Abstract
We have recently shown that p-cyanophenylalanine (Cnf) and a thioamide can be used as a minimally-perturbing Förster resonant energy transfer (FRET) pair to monitor protein conformation. We have also shown that thioamide analogs of natural amino acids can be incorporated into full-sized proteins through native chemical ligation. For intermolecular studies with Cnf/thioamide FRET pairs, Cnf can be incorporated into proteins expressed in E. coli through unnatural amino acid mutagenesis using a Cnf-specific tRNA synthetase. For intramolecular studies, a Cnf-labeled protein fragment can be expressed in E. coli and then ligated to a thioamide-labeled peptide synthesized on solid phase. This combination of methods allows for rapid access to double-labeled proteins with a minimum of unnecessary chemical synthesis. We demonstrate the utility of this approach by studying the binding of peptides to the protein calmodulin and by determining the orientation of the N- and C-termini in the amyloidogenic protein α-synuclein.
INTRODUCTION
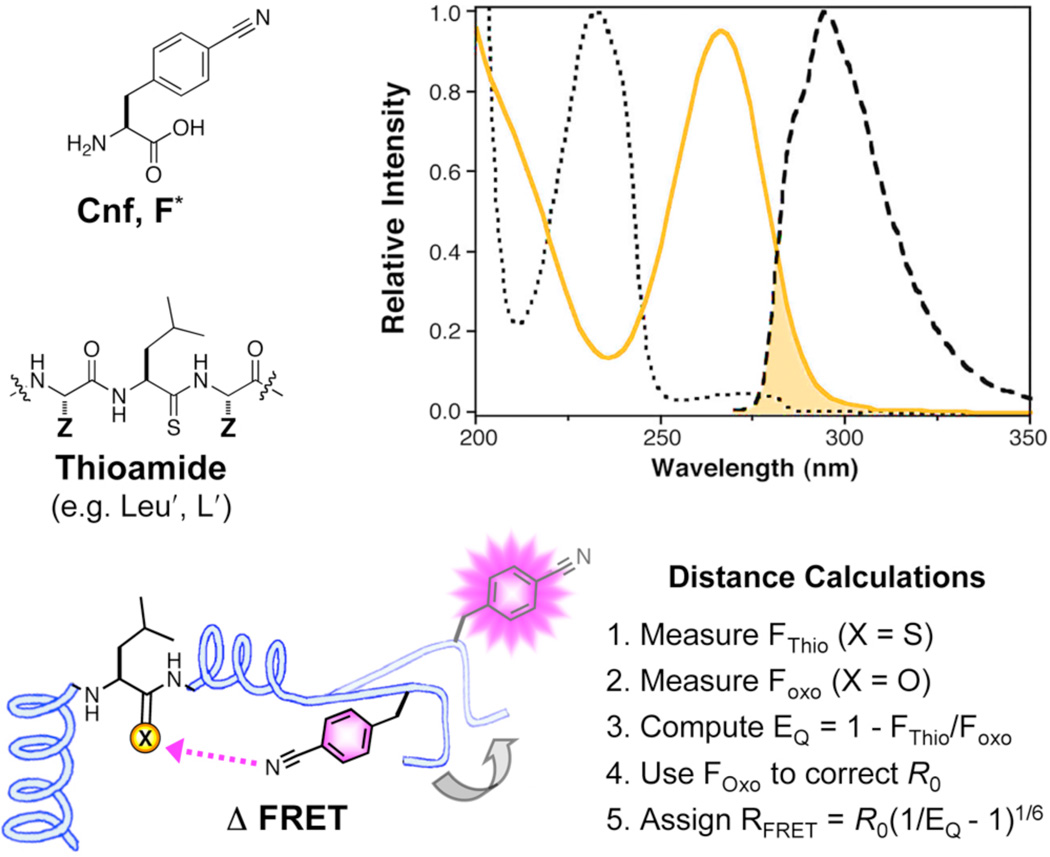
Fluorescence spectroscopy can be used to monitor confor-mational changes in proteins by exploiting distance-dependent interactions such as Förster resonant energy transfer (FRET).1,2 In a FRET experiment, a donor fluorophore is paired with an acceptor fluorophore or quencher. An increase in chromophore separation leads to an increase in donor fluorescence and a decrease in acceptor fluorescence or quenching with an R−6 distance dependence. The utility of the data obtained in these experiments often depends on the ability to label the proteins at appropriate locations. Bulky fluorophores are limited in the number of places that the label can be placed, and are subject to the concern that the label itself alters the observed motion.3 Therefore, our laboratory has developed the thioamide – sulfur replacement of oxygen in the peptide backbone – as an extremely small label compatible with virtually any position in the protein. (Fig. 1)
Figure 1.
Cyanophenylalanine/Thioamide FRET Interactions. Top Left: Cyanophenylalanine (Cnf, F*) and leucyl thioamide (Leu′, L′). Top Right: Normalized absorption (dotted black line) and fluorescence emission (dashed black line) spectrum of Cnf and absorption spectrum of Leu′Ala (yellow line). The shaded area indicates the spectral overlap that contributes to FRET. Bottom: Distance (RFRET) calculations are made using Förster theory as described in text.
Thioamides can quench fluorophores that have spectral emission overlap with thioamide absorption, such as cyanophenylalanine (Cnf) and tyrosine, through a FRET mechanism.4,5 Other fluorophores are quenched through a photoinduced electron transfer (PET) mechanism, like tryptophan, coumarin, and acridone.5,6 Here we will focus on thioamide FRET partners, for which more reliable distance information can be obtained by applying Förster theory. To use thioamide FRET quenching to study protein conformation, we generate constructs in which we label the protein with a Cnf donor fluorophore and a thioamide analog of one of the natural amino acids. We denote these analogs using either the three or one letter amino acid code with a prime symbol indicating the thiocarbonyl (thioleucine is shown in Figure 1). Distance constraints are assigned by computing quenching efficiency (EQ = 1 – FThio/FOxo) from the fluorescence of equimolar concentrations of Cnf/thioamide-labeled protein (FThio) and a Cnflabeled control (FOxo). After appropriate corrections to the Förster radius (R0) for each Cnf location, the Cnf/thioamide separation (RFRET) is determined according to Förster theory. (Fig. 1, and discussion below) These distance measurements are used to monitor conformational changes in the protein.
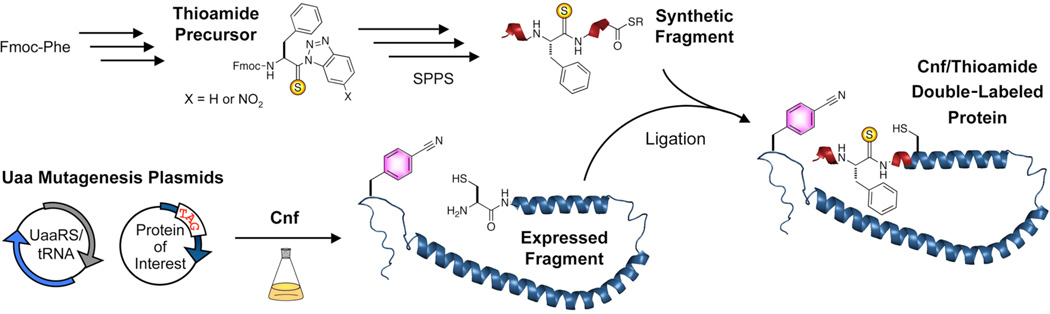
All of our initial studies used small peptides and proteins in which the thioamide was incorporated by solid phase peptide synthesis (SPPS). In order to use fluorophore/thioamide pairs to monitor structural changes in full-sized proteins, we needed to identify methods for incorporating these two modifications with high efficiency. We have recently published an account of our studies of the compatibility of thioamides with native chemical ligation (NCL) reactions.7 NCL is a valuable method for synthesizing proteins by fragment condensation of a C-terminal thioester peptide and an N-terminal Cys peptide in buffered aqueous media.8,9 We found that thioamides could be placed at nearly any position in the thioester or Cys peptide fragments. Furthermore, we found that thioamides could be used in NCL reactions in which the N-terminal Cys fragment was expressed as a protein construct in E. coli (expressed protein ligations). In general, we wish to minimize the need for manual synthesis by maximizing the role of biosynthesis in preparing labeled proteins. These protein ligations can limit the portion that must be prepared by SPPS to a few amino acids around the thioamide. SPPS is deemed necessary for the incorporation of backbone thioamide modifications, but sidechain fluorophores like Cnf can be incorporated during ribosomal biosynthesis.
To easily incorporate Cnf, we have employed unnatural amino acid (Uaa) mutagenesis methods pioneered by Schultz and coworkers.10,11 These methods allow one to insert a Uaa at a specific site in the protein by using a so-called orthogonal aminoacyl tRNA synthetase (RS) that is selective for the Uaa. The UaaRS charges a tRNA that recognizes the UAG stop codon. The UAG stop codon is mutated into the protein coding sequence at the site of interest. Plasmids coding for the UaaRS, tRNA, and protein of interest are transformed into E. coli. After growth in the presence of the Uaa, protein containing the unnatural amino acid is purified for use in biochemical experiments or subsequent NCL reactions.
Here, we made use of Uaa mutagenesis to incorporate donor fluorophores for thioamide FRET experiments examining both protein/protein interactions and intramolecular measurements of protein folding. In intermolecular experiments, no manipulation of the Uaa-containing protein is required following purification. The protein is simply mixed with a thioamide-labeled peptide or protein and quenching observed. We use this strategy to examine a series of pairwise interactions in which Cnf is incorporated in the calcium sensor protein calmodulin (CaM) and the thioamide is incorporated in a CaM-binding peptide. The combinatorial nature of these experiments allows one to rapidly generate a matrix of m × n FRET measurements from m Cnf-labeled proteins and n thioamide-labeled peptides. To carry out intramolecular experiments, we combined Uaa mutagenesis with NCL. Cnf was incorporated by Uaa mutagenesis into protein constructs in which the termini could be used in ligation to thiopeptides. Strategies for ligating to both the C- and N-termini are described. The resulting double-labeled proteins can then be used in intramolecular folding experiments. Again, combinatorial assembly can be used to make m × n double-labeled proteins from m N-terminal fragments and n C-terminal fragments. We applied our double-labeling strategy to study conformational changes in α-synuclein (αS), an amyloidogenic protein found to aggregate in Parkinson’s Disease.12–14
Our methods should be applicable to a wide variety of proteins: essentially any proteins that express well in E. coli and are amenable to refolding after ligation. Moreover, since this combination of Uaa mutagenesis and NCL to label proteins on the backbone and sidechain is unprecedented (examples of sidechain ligation using Uaa mutagenesis exist),15,16 the considerations here should be useful to those who wish to combine these two methods for other reasons, such as incorporating olefin or D-amino acid modifications in combination with unnatural sidechains.17–20
RESULTS AND DISCUSSION
To begin combining Uaa mutagenesis with thioamides, we used CaM as a model system. We chose CaM because it expresses well in E. coli and it has been thoroughly studied from a structural and biophysical standpoint.21–27 CaM has been shown to bind a variety of helical peptides derived from its endogenous protein interaction partners.28,29 These peptides bind with high affinity in a central groove formed by the folding of the N- and C-terminal CaM lobes. In particular, we studied the binding of a peptide derived from the intracellular domain of an olfactory cyclic nucleotide-gated ion channel (pOCNC).30 This peptide binds with a low nM affinity, and its CaM-bound structure has been determined previously by NMR.30,31 Therefore, we expected that 1:1 mixtures of 10 µM each CaM and pOCNC would be stably bound and that the NMR structures could be used to determine interatomic distances. These measurements would then be compared to the corresponding distances derived from our FRET measurements.
