouse 10 weeks post tamoxifen. KN: tumor from KrasLSL-G12D; Nkx2-1F/F mice 1-2 months after initiation. LungΔN: lung from RosaCreERT2; Nkx2-1F/F mouse 10 weeks post tamoxifen. Scale bar, 200 μm (GI Jxn), 100 μm (all others).
(D) IHC for GKN1 and CTSE in human lung adenocarcinomas. Scale bar, 100 μm.
We also harvested mRNA from tumors in KrasLSL-G12D; Nkx2-1F/F mice (designated “KN”) 3-4 months after initiation by Ad-Cre. 1828 genes were differentially expressed between KN tumors and Nkx2-1-positive tumors (Table S1), including 1137 upregulated and 691 downregulated genes. Surprisingly, Metacore analysis for disease-specific gene collections and networks identified the categories “Stomach Neoplasms” and “Stomach Diseases” as enriched in KN tumors (Table S2). Evaluation of individual genes revealed that Nkx2-1 deletion led to the de-repression of gastrointestinal (GI) transcripts in both TM and KN groups (Figure 4A, 4B and S4A). These included some genes that are expressed in both the proximal lung and the GI tract, as well as others that are never expressed in the lung (Su et al., 2009). These changes in mRNA levels occurred rapidly, with Nkx2-1 levels reaching their nadir by day 3 and Nkx2-1 target genes (Sftpa1, Sftpb and Sftpc) declining from day 3 to day 6 (Figure 4B). Several GI transcripts also showed induction at day 3.
We next asked whether Nkx2-1 deletion caused a differentiation state change mimicking a specific tissue or cell-type within the GI tract. We therefore evaluated the levels of 1137 genes upregulated in KN tumors across a large panel of normal murine tissues (Su et al., 2009). We found that this gene set is highly correlated with genes expressed in the stomach, and to a lesser extent in the small and large intestines, but not with expression patterns in any other tissues (Figure S4B). Several de-repressed genes are expressed at much greater levels in the stomach than the intestines, including Gkn1, Gkn3, Vsig1, Ctse and Muc5AC (Menheniott et al., 2010; Oien et al., 2004; Scanlan et al., 2006; Su et al., 2009). In addition, Nkx2-1-negative tumors and normal stomach express only the isoforms of Hnf4a driven by the P2 promoter (data not shown), whereas both the P1 and P2 promoters are active in the intestine (Tanaka et al., 2006). Nkx2-1-deleted tumors did not express Muc2, a type of mucin expressed in the intestinal but not the gastric mucosa (Figure S4C). These data suggest that Nkx2-1 loss causes tumors to adopt a new identity that is closest overall to the gastric epithelium.
We used IHC to evaluate the expression pattern of four gastric proteins in tumors and normal lung after Nkx2-1 deletion (Figure 4C). Hnf4α and Pdx1 are transcription factors that regulate gastrointestinal differentiation (Hayhurst et al., 2001; Offield et al., 1996). Cathepsin E (Ctse) and Gastrokine 1 (Gkn1, a stomach-specific protein), are secreted proteins (Chlabicz et al., 2011; Oien et al., 2004). These four proteins were undetectable in normal lung and Nkx2-1-positive control tumors, but were expressed KrasLA2 tumors 10 weeks after Nkx2-1 deletion (LA2ΔN) and in KN tumors. Hnf4α was expressed in essentially all Nkx2-1-deleted tumor cells, whereas expression of the other proteins was more heterogeneous from cell to cell.
In normal (Kras-wild type) lung, Nkx2-1 deletion led to the expression of Hnf4α and Cathepsin E in hyperplastic alveolar cells, but not Gastrokine1 or Pdx1 (Figure 4C). None of these proteins was detectable in Nkx2-1-deleted bronchiolar epithelium. Thus, a specific cell type in the distal adult lung is primed both to proliferate and adopt a gastric cell fate upon Nkx2-1 deletion. The capacity of these cells to de-repress gastric markers appears more limited than Kras-driven tumors, suggesting that KrasG12D may activate specific signaling pathways that augment the change in differentiation state.
Finally, we determined whether human mucinous lung adenocarcinomas also exhibit evidence of gastric differentiation. A strong inverse correlation between NKX2-1 and HNF4α expression was recently demonstrated in human lung adenocarcinomas (Kunii et al., 2011). We therefore evaluated the expression of two stomach-restricted proteins (GKN1 and CTSE) in 37 human lung adenocarcinomas. In NKX2-1-negative, mucinous human lung adenocarcinomas (n=11), we found that GKN1 was expressed in 6/11 cases and that CTSE was strongly and diffusely expressed in all eleven tumors (Figure 4D and S4D). In contrast, NKX2-1-positive lung adenocarcinomas (n=26) were entirely negative for GKN1. Most NKX2-1-positive lung tumors were either negative (n=14) or only focally positive (n=10) for CTSE. These results suggest that NKX2-1 has an evolutionarily conserved ability to prevent gastric differentiation in the adult pulmonary epithelium.
