Abstract
The Escherichia coli replisome contains three polymerases, one more than necessary to duplicate the two parental strands. Using single-molecule studies, we reveal two advantages conferred by the third polymerase. First, dipolymerase replisomes are inefficient at synthesizing lagging strands, leaving single-strand gaps, whereas tripolymerase replisomes fill strands almost to completion. Second, tripolymerase replisomes are much more processive than dipolymerase replisomes. These features account for the unexpected three-polymerase-structure of bacterial replisomes.
For 40 years, it has been assumed that replisomes contain two DNA polymerases, one for each strand at a replication fork1,2. However, the E. coli replicase Pol III* has recently been shown to contain three Pol III cores3. Furthermore, recent in vivo support for a tripolymerase (TriPol) replisome has been provided by slimfield microscopy, which demonstrated that E. coli replication forks contain three Pol III cores, not two4.
The failure to foresee the three-polymerase structure of the bacterial replisome is attributable to the peculiarities of the E. coli dnaX gene, which encodes two proteins, τ and γ. The protein τ is the full-length product and γ is truncated by a translational frameshift5. The C-terminal sequences of τ bind the Pol III core (and DnaB helicase), but the truncated γ binds neither protein. The E. coli clamp loader contains three copies of the DnaX protein and thus was presumed to contain two τ and one γ to yield the `classic' dipolymerase replisome, which also explains why E. coli produces γ (that is, to prevent the binding of an unnecessary third Pol III core). However, mixing all the subunits simultaneously yields a Pol III* with three τ and three Pol III cores, while excluding γ3. Furthermore, mutation of dnaX shows that γ is not required for E. coli viability, whereas τ is essential6. We presume E. coli produces γ to form a γ-only clamp loader that assembles β-clamps onto DNA for use by other enzymes besides Pol III.
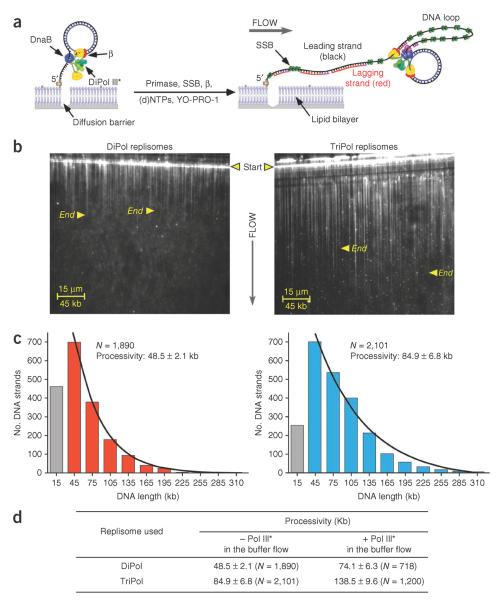
Why would a replisome contain three polymerases when there are only two DNA strands to replicate? To address this question, we used single-molecule total internal reflection fluorescence microscopy to monitor the behavior of E. coli replisomes that contain either two or three polymerases (DiPol III* can be assembled by forcing one γ to exchange with one τ). We assembled DnaB helicase, DiPol III* or TriPol III*, and the β clamp on a 5′-biotinylated 100-mer rolling circle DNA to form TriPol and DiPol replisomes (see Fig. 1a and Supplementary Methods)7. We then attached the replisome–DNA complex to a lipid bilayer within a flow cell. Unbound proteins were washed away and the hydrodynamic flow enabled the replisome–DNA complexes to migrate in the bilayer and accumulate along a diffusion barrier7,8. Replication was initiated by flowing a buffer that included dNTPs and NTPs.
Figure 1.
TriPol replisomes are more processive than DiPol replisomes. (a) Scheme of single-molecule experiments. For clarity, only the DiPol replisome is illustrated. (b) DNA products from either the DiPol (left) or TriPol (right) replisome, using 250 nM primase. The endpoints of two representative DNA products are marked with arrowheads. (c) DNA length distribution histograms. Numbers represent the single-exponential fit ± s.e.m. of the total number (N) of molecules analyzed. Gray bars represent DNA strand lengths below 15 kb that were undersampled because they were obscured by the width of the diffusion barrier. Left, DiPol replisomes, right, TriPol replisomes. (d) Processivity of DiPol and TriPol replisomes, where the indicated polymerase is present or absent from the buffer flow.