Since Cnf is excited at 240 nm and fluoresces with a maximum at 295 nm, we began by mutating the two endogenous Tyr residues in CaM to Phe, generating a construct hereafter referred to as CaMF.32 . This all Phe background” ensures that Tyr fluorescence does not confound our ability to interpret Cnf data. Both the extinction coefficient and quantum yield of Phe (ε240 = 65 cm−1 •M−1 Φ = 0.022) are substantially smaller than the values for Cnf (ε240 = 10,333 cm−1 •M−1 , Φ = 0.110), so in these proteins all fluorescence emission at 295nm should derive from Cnf.32,33
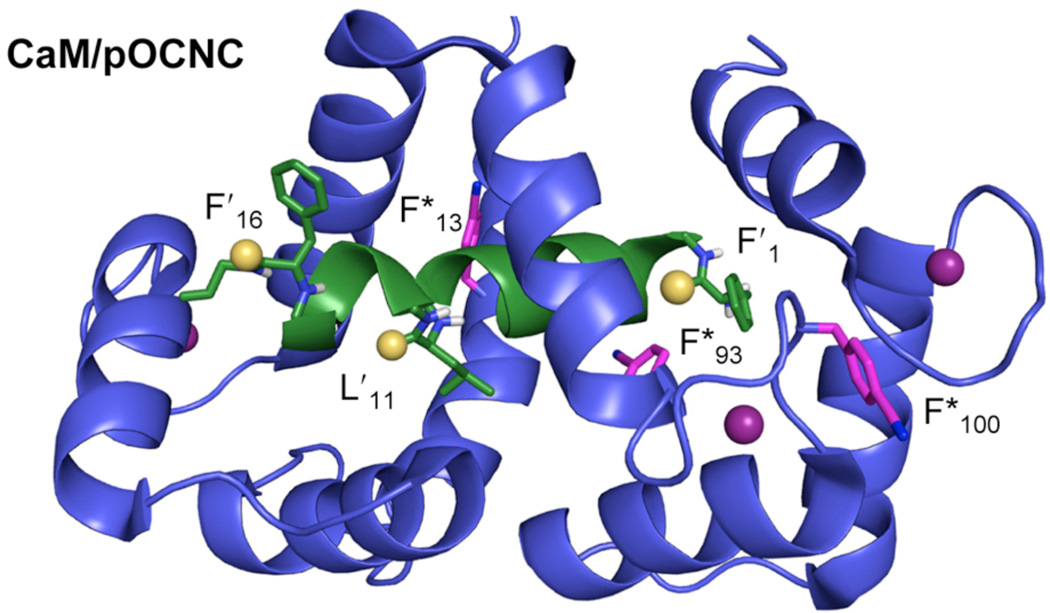
To incorporate Cnf, we used a plasmid containing a UaaRS/tRNA pair previously selected for Cnf incorporation by Mehl and coworkers.34 This was paired with a plasmid encoding CaM under IPTG control. We expressed CaMF in M9 media supplemented with Cnf and purified the full-length protein by two rounds of phenyl sepharose chromatography. Four positions were chosen for incorporation of Cnf: F13, F17, F93, and Y100. Four pOCNC peptides were synthesized: an all Phe oxopeptide (pOCNC), and three thiopeptides – pOCNC-F′1, pOCNC-L′11, and pOCNC-F′16. Before performing fluorescence titration experiments, native PAGE gel analysis was used to ensure that each CaMF /pOCNC combination formed stable complexes. This allowed us to eliminate CaMFF*17 from further binding experiments (see Supporting Information).
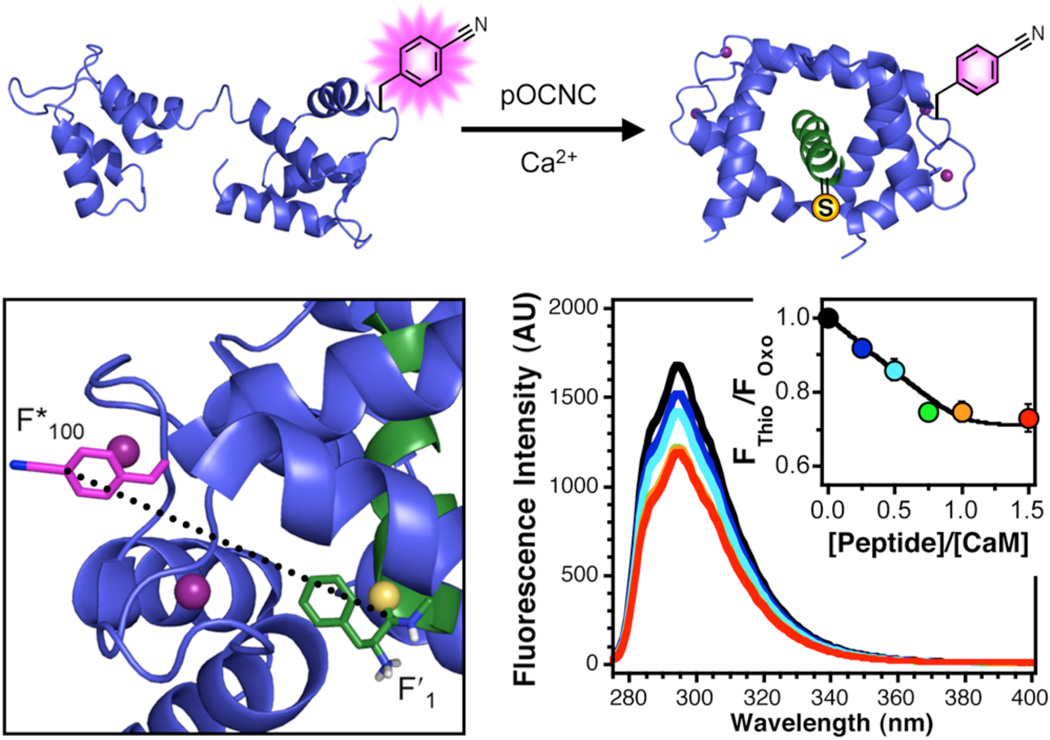
For each of the other Cnf mutants, we performed the same set of experiments, which are described using CaMFF*100 as an example. Fluorescence wavelength scans were acquired for mixtures of 10 µM CaMFF*100 with between 0 and 15 µM pOCNC or pOCNC-F′1. For each concentration, the fluorescence emission from the pOCNC-F′1 complex (FThio) was normalized to the fluorescence emission from the pOCNC complex (FOxo). For F*100 a concentration-dependent decrease was observed for FThio, while no change was seen FOxo. Plotting FThio/FOxo (or 1 – EQ) as a function of concentration shows that binding reaches saturation at 1:1 CaM FF*100/pOCNC-F′1 (Fig. 3) The quenching (FRET) efficiency can then be used to calculate a separation according to RFRET = R0(1/EQ - 1). We observed 25% quenching at saturating concentrations of pOCNC-F’1, which translates into a 20 Å separation using Förster theory. This is in excellent agreement with the 19 Å average separation computed from the 20 lowest energy structures deposited from the NMR structure determination by Contessa et al.31 These calculations were performed using Φ = 0.110 and an orientation factor (κ2 ) of 2/3.
Figure 3.
FRET in CaM/Peptide Complexes. Top: Binding of thioamide-labeled peptides (green) to Cnf-labeled CaM (blue) in the presence of Ca2+. Bottom Left: An image showing Cnf (pink) at position 100 in calmodulin (CaMFF*100) and thiophenylalanine (yellow sulfur atom) at position 1 in the pOCNC peptide (pOCNC-F′1). Ca2+ ions are shown as purple spheres. Adapted from PDB ID 1SYD using PyMol. The 18.9 Å separation shown represents an average of the separation of the center of the thioamide from the center of the ε-carbons of the Cnf ring. Bottom Right: Fluorescence emission spectra of solutions of 10 µM CaMFF*100 in the presence of increasing concentrations of pOCNC-F′1. Inset: Relative fluorescence of complexes of CaMFF*100/pOCNC-F′1 (FThio) to an oxoamide control CaMFF*100/pOCNC (FOxo). See Supporting Information for details.
We completed comparable experiments for each pair of Cnf mutant and thiopeptide. The separation in the seven pairs spanned a distance range from 8 to 31 Å, covering the range that is accessible with the Cnf/thioamide FRET pair. In the case of CaM FF*100/pOCNC-F′1, no change in fluorescence was observed in the “oxoamide control” experiment. For other mutants, some changes in fluorescence were observed for pOCNC binding. (See Supporting Information) Regardless, these effects can be deconvoluted by calculating EQ relative to the oxoamide control as we do. Such oxoamide control experiments are essential as many factors can influence fluorescence. 35–37Therefore, we always compare any thioamide quenching measurement to the most relevant control, in this case the CaM/pOCNC oxopeptide complex.
The oxopeptide complex is also valuable in determining the appropriate value of Φ to be used in FRET distance calculations. The changes in fluorescence intensity upon pOCNC binding represent a change in Cnf quantum yield as it moves into a new environment. For example, the emission of CaM FF*93 starts out at 3.8% of the emission of an equimolar concentration of CaM FF*100, but it increases 3.2-fold upon pOCNC binding. Therefore, the relevant quantum yield for the CaMFF*93/pOCNC complex is 0.014. We use this value in our calculation of Rfret for all thiopeptide complexes with CaM FF*93. Similar corrections to the quantum yield were performed for CaMFF*13. Correction of the quantum yield of CaMFF*100 was not necessary, presumably because Cnf is solvent-exposed at this position.
The distances calculated from our FRET measurements are collected in Table 1. All measurements are within 3 Å of the average value observed in the NMR structure except for F*100/F′16. Moreover, our measurements are always correct in predicting the relative order of thioamide proximity for a given Cnf mutant. While it may be surprising that we achieve reasonable agreement between theory and experiment using a value of 2/3 for κ2 , deriving κ2 values from the 20 lowest energy NMR structures has only a small effect on this agreement. (See Supporting Information) The RFRET values obtained using κ2 = 2/3 are more representative of the application of thioamide FRET in a structure determination experiment (i.e. one in which the target structure is unknown and κ2 would be difficult to fit accurately).
Table 1.
Distances in CaM/pOCNC Complexes
| CaM/pOCNC Mutants (Φ)a | % EQ Obsvdb |
RFRET (Å)c |
RNMR (Å)d |
|---|---|---|---|
| All Phe control | 0 | — | — |
| F*13/F′1 (0.003) | 11 ± 1 | 13 | 14 |
| F*13/L′11 (0.003) | 37 ± 2 | 11 | 14 |
| F*17 | NBe | — | — |
| F*93/F′1 (0.014) | 79 ± 2 | 12 | 9 |
| F*93/L′11 (0.014) | 43 ± 4 | 13 | 12 |
| F*100/F′1 (0.110) | 25 ± 3 | 20 | 19 |
| F*100/L′11 (0.110) | 17 ± 2 | 21 | 24 |
| F*100/F′16 (0.110) | 10 ± 3f | 23 | 31 |
Φ determined by comparison to fluorescence emission of 10 µM Cnf, Φ = 0.110.
EQ determined for 1:1 CaM/pOCNC complex.
RFRET calculated from EQ as described in Supporting In formation.
RNMR is an average value calculated from the twenty lowest energy structures in PDB ID 1SYD.
NB indicates no binding by native PAGE gel analysis.
Mixture of R17 D- and L epimers of pOCNC-F′16 peptide. Both bind with high affinity by native PAGE gel analysis.