Nkx2-1 functions as a transcriptional activator in lung adenocarcinoma
In the hematopoietic system, master regulators of differentiation can activate one differentiation state and repress alternative fates by directly binding to genes characteristic of each state (Wontakal et al., 2012). Although Nkx2-1 binds numerous pulmonary genes, it is not known whether it can also directly bind and repress gastric genes. We therefore used chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) to map Nkx2-1 binding sites in lung tumors from KrasLA2 mice and determine which differentially expressed genes are associated with Nkx2-1 binding sites. Nkx2-1 binding sites were enriched at promoters relative to other locations in the genome (15.9% percent of sites occurred between 3 kb upstream and 1 kb downstream of transcriptional start sites), consistent with a role in transcriptional activation (Figure 5A and S5A). Nkx2-1 binding sites were significantly associated with promoters of genes that decrease after acute Nkx2-1 deletion (Fisher’s exact test, p<10−17) and depleted at genes that are de-repressed (p<10−5). Specifically, Nkx2-1 binds 58% of genes with decreased expression after deletion but only 23% of de-repressed genes (Figure 5B and S5B). We also quantitated Nkx2-1 binding at a set of 158 genes that were both increased in KN tumors and also expressed at least four fold higher in normal stomach than normal lung. Only 12.7% of these “stomach high” genes were bound by Nkx2-1 (Figure 5B). Selected binding sites were confirmed with qPCR and an independent Nkx2-1 antibody (Figure S5C). Taken together, these data demonstrate the role of Nkx2-1 as a direct transcriptional activator in lung tumors but suggest that its ability to repress gastric differentiation may occur by mechanisms other than direct promoter binding.
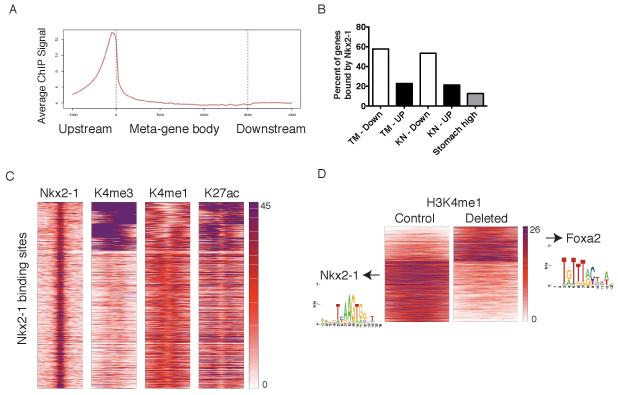
Figure 5. Global analysis of Nkx2-1 binding sites in lung adenocarcinoma and correlation with gene expression.
(A) Average Nkx2-1 ChIP signal on normalized gene structure across all genes (gene bodies adjusted to 3 kb).
(B) Percent of differentially expressed genes with Nkx2-1 binding sites in their promoters (from 3 kb upstream to 1 kb downstream of the TSS). TM: tumors from KrasFSF-G12D; RosaCreERT2; Nkx2-1F/F mice six days after treatment with tamoxifen. KN: tumors from KrasLSL-G12D; Nkx2-1F/F mice 3-4 months after initiation by Ad-Cre. “Down”: genes whose expression was significantly lower in Nkx2-1-deleted group vs. control tumors. “UP”: genes whose expression was significantly higher in Nkx2-1-deleted group than in control tumors.
(C) Heatmaps displaying k means clustering (n=7 groups) of histone modifications at each Nkx2-1 binding site (n= 29,782) in the lung adenocarcinoma genome. 1 kb regions around the center of each Nkx2-1 binding site are depicted (5 kb for H3K27me3).
(D) Heatmap centered on H3K4me1 peaks (n=15,089) depicting significant changes in enrichment six days after Nkx2-1 deletion. Heat maps are centered on H3K4me1 peaks and display 1 kb regions around the center of each peak. Arrows point to motifs enriched in each subgroup.
See also Figure S5.
To characterize the epigenetic state of Nkx2-1 binding sites in lung tumors, we performed ChIP-Seq for four histone post-translational modifications: H3K4me1 (a marker of enhancers and promoters), H3K4me3 (a marker of promoters), H3K27ac (a marker of activity at promoters and enhancers) and H3K27me3 (a marker of Polycomb-mediated gene repression) (reviewed in Zhou et al., 2011). H3K4me1 and H3K27ac modifications were detectable at most Nkx2-1 binding sites, a subset of which also exhibited high levels of H3K4me3 (Figure 5C). Thus, Nkx2-1 binding sites have epigenetic features of active enhancers and promoters. We also evaluated Nkx2-1 and H3K27me3 levels at the transcription start sites (TSS) of the genes de-repressed by acute Nkx2-1 deletion (Figure S5C). Genes without Nkx2-1 binding had higher levels of H3K27me3 near their TSS than genes bound by Nkx2-1.
These results raise the question of how Nkx2-1 loss indirectly de-represses genes that have undergone polycomb-mediated silencing. The greatest number of epigenetic changes induced by Nkx2-1 deletion occurred in the H3K4me1 and H3K27ac modifications (Figure 5D and S5D), consistent with reports that these modifications correlate more closely with differentiation state than others (Creyghton et al., 2010; Shen et al., 2012). Motif analysis revealed that in the H3K4me1 dataset, Nkx2-1 binding motifs were enriched in the peaks that decreased upon deletion, but not in peaks that increased (Figure 5D, left). In contrast, H3K4me1 peaks that increase with Nkx2-1 deletion were enriched for the motif bound by Foxa1 and Foxa2 (Figure 5D, right). We therefore investigated whether Foxa1/2 might play a role in the induction of gastric differentiation by Nkx2-1 deletion.
Nkx2-1 regulates global Foxa1/2 binding in lung adenocarcinoma
NKX2-1 physically interacts with FOXA1/2 in human lung adenocarcinoma cells, and the two proteins bind to adjacent sites at several genes expressed in the lung to cooperatively activate transcription (Costa et al., 2001; Minoo et al., 2007). Foxa1/2 are also expressed in the GI tract and liver, where they activate several genes de-repressed by Nkx2-1 deletion, including Hnf4a and Pdx1 (Gao et al., 2008; Wederell et al., 2008). Foxa1/2 are expressed in both Nkx2-1-positive and deleted murine lung tumors (Figure S6A). We hypothesized that physical interaction with Nkx2-1 is required for Foxa1/2 binding to pulmonary genes in lung adenocarcinoma, and that loss of Nkx2-1 releases Foxa1/2 from these loci, enabling them to activate non-pulmonary genes.
