Abstract
The enteric nervous system (ENS), the intrinsic innervation of the gastrointestinal tract, is an essential component of the gut neuromusculature and controls many aspects of gut function, including coordinated muscular peristalsis. The ENS is entirely derived from neural crest cells (NCC) which undergo a number of key processes, including extensive migration into and along the gut, proliferation, and differentiation into enteric neurons and glia, during embryogenesis and fetal life. These mechanisms are under the molecular control of numerous signaling pathways, transcription factors, neurotrophic factors and extracellular matrix components. Failure in these processes and consequent abnormal ENS development can result in so-called enteric neuropathies, arguably the best characterized of which is the congenital disorder Hirschsprung disease (HSCR), or aganglionic megacolon. This review focuses on the molecular and genetic factors regulating ENS development from NCC, the clinical genetics of HSCR and its associated syndromes, and recent advances aimed at improving our understanding and treatment of enteric neuropathies.
Keywords: enteric nervous system, Hirschsprung disease, enteric neuropathies, neural crest cells, RET
A functional gastrointestinal (GI) tract is essential for transporting, absorbing, digesting, and excreting food and waste, for protecting the host from ingested pathogens, allergens, and toxins, and for continuously monitoring and responding to the state of the intestinal lumen. The principal conductor of this highly orchestrated symphony is the enteric nervous system (ENS), a vast and complex network of neurons and glial cells that is located along the length of the GI tract and represents an important component of the autonomic nervous system (1). While extrinsic innervation from the central nervous system can modulate ENS function, the ENS serves as an intrinsic nervous system for the gut, capable of controlling most of the functions of the intestine independently. It is because of its complexity and autonomous function that the ENS is often referred to as the `second brain' (2).
The ENS contains approximately 100 million neurons (3) within at least 18 functional classes (4). It is organized in two concentric rings of interconnected ganglia, consisting of both enteric neurons and enteroglial cells, interconnected by interganglionic fibers (Fig. 1). The outer ring is the myenteric (Auerbach's) plexus, located between the circular and longitudinal muscle layers, and the inner ring is the submucosal (Meissner's) plexus, which is absent in the esophagus. The ENS contains many different types of neurons, including primary afferent neurons that sense chemical or mechanical stimuli in the lumen and then signal via ascending and descending interneurons to excitatory and inhibitory motoneurons that control effector cell function (4). The reflex circuits produced by these synaptically interconnected neurons are responsible for coordinating the major functions of the ENS, including the regulation of intestinal motility, absorption, secretion, and blood flow.
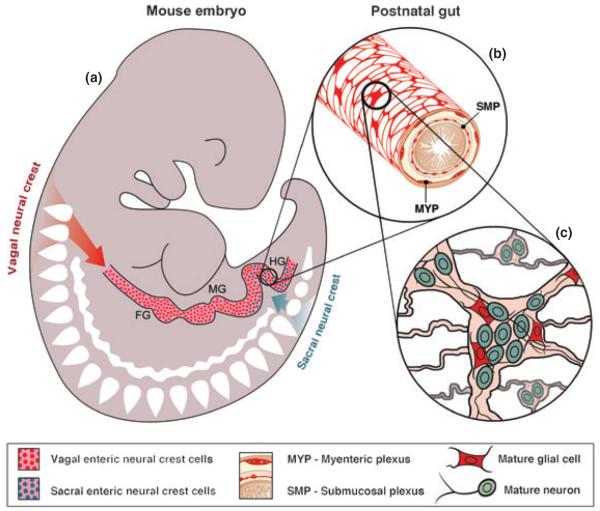
Fig. 1.
Schematic showing formation of enteric nervous system (ENS) from neural crest-derived precursors. (a) Vagal neural crest-derived cells enter the proximal (oral) end of the embryonic gut and migrate along its entire length giving rise to the majority of the neurons and glia of the ENS. Sacral neural crest-derived cells enter the hindgut and migrate in an oral direction to form neurons and glia in the distal portion of the gut. (b) In the post-natal gut, the ENS is organized in web-like plexuses that are located between the muscle layers: the myenteric plexus is situated between the longitudinal and circular muscle layers, and the submucosal plexus between the circular muscle and the mucosa. (c) The enteric plexuses contain small groupings of enteric neurons and glia. FG, foregut; MD, midgut; HD, hindgut.
Congenital and acquired enteric neuropathies can lead to serious health consequences, typically manifesting with abnormal gut motor function (5, 6). For example, inflammatory enteric neuropathies, which can be caused by paraneoplastic, infectious, or immune-mediated diseases, can lead to gastroparesis, intestinal pseudo-obstruction, or colonic inertia. Deficiencies of selective neurotransmitters have been described in esophageal achalasia and congenital hypertrophic pyloric stenosis, both associated with abnormal intestinal sphincter function. Mitochondrial dysfunction can lead to intestinal dysmotility from neuronal cell injury, as in mitochondrial neurogastrointestinal encephalopathy. Enteric ganglioneuromas, occurring in multiple endocrine neoplasia type 2B, are associated with severe colonic dysmotility (7).
The classic enteric neuropathy is Hirschsprung disease (HSCR), a congenital disease characterized by the absence of enteric ganglia along variable lengths of distal colon (8, 9). Congenital aganglionosis, which occurs in 1 in 5000 live-births, is limited to the rectosigmoid colon in 80% of cases, referred to as short-segment HSCR (S-HSCR), and most commonly presents with the failure of a newborn to pass meconium within 48 h of life, often with abdominal distension and vomiting. A contrast enema X-ray showing narrowing of the distal colorectum with dilation proximally supports the diagnosis of HSCR, and a rectal biopsy revealing the absence of enteric ganglia makes the definitive diagnosis. Treatment consists of surgical resection of the aganglionic segment and anastomosis of normally ganglionated bowel to the anus.
This review focuses on the molecular and cellular factors regulating ENS morphogenesis, the clinical genetics of HSCR and its associated syndromes, and recent advances aimed at improving our understanding and treatment of enteric neuropathies.
Development of the ENS
The ENS is entirely derived from the neural crest, a transient structure that arises early in development during formation of the neural tube, the precursor of the brain and spinal cord. The progeny of the neural crest, neural crest cells (NCC), migrate extensively throughout the embryo, proliferate, and differentiate into a wide variety of cell types including melanocytes, craniofacial cartilage and bone, neurons and glia of the peripheral and ENS, and smooth muscle (10). Failure in key processes underlying NCC development can result in a number of wide ranging clinically important neural crest disorders (neurocristopathies) that affect pigmentation, alter craniofacial formation, result in deafness, give rise to tumors, or impact the innervation of the gut (i.e. HSCR) (11).