These CaM experiments involved intermolecular quenching, but our method, like FRET in general, is equally applicable to intramolecular conformational changes. However, obtaining Cnf/thioamide double-labeled proteins represents a biosynthetic challenge, as one ideally wishes to have complete freedom in placing the chromophores within the protein sequence. While this is easily accomplished in small peptides made by SPPS, and conceivably could be done by NCL, we wished to use a combination of NCL and Uaa mutagenesis to minimize unnecessary peptide synthesis. Therefore, we assessed the compatibility of these two methods. We chose αS as a model system because we had previously shown that thioamides could be incorporated into αS, and we wished to compare PET quenching of Trp in αS to FRET quenching of Cnf at similar locations.7
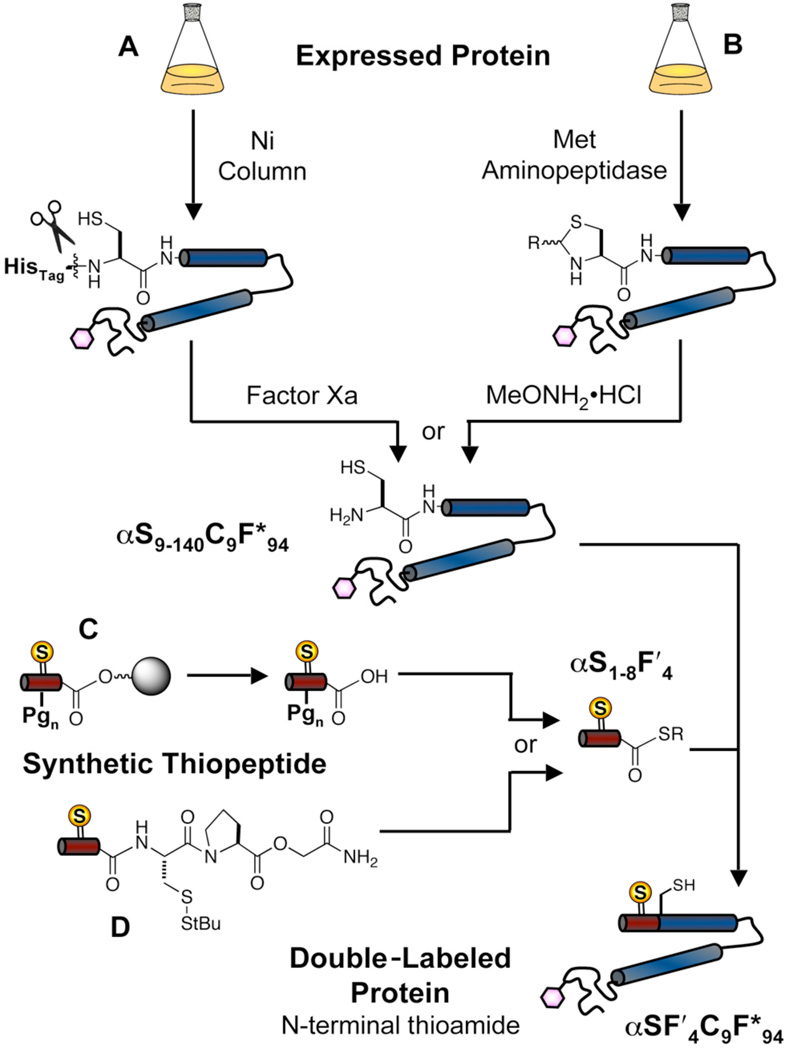
First, we examined thioamide incorporation at the αS N-terminus. For protein expression, we used the UaaRS/tRNA plasmid pDULE2-Cnf and a plasmid encoding αS with an N-terminal truncation that could be revealed after proteolysis of a His10 purification sequence using Factor Xa (Fig. 5A).38 The proteolyzed protein contains an N-terminal Cys for use in NCL with the synthetic thioester. The Factor Xa method provides a general method for generating NCL constructs containing unnatural amino acids, but in the specific context of αS, prolonged incubation with Factor Xa can lead to protein loss via aggregation. Therefore, we have also used constructs directly encoding αS with an N-terminal truncation plus Met in a fashion similar to that described by Hejjaoui et al.39 The N-terminal Met residue is removed in vivo by Met aminopepti-dase to reveal the requisite Cys.40 Protein harvested from E. coli contains an N-terminal thiazolidine ring formed by the condensation of Cys with either pyruvate or acetaldehyde. This is treated with methoxylamine hydrochloride to reveal free Cys for NCL reactions. Regardless of the method used in producing the expressed fragment, once purified, usage in subsequent NCL reactions is identical.
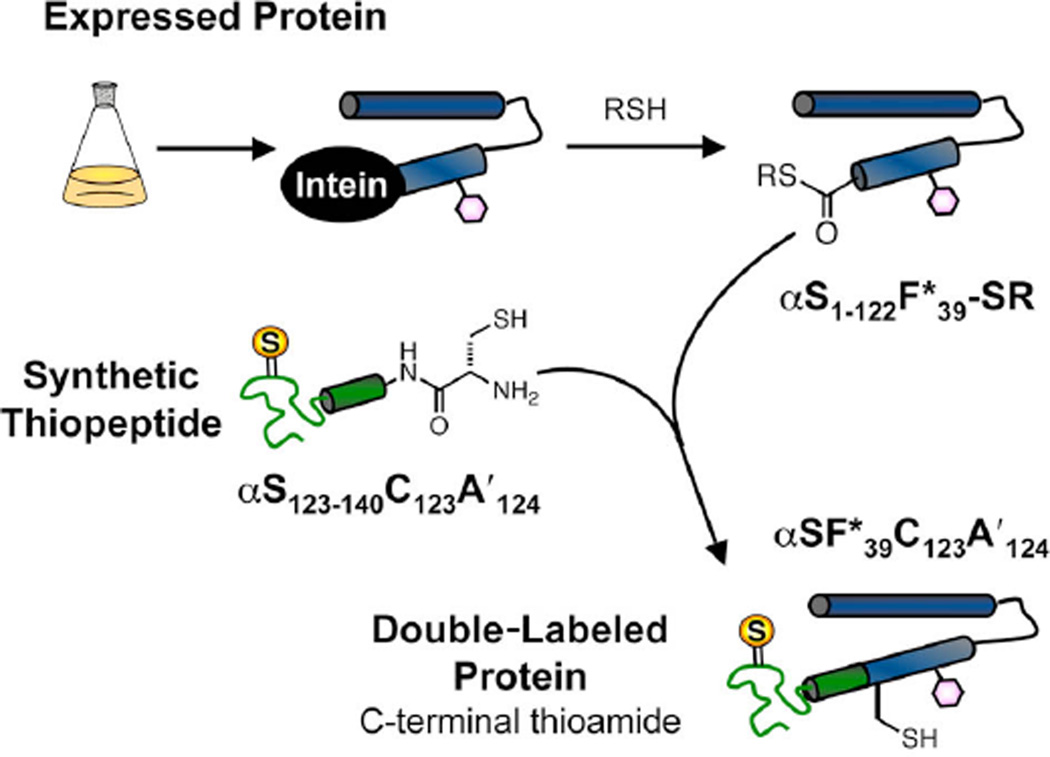
Figure 5.
Semi-synthesis of Double-Labeled Proteins with N-terminal Thioamides. Expressed C-terminal protein fragment: A) Using a plasmid encoding an N-terminal HisTag with a Factor Xa site; B) Using a plasmid encoding the truncated protein where N-terminal Met is removed by Met aminopeptidase. MeONH2•HCl used to cleave the thiazolidine which forms with intracellular aldehydes. (R = Me or CH2CO2H) Synthetic N-terminal thiopep-tide thioester: C) Synthesis on Cl-Trt resin and activation to form thioester with PyBOP, followed by deprotection with TFA; D) Synthesis on Rink amide resin with CbPGo masked thioester. The expressed protein fragment and synthetic thiopeptide are combined in the presence of thiophenol and the ligated, full-length protein isolated.
Synthesis of a thioamide-containing thioester peptide using Fmoc chemistry could be accomplished by one of two methods. The thiopeptide can be synthesized on Cl-Trt resin, cleaved under mild conditions (10% AcOH) to retain the sidechain protecting groups, and then activated using PyBOP to form the thioester (Fig. 5C).41,42 This method can lead to epimerization of the α-carbon of the C-terminal amino acid, but this can be avoided by using short (< 30 min) reaction times during PyBOP activation.43 A more significant inherent limitation is the insolubility of protected peptides > 15 amino acids in most solvents.41 This limits use of the PyBOP method to short peptides.
The other method makes use of a thioester that is masked during synthesis as a disulfide (Fig. 5D). Several variations of masked thioester exist. In our previous work, we used a three residue CbPGo sequence where Cb is a t-Bu-protected Cys and Go is glycolic acid.7,44,45 Deprotection of the Cys thiol initiates a cascade reaction generating a diketopiperazine thioester that is competent in NCL reactions. Synthesis of peptides with the CbPGo linker is inefficient because of ester linkage instability. Therefore we reserve its use for longer peptides where it is necessary.
All αS protein constructs tested contain Tyr to Phe mutations at positions 39, 125, 133, and 136 to generate an “all Phe” background construct, αSF. As was the case for CaMF, these mutations allow us to more easily interpret Cnf fluorescence data. Combinations of methods 5a or 5b with 5c or 5d were used to synthesize double-labeled αS. While all combinations were viable, for αSFF′4C9F*94, our highest yields were obtained with a combination of methods 5b and 5c. Protein expression yields were typically 3 mg/L of culture after methoxylamine treatment. Ligations were typically carried out on the 0.1 µmol scale at pH 7.2 with 1% thiophenol. After 24 hours, 60 – 70% yields of ligated product were typically seen, and the products purified by C4 reverse phase HPLC, giving isolated yields of about 0.4 mg for a given ligation. PAGE gel, spectroscopic, and MALDI MS analyses of the ligation confirm production of homogeneous, double-labeled αS. Data for other reactions are shown in Supporting Information.
The methods shown in Figure 5 permit protein double-labeling with thioamides located at the N-terminus. To insert thioamides at the C-terminus, we made use of intein fusion constructs popularized by Muir and others.9 These self-splicing domains have been engineered to stall at a thioester intermediate and can be intercepted by treatment with a small molecule thiol to give a thioester suitable for NCL. We have found that the expression of intein fusion constructs is compatible with Uaa mutagenesis. In fact, yields of intein fusions of αS Cnf mutants (e.g. αSF1–122F*94-Int) are higher (20 mg/L culture) than comparable expressions of the full-length protein using either T7 or PRK vectors previously used to express αS mutants (3 – 5 mg/L). The synthetic thiopeptide with N-terminal Cys (αSF123–140C123A′124) was prepared by standard SPPS.
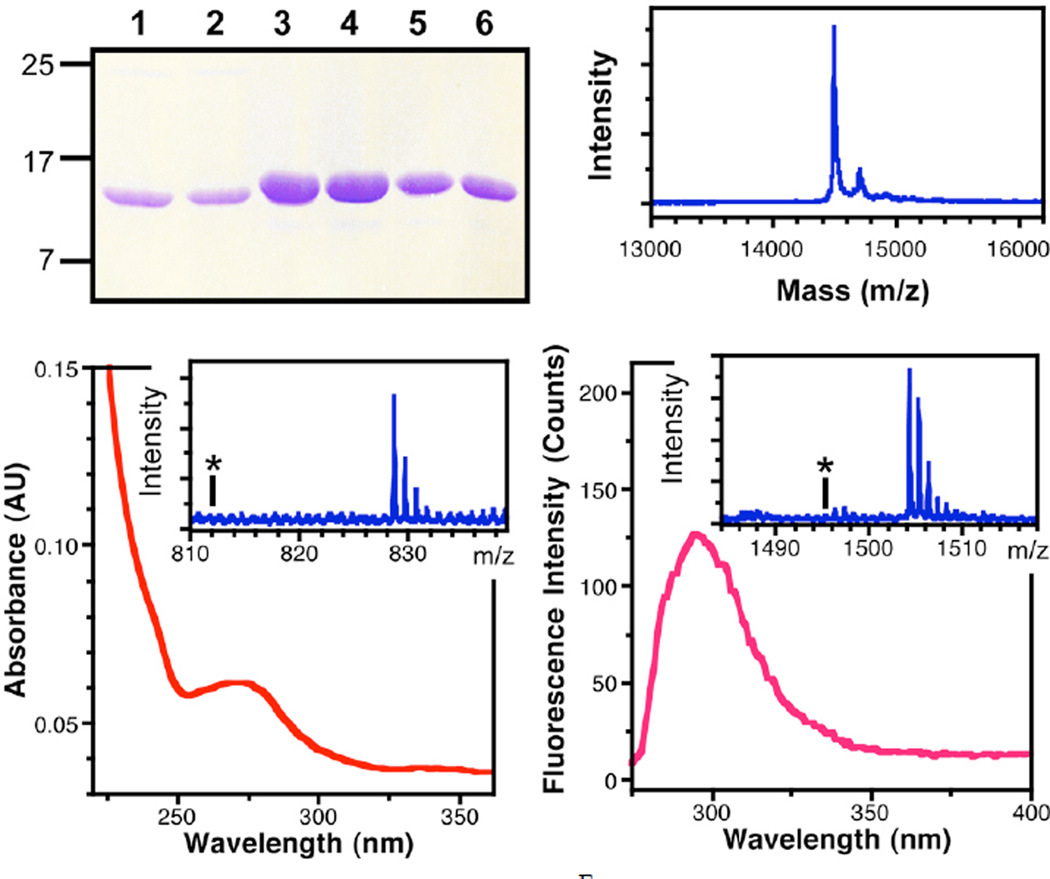
The thioester αSF1–122F*39-SR was prepared from αSF1–122F*39-Int using the thiol mercaptoethane sulfonate (MESNA). This was combined with αSF123–140C123A′124 in a buffer containing thiophenol. Although some C-terminal Asn cyclization (~ 25% based on crude MALDI MS estimates, see Supporting Information) of the intein thioester occurred in competition with the desired MESNA thiolysis, this was not a major problem since the resulting cyclic succinimide was not competent for ligation. Ligations were typically carried out on the 0.3 µmol scale. After 16 hours, 50% yields were obtained, and the products purified by FPLC using an ion-exchange column, giving isolated yields of 0.6 – 0.7 mg for a given ligation reaction. Once again PAGE gels, spectroscopy, and MALDI MS were used to analyze the ligation reaction. These data are shown in Figure 8. The data confirm that we have again produced homogeneous, full-length, double-labeled αS.