To test this hypothesis, we confirmed that Nkx2-1 physically interacts with Foxa1/2 in murine lung adenocarcinoma cells (Figure 6A). We then performed ChIP-Seq for Foxa1/2 from tumors from KrasLA2 mice to determine whether loss of Nkx2-1 would alter the distribution of Foxa1/2 binding sites. We identified 23,039 significantly enriched peaks in control tumors and 14,526 in tumors 6 days after Nkx2-1 deletion (p<10−6) (Figure S6B). Comparison of Foxa1/2 peaks in each dataset revealed numerous changes in binding near genes that are differentially expressed upon Nkx2-1 deletion (Figure 6B-D, S6C and data not shown).
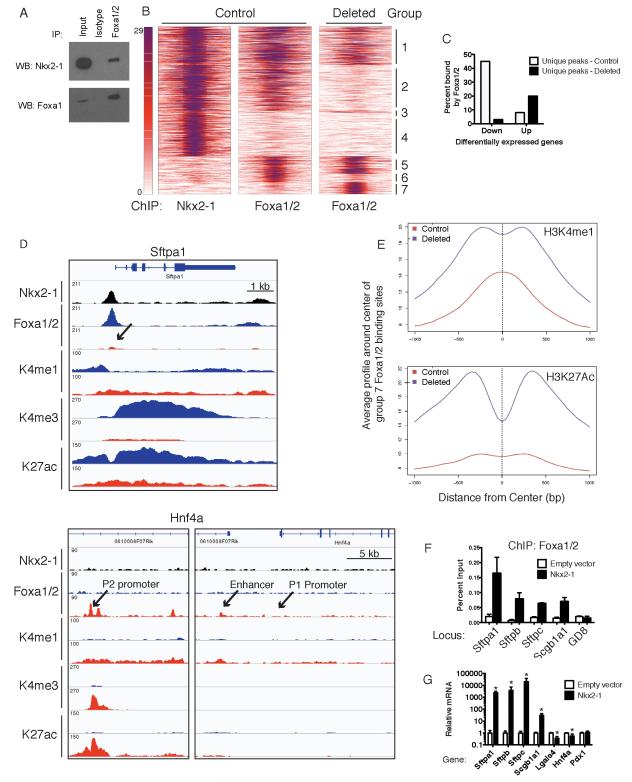
Figure 6. Nkx2-1 regulates global Foxa1/2 binding in lung adenocarcinoma.
(A) Immunoprecipitation of Foxa1/2 from an Nkx2-1-deleted adenocarcinoma cell line transduced with MSCV-Nkx2-1 expression vector followed by immunoblot for Nkx2-1 (top) or Foxa1 (bottom).
(B) Heatmaps correlating Nkx2-1 binding sites in control KrasLA2 lung tumors with Foxa1/2 binding sites in control and Nkx2-1-deleted tumors as determined by ChIP-Seq analysis. The Y-axis contains all peaks that are bound by either Nkx2-1 or Foxa1/2 in control and deleted tumors (n= 42,762). Peaks are divided into 7 groups based on whether they are unique to one dataset or shared between 2 or 3 datasets. Heat maps depict the signal of each transcription factor and extend 500 bp on each side of the peak center.
(C) Percent of differentially expressed genes six days after Nkx2-1 deletion (tamoxifen group) that are associated with Foxa1/2 peaks unique to control or deleted tumors. Unique peaks were defined as those present at p<10−6 in control tumors but not p<10−3 in Nkx2-1-deleted tumors.
(D) Representative signal of Nkx2-1, Foxa1/2 and histone modifications at Sftpa1 and Hnf4a. Black: Nkx2-1 ChIP. Blue: control tumors. Red: Nkx2-1 deleted tumors.
(E) Average signal profile of H3K4me1 (left) and H3K27ac (right) around group 7 Foxa1/2 binding sites in control and Nkx2-1-deleted tumors.
(F) ChIP-qPCR for Foxa1/2 at pulmonary gene promoters in an Nkx2-1-negative lung adenocarcinoma cell line derived from a KrasLSL-G12D; p53F/F; Nkx2-1F/F mouse (n=3-4 ChIPs/locus). Cells were stably transduced with MSCV-Nkx2-1 retrovirus or empty vector control. Data are represented as mean +/− SEM. *p<0.03 vs. empty vector cells for each locus. GD8: negative control region.
(G) qRT-PCR analysis of Nkx2-1-deleted lung adenocarcinoma cell lines after stable transduction with MSCV-Nkx2-1 or empty vector. Data are pooled from three independent experiments and represented as mean +/− SD. *p<0.02.
We assembled a set of all sites bound by Nkx2-1 and/or Foxa1/2 (in both control and Nkx2-1-deleted tumors), then classified each site as unique to one dataset or shared between datasets (see Extended Experimental Procedures and Table S4). We identified 21,638 sites with evidence of binding by both Nkx2-1 and Foxa1/2 in control tumors (groups 1-2) (Figure 6B and Table S3-4). Strikingly, Foxa1/2 binding was not detectable at more than half of these sites in Nkx2-1-deleted tumors (group 2), consistent with the possibility that Foxa1/2 are recruited to those sites by physical interaction with Nkx2-1. Loss of Foxa1/2 binding could be identified at canonical Nkx2-1 target genes, including Sftpa1 and Sftpb (Figure 6D and data not shown). We also identified sites that were bound only by Nkx2-1 (group 4) or Foxa1/2 (groups 5-6) in control tumors. Our analysis further revealed 3513 Foxa1/2 binding sites in deleted tumors that were not detectable in control tumors (groups 3 and 7), most of which did not overlap with Nkx2-1 binding sites (group 7, n=2903). Many of these de novo sites could be found near genes de-repressed by Nkx2-1 deletion, including the enhancer and the P2 promoter, but not the P1 promoter, of Hnf4a (Figure 6D).