Studies in the 1950s and 1970s on chick embryos demonstrated that the vagal (hindbrain) region of the neural crest, adjacent to somites 1–7, gives rise to the majority of ENS cells along the entire length of the gut. In the last few decades, numerous aspects of ENS development have been extensively studied, with remarkable conservation of cell behavior and molecular control described in species such as fish, avians, mice and humans. In the mouse, the most widely studied animal model of ENS development, vagal NCC emerge from the neural tube around embryonic day 8.5 (E8.5), reach the foregut at E9–E9.5 (12, 13), and migrate rostrocaudally to colonize the entire length of the gut by E13.5–14 (14). In the human, this journey by vagal NCC along the gut begins at week 4 of gestation and is completed by approximately week 7 (15). In addition to this principal vagal NCC contribution to the gut, a second, more caudal region of the neuraxis, the sacral neural crest, also contributes a smaller number of cells that mainly colonize the terminal region of the mouse and avian hindgut (16, 17). Sacral neural crest-derived cells migrate rostrocaudally along the hindgut, opposite to the direction of migration of the vagal neural crest-derived cells. The neuronal subtypes and role(s) of sacral NCC remain unclear, and whether the sacral neural crest contributes to human ENS formation is still unknown.
In order to form a functional ENS along the entire length of the gut, vagal and sacral NCC must undergo a number of key processes including migration, survival, proliferation, neuronal and glial differentiation, and axon formation (Figure 1). Numerous transcription factors, signaling pathways, and neurotrophic factors have been shown to be involved in regulating each of these critical processes, a number of which are summarized in Table 1, elaborated upon below, and reviewed previously (18, 19). These advances in our understanding of ENS development have allowed a number of markers to be used to identify undifferentiated NCC prior to their entry into the gut (termed pre-enteric NCC), including the transcription factors Sox10 and Phox2b, the G protein-coupled receptor endothelin receptor B (EdnrB), the low-affinity nerve growth receptor p75, and the receptor tyrosine kinase Ret (Table 1).
Table 1.
Molecular markers expressed by NCC and their cellular derivatives at key stages of ENS development in the mouse [modified from (19)].
| Stage of development | Key cellular events | Cell types | Markers expressed by NCC and their progeny |
|---|---|---|---|
| E8.5 | Delamination from vagal region of neural tube | Vagal NCCs | Sox10/p75 |
| E9.5 | Invasion of the embryonic foregut by vagal progenitor cells | Pre-enteric NCCs | Sox10/p75±RET/Phox2b |
| E10.5 | Rostrocaudal migration of progenitor cells | ENCCs (progenitor cells) | Sox10/p75/RET/Phox2b; EDNRB; Mash1 |
| Proliferation of progenitor cells | Immature neurons | RET/Phox2b/PGP9.5/HuC-D/TuJ1±Mash1/TH | |
| Start of neuronal differentiation | |||
| E11.5 | Rostrocaudal migration of progenitor cells | ENCCs (progenitor cells) | Sox10/p75/RET/Phox2b; EDNRB; Mash1 |
| Proliferation of progenitor cells | Immature neurons | RET/Phox2b/PGP9.5/HuC-D/TuJ1±Mash1/TH; ± NOS; ±Calb | |
| Neuronal differentiation (appearance of first neurotransmitters) | Immature glial cells | Sox10/p75/B-FABP | |
| Start of glial differentiation | |||
| E13.5 | Completion of rostrocaudal migration of vagal progenitor cells | Sacral NCCs | Sox10/p75±RET/Phox2b |
| Invasion of the embryonic hindgut and caudo-rostral migration of sacral progenitor cells | ENCCs (progenitor cells) | Sox10/p75/RET/Phox2b; EDNRB | |
| Proliferation of progenitor cells | Immature neurons | RET/Phox2b/PGP9.5/HuC-D/TuJ1±NOS; ± Calb; ± VIP; ± NPY | |
| Neuronal differentiation | Immature glial cells | Sox10/p75/B-FABP | |
| Glial differentiation | |||
| P0 (to adult) | Proliferation of progenitor cells | ENCCs (progenitor cells) | Sox10/p75±RET/Phox2b |
| Differentiation of mature neuronal phenotypes | Neurons | RET/Phox2b/PGP9.5/HuC-D/TuJ1±NOS; ± Calb; ± VIP; ± NPY; ± SubP; ± CGRP; ± 5HT; ± ChAT; ±Calret | |
| Differentiation of mature glial phenotype/s | |||
| Gangliogenesis | Glial cells | Sox10/p75/Phox2b/B-FABP/S100β/GFAP | |
| Formation of functional neuronal circuits (i.e. onset of co-ordinated intestinal motility) |
E, embryonic day; ENCC, enteric neural crest cells; ENS, enteric nervous system; NCC, neural crest cells.
Enteric NCC (ENCCs), as they are termed upon entering the gut, are undifferentiated at the wavefront of migration and express Sox10, Ret, p75, Phox2b, Ednrb and the transcriptional regulator Mash1. Behind the migratory wavefront, ENCCs are at different stages of maturation, with neuronal differentiation, which begins before glial differentiation, commencing shortly after they invade the foregut. This commitment to the neuronal lineage is associated with downregulation of Sox10 and p75, maintenance of Ret and Phox2b expression, and upregulation of pan-neuronal markers, including PGP9.5, neurofilament, neuronal class III β-tubulin (Tuj1), HuC and HuD. Although committed to a neuronal fate, these cells are still considered to be progenitors as they lack neuron-subtype-specific markers (e.g. NOS, VIP, NPY, SubP, ChAT) and remain mitotically active. To commit to the glial lineage, ENCCs maintain Sox10 and p75 expression, downregulate Ret, and upregulate B-FABP first and S100 and GFAP later in their differentiation (Table 1).
Molecular mechanisms in ENS development
ENS development is a highly dynamic process in which ENCC migration, proliferation, and differentiation all occur simultaneously at different positions along the gut. While most cells at the migratory wavefront continue to proliferate and invade regions lacking ENCCs, those behind the wavefront establish themselves and begin to differentiate into neurons or glia, although some continue to proliferate to fill gaps created by the growing intestine. Several signaling pathways have essential roles in coordinating these processes and the timing and location of their expression is critical.
Ret and EdnrB signaling: balancing cell proliferation and differentiation in the ENS
Glial cell-derived neurotrophic factor (GDNF) is expressed in the mesoderm of the embryonic gut and activates a receptor complex on migrating ENCCs. This complex consists of the transmembrane receptor tyrosine kinase, Ret, and a co-receptor, Gdnf family receptor α1 (GFRα1) (20). Null mutations of Ret, Gdnf, or GFRα1 result in aganglionosis of the small and large intestine (21–24), and GDNF haploinsufficiency leads to severe hypoganglionosis (25) (Table 2). At early stages, Ret signaling supports the survival of ENCCs, with its loss leading to significant apoptosis in the foregut (26). Ret is also strongly mitogenic (27–31), an essential function for expanding the pool of ENCCs so that sufficient numbers are available to colonize the entire GI tract. The proliferative effect of Gdnf-Ret signaling predominates during early stages when ENCCs are still migrating along the gut (27, 31), whereas at later developmental stages, Gdnf-Ret promotes neuronal differentiation (26–28). GDNF is also a potent chemoattractive factor for ENCCs, expressed most highly in the intestinal mesenchyme ahead of the wavefront and thereby possibly drawing them distally down the gut (31–33). A neurotrophic factor closely related to GDNF, neurturin, also activates the Ret receptor. Mouse mutations in this gene result in hypoganglionosis specifically affecting the myenteric plexus (34).