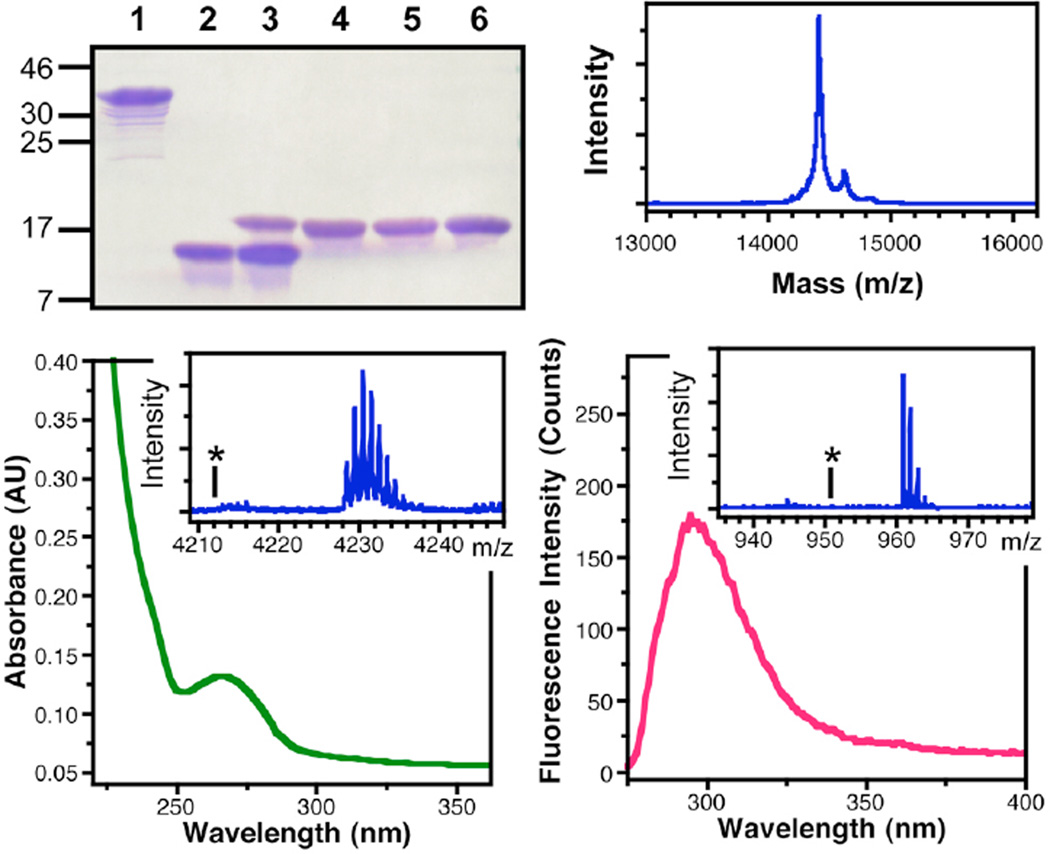
Figure 8.
Characterization of αSFF*39C123A′124 Ligation Product. Top Left: PAGE gel showing: αSF1–122F*39-Int before (1) and after thiolysis of the intein to generate αSF1–122F*39-SR thioester (2), ligation of αSF123–140C123A′124 to αSF1–122F*39-SR thioester (3), purified αSFF*39C123A′124 (4), αSFF*39C123 (5), and αSF (6). Top Right: MALDI MS analysis of full-length αSFF*39C123A′124. Bottom Left: UV/Vis absorption spectrum of αSFF*39C123A′124 showing thioamide absorption at 266 nm. Inset: MALDI MS analysis of trypsinized 103–140 fragment, confirming the presence of the thioamide at A′124; Calcd m/z (M+H): 4228.69, Obsvd: 4228.47. The asterisk indicates the absence of peaks corresponding an oxoamide at Ala124 in the 103–140 fragment; Calcd m/z (M+H): 4212.72. Bottom Right: Fluorescence emission spectrum of αSFF*39C123A′124 showing Cnf emission at 295 nm. Inset: MALDI MS analysis of trypsinized 35–43 fragment, confirming the presence of Cnf at position 39; Calcd m/z (M+H): 960.52, Obsvd: 960.77. The asterisk indicates the absence of peaks corresponding Tyr at position 39 in the 35–43 fragment; Calcd m/z (M+H): 951.51.
Using these two methods, we should be able to label a variety of proteins with thioamides at the N- or C-terminus, with complete freedom in placement of the donor fluorophore. Since we have previously shown that thioamides are compatible with the MeONH2•HCl conditions used to deprotect N-terminal thiazolidines, the methods used here should also be compatible with internal thioamide-labeling through three component ligations. In addition, we note that the use of C-terminal inteins may be generally useful to those employing Uaa mutagenesis. One major limitation of the method is that the inherent competition with release factors at the UAG stop codon can lead to protein truncation.46 Purifying the full-length protein containing the Uaa can be difficult for sites near the C-terminus, where the truncated form is only slightly shorter. The His-tagged intein provides a convenient handle that will only be present in the untruncated protein. Since the intein can be cleaved by thiolysis or hydrolysis in a traceless manner, the Uaa-labeled fragment or protein can be obtained in pure form in an efficient manner.
In order to demonstrate the value of double-labeling a protein such as αS, we carried out experiments in which intramolecular conformational changes were monitored. αS aggregates by first forming soluble oligomers which eventually convert into larger, insoluble fibrils.13,14,47,48 Both the oligomers and fibrils have been shown to be cytotoxic, and recent evidence has shown that extracellular αS can enter neurons and catalyze misfolding of αS within healthy neurons.49–52 Therefore, there is substantial current interest in understanding the structural dynamics of the meta-stable αS monomers and the rearrangements that occur on formation of oligomers and subsequent fibrillization. Several researchers have recently shown the value of being able to construct semi-synthetic αS to study the effects of post-translational modification on its folding, our modification should provide a uniquely non-perturbing method for tracking its dynamics.7,39,53–55
We envision thioamide quenching as a method for site-selectively monitoring conformational changes during αS refolding. While a number of researchers have studied αS conformation using other techniques such as NMR, IR, and EPR, we find fluorescence to be particularly useful because the sensitivity of the technique permits one to use low concentrations of double-labeled αS which should not undergo aggregation on the timescale of our experiments.47,48,52,56–60 This strategy allows us to monitor long-range (25 – 30 Å) interactions that are difficult to observe with other techniques. Moreover, FRET is easily applied to interactions involving residues in dynamic regions of proteins, such as the termini of αS.
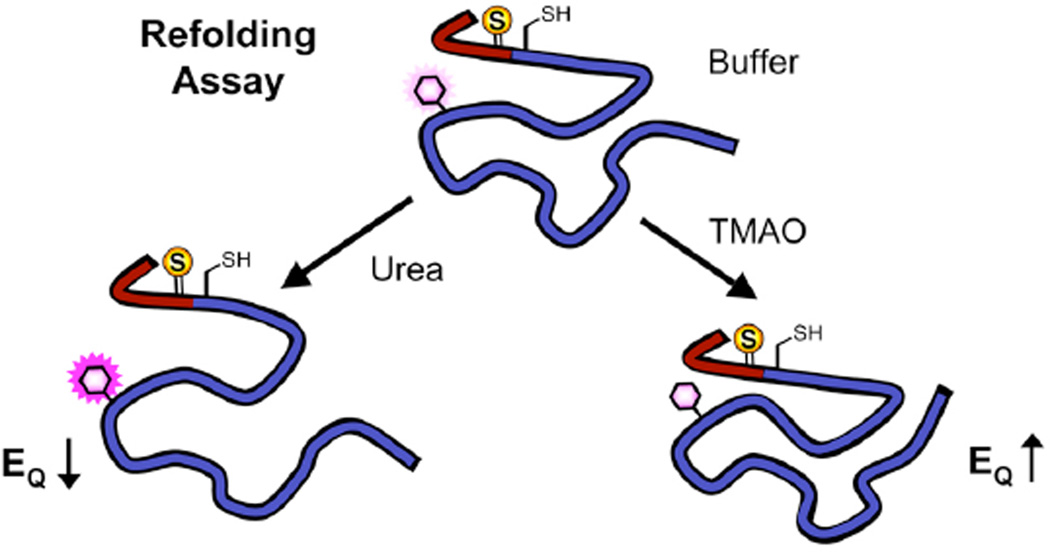
We have previously shown that conformational dynamics during aggregation can be monitored by intramolecular quenching of Trp by a thioamide.7 Here, we demonstrate that we can monitor conformational changes in monomeric αS, using urea or trimethylamine oxide (TMAO) to denature or compact αS, respectively. Performing oxoamide control experiments allows us to correct for any changes in fluorescence not due to the thioamide, and correct the Cnf quantum yield in order to properly interpret the observed quenching in terms of interchromophore distance.