Motif analysis revealed that sites in groups 1-2 were enriched in both Nkx2-1 and Foxa1/2 binding motifs, whereas group 3 sites (Nkx2-1 unique) contained Nkx2-1 but not Foxa1/2 binding motifs (Table S3). In contrast, de novo Foxa1/2 sites (group 7) were enriched not only for Foxa1/2 but also for the Hnf4α binding motif, which was not enriched in any other subgroup. Foxa1/2 and Hnf4α physically interact and coordinately regulate gene expression in non-pulmonary tissue (Hoffman et al., 2010).
We observed a highly significant association between dynamic Foxa1/2 peaks and differentially expressed genes (p<10−25, Fisher’s exact test). In order to capture peaks bound to enhancers as well as promoters, we assigned Foxa1/2 peaks to genes whose TSS was up to 10 kb from the peak (see Extended Experimental Procedures). 45% of genes whose levels declined six days after Nkx2-1 deletion were associated with Foxa1/2 peaks unique to control tumors (Figure 6C and S6C), whereas only 3% of these genes were associated with de novo Foxa1/2 peaks. In addition, 20% of genes that were de-repressed by Nkx2-1 deletion were associated with de novo Foxa1/2 peaks, whereas only 8% were associated with peaks unique to control tumors. Five of the top ten genes de-repressed by acute Nkx2-1 deletion were associated with de novo Foxa1/2 peaks (data not shown). Foxa1/2 inhibition by RNA interference in an Nkx2-1-deleted lung adenocarcinoma cell line reduced the levels of several gastrointestinal transcripts that are de-repressed by Nkx2-1 deletion in vivo (Figure S6D-E).
We next evaluated levels of histone modifications in each group of Nkx2-1 and Foxa1/2 binding sites (Figure 6E, S6F and Table S4). In control tumors, sites with evidence of Nkx2-1 binding (groups 1-4) had the highest levels of H3K4me1, H3K4me3 and H3K27ac and lowest levels of H3K27me3. Upon Nkx2-1 deletion, group 2 sites, which lose Foxa1/2 binding, also exhibit a decline in H3K4me1 and H3K27ac signal (Figure S6F). This suggests that combinatorial binding of Nkx2-1 and Foxa1/2 is critical for maintaining an active chromatin state at these loci. De novo Foxa1/2 binding sites (group 7) exhibited the highest levels of H3K27me3 and lowest levels of H3K4me3 and H3K27ac in control tumors (Figure S6F). The H3K4me1 modification exhibited a clear peak at de novo Foxa1/2 binding sites in control tumors, albeit of lower intensity than in other subgroups. Upon Nkx2-1 deletion, the levels of H3K4me1 and H3K27ac modifications at de novo Foxa1/2 sites increased and their distribution shifted from unimodal to bimodal (Figure 6E and S6G). Bimodal distribution of H3K4me1 at Foxa2 binding sites has previously been correlated with tissue-specific gene activity and nucleosome displacement (Hoffman et al., 2010). Thus, de novo Foxa1/2 binding sites exhibit an increase in histone modifications associated with an active chromatin state in Nkx2-1-deleted tumors.
We next asked whether Nkx2-1 re-expression was sufficient to modulate Foxa1/2 binding in an Nkx2-1-deleted lung adenocarcinoma cell line. Control cells exhibited no evidence of Foxa1/2 binding near several pulmonary genes that are bound by Foxa1/2 in Nkx2-1-positive tumors (Figure 6F). Re-expression of Nkx2-1 restored Foxa1/2 binding to all of these loci (Figure 6F) and induced expression of the corresponding genes (Figure 6G). Thus, Nkx2-1 appears to be both necessary and sufficient for Foxa1/2 binding at these genes. In contrast, Nkx2-1 re-expression caused a partial decrease in Hnf4α and Lgals4 mRNA levels (Figure 6G), but Foxa1/2 remained bound to sites near these genes (Figure S6H). Furthermore, Nkx2-1 affected neither Pdx1 expression (Figure 6G) nor Foxa1/2 binding at the Pdx1 area IV enhancer (Figure S6H). Thus, the epigenetic changes induced by Nkx2-1 deletion are not completely reversed by Nkx2-1 re-expression in vitro.
These results demonstrate that Nkx2-1 is required for Foxa1/2 binding to a large number of sites in the lung adenocarcinoma genome, including many genes highly expressed in distal lung. Furthermore, the loss of Nkx2-1 enables Foxa1/2 to bind de novo sites, many of which are near genes whose expression is normally restricted to the GI tract.
Initiation of Nkx2-1-negative lung adenocarcinomas is dependent upon Hnf4α expression
Finally, we sought to determine whether the gastric differentiation program regulates the growth of Nkx2-1-negative lung adenocarcinomas. We focused our studies on the transcription factor Hnf4α because it promotes epithelial differentiation (Cattin et al., 2009; Hayhurst et al., 2001) and regulates cancer progression in the murine GI tract and liver (Darsigny et al., 2010; Hatziapostolou et al., 2011). Hnf4α also represses Hmga2 in human liver cancer cells (Santangelo et al., 2011). Furthermore, Hnf4α is expressed in human mucinous lung adenocarcinomas (Kunii et al., 2011). Given that lung tumors in KrasLSL-G12D; Nkx2-1F/F mice are well-differentiated and non-metastatic, we hypothesized that Hnf4α regulates a gastric differentiation program that prevents Nkx2-1-negative tumors from reaching an embryonal and/or metastatic differentiation state.