Table 2.
Mouse models of intestinal aganglionosis and gut phenotypes in homozygous null and heterozygous mutants
| Gene | Homozygous phenotype | Heterozygous phenotype |
|---|---|---|
| Ret | AG of small and large intestine (21) | Normal (21) |
| Gdnf | AG of small and large intestine (22,23,24) | Hypoganglionosis (25) |
| Gfr α 1 | AG of small and large intestine (107,108) | Normal (107) |
| Et3ls (lethal spotting) | Distal colon AG (36) | Normal (36) |
| Ednrbsl (piebald lethal) | Distal colon AG (35) | Hyperganglionosis in submucosal plexus (109) |
| Ece-1 | Distal colon AG (37) | ND |
| Sox10 | Total intestinal AG (50) | Distal colon AG (50,51,110) |
| b1-integrin | Distal colon AG (59) | ND |
| Phox2b | Total intestinal AG (56) | Normal |
| Mash1 | AG of esophagus (111) | ND |
| Pax3 | AG of small and large intestine (55) | Normal |
| Ihh | Patchy AG in small and large intestine (112) | ND |
AG, aganglionosis; ND, not done.
Another major signaling pathway in ENS development is endothelin-3 (ET3) – EDNRB. ET3 is a 21 amino acid peptide expressed in the gut mesoderm, while its G protein-coupled receptor, EDNRB, is present on the ENCC. Targeted mutation of either gene in mice, or of the endothelin converting enzyme-1 (ECE-1) that cleaves ET3 from its larger precursor, leads to aganglionosis of the distal colon and pigmentation defects due to deficient melanocytes (35–37). The primary role of EDNRB signaling is to inhibit the differentiation of ENCCs (28, 38, 39) and to keep them in a proliferative state (30, 39), thereby maintaining a pool of uncommitted progenitors. Whereas these undifferentiated ENCCs can continue migrating, once they differentiate into neurons they become post-mitotic and unable to migrate any farther, leading to the distal aganglionosis seen in ET3 and EDNRB mutant animals (40) (Table 2).
A critical determinant of normal ENS development is the size of the ENCC population available to colonize the gut. Experimentally reducing the size of the vagal neural crest in avians leads to distal intestinal aganglionosis (41–43). It has been shown that a critical density of cells at the wavefront is necessary to form the cellular strands that drive ENCC migration (44, 45). A low ENCC density delays their rate of migration, which may leave the cells unable to colonize a distal environment that has changed by the time they arrive (46, 47), thus coupling cell numbers, rate of migration, and maturation of the microenvironment. ENCC proliferation is thus critically important to drive their colonization, as shown by both experimental and mathematical modeling (48, 49). Balancing proliferation and differentiation are therefore vital to ENS development. Whereas ET3 and GDNF act synergistically to enhance ENCC proliferation (30), they have antagonistic roles with respect to ENCC differentiation and migration, with ET3 inhibiting both of these GDNF-mediated processes (28, 31, 38, 39). This coordinated activity is essential, particularly in the cecal region, where GDNF expression is strongest at the stage when ENCCs are arriving there (31, 33).
Additional signals involved in ENS formation
Haploinsufficiency of Sox10, a SRY-related HMG transcription factor expressed by undifferentiated ENCC progenitors, leads to distal colonic aganglionosis in mice (50, 51). Sox10 supports the survival of ENCC progenitor cells, with its loss leading to NCC apoptosis prior to their arrival in the foregut (52). Acting together with EdnrB, whose enhancer has SOX10-binding sites (53), Sox10 also acts by maintaining ENCCs in an undifferentiated state (54). Similar to ET3 signaling, Sox10 appears to promote maintenance of a pool of progenitor cells for ENS colonization. Sox10 has also been implicated in the activation of Ret transcription (55) and may influence ENS development via that pathway as well. Mice lacking Phox2B, a transcription factor expressed by ENCC progenitor cells (56), develop aganglionosis below the stomach, similar to the Ret knockout phenotype (Table 2). This may be explained by the finding that Phox2B is required for Ret expression (56).
NCC migration, survival, and proliferation rely not only on signaling molecules acting on their respective receptors, but also on the interactions between NCC and the extracellular matrix (57). The aganglionic colon of ET3 mutant mice is rich in laminin, which promotes enteric neurogenesis and may contribute to the premature differentiation and distal aganglionosis occurring in this model (58). Studies in EDNRB mutant mice suggest that the observed delay in ENCC migration results in ENCCs reaching the colon at a time when it has become non-permissive, possibly due to increased laminin expression in the more mature colon (46). ENCC-specific deletion of β1 integrin, which is expressed by ENCCs and required for their interaction with the extracellular matrix, leads to abnormal cellular adhesion, delayed migration, and distal aganglionosis (59) (Table 2). The migratory defect occurs specifically in the cecum/proximal hindgut and is thought to be due to a requirement for β1 integrin-mediated interactions between ENCCs with tenascin-C and fibronectin, which are both highly expressed in the mouse cecum when ENCCs arrive there (60). In avians, ENS formation requires the presence of endothelial cells, which ENCCs appear to use as a scaffold to guide their migration via a β1 integrin-dependent interaction between ENCCs and the endothelial cell basement membrane (61).
Recently, retinoic acid was shown to be required for ENS development, supporting a non-genetic factor as a possible contributor to the pathogenesis of HSCR. Mice depleted of Vitamin A show colorectal aganglionosis due to impaired lamellipodia formation and reduced ENCC migration in response to GDNF (62).
Hirschsprung disease
HSCR and the challenge of colonizing the distal bowel
Why the distal end of the colon is particularly susceptible to aganglionosis, as occurs in HSCR, is not entirely clear. Reduction of Ret dosage to one third of normal levels leads to colonic aganglionosis (63). Other mouse models also display aganglionosis limited to the colorectal region, including mice expressing only the Ret51 allele (64), Sox10 heterozygotes (51), and targeted mutants of the ET3-EdnrB pathway (35, 36). As the majority of ENS colonization occurs rostrocaudally, distal aganglionosis may simply reflect the distance ENCCs need to migrate to reach the end of the gut. Given the importance of ENCC proliferation for generating an adequate pool of progenitor cells to populate the intestine, inadequate cell numbers could account for the distal aganglionosis. However, conditional inactivation of Ret in late development, after ENS migration has completed, leads to enteric neuronal cell death specifically in the colon (63), suggesting that certain aspects of ENCC development may be unique to the distal gut and that other etiologies may account for the HSCR phenotype.