The NCL procedures that we use to synthesize our thioamide-labeled proteins introduce a Cys into the αS sequence at position 9 or 123. However, we have previously shown that the presence of non-native Cys in the sequence does not cause aberrant aggregation when incubated with β-mercaptoethanol (BME) to prevent disulfide formation.7 While Cys can have an effect on Cnf emission, since we include Cys mutations in our oxoamide control experiments, we can account for these changes as well.34,61
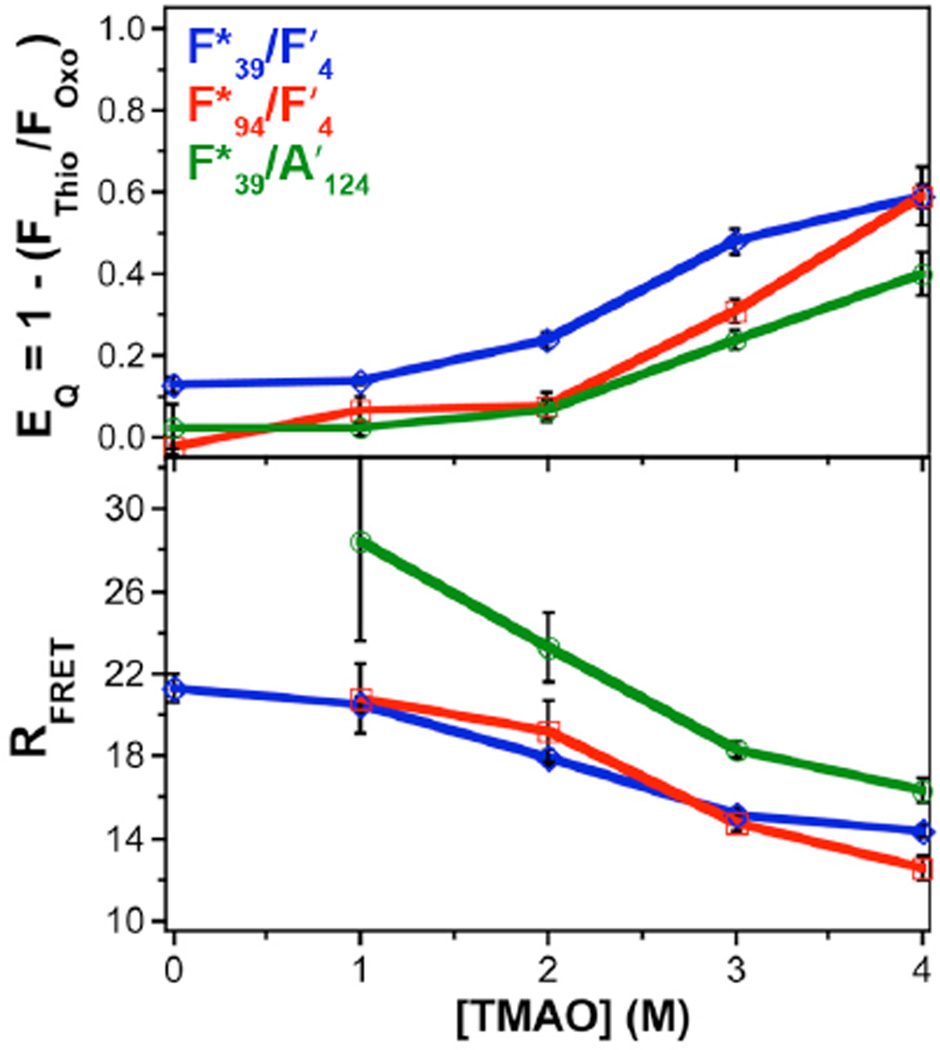
We carried out refolding experiments with the constructs described above, αSFF′4C9F*94 and αSFF*39C123A′124, as well as αSFF′4C9F*39. In the “native” state, when our constructs were incubated in buffer alone, we observed varying levels of quenching relative to the equivalent oxoprotein control. (Fig. 10) For F*94/F′4 and F*94/A′124 we observe no quenching. For F*39/F′4, we observe some quenching, which we interpret as resulting from local compaction of the N-terminus. All of these observations are consistent with previous FRET experiments performed on αS. The observed N-terminal compaction is similar to the roughly 30 Å separation observed by Rhoades and coworkers using Alexa Fluor 488 and Alexa Fluor 594 dyes in single molecule FRET experiments.62 Ferreon et al. observed FRET efficiencies consistent with separations of ~ 70 Å in single molecule FRET experiments with αS labeled at positions 7 and 84 with the same dye pair.63,64 This is well outside the useful range of the Cnf/thioamide FRET pair, so it is not surprising that we see no quenching for F*94/F′4. Our results for F*39/F′4 are consistent with experiments carried out by Lee, Gray, and coworkers using Trp and nitrotyrosine as a FRET pair.65,66 Our F*39/A′124 data are consistent with reports by Nath et al. showing a ~ 45 Å interaction between probes at positions 33 and 130, which is beyond the range of the Cnf/thioamide pair.62
Figure 10.
Refolding of αS in TMAO. Top: Quenching efficiency of αSFF′4C9F*94 (red), αSFF*39C123A′124 (green), or αSFF′4C9F*39 (blue) determined in phosphate buffer at 25 °C at varying concentrations of TMAO. Quenching efficiency determined by comparison to the equivalent oxoprotein: αSFC9F*94, αSFF*39C123, or αSFC9F*39, respectively. Bottom: Interchromophore distances computed using Förster theory as described in Supporting Information.
When 6 M urea was used as an additive, an increase in FThio/FOxo was observed for F*39/F′4, consistent with greater unfolding of the protein. For the other two mutants, no change in FThio/FOxo was observed, indicating that these regions were indeed less compacted. Control experiments using Cnf itself indicate that although an inherent change in Cnf quantum yield plays a role in the observed change in fluorescence, there is nonetheless an observable expansion of αS once one corrects for inherent effects on Cnf fluorescence. (See Supporting Information) In contrast, an increase in quenching (i.e. FRET) was observed as a function of TMAO concentration for all three mutants. The changes in EQ as a function of TMAO concentration can be seen in Figure 10. This indicates increased compaction of both N- and C- termini, as well as folding of the N-terminal region on itself.
To assign distance constraints based on these measurements, we converted the measured EQ values to distances using Förster theory. As with CaM, we computed a specific R0 value for each construct using the quantum yields of Cnf for the oxoamide control proteins αSFC9F*39 (R0 = 15.6 Å in 0 M TMAO), αSFC9F*94 (R0 = 13.6 in Å in 0 M TMAO), or αSFF*39C123 (R0 = 13.5 Å in 0 M TMAO). Förster radii for each mutant in other TMAO concentrations are given in Supporting Information. We use these rather than simply using R0 values calculated from free Cnf because the variation among the different mutants demonstrates that the protein context influences quantum yield. The RFRET values that result from our calculations indicate that Cnf and the thioamide come into very close proximity in TMAO. While these results are consistent with the single molecule studies of Deniz and coworkers, it is surprising that our RFRET values are so small.64 This may be the result of a dominant contribution from very close contact. Ferreon et al. observe subpopulations with near unity FRET efficiencies, but the Alexa Fluor FRET pair that they use has high FRET efficiency for any separation less than 30 Å.63,67 Our probes have a shorter effective range (8 – 31 Å) than theirs, and therefore provide a more precise report on the close contact induced by TMAO. We will continue to study this phenomenon, not only with thioamides, but also with other short range FRET pairs such as Cnf and Trp.68,69 Finally, it may be more appropriate to use polymer models rather than simple Förster theory to interpret our FRET data.70,71 These models have clear value when interpreting single molecule FRET data, which may have broad and asymmetric FRET efficiency distributions. Although the value is less clear for ensemble measurements, we will explore the interpretation of our FRET data using models such as those used by Lee, Gray, Rhoades, and Plaxco.62,65,66,72,73
CONCLUSIONS
The application of our recently developed Cnf/thioamide FRET probe pair to studies of the folding of full-sized proteins requires the ability to synthesize those labeled proteins in an efficient manner. Although we could, in theory, construct Cnf- and thioamide-labeled proteins purely through ligation, we have developed a far more expedient approach by melding the methods of unnatural amino acid mutagenesis and protein ligation. Intermolecular studies simply require that Cnf be incorporated into proteins expressed in E. coli through established unnatural amino acid mutagenesis techniques. Intramolecular studies require more complex semi-synthesis to obtain doubly labeled proteins, wherein a Cnf-labeled protein fragment can be expressed in E. coli and then ligated to a synthetic thiopeptide. This combination of methods minimizes SPPS of natural amino acid segments. The same methods can be used to synthesize proteins labeled with other donor fluorophores such as coumarin, or any fluorophore conjugated to a bioorthogonal handle, such as p-azidophenylalanine.74,75 In fact, we have identified a synthetase for the incorporation of the fluorescent amino acid acridonylalanine (Acd), which we have previously shown to be quenched by thioamides.6,76 We are currently working to optimize yields of Acd/thioamide labeled proteins using methods analogous to those described here.
The doubly labeled proteins produced using these methods can be applied to monitor protein folding and dynamics. For example, here we have studied the structure of a CaM/peptide complex and determined the average separation of residues in the disordered protein αS in various buffer conditions. The benchmarking results using CaM help to validate our method and provide a foundation for the application of thioamide FRET to other systems. These experiments show clearly that meaningful interpretations of the interchromopore distances require determining changes in the donor quantum yield. This is easily accomplished using the oxoamide control experiments that we always include in order to determine quenching efficiency. Moreover, the dependence of quenching efficiency on Cnf quantum yield supports a FRET mechanism for quenching as opposed to Dexter or PET mechanisms, which were difficult to discount based on previous data.4,77 The confirmation of a FRET mechanism for Cnf/thioamide interactions allows one to confidently interpret thioamide quenching using Förster’s equations for future applications. Nonetheless, it is worth noting that there was an imperfect match of observed distances (RFRET) and predicted distances (RNMR) that was not resolved by considering the orientation parameter κ2. This may be because simply using the 20 structures developed in solving the NMR structure does not fully account for intermediate geometries between these valleys on the potential energy surface. Calculating RFRET by sampling κ2 over extended molecular dynamics trajectories may give improved agreement with the FRET data. Additionally, fluorescence lifetime measurements (which are concentration-independent) would remove the influence of small deviations in concentration on our determination of EQ. We will explore both of these aspects in subsequent work.
The αS experiments show how one might use our method in an intramolecular sense. The addition of TMAO results in compaction of monomeric αS, consistent with previous independent observations by Deniz as well as small angle X-ray scattering experiments by Fink.64,78 The short effective range of our probes allows us to obtain more precise information about close contact of regions of the protein than similar experiments with larger fluorophores. Moreover, the small size of our chromophores leaves less uncertainty in their location relative to the protein structure. Thus, the thioamide probes provide new short-range information on TMAO-induced conformations of αS, complementing data obtained in single molecule experiments. We continue to apply fluorophore/thioamide pairs to the study of folding transitions in αS monomers as well as to monitoring conformational changes during aggregation.
Of course, our labeling strategy is not restricted to CaM or αS. Any protein which can be expressed at high levels in E. coli and reversibly unfolded should be amenable to double-labeling using a combination of Uaa mutagenesis and NCL. This strategy should be particularly appealing when one of the modifications cannot be incorporated ribosomally (i.e. backbone modifications such as olefin isosteres). For larger dyes that are attached post-translationally, Uaa mutagenesis at two sites, or used in combination with Cys modification is probably more efficient.79–82 However, even in those cases, it can be difficult to separate labeled from unlabeled protein, whereas the ligated products of our reactions differ substantially in size and can easily be separated by HPLC or FPLC. The major current limitations of our labeling methods are that Uaa mutagenesis expression yields can be low (3 – 5 mg/L), and if these are coupled to inconsistent NCL yields, it can be difficult to obtain sufficient quantities of protein for biophysical experiments. We will continue methodological studies to improve both aspects to make these methods as accessible as possible to the general biochemical community.
EXPERIMENTAL PROCEDURES
General Information
Fmoc-L-4-cyanophenylalanine (Fmoc-Cnf-OH) was purchased from Peptech (Burlington, MA). Boc-L-thionophenylalanine-1-(6-nitro)benzotriazolide, Fmoc-Gln(Trt)-OH, Fmoc-Asn(Trt)-OHwere purchased from Bachem (Torrance, CA) or EMD Chemicals (Philadelphia, PA). Benzotriazol-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate (PyBOP), Fmoc-Ala-OH, Fmoc-Leu-OH, Fmoc-Glu(OtBu)-OH, Fmoc-Lys(Boc)-OH, Fmoc-Trp(Boc)-OH, Fmoc-Arg(Pbf)-OH, Fmoc-Pro-OH, Fmoc-Thr(tBu)-OH, Fmoc-Met-OH, Fmoc-Gly-OH, Fmoc-Val-OH, Fmoc-Phe-OH, Fmoc-Asp(OtBu)-OH, Fmoc-Ser(tBu)-OH, Fmoc-Cys(Trt)-OH, 2-chlorotrityl chloride resin, and Rink amide resin were purchased from Novabiochem (San Diego, CA). Piperidine and 2-(1H–benzotriazol-1-yl)-1,1,3,3- tetramethyluronium hexafluorophosphate (HBTU) were purchased from American Bioanalytical (Natick, MA). Sigma-cote, N,N-diisopropyl ethylamine (DIPEA), thiophenol, tri-fluoroacetic acid (TFA), tris(2-carboxyethyl)phosphine hydro-chloride (TCEP), were purchased from Sigma-Aldrich (St. Louis, MO). Ni-NTA resin was from Qiagen (Valencia, CA). E. coli BL21(DE3) cells were purchased from Stratagene (La Jolla, CA). Sequencing-grade trypsin was purchased from Promega (Madison, WI). Restriction Grade Factor Xa protease was purchased from Novagen (San Diego, CA). All other reagents were purchased from Fisher Scientific (Pittsburgh, PA).