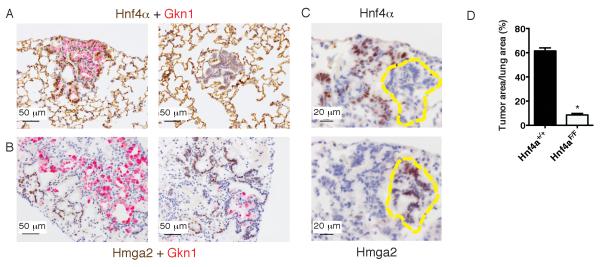
To test this hypothesis, we utilized a conditional allele of Hnf4a (Hayhurst et al., 2001) to generate KrasLSL-G12D; Nkx2-1F/F; Hnf4aF/F mice. We infected mice with a lentivirus (Lenti-Cre) that integrates into the genome and stably expresses Cre, thereby increasing the probability that all conditional alleles are recombined in an infected cell. By IHC, ~50% of lesions exhibited a complete loss of Hnf4α protein expression (Figure 7A-C and data not shown). Hnf4α–negative cells did not exhibit major morphologic differences from adjacent Hnf4α-positive cells and were classified as mucinous, well-differentiated adenocarcinoma. Hnf4α was required for the expression of Gkn1, but not Ctse (Figure 7A and S7A). Furthermore, combined deletion of Hnf4a and Nkx2-1 was sufficient to de-repress the embryonal protein Hmga2 in neoplastic cells (Figure 7B-C and S7A). We also found that concomitant deletion of Nkx2-1 and Hnf4a is sufficient to de-repress Hmga2 in the distal lung epithelium in the absence of oncogenic Kras (Figure S7B). Thus, Hnf4α activates a portion of the gastric differentiation program and represses the embryonal proto-oncogene Hmga2 in the absence of Nkx2-1.
Figure 7. Nkx2-1 negative lung adenocarcinoma initiation is dependent on Hnf4α.
(A) Dual color IHC for total Hnf4α (brown) and Gkn1 (red) in tumors arising after Lenti-Cre infection in a KrasLSL-G12D; Nkx2-1F/F; Hnf4aF/F mouse. Scale bar, 50 μm.
(B) Dual color IHC for Hmga2 (brown) and Gkn1 (red) in tumors arising after Lenti-Cre infection in a KrasLSL-G12D; Nkx2-1F/F; Hnf4aF/F mouse. Scale bar, 50 μm.
(C) IHC for Hnf4α (P2 isoform) (top) and Hmga2 (bottom) on serial sections of tumors arising four weeks after Lenti-Cre infection in a KrasLSL-G12D; Nkx2-1F/F; Hnf4aF/F mouse. Scale bar, 20 μm.
(D) Quantitation of tumor burden four weeks after Lenti-Cre infection (5 × 105 pfu/mouse) in KrasLSL-G12D; Nkx2-1F/F (n=6) mice and KrasLSL-G12D; Nkx2-1F/F; Hnf4aF/F (n=5) mice. *p<0.0001.
Data are represented as mean +/− SEM.
See also Figure S7.
Surprisingly, we observed a dramatic reduction in tumor burden in KrasLSL-G12D; Nkx2-1F/F; Hnf4aF/F mice compared to controls (Figure 7D). This suggests that most nascent lung tumor cells cannot tolerate the combined loss of these two transcription factors. Deletion of Hnf4a did not impact apoptosis at 5-7 days after tumor initiation, suggesting other mechanisms may be limiting tumor initiation (Figure S7C). Hnf4a deletion had no effect on the number of hyperplastic lesions induced by Nkx2-1 deletion in the context of wild-type Kras (Figure S7D). Thus, partial loss of gastric differentiation and de-repression of Hmga2 is not sufficient to enhance tumor growth or cause tumors to adopt a poorly differentiated state, at least not at this stage of tumor evolution. Instead, these data suggest that Hnf4a deletion puts tumor cells at a growth disadvantage or prevents transformation altogether.
DISCUSSION
Nkx2-1 and Hnf4a regulate active and latent differentiation programs in lung adenocarcinoma
We have demonstrated that Nkx2-1 not only promotes pulmonary differentiation and homeostasis, but also inhibits gastric differentiation in adult alveolar epithelium and in peripheral lung adenocarcinomas. We speculate that the ability of lung epithelial cells to assume a gastric identity after Nkx2-1 loss is a consequence of the lung’s embryologic origins in the developing foregut (Costa et al., 2001). We have also shown that concomitant loss of Nkx2-1 and Hnf4a, a master regulator of GI differentiation, results in the loss of Gastrokine1 expression and de-represses the embryonal protein Hmga2. These data suggest that lung epithelial cells have multiple layers of differentiation, including an active pulmonary differentiation program and a latent gastric differentiation program, and that inactivation of both programs may be required for the de-repression of embryonal genes such as Hmga2 (Figure S7E). Even combined deletion of Nkx2-1 and Hnf4a did not drive tumors directly into a poorly differentiated state, suggesting that other mediators of adult-type differentiation remain active in Nkx2-1/Hnf4α double-negative tumor cells. It is notable that in a previous study, we did not observe high levels of gastric transcripts in Nkx2-1-negative cell lines that were derived from poorly differentiated tumors (Winslow et al., 2011), consistent with the concept that tumor cells may downregulate multiple differentiation programs to reach a highly primitive, metastatic state.
There is evidence that latent differentiation programs may be commonly de-repressed during the evolution of human cancer. Foregut genes are upregulated in human pancreatic intraepithelial neoplasia (PanIN) relative to normal pancreas (Prasad et al., 2005), including some genes (e.g., Ctse) that are also expressed in both human and murine mucinous lung adenocarcinoma. In contrast, precursor lesions for esophageal and gastric carcinomas often adopt an intestinal identity (Yuasa, 2003). These observations suggest that acquisition of an alternative adult cell fate may be a common, and even advantageous, event during cancer evolution.