Genes involved in the development of HSCR disease
HSCR is considered an inherited disease which can be transmitted in a Mendelian way, both as a dominant trait and as a recessive trait. The majority of cases are probably polygenic/multifactorial with differences in sex ratio, with a male predominance in S-HSCR (4:1), incomplete penetrance and variable expression. Associations with a large number of syndromes and congenital malformations have been observed (65). Linkage analyses of multiplex HSCR families revealed that the RET gene, located at 10q11.2, is the major risk factor as almost all HSCR families showed linkage with RET (66, 67). Coding sequence mutations in RET are responsible for a dominant form of HSCR (with incomplete penetrance) and coding and splice site mutations have been identified in up to 50% of familial cases and 15–35% of sporadic cases (68). The mutations are scattered throughout the RET-coding sequence, including large and micro-deletions and a variety of point mutations. RET mutations associated with HSCR are believed to cause a loss of function (haploinsufficiency) (69–71). However, RET mutations on their own might not result in aganglionosis, as the penetrance of the RET mutations (in general) is 72% in males and 51% in females. In addition to RET, mutations have been found in 11 other genes (Table 3). Mutations in these genes, namely EDNRB (72), EDN3 (73, 74), ECE1 (75), GDNF (76, 77), NTN (78), SOX10 (79), PHOX2B (80), KIAA1279/KBP (81), ZFHX1B (82, 83), TTF-1 (84) and NRG1 (85), do not account for more than 20% of the cases, supporting genetic heterogeneity for this disorder.
Table 3.
Hirschsprung disease associated genes and their clinical features (modified from (99))
| Gene symbol | Position | Inheritance | Phenotype |
|---|---|---|---|
| RET | 10q11.2 | Dominant, incomplete penetrance | Non-syndromic/MEN2A |
| GDNF | 5p13 | Non-Mendelian | Non-syndromic |
| NTN | 19p13 | Non-Mendelian | Non-syndromic |
| EDNRB | 13q22 | Recessive Dominant (de novo in 80%) | Shah–Waardenburg Non-syndromic |
| EDN3 | 20q13 | Recessive Dominant, incomplete penetrance | Shah–Waardenburg Non-syndromic |
| PH0X2B | 4p12 | Dominant (de novo in 90%) | Haddad syndrome (CCHS) |
| SOX10 | 22q13 | Dominant (de novo in 75%) | Shah–Waardenburg |
| ECE1 | 1p36 | Dominant (de novo) | Congenital heart formation |
| ZFHX1B (SIP1) | 2q22 | Dominant (de novo) | Mowat–Wilson |
| KIA1279 (KBP) | 10q22.1 | Recessive | Goldberg–Shprintzen |
| TTF1 (TITF1) | 14q13 | – | Non-syndromic |
| NRG1 | 8p21 | – | Non-syndromic |
CCHS, congenital central hypoventilation syndrome.
Genes involved in syndromic HSCR
Mutations in many of the genes are found in syndromic HSCR cases (Table 3). Mutations in EDNRB, EDN3 and SOX10 were identified in a patient with Shah–Waardenburg syndrome (WS4), which is characterized by congenital hearing loss, pigmentary abnormalities of the hair, skin and eyes, and HSCR disease (79, 86). Mutations in PHOX2B have been identified in patients with congenital central hypoventilation syndrome (CCHS) and HSCR disease. CCHS is a rare disorder characterized by impairment of autonomic control of spontaneous respiration in the absence of other lung or cardiac disease (80). The coexistence of CCHS and HSCR is known as Haddad syndrome. Mutations in ZFHX1B have been identified in patients with Mowat–Wilson syndrome, an autosomal dominant disorder characterized by mental retardation, epilepsy, delayed motor development, and HSCR disease (82). Mutations in KIAA1279 [now called kinesin-binding protein (KBP)] have been identified in patients with Goldberg–Shprintzen syndrome, a rare autosomal recessive disorder characterized by HSCR, microcephaly, mental retardation, and polymicrogyria (81). A mutation in ECE1 was identified in a single patient with craniofacial and cardiac defects (75). Finally, specific mutations in RET have been found in patients with HSCR in combination with the cancer syndrome multiple endocrine neoplasia type 2A (MEN2A) or familial medullary thyroid carcinoma (FMTC) (87).
Besides mutations in these genes, chromosomal abnormalities are observed in 12% of all syndromic HSCR cases. Trisomy 21 (Down syndrome), which occurs in up to 10% of children with HSCR, is the most frequent, accounting for >90% of all known chromosomal defects associated with this disease (65).
Non-coding RET variants
As mentioned above, RET-coding mutations have been identified in 50% of familial cases. However, regardless of the RET-coding mutation status, almost all familial cases are linked to the RET locus (66, 88). This suggests that non-coding RET mutations must play a major role in the remaining cases. This idea was corroborated in association studies on sporadic (simplex) cases with and without RET-coding mutations, performed on several Caucasian populations and an Asian population. These studies revealed a strong association between a certain haplotype (covering 27 kb in total) and the disease. This haplotype starts 4 kb upstream of the RET transcription start site, going all the way to the beginning of exon 2. This haplotype is present in 56–62% of patients, but only 20% of controls, in the Caucasian population. In the Chinese patient population, the frequency of this haplotype was 85%, and 40% in controls. This finding might partially explain the higher incidence of HSCR in Asians compared to Caucasians. Several groups have focused their studies on fine-mapping the associated region to identify the location of the causative variant (84, 89–96).
Susceptible HSCR loci
Investigators are actively searching for additional susceptibility loci, with or without an associated RET mutation. Linkage studies on multiplex HSCR families identified a new locus at 9q31 in families linked to RET but without a RET-coding mutation (88). Sibpair analysis in nuclear families with S-HSCR resulted in significant allele sharing with markers on 10q11 (RET), and two new loci on 19q12 and on 3p21, respectively (66). A genome-wide scan on 43 Mennonite trios, all belonging to the same large kindred, resulted in three loci, two of which were known loci (13q22.3-q31.1, EDNRB; 10q11.21, RET) and one new locus on 16q23.3 (97). Studying a large multigenerational Dutch family with an isolated HSCR phenotype resulted in the identification of a new susceptibility locus on 4q31-32 (98). Finally, a genome-wide association study on Chinese patients identified NRG1 as a susceptibility locus for HSCR (85).
Genetic testing for HSCR – what and who to test?
Current knowledge on the genetic background of HSCR has brought forward, for the patient, several important new insights. The most significant is perhaps the fact that a few percent (up to 3%) of non-syndromic HSCR patients have a RET mutation that predisposes them not only for HSCR but also for the cancer syndrome MEN2A or FMTC. As the detection of such a RET mutation has profound clinical consequences for the patients and his or her family it is recommended that all non-syndromic HSCR patients are screened for these specific mutations (87, 99). This testing should only be offered in combination with genetic counseling. Genetic counseling is also recommended in all syndromic HSCR cases. Depending on the disease phenotype one could decide to screen for specific gene(s). Another reason for molecular testing of RET could be to give more accurate estimates for recurrence risks to parents of a non-syndromic HSCR patient. For instance, the finding of a pathogenic RET mutation in a male proband with L-HSCR and the exclusion of this mutation in the parents may allow lowering of the recurrence risk of 13–17% to less than 1%, taking into account the theoretical possibility of a mosaic germ line mutation in one of the parents (100).