Milli-Q filtered (18 MΩ) water was used for all solutions (Millipore; Billerica, MA). Matrix-assisted laser desorption ionization (MALDI) mass spectra were collected with a Bruker Ultraflex III MALDI-TOF-TOF mass spectrometer (Billerica, MA). UV/Vis absorbance spectra were obtained with a Hewlett-Packard 8452A diode array spectrophotometer (Agilent Technologies, Santa Clara, CA). Fluorescence spectra were collected with a Varian Cary Eclipse fluorescence spectrophotometer fitted with a Peltier multicell holder (currently Agilent Technologies).
Peptide Synthesis
All peptides were synthesized using a manual, Fmoc-based solid-phase procedure described previ-ously.4,7 Thioamide monomers were synthesized as previously described.4,83
CaM Expression and Purification
Each plasmid containing a CaM mutant (pCaM-TAG13, pCaM-TAG17, pCaM-TAG93, or pCaM-TAG100, see Supporting information for plasmid construction) was transformed into pDULE2-Cnf-containing E. coli BL21(DE3) cells. These cells were cultured and made competent through the Hanahan method.84 First, the pDULE2-Cnf plasmid was transformed into BL21(DE3) cells. Then, a colony was picked based on streptomycin (Strep) resistance to grow cultures of competent cells. Once pCaM was transformed into these cells, selection was based on both am-picillin (Amp) and Strep resistance. Single colonies were used to inoculate 5 mL of LB media containing Amp (100 µg/mL) and Strep (100 µg/mL). This primary 5 mL culture was incubated at 37 °C with shaking at 250 RPM overnight, and then added to 1 L of M9 minimal media containing Amp and Strep at the same concentrations. M9 minimal media was prepared by adding the following autoclaved solutions to 1 L of auto-claved water containing 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl and 1 g NH4Cl: 1 mL of 2 M MgSO4, 1 mL of 15 mg/mL FeCl2 (in 1.0 M HCl), 1 mL of 15 mg/mL ZnCl2 (in acidified H2O), 2 mL of 10% Bacto™ Yeast Extract, 20 mL of 10% glycerol (v/v) and 1 µL of 1 M CaCl2. The 1 L secondary culture was incubated at 37 °C with shaking at 250 RPM until the OD600 reached 0.7 – 0.9 AU. Protein expression was induced with isopropyl D-galactoside (IPTG) and 95 mg (0.5 mM) Cnf, and the culture was incubated at 37 °C for an additional 12 h. The cells were recovered from media at 6000 × g for 10 minutes. The resulting pellet was suspended in 15 mL of CaM resuspension buffer (50 mM 3-(N-morpholino) propanesulfonic acid (MOPS), 100 mM KCl, 1 mM ethylenedia-minetetraacetic acid (EDTA), pH 7.5). Cell were lysed by sonication, alternating between one minute of sonication. CaCl2 was added to the sonicated lysate to a final concentration of 20 mM prior to centrifugation for 10 minutes at 6,000 × g, 4 °C. Following centrifugation, the supernatant was collected for purification. CaM was purified from the cleared cell lysate using a phenyl-sepharose (PS) CL-4B column with EDTA as the eluent. A column with a total resin bed volume of approximately 8 mL was first equilibrated with four 10 mL washes of PS Buffer A (50 mM Tris base, 1 mM CaCl2). Then the cleared cell lysate was added to the column and collected as it ran through to save any protein that did not bind to the column. Next, the column was washed four times with 10 mL PS Buffer A. The column was then washed with four 10 mL portions of high-salt PS Buffer B (50 mM Tris base, 0.5 M NaCl, 0.1 mM CaCl2). Finally, an additional 10 mL wash of PS Buffer A was performed to restore the column to a low-salt condition. CaM was eluted with 18 mL of PS Buffer C (10 mM Tris base, 10 mM EDTA, pH 7.5. This elution solution was then re-saturated with CaCl2 to a concentration of 20 mM. This elution solution was repurified on a second PS column using the same procedure to reduce truncation product. For the second PS column, rather than adding 18 mL of PS Buffer C directly to the column, 1 mL elutions were collected and analyzed by SDS-PAGE gel. Collection stopped when the protein was no longer visible by gel. The elution fractions were dialyzed against CaM Buffer (140 mM KCl, 15 mM HEPES, 6 mM CaCl2, pH 6.7) and stored at −20 °C. Proteins were analyzed by SPS PAGE gel and MALDI MS for purity.
CaM/pOCNC Binding Assays
After synthesis, HPLC purification, and lyophilization, the various pOCNC peptides were redissolved in CaM buffer (140 mM KCl, 10 mM HEPES, 6 mM CaCl2, pH 6.7). The concentrations of the peptides were determined by UV/Vis spectroscopy, using an extinction coefficient of 10, 270 M−1 cm- at 273 nm for the thioamide-containing peptides and 600 M−1 cm−1 at 257 nm for the oxoamide-containing peptide. The concentration of the CaM proteins was determined through the bicinchioninic acid (BCA) assay. For each peptide/protein combination, six solutions were prepared that contained 10 µM CaM protein and 0 to 15 µM pOCNC peptide. In addition, a solution of 10 µM peptide in the absence of protein was made to confirm that no background fluorescence resulted from the peptide. The solutions were prepared three times in order to obtain data in triplicate. EQ was computed from the raw fluorescence data for CaM with thioamide-labeled pOCNC (FThio) or pOCNC (FOxo).
C-Terminal Fragment Expression (Factor Xa)
The plasmid pET16b_HTag-αS Δ1–8C9F*39 (see Supporting Information for plasmid construction) was transformed into E. coli BL21(DE3) cells containing pDULE2-Cnf (see above) and colonies were selected on the basis of Amp and Strep resistance. Following growth of a primary culture in LB media, 1L of M9 minimal media containing glycerol as a carbon source (0.25% v/v) was inoculated and allowed to grow at 37 °C with shaking at 250 RPM. When the OD600 reached 0.8, Cnf was added (150 mg), and the culture was allowed to incubate overnight at 37 °C. The boiled and cleared lysate was prepared as described and incubated with 2 mL Ni-NTA resin for 1 hour at room temperature.7 The resin was washed with 20 mL of 50 mM Tris, 150 mM NaCl, pH 8. The protein was eluted in eight 1.5 mL portions of 50 mM Tris, 150 mM NaCl, 250 mM imidazole, pH 8. Elution fractions were analyzed by SDS-PAGE gel, combined, and dialyzed against Factor Xa cleavage buffer (50 mM Tris, 150 mM NaCl, 5 mM CaCl2, pH 8.0) overnight. Following dialysis, His-tag cleavage was achieved by overnight incubation with Factor Xa protease (10 units per 1 mg of HTag-αS Δ1–8C9F*39). Factor Xa was inactivated by boiling the reaction for 20 minutes and the precipitated enzyme was removed by centrifugation for 20 minutes at 30,000 × g, 4 °C. The supernatant was then subjected to incubation with 1 mL of Ni-NTA resin for 1 hr at room temperature to capture any remaining HTag-αS9–140 C9F*39. The flow-through containing αS Δ1–8C9F*39 was collected, analyzed by SDS-PAGE gel and MALDI MS, dialyzed overnight against water, and stored at − 80 °C.
C-Terminal Fragment Expression (Met Aminopepti-dase)
The plasmids pRK_αSFΔ2–8C9F*39 and pRK_αSFΔ2–8C9F*94 (see Supporting Information for plasmid construction) were transformed into competent E. coli BL21(DE3) cells with pDULE2-Cnf (see above). Transformed cells were selected on the basis of Amp and Strep resistance. Single colonies were used to inoculate 5 mL of LB media supplemented with Amp (100 ug/mL) and Strep (100 ug/mL). The primary 5 mL culture was incubated at 37 °C with shaking at 250 RPM for 5 hours and then used to inoculate 1 L of LB media. When the OD600 of the secondary culture reached 0.8, the cells were isolated by centrifugation at 5000 × g for 15 minutes and gently re-suspended in 1 L of M9 minimal media containing Cnf (100 mg). M9 minimal media was prepared as described above. The cells were harvested at 5000 × g for 15 min and the resulting pellet was resuspended in 20 mM Tris pH 8. Following sonication, the cell lysate was boiled for 20 min prior to centrifugation for 20 min at 30,000 × g, 4 °C. The cleared lysate was loaded onto a Superdex 200 column (25 cm) connected to a BioCad Sprint (FPLC) system and eluted with 20 mM Tris, pH 8. FPLC fractions from size-exclusion chromatography were analyzed by SDS PAGE. The fractions containing αS were then loaded onto a HighTrap™ Q HP column and eluted over a 100 min NaCl gradient (0 to 0.5 M NaCl in 20 mM Tris, pH 8). FPLC fractions from ion-exchange chromatography were dialyzed against 20 mM Tris, 100 mM NaCl, pH 7.4 and analyzed by SDS PAGE gel, MALDI MS, UV/Vis, and fluorescence spectroscopy. Following dialysis, guanidinium hydrochloride and methoxylamine hydrochloride were added to a final concentration of 6 M and 400 mM, respectively. The pH was adjusted to 4.0 with NaOH and the solution was stirred overnight at 4 °C. Removal of N-terminal cysteine adducts was monitored by MALDI MS. Following deprotection, the solution was dialyzed against water and dried in a vacuum centrifuge.
Thioester Synthesis (PyBOP)
Peptide synthesis and selective cleavage from Cl-Trt resin was performed as previously described.7 Cleaved N-terminal peptide was dissolved in THF, and 2-mercaptopropionate (3 equiv) was added to the solution. After stirring for 5 min, DIPEA (6 equiv) and a solution of PyBOP pre-dissolved in warm THF (3 equiv) were added to the reaction mixture. The solution was allowed to stir for 30 min at room temperature, at which point the solvent was removed by rotary evaporation. Sidechain protecting groups were removed by incubating the peptide for 10 min with a cleavage cocktail containing TFA, TIPS, thioanisole, and 1,2-ethanedithiol (47:1:1:1). Following concentration of the cleavage solution by rotary evaporation, the peptide was precipitated with ether and isolated by centrifugation. The peptide thioester was purified by HPLC and dried in a vacuum centrifuge.
Thioester Synthesis (CbPGo)
Ac-MDV′3FMKGLCbPGo (αS1–8V’3-CbPGo,) was synthesized essentially as previously described, with the following modifications.7 Bromoacetic acid (0.0695 g, 500 µmol, 5 equiv) was pre-activated with diisopropyl carbodiimide (78 µL, 500 µmol, 5.0 equiv) for 30 min in dry DMF (6 mL). Also, the peptide (αS1–8V’3-CbPGo) was cleaved from the resin with the treatment of CH2Cl2/TFA/thioanisole (38:1:1 v/v, 5 mL) on a rotisserie for 1 h. The resulting solution was concentrated by rotatory evaporation, precipitated in cold diethyl ether (20 mL), centri-fuged, and decanted ether. The precipitated peptide was dissolved in acetonitrile/water (50:50 v/v) for HPLC purification.