Nkx2-1 regulates tissue-specific binding by Foxa1/2
The Foxa1/2 transcription factors are required for the proper development of the lung (Wan et al., 2005) and several other tissues (Kaestner, 2010). In the adult, Foxa1/2 physically interact with lineage-specific transcription factors (e.g., Nkx2-1 in the lung, Hnf4α in the liver, and estrogen receptor (ER) in the breast), to cooperatively activate tissue-specific gene expression (Hoffman et al., 2010; Lupien et al., 2008; Minoo et al., 2007). However, the mechanisms by which Foxa1/2 are recruited to genes in a tissue-specific manner have not been fully delineated (Kaestner, 2010). Our data demonstrate that Nkx2-1 is required for proper Foxa1/2 localization and implicate a global change in Foxa1/2 binding as a critical step in the transition from a pulmonary to a gastric differentiation state. In contrast, ER is not required to maintain FOXA1 at its target genes in human breast carcinoma cells (Lupien et al., 2008). It remains to be tested whether other tissue-specific transcription factors regulate Foxa1/2 localization in a similar manner.
Loss of Nkx2-1 also results in the emergence of de novo Foxa1/2 binding sites, a subset of which is marked by the H3K4me1 histone modification and exhibits an increase in H3K27ac modification upon Nkx2-1 deletion. These changes in histone modifications are characteristic of poised promoters and enhancers, which regulate genes whose expression increases during changes in differentiation state (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Wamstad et al., 2012; Zentner et al., 2011). In human breast carcinoma cells, tissue-specific FOXA1 binding may be regulated by H3K4me1 and H3K4me2 modifications (Lupien et al., 2008). These data raise the possibility that the H3K4me1 modification mediates Foxa1/2 recruitment to some of its de novo binding sites after Nkx2-1 deletion, and that these poised DNA elements may be part of the epigenetic basis for the latent gastric differentiation program in lung tumors.
Master regulators of differentiation also regulate tumor growth
The dramatic effects of Nkx2-1 and Hnf4a deletion on tumor burden underscore the connection between differentiation state and tumor progression. We hypothesize that Nkx2-1 is required for full transcriptional induction of the feedback inhibitor Spry2 in response to KrasG12D and that lack of Spry2 induction enhances Kras-driven MAPK activity and tumor growth (Figure S3D). Alternatively, Nkx2-1 may also control the transcription of cell cycle regulators such as E2f3, whose promoter is bound by Nkx2-1 in both developing lung and tumors (Tagne et al., 2012 and data not shown). In contrast, it is not clear why Hnf4a deletion impairs the initiation of Nkx2-1-negative tumors. In non-pulmonary tissues, Hnf4α regulates many processes that may impact tumorigenesis, including proliferation (Bonzo et al., 2012), metabolism (Hayhurst et al., 2001), and inflammation (Hatziapostolou et al., 2011).
In conclusion, our results suggest that some normal cells have latent differentiation programs that can be traced to their developmental origins and may be unleashed during cancer progression. We speculate that just as active differentiation programs influence tumor initiation, latent differentiation programs may shape the specific evolutionary course taken by a neoplastic cell during cancer progression. Our data also imply that even though tumors often evolve toward a primitive state, there may be limits to cellular plasticity during the early stages of tumor evolution.
EXPERIMENTAL PROCEDURES
Mice and tumor initiation
Mice harboring KrasLA2 (Johnson et al., 2001), KrasLSL-G12D (Jackson et al., 2001), KrasFSF-G12D (Young et al., 2011), p53F/F (Jonkers et al., 2001), Nkx2-1F/F (Kusakabe et al., 2006), Hnf4aF/F (Hayhurst et al., 2001), RosaCreERT2 (Ventura et al., 2007) alleles have been previously described. Mice were infected intratracheally with adenovirus or lentivirus (DuPage et al., 2009). Animal studies were approved by the Committee for Animal Care (institutional animal welfare assurance no. A-3125-01).
Histology and immunohistochemistry
Tissues were fixed in 10% formalin overnight and paraffin embedded. Immunohistochemistry (IHC) was performed on a Thermo Autostainer 360 machine followed by hematoxylin counterstain.
qRT-PCR and immunoblotting
Quantitative RT-PCR was performed on Trizol-extracted RNA. qPCR reactions were performed using Taqman Fast Universal Master Mix and probes (Applied Biosystems). Taqman probes and antibodies are listed in Extended Experimental Procedures.
Microarray analysis
Mouse lung tumors were snap frozen in liquid nitrogen. RNA was extracted using Trizol (Invitrogen), analyzed for RNA integrity, and prepared with Affymetrix GeneChip WT Sense Target Labeling and Control Reagents kit, followed by hybridization to Affymetrix GeneChip Mouse Exon 1.0 ST arrays.
ChIP-Seq and analysis
Genome-wide localization of Nkx2-1, Foxa1/2 and four histone modifications was performed on tumors from KrasLA2 mice by chromatin immunoprecipitation followed by high throughput sequencing on an Illumina HiSeq 2000. For ChIP qPCR from cell lines, the MAGnify Chromatin Immunoprecipitation kit (Invitrogen) was used according to the manufacturer’s instructions. qPCR was performed using Fast Sybr Green Master Mix (Applied Biosystems) on a StepOne Plus Real Time PCR system. See Extended Experimental Procedures for antibodies and qPCR primers.
Human studies
Formalin fixed, paraffin-embedded (FFPE) tumors were obtained from the archives of the Department of Pathology at the Brigham and Women’s Hospital and US Biomax. Studies were carried out in accordance with protocols approved by the Partners Health Care Institutional Review Board and the MIT Committee on the Use of Humans as Experimental Subjects.
Supplementary Material
HIGHLIGHTS.