Future directions
Better understanding of, and therapies for, congenital diseases affecting the ENS
After over two decades of intense investigations, our understanding of the developmental biology, genetics and etiology of HSCR has significantly advanced. However, the defining characteristics of a number of other enteric neuropathies are still lacking (101, 102). This is partly due to the relatively rare incidence of these diseases, the lack of well defined neuropathological features, and the scarcity of animal models in which specific gut motility defects can be investigated. Future work may be directed towards: (i) identifying phenotype/genotype associations within well-defined patient groupings (which may require the establishment of international consortia to accumulate a `critical mass' of patients and tissues for analysis); (ii) utilizing evolving genetic technologies to identify candidate genes for enteric neuropathies; (iii) using animal models including zebrafish and mice to investigate potential mechanisms underlying enteric neuropathies, including aberrant development of neuronal subtypes, inappropriate neuronal wiring and network formation, and (iv) developing novel therapies for congenital diseases affecting the ENS (Fig. 2).
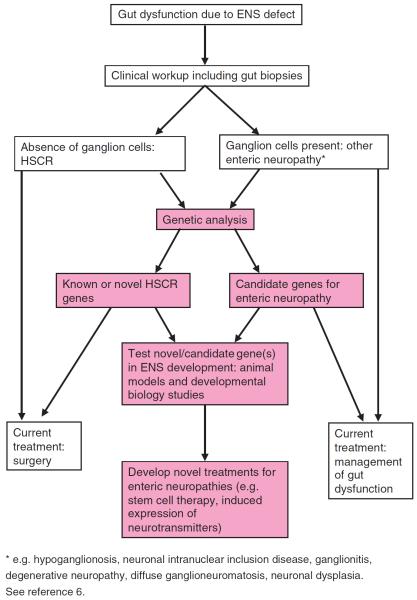
Fig. 2.
Flow diagram showing potential experimental approaches for gaining insight to the molecular mechanisms underlying enteric neuropathies, and development of novel therapies for their treatment.
For this latter point, although gut motility disorders represent relatively rare but clinically challenging conditions with little in the way of definitive cures, a number of groups worldwide are currently focusing on the possibility of utilizing stem cells to replace or restore missing or defective ENS cells, particularly with therapy for HSCR in mind. Typical investigative approaches include isolation and propagation of stem cells from various sources (including the gut or CNS, as well as iPS and embryonic stem cells), transplantation of stem cells into mouse or other models of gut aganglionosis, and assessment of gut function following transplantation (for reviews see (103–106)). The next challenges in this field will be to progress from animal models to the isolation and characterization of stem cells from human sources, and move to `first in man' studies whereby stem cell delivery methods, safety and efficacy can be assessed. With these steps in mind, advances in the understanding and treatment of ENS disorders will continue to advance at a rapid pace.
Acknowledgement
A. M. G. is supported by NIH R01DK080914.
Footnotes
Conflict of interest
The authors have no conflict of interest.
References
- 1.Furness JB. The enteric nervous system. Blackwell Publishing; Oxford: 2006. [Google Scholar]
- 2.Gershon MD. The enteric nervous system: a second brain. Hosp Pract(Off Ed) 1999;34:31–2. 35–8, 41–2. doi: 10.3810/hp.1999.07.153. [DOI] [PubMed] [Google Scholar]
- 3.Schemann M. Control of gastrointestinal motility by the “gut brain”-the enteric nervous system. J Pediatr Gastroenterol Nutr. 2005;41(Suppl. 1):S4–S6. doi: 10.1097/01.scs.0000180285.51365.55. [DOI] [PubMed] [Google Scholar]
- 4.Brookes SJ. Classes of enteric nerve cells in the guinea-pig small intestine. Anat Rec. 2001;262:58–70. doi: 10.1002/1097-0185(20010101)262:1<58::AID-AR1011>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.De Giorgio R, Camilleri M. Human enteric neuropathies: morphology and molecular pathology. Neurogastroenterol Motil. 2004;16:515–531. doi: 10.1111/j.1365-2982.2004.00538.x. [DOI] [PubMed] [Google Scholar]
- 6.Kapur RP. Pathology of intestinal motor disorders in children. Surg Pathol Clin. 2010;3:711–741. doi: 10.1016/j.path.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Hofstra RM, Landsvater RM, Ceccherini I, et al. A mutation in the RET proto-oncogene associated with multiple endocrine neoplasia type 2B and sporadic medullary thyroid carcinoma. Nature. 1994;367:375–376. doi: 10.1038/367375a0. [DOI] [PubMed] [Google Scholar]
- 8.Amiel J, Sproat-Emison E, Garcia-Barcelo M, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 9.Kenny SE, Tam PK, Garcia-Barcelo M. Hirschsprung's disease. Semin Pediatr Surg. 2010;19:194–200. doi: 10.1053/j.sempedsurg.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Kalcheim C, Le Douarin NM. The Neural Crest. Cambridge University Press; Cambridge: 1999. [Google Scholar]
- 11.Etchevers HC, Amiel J, Lyonnet S. Molecular bases of human neurocristopathies. Adv Exp Med Biol. 2006;589:213–234. doi: 10.1007/978-0-387-46954-6_14. [DOI] [PubMed] [Google Scholar]
- 12.Durbec PL, Larsson-Blomberg LB, Schuchardt A, et al. Common origin and developmental dependence on c-ret of subsets of enteric and sympathetic neuroblasts. Development. 1996;122:349–358. doi: 10.1242/dev.122.1.349. [DOI] [PubMed] [Google Scholar]
- 13.Anderson RB, Stewart AL, Young HM. Phenotypes of neural-crest-derived cells in vagal and sacral pathways. Cell Tissue Res. 2006;323:11–25. doi: 10.1007/s00441-005-0047-6. [DOI] [PubMed] [Google Scholar]
- 14.Young HM, Hearn CJ, Ciampoli D, et al. A single rostrocaudal colonization of the rodent intestine by enteric neuron precursors is revealed by the expression of Phox2b, Ret, and p75 and by explants grown under the kidney capsule or in organ culture. Dev Biol. 1998;202:67–84. doi: 10.1006/dbio.1998.8987. [DOI] [PubMed] [Google Scholar]
- 15.Wallace AS, Burns AJ. Development of the enteric nervous system, smooth muscle and interstitial cells of Cajal in the human gastrointestinal tract. Cell Tissue Res. 2005;319:367–382. doi: 10.1007/s00441-004-1023-2. [DOI] [PubMed] [Google Scholar]
- 16.Burns AJ, Le Douarin NM. The sacral neural crest contributes neurons and glia to the post- umbilical gut: spatiotemporal analysis of the development of the enteric nervous system. Development. 1998;125:4335–4347. doi: 10.1242/dev.125.21.4335. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Chan AK, Sham MH, et al. Analysis of the sacral neural crest cell contribution to the hindgut enteric nervous system in the mouse embryo. Gastroenterology. 2011;141:992–1002. e1001–1006. doi: 10.1053/j.gastro.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Heanue TA, Pachnis V. Enteric nervous system development and Hirschsprung's disease: advances in genetic and stem cell studies. Nat Rev Neurosci. 2007;8:466–479. doi: 10.1038/nrn2137. [DOI] [PubMed] [Google Scholar]