Ligation to form αSFF′4C9F*39, αSFV′3C9F*39, and αSFF′4C9F*94
The expressed C-terminal protein fragment (1 equiv, 0.1 µmol) was dissolved in 100 µL of freshly prepared ligation buffer (6 M GdnHCl, 200 mM Na2HPO4, 20 mM TCEP, 1% v/v thiophenol, pH 7.2 for PyBOP-based thioesters or pH 8.0 for C PGo thioesters). The dissolved protein fragment was transferred to a microcentrifuge tube containing 1.2 equiv of the dried N-terminal peptide thioester, purged with argon, and allowed to incubate overnight at 37 °C with shaking at 1000 RPM. The ligation solution was diluted to 3 mL and dialyzed against water prior to HPLC purification (see Supporting Information). Following HPLC purification, the product fractions were combined, concentrated, and exchanged into 20 mM Tris, 100 mM NaCl, pH 7.4 using an Amicon (Millipore) Ultra 0.5 mL 3 kDa spin column.
Expression and purification of αS 1-122F*39-Int
A plasmid encoding αS1–122F*39-Int-HTag was transformed into E. coli BL21(DE3) cells containing the pDULE2-Cnf gene (prepared and made competent using the Hanahan method).84 A starter culture of 4 mL LB media was inoculated with a single colony and was grown at 37 °C in the presence of Amp (100 mg/L) and Strep (100 mg/L) for approximately 4 h. A secondary culture of 1 L M9 media was inoculated with 4 mL of the starter culture and grown at 37 °C with the same concentrations of Amp and Strep. M9 minimal media was prepared as described above. The culture was induced by the addition of Cnf (150 mg) followed by 0.5 mM of IPTG when it reached an OD600 of 0.8 and allowed to grow at 37 °C overnight. Cells were harvested at 5, 000 RPM using a GS3 rotor and Sorvall RC-5 centrifuge for 20 min at 4 °C. After discarding the supernatant, the pellet was resuspended in 30 mL of resuspension, pH 8.3, protease inhibitor cocktail, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 10 units/mL DNAse1–Grade II. The buffer: 40 mM Tris, 5 mM EDTA cells were lysed by sonication, and the lysate was centrifuged at 13, 900 RPM for 30 min at 4 °C. The supernatant was then incubated with Ni-NTA resin for 1 h on ice. This slurry was loaded into a column and the liquid allowed to flow through. The resin was washed extensively, first with 15 mL of Wash Buffer A (50 mM HEPES, pH 7.0), then twice with 10 mL of Wash Buffer B (50 mM HEPES, 10 mM imidazole, pH 7.0. The protein was eluted with five 2 mL portions of elution buffer (50 mM HEPES, 300 mM imidazole, pH 7.0). The eluted fractions were combined and incubated with 400 mM sodium 2-mercaptoethanesulfonate (MESNA) for thiolysis for 48 h, stirring at 4 °C. The protein thioester was dialyzed against 20 mM Tris, pH 8.0 overnight, and incubated with Ni-NTA resin for 1 h on ice. The flow-through was collected, dialyzed against 20 mM sodium citrate, pH 5.0, purified over HiTrap SP column using a 100 min NaCl gradient (0 to 0.5 M NaCl in sodium citrate, pH 5). The product fractions were pooled and dialyzed against Milli-Q water overnight, dried in a vacuum centrifuge, and stored at − 80 °C until further use.
Ligation to Form aS FF*94C123A’124
Protein thioester αS 1–122F*39 (1.5 equiv, 0.45 µmol, 1.5 mM) and C-terminal protein fragment αS 123–140C123A´124 (1 equiv, 0.3 µmol, 1 mM) were dissolved in 300 µL of a freshly made, degassed ligation buffer (6 M Gdn•HCl, 200 mM Na2HPO4, 20 mM TCEP, and 1% v/v thiophenol, pH 7.0). The reaction solution was placed in an incubator at 37 °C, shaking at 1000 RPM. The ligation progress was monitored by MALDI MS, and allowed to react until all the protein thioester was either consumed or hydro-lyzed. Upon completion, the ligation solution was brought up to 1.5 mL with water and dialyzed against 20 mM Tris, pH 8.0 for 4 h. Then, the ligated product, αS FF*39C123A´124, was purified over a HiTrap™ Q HP column using a 100 min NaCl gradient (0 to 0.5 M NaCl in 20 mM Tris, pH 8). The product fractions were pooled, concentrated, and the buffer was ex- changed for 20 mM Tris, 100 mM NaCl, pH 7.4 using an Amicon (Millipore) Ultra 0.5 mL 10 kDa spin column.
Trypsin Digest
Proteins (10 – 50 µg) were incubated with 5 – 10 µL aliquots of sequencing-grade modified trypsin (0.1 mg/mL) in 20 mM Tris, 100 mM NaCl, pH 7.5 at 37 °C for 4 −8 hours. An aliquot (1.0 µL) of the digestion reaction was taken and analyzed by MALDI MS (Figure 6, Figure 8, and Supporting Information).
Figure 6.
Characterization of αSFF′4C9F*94 Ligation Product. Top Left: PAGE gel showing: αSF9–140C9F*94 before (1) and after (2) MeONH2•HCl treatment, ligation of αSF9–140C9F*94 to αSF1–8F′4-SR thioester (3), purified αSF-F′4C9F*94 (4), αSFC9F*94 (5), and αSWT (6). See HPLC chromatogram in Supporting Information for separation of ligated product. Top Right: MALDI MS analysis of full-length αSFF′4C9F*94. Bottom Left: UV/Vis absorption spectrum of αSFF′4C9F*94 showing thioamide absorption at 266 nm. Inset: MALDI MS analysis of trypsinized 1–6 fragment, confirming the presence of the thioamide at F′4; Calcd m/z (M+H): 828.35, Obsvd: 828.65. The asterisk indicates the absence of peaks corresponding an oxoamide at Phe4 in the 1–6 fragment; Calcd m/z (M+H): 812.38. Bottom Right: Fluorescence emission spectra of αSFF′4C9F*94 showing Cnf emission at 295 nm. Inset: MALDI MS analysis of trypsinized 81–96 fragment, confirming the presence of the Cnf at position 94; Calcd m/z (M+H): 1503.78, Obsvd: 1504.30. The asterisk indicates the absence of peaks corresponding Tyr at position 94 in the 81–96 fragment; Calcd m/z (M+H): 1494.78.
Refolding Assays
Prior to performing the experiment, the concentrations of all double-labeled proteins were adjusted using the BCA assay to match their respective oxoamide (donor only) control proteins. Tris buffers (20 mM Tris, 100 mM NaCl, 1 mM βME, pH 7.5) containing trimethylamine oxide (TMAO) were prepared such that upon addition of protein (to a final concentration of 1 µM), the final TMAO concentrations were 0 M, 1 M, 2 M, 3 M, and 4 M. Buffers containing urea were similarly prepared such that upon addition of protein, the final urea concentration was 6 M. Fluorescence spectra were measured immediately following sample dilution in TMAO or urea buffer (to a final volume of 130 µL). All samples were prepared and measured in triplicate.
Supplementary Material
Figure 2.
Combining Unnatural Amino Acid Mutagenesis and Native Chemical Ligation. Thioamide-bearing peptides are synthesized on solid phase starting from activated benzotriazole precursors and standard Fmoc-protected amino acids. For N-terminal thioamides, a thioester is installed as a masked group or formed by post-synthetic activation. To generate the Cnf-labeled protein fragment, E. coli cells are transformed with two DNA plasmids; one encoding the protein with a UAG stop codon at the site of interest, and one encoding an aminoacyl tRNA synthetase that is selective for an unnatural amino acid (UaaRS) with a tRNA that recognizes the UAG stop codon. Protein is expressed under standard conditions and the N-terminal Cys generated by proteolysis. Ligation of the two fragments gives the full length protein, labeled with both Cnf and the thioamide.
Figure 4.
CaM/pOCNC Label Sites. Cnf shown at three positions in CaM with three possible pOCNC thiopeptides (coloring as in Fig. 3). Images based on one of the 20 low energy structures in PDB ID 1SYD, modified using PyMol.
Figure 7.
Semi-synthesis of Double-Labeled Proteins with C-terminal Thioamides. An expressed N-terminal protein fragment is generated using a plasmid encoding a truncated version of the protein with a C-terminal intein. Thiolysis of the stalled intein generates a thioester suitable for NCL. The synthetic thiopeptide with an N-terminal Cys is made by standard SPPS. The expressed protein fragment and synthetic thiopeptide are combined in the presence of thiophenol and ligated, full-length protein isolated.
Figure 9.
Refolding Assay. Monomeric double-labeled αS (e.g. αSFF′4C9F*94) is diluted to 1 µM in either Tris buffer with 1 mM BME, or the same buffer with urea or TMAO added. Cnf fluorescence is measured at 295 nm (FThio). Identical experiments are carried out with donor-only labeled αS (e.g. αSFC9F*94) to determine FOxo. EQ = 1 – FThio/FOxo.
ACKNOWLEDGMENT
This work was supported by funding from the University of Pennsylvania, including a grant from the Institute on Aging, the Searle Scholars Program (10-SSP-214 to EJP), the National Science Foundation (NSF CHE-1150351) and the National Institutes of Health (NIH NS081033 to EJP). Instruments supported by the NSF and NIH include: HRMS (NIH RR-023444) and MALDI-MS (NSF MRI-0820996). We thank Prof. Ryan Mehl for the gift of the pDULE2-Cnf plasmid. We also thank Dr. Christopher Lanci of the UPenn Biological Chemistry Resource Center for assistance with circular dichroism spectroscopy (supported by UPenn Laboratory for Research on the Structure of Matter with NSF MRSEC DMR 0520020, 1120901). RFW thanks the NIH for funding through the Chemistry-Biology Interface Training Program (T32 GM07133). CMF thanks the Roy and Diana Vagelos Scholars Program in the Molecular Life Sciences for support.
ABBREVIATIONS
- FRET
Förster resonant energy transfer Cnf or F*, p-cyanophenylalanine
- PET
photoinduced electron transfer
- SPPS
solid phase peptide synthesis
- NCL
native chemical ligation
- Uaa
unnatural amino acid
- RS
aminoacyl tRNA synthetase
- CaM
calmodulin
- αS
α-synuclein
- OCNC
olfactory cyclic nucleotide-gated channel
- IPTG
isopropyl β-d-thiogalactopyranoside
- PAGE
polyacrylamide gel electrophoresis
- MPAA
4-mercaptophenylacetic acid
- Fmoc
9-fluorenylmethoxycarbonyl
- Cl-Trt
2-chlorotrityl
- TFA
trifluoroacetic acid
- PyBOP
benzotri-azol-1-yl-oxytri-pyrrolidinophosphonium hexafluoro-phosphate
- Cb
S-tert-Butyl-L-cysteine
- Go
glycolic acid
- MESNA
mercap-toethane sulfonate
- TMAO
trimethylamine oxide
- BME
β-mercaptoethanol
- Acd
acridonylalanine thioamide residues are represented by the one or three letter code of the equivalent oxo-amide amino acids with a prime symbol (e.g. L′ or Leu′ represent thioleucine)
Footnotes
ASSOCIATED CONTENT
Supporting Information. Characterization of all peptides and proteins, assessment of CaM mutants by native PAGE gel and circular dichroism (CD), primary fluorescence data for CaM experiments, analysis of fluorescence data using Förster theory, HPLC analysis of NCL reactions, details of UV/Vis, fluorescence, and trypsin/MALDI MS characterization of NCL products, primary fluorescence data and PAGE gel analysis of αS refolding experiments. This material is available free of charge via the Internet at http://pubs.acs.org. Primary data used to generate figures and tables have been digitally archived and can be obtained by emailing the corresponding author.