Lung adenocarcinomas adopt a gastric identity upon Nkx2-1 loss
Nkx2-1 regulates tissue-specific Foxa1/2 binding in lung tumors
Nkx2-1-negative human lung adenocarcinomas exhibit gastric differentiation
Loss of Hnf4α de-represses Hmga2 in the absence of Nkx2-1
ACKNOWLEDGMENTS
We thank N. Dimitrova, D. Feldser and M. Mendoza for critical reading of the manuscript; the Swanson Biotechnology Center for histology, Illumina sequencing and bioinformatic support; the BioMicro Center for library preparation and Illumina sequencing; L. Sholl and L. Chirieac for assistance with human lung adenocarcinoma collection; M. Winslow and the entire Jacks lab for helpful discussions. E.L.S. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. This work was supported by National Institutes of Health grants U01-CA84306 (to T.J.) and K08-CA154784-01 (to E.L.S.), the Howard Hughes Medical Institute, the Ludwig Center for Molecular Oncology at MIT, and in part by the Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. T.J. is the David H. Koch Professor of Biology and a Daniel K. Ludwig Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Gene expression data is accessible in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE36473 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ptoffussocsmytc&acc=GSE36473). ChIP-Seq data is accessible via GEO and in the NCBI Sequence Read Archive database under accession numbers GSE43252/SRP017753. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43252)
REFERENCES
- Abate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 2008;76:717–727. doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta JA, Perner S, Iafrate AJ, Yeap BY, Weir BA, Johnson LA, Johnson BE, Meyerson M, Rubin MA, Travis WD, et al. Clinical Significance of TTF-1 Protein Expression and TTF-1 Gene Amplification in Lung Adenocarcinoma. J. Cell. Mol. Med. 2008;13:1977–1986. doi: 10.1111/j.1582-4934.2008.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans T, Paesmans M, Mascaux C, Martin B, Meert AP, Haller A, Lafitte JJ, Sculier JP. Thyroid transcription factor 1--a new prognostic factor in lung cancer: a meta-analysis. Ann. Oncol. 2006;17:1673–1676. doi: 10.1093/annonc/mdl287. [DOI] [PubMed] [Google Scholar]
- Boggaram V. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin. Sci. (Lond.) 2009;116:27–35. doi: 10.1042/CS20080068. [DOI] [PubMed] [Google Scholar]
- Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of Hepatocyte Proliferation by Hepatocyte Nuclear Factor 4alpha in Adult Mice. J. Biol. Chem. 2012;286:29635–29643. doi: 10.1074/jbc.M111.334599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin AL, Le Beyec J, Barreau F, Saint-Just S, Houllier A, Gonzalez FJ, Robine S, Pincon-Raymond M, Cardot P, Lacasa M, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell. Bio. 2009;29:6294–6308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, Whitsett JA. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J. Clin. Invest. 2009;119:2914–2924. doi: 10.1172/JCI39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlabicz M, Gacko M, Worowska A, Lapinski R. Cathepsin E (EC 3.4.23.34) - a review. Folia Histochem. Cytobiol. 2011;49:547–557. doi: 10.5603/fhc.2011.0078. [DOI] [PubMed] [Google Scholar]
- Copin MC, Buisine MP, Leteurtre E, Marquette CH, Porte H, Aubert JP, Gosselin B, Porchet N. Mucinous bronchioloalveolar carcinomas display a specific pattern of mucin gene expression among primary lung adenocarcinomas. Hum. Pathol. 2001;32:274–281. doi: 10.1053/hupa.2001.22752. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Lim L. Transcription factors in mouse lung development and function. Am. J. Physio.l Lung Cell. Mol. Physiol. 2001;280:L823–838. doi: 10.1152/ajplung.2001.280.5.L823. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsigny M, Babeu JP, Seidman EG, Gendron FP, Levy E, Carrier J, Perreault N, Boudreau F. Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res. 2010;70:9423–9433. doi: 10.1158/0008-5472.CAN-10-1697. [DOI] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Prot. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finberg KE, Sequist LV, Joshi VA, Muzikansky A, Miller JM, Han M, Beheshti J, Chirieac LR, Mark EJ, Iafrate AJ. Mucinous differentiation correlates with absence of EGFR mutation and presence of KRAS mutation in lung adenocarcinomas with bronchioloalveolar features. J. Mol. Diagn. 2007;9:320–326. doi: 10.2353/jmoldx.2007.060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nature reviews. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfield D. Can K-ras-mutated atypical adenomatous hyperplasia be another precursor lesion for mucinous bronchioloalveolar carcinoma? Am. J. Clin. Path. 2008;130:315–316. [PubMed] [Google Scholar]
- Hata A, Katakami N, Fujita S, Kaji R, Imai Y, Takahashi Y, Nishimura T, Tomii K, Ishihara K. Frequency of EGFR and KRAS mutations in Japanese patients with lung adenocarcinoma with features of the mucinous subtype of bronchioloalveolar carcinoma. J. Thorac. Oncol. 2010;5:1197–1200. doi: 10.1097/JTO.0b013e3181e2a2bc. [DOI] [PubMed] [Google Scholar]
- Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Hoshi S, Hoshi N, Okamoto M, Paiz J, Kusakabe T, Ward JM, Kimura S. Role of NKX2-1 in N-bis(2-hydroxypropyl)-nitrosamine-induced thyroid adenoma in mice. Carcinogenesis. 2009;30:1614–1619. doi: 10.1093/carcin/bgp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Gen. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kaestner KH. The FoxA factors in organogenesis and differentiation. Curr. Opin. Genet. Dev. 2010;20:527–532. doi: 10.1016/j.gde.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc. Natl. Acad. Sci. U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Kunii R, Jiang S, Hasegawa G, Yamamoto T, Umezu H, Watanabe T, Tsuchida M, Hashimoto T, Hamakubo T, Kodama T, et al. The predominant expression of hepatocyte nuclear factor 4alpha (HNF4alpha) in thyroid transcription factor-1 (TTF-1)-negative pulmonary adenocarcinoma. Histopathology. 2011;58:467–476. doi: 10.1111/j.1365-2559.2011.03764.x. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Kawaguchi A, Hoshi N, Kawaguchi R, Hoshi S, Kimura S. Thyroid-specific enhancer-binding protein/NKX2. 1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol. Endocrinol. 2006;20:1796–1809. doi: 10.1210/me.2005-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Chen G, Xu Y, Haitchi HM, Du L, Keiser AR, Howarth PH, Davies DE, Holgate ST, Whitsett JA. Airway epithelial transcription factor NK2 homeobox 1 inhibits mucous cell metaplasia and Th2 inflammation. Am. J. Respir. Crit. Care Med. 2011;184:421–429. doi: 10.1164/rccm.201101-0106OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Tsuchiya T, Hao H, Tompkins DH, Xu Y, Mucenski ML, Du L, Keiser AR, Fukazawa T, et al. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J. Clin. Invest. 2012;122:4388–4400. doi: 10.1172/JCI64048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menheniott TR, Peterson AJ, O’Connor L, Lee KS, Kalantzis A, Kondova I, Bontrop RE, Bell KM, Giraud AS. A novel gastrokine, Gkn3, marks gastric atrophy and shows evidence of adaptive gene loss in humans. Gastroenterology. 2010;138:1823–1835. doi: 10.1053/j.gastro.2010.01.050. [DOI] [PubMed] [Google Scholar]
- Minoo P, Hu L, Xing Y, Zhu NL, Chen H, Li M, Borok Z, Li C. Physical and functional interactions between homeodomain NKX2. 1 and winged helix/forkhead FOXA1 in lung epithelial cells. Mol. Cell. Biol. 2007;27:2155–2165. doi: 10.1128/MCB.01133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev. Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Oien KA, McGregor F, Butler S, Ferrier RK, Downie I, Bryce S, Burns S, Keith WN. Gastrokine 1 is abundantly and specifically expressed in superficial gastric epithelium, down-regulated in gastric carcinoma, and shows high evolutionary conservation. J. Pathol. 2004;203:789–797. doi: 10.1002/path.1583. [DOI] [PubMed] [Google Scholar]
- Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, Hruban RH, Goggins M, Leach SD. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619–1626. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell PA, Wainer Z, Wright GM, Daniels M, Conron M, Williams RA. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J. Thorac. Oncol. 2011;6:1496–1504. doi: 10.1097/JTO.0b013e318221f701. [DOI] [PubMed] [Google Scholar]
- Santangelo L, Marchetti A, Cicchini C, Conigliaro A, Conti B, Mancone C, Bonzo JA, Gonzalez FJ, Alonzi T, Amicone L, et al. The stable repression of mesenchymal program is required for hepatocyte identity: a novel role for hepatocyte nuclear factor 4alpha. Hepatology. 2011;53:2063–2074. doi: 10.1002/hep.24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan MJ, Ritter G, Yin BW, Williams C, Jr., Cohen LS, Coplan KA, Fortunato SR, Frosina D, Lee SY, Murray AE, et al. Glycoprotein A34, a novel target for antibody-based cancer immunotherapy. Cancer Immunity. 2006;6:2. [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenhouse G, Fyfe N, King G, Chapman A, Kerr KM. Thyroid transcription factor 1 in pulmonary adenocarcinoma. J. Clin. Pathol. 2004;57:383–387. doi: 10.1136/jcp.2003.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A, Walker JR, Zhang J. Gene Expression Omnibus GSE 15998. 2009. Mouse Exon Atlas. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15998. [Google Scholar]
- Tagne J-B, Gupta S, Gower AC, Shen SS, Varma S, Lakshminarayanan M, Cao Y, Spira A, Volkert TL, et al. Genome-wide analyses of Nkx2-1 binding to transcriptional target genes uncover novel regulatory patterns conserved in lung development and tumors. PLoS One. 2012;7:e29907. doi: 10.1371/journal.pone.0029907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Jiang S, Hotta H, Takano K, Iwanari H, Sumi K, Daigo K, Ohashi R, Sugai M, Ikegame C, et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J. Pathol. 2006;208:662–672. doi: 10.1002/path.1928. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J. Biol. Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Francis JM, Woo MS, Etemad B, Lin W, Fries DF, Peng S, Snyder EL, Tata PR, Izzo F, et al. Integrated cistromic and expression analysis of amplified NKX2-1 in lung adenocarcinoma identifies LMO3 as a functional transcriptional target. Genes Dev. 2013;27:197–210. doi: 10.1101/gad.203208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nuc. Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wontakal SN, Guo X, Smith C, MacCarthy T, Bresnick EH, Bergman A, Snyder MP, Weissman SM, Zheng D, Skoultchi AI. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3832–3837. doi: 10.1073/pnas.1121019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Yanagisawa K, Sugiyama R, Hosono Y, Shimada Y, Arima C, Kato S, Tomida S, Suzuki M, Osada H, Takahashi T. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell. 2012;20:348–361. doi: 10.1016/j.ccr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG, Park BJ, Rusch VW, Travis WD. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod. Pathol. 2011;24:653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71:4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zhang Z, Liao W, Zhao X, Liu L, Wu Y, Liu Z, Li Y, Zhong Y, Chen K, et al. The tumor-suppressor gene Nkx2. 8 suppresses bladder cancer proliferation through upregulation of FOXO3a and inhibition of the MEK/ERK signaling pathway. Carcinogenesis. 2012;33:678–686. doi: 10.1093/carcin/bgr321. [DOI] [PubMed] [Google Scholar]
- Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat. Rev. Cancer. 2003;3:592–600. doi: 10.1038/nrc1141. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.