- 19.Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol. 2012;366:64–73. doi: 10.1016/j.ydbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Manie S, Santoro M, Fusco A, et al. The RET receptor: function in development and dysfunction in congenital malformation. Trends Genet. 2001;17:580–589. doi: 10.1016/s0168-9525(01)02420-9. [DOI] [PubMed] [Google Scholar]
- 21.Schuchardt A, D'Agati V, Larsson-Blomberg L, et al. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 22.Pichel JG, Shen L, Sheng HZ, et al. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature. 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- 23.Moore MW, Klein RD, Farinas I, et al. Renal and neuronal abnormalities in mice lacking GDNF. Nature. 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez MP, Silos-Santiago I, Frisen J, et al. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- 25.Shen L, Pichel JG, Mayeli T, et al. Gdnf haploinsufficiency causes Hirschsprung-like intestinal obstruction and early-onset lethality in mice. Am J Hum Genet. 2002;70:435–447. doi: 10.1086/338712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taraviras S, Marcos-Gutierrez CV, Durbec P, et al. Signalling by the RET receptor tyrosine kinase and its role in the development of the mammalian enteric nervous system. Development. 1999;126:2785–2797. doi: 10.1242/dev.126.12.2785. [DOI] [PubMed] [Google Scholar]
- 27.Chalazonitis A, Rothman TP, Chen J, et al. Age-dependent differences in the effects of GDNF and NT-3 on the development of neurons and glia from neural crest-derived precursors immunoselected from the fetal rat gut: expression of GFRalpha-1 in vitro and in vivo. Dev Biol. 1998;204:385–406. doi: 10.1006/dbio.1998.9090. [DOI] [PubMed] [Google Scholar]
- 28.Hearn CJ, Murphy M, Newgreen D. GDNF and ET-3 differentially modulate the numbers of avian enteric neural crest cells and enteric neurons in vitro. Dev Biol. 1998;197:93–105. doi: 10.1006/dbio.1998.8876. [DOI] [PubMed] [Google Scholar]
- 29.Heuckeroth RO, Lampe PA, Johnson EM, et al. Neurturin and GDNF promote proliferation and survival of enteric neuron and glial progenitors in vitro. Dev Biol. 1998;200:116–129. doi: 10.1006/dbio.1998.8955. [DOI] [PubMed] [Google Scholar]
- 30.Barlow A, de Graaff E, Pachnis V. Enteric nervous system progenitors are coordinately controlled by the G protein-coupled receptor EDNRB and the receptor tyrosine kinase RET. Neuron. 2003;40:905–916. doi: 10.1016/s0896-6273(03)00730-x. [DOI] [PubMed] [Google Scholar]
- 31.Mwizerwa O, Das P, Nagy N, et al. Gdnf is mitogenic, neurotrophic, and chemoattractive to enteric neural crest cells in the embryonic colon. Dev Dyn. 2011;240:1402–1411. doi: 10.1002/dvdy.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young HM, Hearn CJ, Farlie PG, et al. GDNF is a chemoattractant for enteric neural cells. Dev Biol. 2001;229:503–516. doi: 10.1006/dbio.2000.0100. [DOI] [PubMed] [Google Scholar]
- 33.Natarajan D, Marcos-Gutierrez C, Pachnis V, et al. Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 2002;129:5151–5160. doi: 10.1242/dev.129.22.5151. [DOI] [PubMed] [Google Scholar]
- 34.Heuckeroth RO, Enomoto H, Grider JR, et al. Gene targeting reveals a critical role for neurturin in the development and maintenance of enteric, sensory, and parasympathetic neurons. Neuron. 1999;22:253–263. doi: 10.1016/s0896-6273(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 35.Hosoda K, Hammer RE, Richardson JA, et al. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 36.Baynash AG, Hosoda K, Giaid A, et al. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 37.Yanagisawa H, Yanagisawa M, Kapur RP, et al. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- 38.Wu JJ, Chen JX, Rothman TP, et al. Inhibition of in vitro enteric neuronal development by endothelin-3: mediation by endothelin B receptors. Development. 1999;126:1161–1173. doi: 10.1242/dev.126.6.1161. [DOI] [PubMed] [Google Scholar]
- 39.Nagy N, Goldstein AM. Endothelin-3 regulates neural crest cell proliferation and differentiation in the hindgut enteric nervous system. Dev Biol. 2006;293:203–217. doi: 10.1016/j.ydbio.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 40.Gershon MD. Endothelin and the development of the enteric nervous system. Clin Exp Pharmacol Physiol. 1999;26:985–988. doi: 10.1046/j.1440-1681.1999.03176.x. [DOI] [PubMed] [Google Scholar]
- 41.der Sanden MJ P-v, Kirby ML, Gittenberger-de Groot A, et al. Ablation of various regions within the avian vagal neural crest has differential effects on ganglion formation in the fore-, mid- and hindgut. Dev Dyn. 1993;196:183–194. doi: 10.1002/aja.1001960305. [DOI] [PubMed] [Google Scholar]
- 42.Burns AJ, Champeval D, Le Douarin NM. Sacral neural crest cells colonise aganglionic hindgut in vivo but fail to compensate for lack of enteric ganglia. Dev Biol. 2000;219:30–43. doi: 10.1006/dbio.1999.9592. [DOI] [PubMed] [Google Scholar]
- 43.Barlow AJ, Wallace AS, Thapar N, et al. Critical numbers of neural crest cells are required in the pathways from the neural tube to the foregut to ensure complete enteric nervous system formation. Development. 2008;135:1681–1691. doi: 10.1242/dev.017418. [DOI] [PubMed] [Google Scholar]
- 44.Young HM, Bergner AJ, Anderson RB, et al. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev Biol. 2004;270:455–473. doi: 10.1016/j.ydbio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Druckenbrod NR, Epstein ML. Behavior of enteric neural crest-derived cells varies with respect to the migratory wavefront. Dev Dyn. 2007;236:84–92. doi: 10.1002/dvdy.20974. [DOI] [PubMed] [Google Scholar]
- 46.Druckenbrod NR, Epstein ML. Age-dependent changes in the gut environment restrict the invasion of the hindgut by enteric neural progenitors. Development. 2009;136:3195–3203. doi: 10.1242/dev.031302. [DOI] [PubMed] [Google Scholar]
- 47.Hotta R, Natarajan D, Thapar N. Potential of cell therapy to treat pediatric motility disorders. Semin Pediatr Surg. 2009;18:263–273. doi: 10.1053/j.sempedsurg.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Landman KA, Simpson MJ, Newgreen DF. Mathematical and experimental insights into the development of the enteric nervous system and Hirschsprung's disease. Dev Growth Differ. 2007;49:277–286. doi: 10.1111/j.1440-169X.2007.00929.x. [DOI] [PubMed] [Google Scholar]