REFERENCES
- 1.Forster T. Discuss. Faraday Soc. 1959;No. 27:7–17. [Google Scholar]
- 2.Royer CA. Chem. Rev. 2006;106:1769–1784. doi: 10.1021/cr0404390. [DOI] [PubMed] [Google Scholar]
- 3.Piston DW, Kremers G-J. Trends Biochem. Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg JM, Batjargal S, Petersson EJ. J. Am. Chem. Soc. 2010;132:14718–14720. doi: 10.1021/ja1044924. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg JM, Wissner RF, Klein AM, Petersson EJ. Chem. Commun. 2012;48:1550–1552. doi: 10.1039/c1cc14708k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg JM, Speight LC, Fegley MW, Petersson EJ. J. Am. Chem. Soc. 2012;134:6088–6091. doi: 10.1021/ja3005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batjargal S, Wang YJ, Goldberg JM, Wissner RF, Petersson EJ. J. Am. Chem. Soc. 2012;134:9172–9182. doi: 10.1021/ja2113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alewood P, Engelhard M, Kent SBH. J. Peptide Sci. 2010;16:513–513. doi: 10.1002/psc.1291. [DOI] [PubMed] [Google Scholar]
- 9.Muralidharan V, Muir TW. Nature Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Schultz PG. Angew. Chem. Int. Ed. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 12.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 13.Wood SJ, Wypych J, Steavenson S, Louis JC, Citron M, Biere AL. J. Biol. Chem. 1999;274:19509–19512. doi: 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- 14.Auluck PK, Caraveo G, Lindquist S. Ann. Rev. Cell Dev. Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen DP, Elliott T, Holt M, Muir TW, Chin JW. J. Am. Chem. Soc. 2011;133:11418–11421. doi: 10.1021/ja203111c. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Fekner T, Ottesen JJ, Chan MK. Angew. Chem. Int. Ed. 2009;48:9184–9187. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary A, Raines RT. ChemBioChem. 2012;12:1801–1807. doi: 10.1002/cbic.201100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valiyaveetil FI, Sekedat M, MacKinnon R, Muir TW. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17045–17049. doi: 10.1073/pnas.0407820101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu YW, Bieschke J, Kelly JW. J. Am. Chem. Soc. 2005;127:15366–15367. doi: 10.1021/ja0551382. [DOI] [PubMed] [Google Scholar]
- 20.Fu YW, Gao JM, Bieschke J, Dendle MA, Kelly JW. J. Am. Chem. Soc. 2006;128:15948–15949. doi: 10.1021/ja065303t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Autric F, Ferraz C, Kilhoffer MC, Cavadore JC, Demaille JG. Biochim. Biophys. Acta. 1980;631:139–147. doi: 10.1016/0304-4165(80)90062-8. [DOI] [PubMed] [Google Scholar]
- 22.Brzeska H, Venyaminov SV, Grabarek Z, Drabikowski W. FEBS Lett. 1983;153:169–173. doi: 10.1016/0014-5793(83)80141-0. [DOI] [PubMed] [Google Scholar]
- 23.Chabbert M, Kilhoffer MC, Watterson DM, Haiech J, Lami H. Biochemistry. 1989;28:6093–6098. doi: 10.1021/bi00440a054. [DOI] [PubMed] [Google Scholar]
- 24.Chapman ER, Alexander K, Vorherr T, Carafoli E, Storm DR. Biochemistry. 1992;31:12819–12825. doi: 10.1021/bi00166a016. [DOI] [PubMed] [Google Scholar]
- 25.Gilli R, Lafitte D, Lopez C, Kilhoffer MC, Makarov A, Briand C, Haiech J. Biochemistry. 1998;37:5450–5456. doi: 10.1021/bi972083a. [DOI] [PubMed] [Google Scholar]
- 26.Kilhoffer MC, Demaille JG, Gerard D. Biochemistry. 1981;20:4407–4414. doi: 10.1021/bi00518a027. [DOI] [PubMed] [Google Scholar]
- 27.Moorthy AK, Gopal B, Satish PR, Bhattacharya S, Bhattacharya A, Murthy MRN, Surolia A. FEBS Lett. 1999;461:19–24. doi: 10.1016/s0014-5793(99)01380-0. [DOI] [PubMed] [Google Scholar]
- 28.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. Nat. Chem. Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vetter SW, Leclerc E. Eur. J. Biochem. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu MY, Chen TY, Ahamed B, Li J, Yau KW. Science. 1994;266:1348–1354. doi: 10.1126/science.266.5189.1348. [DOI] [PubMed] [Google Scholar]
- 31.Contessa GM, Orsale M, Melino S, Torre V, Paci M, Desideri A, Cicero DO. J. Biomol. NMR. 2005;31:185–199. doi: 10.1007/s10858-005-0165-1. [DOI] [PubMed] [Google Scholar]
- 32.Serrano AL, Troxler T, Tucker MJ, Gai F. Chem. Phys. Lett. 2010;487:303–306. doi: 10.1016/j.cplett.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du H, Fuh RCA, Li JZ, Corkan LA, Lindsey JS. Photochem. Photobiol. 1998;68:141–142. [Google Scholar]
- 34.Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Biochemistry. 2009;48:9040–9046. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Liu B, Yu HT, Barkley MD. J. Am. Chem. Soc. 1996;118:9271–9278. [Google Scholar]
- 36.Lakowicz JR. Principles of fluorescence spectroscopy. Third ed. New York, NY: Springer; 2006. [Google Scholar]
- 37.Chen H, Ahsan SS, Santiago-Berrios MEB, Abruña HD, Webb WW. J. Am. Chem. Soc. 2010;132:7244–7245. doi: 10.1021/ja100500k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muir TW. Ann. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 39.Hejjaoui M, Haj-Yahya M, Kumar KSA, Brik A, Lashuel HA. Angew. Chem. Int. Ed. 2011;50:405–409. doi: 10.1002/anie.201005546. [DOI] [PubMed] [Google Scholar]
- 40.Gentle IE, De Souza DP, Baca M. Bioconj. Chem. 2004;15:658–663. doi: 10.1021/bc049965o. [DOI] [PubMed] [Google Scholar]
- 41.Flemer S. J. Peptide Sci. 2009;15:693–696. doi: 10.1002/psc.1181. [DOI] [PubMed] [Google Scholar]
- 42.Camarero JA, Mitchell AR. Protein Peptide Lett. 2005;12:723–728. doi: 10.2174/0929866054864166. [DOI] [PubMed] [Google Scholar]
- 43.Nagalingam AC, Radford SE, Warriner SL. Synlett. 2007:2517–2520. [Google Scholar]
- 44.Kang J, Macmillan D. Organic & Biomolecular Chemistry. 2010;8:1993–2002. doi: 10.1039/b925075a. [DOI] [PubMed] [Google Scholar]
- 45.Kawakami T, Aimoto S. Tetrahedron. 2009;65:3871–3877. [Google Scholar]
- 46.Young TS, Ahmad I, Yin JA, Schultz PG. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 47.Uversky VN, Li J, Fink AL. J. Biol. Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 48.Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volpicelli-Daley LA, Luk KC, Patel TP, Tanik SA, Riddle DM, Stieber A, Meaney DF, Trojanowski JQ, Lee VMY. Neuron. 2011;72:57–71. doi: 10.1016/j.neuron.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winner B, Jappelli R, Maji SK, Desplats PA, Boyer L, Aigner S, Hetzer C, Loher T, Vilar M, Campionic S, Tzitzilonis C, Soragni A, Jessberger S, Mira H, Consiglio A, Pham E, Masliah E, Gage FH, Riek R. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conway KA, Harper JD, Lansbury PT. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 53.Hejjaoui M, Butterfield S, Fauvet B, Vercruysse F, Cui J, Dikiy I, Prudent M, Olschewski D, Zhang Y, Eliezer D, Lashuel HA. J. Am. Chem. Soc. 2012;134:5196–5210. doi: 10.1021/ja210866j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meier F, Abeywardana T, Dhall A, Marotta NP, Varkey J, Langen R, Chatterjee C, Pratt MR. J. Am. Chem. Soc. 2012;134:5468–5471. doi: 10.1021/ja300094r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fauvet B, Fares MB, Samuel F, Dikiy I, Tandon A, Eliezer D, Lashuel HA. J. Biol. Chem. 2012 doi: 10.1074/jbc.M112.383711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allison JR, Varnai P, Dobson CM, Vendruscolo M. J. Am. Chem. Soc. 2009;131:18314–18326. doi: 10.1021/ja904716h. [DOI] [PubMed] [Google Scholar]
- 57.Celej MS, Sarroukh R, Goormaghtigh E, Fidelio GD, Ruysschaert JM, Raussens V. Biochem. J. 2012;443:719–726. doi: 10.1042/BJ20111924. [DOI] [PubMed] [Google Scholar]
- 58.Karyagina I, Becker S, Giller K, Riedel D, Jovin TM, Griesinger C, Bennati M. Biophys. J. 2011;101:L1–L3. doi: 10.1016/j.bpj.2011.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramakrishnan M, Jensen PH, Marsh D. Biochemistry. 2006;45:3386–3395. doi: 10.1021/bi052344d. [DOI] [PubMed] [Google Scholar]
- 60.Wu KP, Baum J. J. Am. Chem. Soc. 2010;132:5546–5547. doi: 10.1021/ja9105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taskent-Sezgin H, Marek P, Thomas R, Goldberg D, Chung J, Carrico I, Raleigh DP. Biochemistry. 2010;49:6290–6295. doi: 10.1021/bi100932p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nath A, Sammalkorpi M, DeWitt DC, Trexler AJ, Elbaum-Garfinkle S, O'Hern CS, Rhoades E. Biophys. J. 2012;103:1940–1949. doi: 10.1016/j.bpj.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferreon ACM, Gambin Y, Lemke EA, Deniz AA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferreon ACM, Moosa MM, Gambin Y, Deniz AA. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17826–17831. doi: 10.1073/pnas.1201802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee JC, Gray HB, Winkler JR. J. Am. Chem. Soc. 2005;127:16388–16389. doi: 10.1021/ja0561901. [DOI] [PubMed] [Google Scholar]
- 66.Lee JC, Langen R, Hummel PA, Gray HB, Winkler JR. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16466–16471. doi: 10.1073/pnas.0407307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deniz AA, Dahan M, Grunwell JR, Ha TJ, Faulhaber AE, Chemla DS, Weiss S, Schultz PG. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3670–3675. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyake-Stoner SJ, Miller AM, Hammill JT, Peeler JC, Hess KR, Mehl RA, Brewer SH. Biochemistry. 2009;48:5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- 69.Tucker MJ, Oyola R, Gai F. J. Phys. Chem. B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 70.Makarov DE, Plaxco KW. J. Chem. Phys. 2009:131. doi: 10.1063/1.3212602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Brien EP, Morrison G, Brooks BR, Thirumalai D. J. Chem. Phys. 2009:130. doi: 10.1063/1.3082151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarney ER, Werner JH, Bernstein SL, Ruczinski I, Makarov DE, Goodwin PM, Plaxco KW. J. Mol. Biol. 2005;352:672–682. doi: 10.1016/j.jmb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 73.Trexler AJ, Rhoades E. Biophys. J. 2010;99:3048–3055. doi: 10.1016/j.bpj.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang JY, Xie JM, Schultz PG. J. Am. Chem. Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 75.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. J. Am. Chem. Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 76.Speight LC, Goldberg JM, Warner JB, Mehl RA, Petersson EJ. Unpublished results. [Google Scholar]
- 77.Speiser S. Chem. Rev. 1996;96:1953–1976. doi: 10.1021/cr941193+. [DOI] [PubMed] [Google Scholar]
- 78.Uversky VN, Li J, Fink AL. FEBS Lett. 2001;509:31–35. doi: 10.1016/s0014-5793(01)03121-0. [DOI] [PubMed] [Google Scholar]
- 79.Brustad EM, Lemke EA, Schultz PG, Deniz AA. J. Am. Chem. Soc. 2008;130:17664-+. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang Y, Russell WK, Wan W, Pai PJ, Russell DH, Liu WS. Mol. Biosys. 2010;6:683–686. doi: 10.1039/b920120c. [DOI] [PubMed] [Google Scholar]
- 81.Wan W, Huang Y, Wang ZY, Russell WK, Pai PJ, Russell DH, Liu WR. Angew. Chem. Int. Ed. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 82.Neumann H, Wang KH, Davis L, Garcia-Alai M, Chin JW. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 83.Shalaby MA, Grote CW, Rapoport H. J. Org. Chem. 1996;61:9045–9048. doi: 10.1021/jo961245q. [DOI] [PubMed] [Google Scholar]
- 84.Hanahan D, Jessee J, Bloom FR. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.