- 49.Simpson MJ, Zhang DC, Mariani M, et al. Cell proliferation drives neural crest cell invasion of the intestine. Dev Biol. 2007;302:553–568. doi: 10.1016/j.ydbio.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 50.Herbarth B, Pingault V, Bondurand N, et al. Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc Natl Acad Sci U S A. 1998;95:5161–5165. doi: 10.1073/pnas.95.9.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 52.Kapur RP. Early death of neural crest cells is responsible for total enteric aganglionosis in Sox10(Dom)/Sox10(Dom) mouse embryos. Pediatr Dev Pathol. 1999;2:559–569. doi: 10.1007/s100249900162. [DOI] [PubMed] [Google Scholar]
- 53.Zhu L, Lee HO, Jordan CS, et al. Spatiotemporal regulation of endothelin receptor-B by SOX10 in neural crest-derived enteric neuron precursors. Nat Genet. 2004;36:732–737. doi: 10.1038/ng1371. [DOI] [PubMed] [Google Scholar]
- 54.Bondurand N, Natarajan D, Barlow A, et al. Maintenance of mammalian enteric nervous system progenitors by SOX10 and endothelin 3 signalling. Development. 2006;133:2075–2086. doi: 10.1242/dev.02375. [DOI] [PubMed] [Google Scholar]
- 55.Lang D, Chen F, Milewski R, et al. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pattyn A, Morin X, Cremer H, et al. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 57.Perris R, Perissinotto D. Role of the extracellular matrix during neural crest cell migration. Mech Dev. 2000;95:3–21. doi: 10.1016/s0925-4773(00)00365-8. [DOI] [PubMed] [Google Scholar]
- 58.Gershon MD. Developmental determinants of the independence and complexity of the enteric nervous system. Trends Neurosci. 2010;33:446–456. doi: 10.1016/j.tins.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Breau MA, Pietri T, Eder O, et al. Lack of beta1 integrins in enteric neural crest cells leads to a Hirschsprung-like phenotype. Development. 2006;133:1725–1734. doi: 10.1242/dev.02346. [DOI] [PubMed] [Google Scholar]
- 60.Breau MA, Dahmani A, Broders-Bondon F, et al. Beta1 integrins are required for the invasion of the caecum and proximal hindgut by enteric neural crest cells. Development. 2009;136:2791–2801. doi: 10.1242/dev.031419. [DOI] [PubMed] [Google Scholar]
- 61.Nagy N, Mwizerwa O, Yaniv K, et al. Endothelial cells promote migration and proliferation of enteric neural crest cells via beta1 integrin signaling. Dev Biol. 2009;330:263–272. doi: 10.1016/j.ydbio.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu M, Sato Y, Lyons-Warren A, et al. Vitamin A facilitates enteric nervous system precursor migration by reducing Pten accumulation. Development. 2010;137:631–640. doi: 10.1242/dev.040550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uesaka T, Nagashimada M, Yonemura S, et al. Diminished Ret expression compromises neuronal survival in the colon and causes intestinal aganglionosis in mice. J Clin Invest. 2008;118:1890–1898. doi: 10.1172/JCI34425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Graaff E, Srinivas S, Kilkenny C, et al. Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev. 2001;15:2433–2444. doi: 10.1101/gad.205001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amiel J, Lyonnet S. Hirschsprung disease, associated syndromes, and genetics: a review. J Med Genet. 2001;38:729–739. doi: 10.1136/jmg.38.11.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gabriel SB, Salomon R, Pelet A, et al. Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet. 2002;31:89–93. doi: 10.1038/ng868. [DOI] [PubMed] [Google Scholar]
- 67.Lyonnet S, Bolino A, Pelet A, et al. A gene for Hirschsprung disease maps to the proximal long arm of chromosome 10. Nat Genet. 1993;4:346–350. doi: 10.1038/ng0893-346. [DOI] [PubMed] [Google Scholar]
- 68.Hofstra RM, Wu Y, Stulp RP, et al. RET and GDNF gene scanning in Hirschsprung patients using two dual denaturing gel systems. Hum Mutat. 2000;15:418–429. doi: 10.1002/(SICI)1098-1004(200005)15:5<418::AID-HUMU3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 69.Edery P, Lyonnet S, Mulligan LM, et al. Mutations of the RET protooncogene in Hirschsprung's disease. Nature. 1994;367:378–380. doi: 10.1038/367378a0. [DOI] [PubMed] [Google Scholar]
- 70.Romeo G, Ronchetto P, Luo Y, et al. Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung's disease. Nature. 1994;367:377–378. doi: 10.1038/367377a0. [DOI] [PubMed] [Google Scholar]
- 71.Pasini B, Borrello MG, Greco A, et al. Loss of function effect of RET mutations causing Hirschsprung disease. Nat Genet. 1995;10:35–40. doi: 10.1038/ng0595-35. [DOI] [PubMed] [Google Scholar]
- 72.Puffenberger EG, Hosoda K, Washington SS, et al. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 73.Edery P, Attie T, Amiel J, et al. Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome) Nat Genet. 1996;12:442–444. doi: 10.1038/ng0496-442. [DOI] [PubMed] [Google Scholar]
- 74.Hofstra RM, Osinga J, Tan-Sindhunata G, et al. A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome) Nat Genet. 1996;12:445–447. doi: 10.1038/ng0496-445. [DOI] [PubMed] [Google Scholar]
- 75.Hofstra RM, Valdenaire O, Arch E, et al. A loss-of-function mutation in the endothelin-converting enzyme 1 (ECE-1) associated with Hirschsprung disease, cardiac defects, and autonomic dysfunction. Am J Hum Genet. 1999;64:304–308. doi: 10.1086/302184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Angrist M, Bolk S, Halushka M, et al. Germline mutations in glial cell line-derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet. 1996;14:341–344. doi: 10.1038/ng1196-341. [DOI] [PubMed] [Google Scholar]
- 77.Salomon R, Attie T, Pelet A, et al. Germline mutations of the RET ligand GDNF are not sufficient to cause Hirschsprung disease. Nat Genet. 1996;14:345–347. doi: 10.1038/ng1196-345. [DOI] [PubMed] [Google Scholar]
- 78.Doray B, Salomon R, Amiel J, et al. Mutation of the RET ligand, neurturin, supports multigenic inheritance in Hirschsprung disease. Hum Mol Genet. 1998;7:1449–1452. doi: 10.1093/hmg/7.9.1449. [DOI] [PubMed] [Google Scholar]
- 79.Pingault V, Bondurand N, Kuhlbrodt K, et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 80.Amiel J, Laudier B, Attie-Bitach T, et al. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat Genet. 2003;33:459–461. doi: 10.1038/ng1130. [DOI] [PubMed] [Google Scholar]
- 81.Brooks AS, Bertoli-Avella AM, Burzynski GM, et al. Homozygous nonsense mutations in KIAA1279 are associated with malformations of the central and enteric nervous systems. Am J Hum Genet. 2005;77:120–126. doi: 10.1086/431244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cacheux V, Dastot-Le Moal F, Kaariainen H, et al. Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum Mol Genet. 2001;10:1503–1510. doi: 10.1093/hmg/10.14.1503. [DOI] [PubMed] [Google Scholar]
- 83.Wakamatsu N, Yamada Y, Yamada K, et al. Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat Genet. 2001;27:369–370. doi: 10.1038/86860. [DOI] [PubMed] [Google Scholar]
- 84.Garcia-Barcelo M, Ganster RW, Lui VC, et al. TTF-1 and RET promoter SNPs: regulation of RET transcription in Hirschsprung's disease. Hum Mol Genet. 2005;14:191–204. doi: 10.1093/hmg/ddi015. [DOI] [PubMed] [Google Scholar]
- 85.Garcia-Barcelo MM, Tang CS, Ngan ES, et al. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease. Proc Natl Acad Sci U S A. 2009;106:2694–2699. doi: 10.1073/pnas.0809630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pingault V, Bondurand N, Lemort N, et al. A heterozygous endothelin 3 mutation in Waardenburg-Hirschsprung disease: is there a dosage effect of EDN3/EDNRB gene mutations on neurocristopathy phenotypes? J Med Genet. 2001;38:205–209. doi: 10.1136/jmg.38.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sijmons RH, Hofstra RM, Wijburg FA, et al. Oncological implications of RET gene mutations in Hirschsprung's disease. Gut. 1998;43:542–547. doi: 10.1136/gut.43.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bolk S, Pelet A, Hofstra RM, et al. A human model for multigenic inheritance: phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci U S A. 2000;97:268–273. doi: 10.1073/pnas.97.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Emison ES, McCallion AS, Kashuk CS, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 90.Fitze G, Appelt H, Konig IR, et al. Functional haplotypes of the RET proto-oncogene promoter are associated with Hirschsprung disease (HSCR) Hum Mol Genet. 2003;12:3207–3214. doi: 10.1093/hmg/ddg354. [DOI] [PubMed] [Google Scholar]
- 91.Fernandez RM, Boru G, Pecina A, et al. Ancestral RET haplotype associated with Hirschsprung's disease shows linkage disequilibrium breakpoint at —1249. J Med Genet. 2005;42:322–327. doi: 10.1136/jmg.2004.023960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burzynski GM, Nolte IM, Bronda A, et al. Identifying candidate Hirschsprung disease-associated RET variants. Am J Hum Genet. 2005;76:850–858. doi: 10.1086/429589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Griseri P, Bachetti T, Puppo F, et al. A common haplotype at the 5' end of the RET proto-oncogene, overrepresented in Hirschsprung patients, is associated with reduced gene expression. Hum Mutat. 2005;25:189–195. doi: 10.1002/humu.20135. [DOI] [PubMed] [Google Scholar]
- 94.Emison ES, Garcia-Barcelo M, Grice EA, et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet. 2010;87:60–74. doi: 10.1016/j.ajhg.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grice EA, Rochelle ES, Green ED, et al. Evaluation of the RET regulatory landscape reveals the biological relevance of a HSCR-implicated enhancer. Hum Mol Genet. 2005;14:3837–3845. doi: 10.1093/hmg/ddi408. [DOI] [PubMed] [Google Scholar]
- 96.Sribudiani Y, Metzger M, Osinga J, et al. Variants in RET associated with Hirschsprung's disease affect binding of transcription factors and gene expression. Gastroenterology. 2011;140:572–582. e572. doi: 10.1053/j.gastro.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 97.Carrasquillo MM, McCallion AS, Puffenberger EG, et al. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet. 2002;32:237–244. doi: 10.1038/ng998. [DOI] [PubMed] [Google Scholar]
- 98.Brooks AS, Leegwater PA, Burzynski GM, et al. A novel susceptibility locus for Hirschsprung's disease maps to 4q31.3-q32.3. J Med Genet. 2006;43:e35. doi: 10.1136/jmg.2005.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brooks AS, Oostra BA, Hofstra RM. Studying the genetics of Hirschsprung's disease: unraveling an oligogenic disorder. Clin Genet. 2005;67:6–14. doi: 10.1111/j.1399-0004.2004.00319.x. [DOI] [PubMed] [Google Scholar]
- 100.Badner JA, Sieber WK, Garver KL, et al. A genetic study of Hirschsprung disease. Am J Hum Genet. 1990;46:568–580. [PMC free article] [PubMed] [Google Scholar]
- 101.Panza E, Knowles CH, Graziano C, et al. Genetics of human enteric neuropathies. Prog Neurobiol. 2012;96:176–189. doi: 10.1016/j.pneurobio.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 102.Di Nardo G, Blandizzi C, Volta U, et al. Review article: molecular, pathological and therapeutic features of human enteric neuropathies. Aliment Pharmacol Ther. 2008;28:25–42. doi: 10.1111/j.1365-2036.2008.03707.x. [DOI] [PubMed] [Google Scholar]
- 103.Hotta R, Natarajan D, Burns AJ, et al. Stem cells for GI motility disorders. Curr Opin Pharmacol. 2011;11:617–623. doi: 10.1016/j.coph.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 104.Wood JD. Enteric nervous system neuropathy: repair and restoration. Curr Opin Gastroenterol. 2011;27:106–111. doi: 10.1097/MOG.0b013e328342a6ea. [DOI] [PubMed] [Google Scholar]
- 105.Schafer KH, Micci MA, Pasricha PJ. Neural stem cell transplantation in the enteric nervous system: roadmaps and roadblocks. Neurogastroenterol Motil. 2009;21:103–112. doi: 10.1111/j.1365-2982.2008.01257.x. [DOI] [PubMed] [Google Scholar]
- 106.Kulkarni S, Becker L, Pasricha PJ. Stem cell transplantation in neurodegenerative disorders of the gastrointestinal tract: future or fiction? Gut. 2012;61:613–621. doi: 10.1136/gut.2010.235614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cacalano G, Farinas I, Wang LC, et al. GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron. 1998;21:53–62. doi: 10.1016/s0896-6273(00)80514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Enomoto H, Araki T, Jackman A, et al. GFRalpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron. 1998;21:317–324. doi: 10.1016/s0896-6273(00)80541-3. [DOI] [PubMed] [Google Scholar]
- 109.von Boyen GB, Krammer HJ, Süss A, et al. Abnormalities of the enteric nervous system in heterozygous endothelin B receptor deficient (spotting lethal) rats resembling intestinal neuronal dysplasia. Gut. 2002;51:414–419. doi: 10.1136/gut.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lane PW, Liu HM. Association of megacolon with a new dominant spotting gene (Dom) in the mouse. J Hered. 1984;75:435–439. doi: 10.1093/oxfordjournals.jhered.a109980. [DOI] [PubMed] [Google Scholar]
- 111.Guillemot F, Lo LC, Johnson JE, et al. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 112.Ramalho-Santos M, Melton DA, McMahon AP. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]