In rolling circle replication, the newly formed leading strand becomes the template for lagging-strand synthesis, producing a duplex `tail' composed of the newly synthesized leading and lagging strands. In our study, the duplex tail was tethered at the diffusion barrier, and replication yielded a growing `curtain' of DNA that was visualized in real time using YO-PRO-1, a fluorescent intercalator (Fig. 1b and Supplementary Videos 1 and 2). Primase, β and SSB are included in the buffer because they are needed repeatedly during lagging-strand synthesis9. However, we omitted Pol III* and DnaB helicase from the buffer flow, so if either of these proteins dissociated from the immobilized DNA, they would be removed by the flow and growth of that particular DNA molecule would terminate.
Comparing the DNA products of DiPol and TriPol replisomes revealed that the TriPol replisome synthesizes much longer DNA products than does the DiPol replisome (Fig. 1b). Quantitation of nearly 2,000 DNA molecules in each reaction demonstrated that the products of TriPol replisomes are nearly twice as long as those of DiPol replisomes (84.9 ± 6.8 kilobases (kb) versus 48.5 ± 2.1 kb, respectively, Fig. 1c). We conclude that the presence of a third polymerase substantially enhances the processivity of the replisome.
We considered whether the increase in TriPol replisome processivity compared to DiPol processivity is due to the three τ subunits in TriPol (versus two τ subunits in DiPol), which bind DnaB helicase10 and thus may stabilize Pol III* and/or DnaB helicase at the fork. To determine this, we repeated the experiments but included either TriPol III* or DiPol III* in the buffer flow. With Pol III* in the buffer flow, only DnaB dissociation should terminate replication because any Pol III* that dissociates can be replaced by another Pol III* from the solution. Thus, if DnaB is stabilized by TriPol III*, longer products should be observed compared to DiPol III*. In fact, inclusion of either TriPol III* or DiPol III* in the buffer flow yielded longer DNA products, but TriPol replisome products were still much longer than DiPol products, indicating that TriPol III* stabilizes DnaB more than it stablilizes DiPol III* (Fig. 1d). Considering that the third polymerase raises the effective concentration of the Pol III core at the fork, the enhanced processivity of TriPil over DiPol could be aided by the spare polymerase replacing a dissociated polymerase (on the leading or lagging strand) and resuming fork progression.
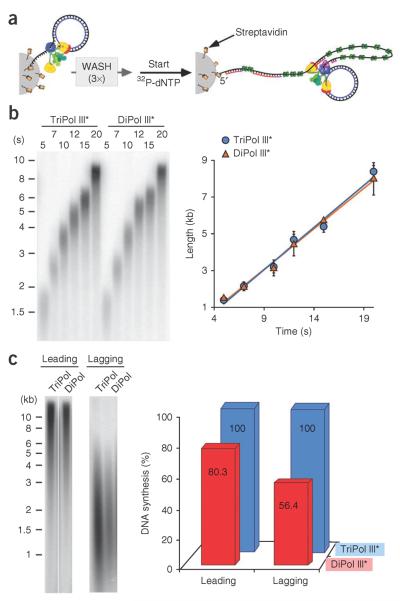
Longer DNA products could also result if the TriPol replisome moves faster than the DiPol replisome. To determine the rate of fork progression, we developed a bead-based assay using a mini–rolling circle that contains only three dNMPs on either strand, allowing us to specifically follow the growth of the leading strand and determine the speed of the replisome. We assembled either DiPol or TriPol replisomes on 5′ biotinylated mini–rolling circle DNA, attached the DNA to streptavidin magnetic beads, then washed the beads to remove unbound proteins before initiating replication (see scheme in Fig. 2a). Our product analysis showed that the DiPol and TriPol replisomes advanced at the same rate (Fig. 2b). Thus, the longer DNA molecules made by TriPol replisomes result from the greater processivity of TriPol over DiPol replisomes and not from a higher rate of fork progression. While this report was in progress, another study observed a difference in product length between DiPol and TriPol replisomes, as a corollary to investigating a different question11.
Figure 2.
TriPol replisomes are more efficient on the lagging strand than DiPol replisomes are. (a) Scheme of the bead-based assay; the DiPol replisome is illustrated for simplicity. (b) Dipol and Tripol replisomes replicate DNA with similar rates. Left, autoradiogram of 0.8% alkaline agarose gel analysis of reactions, using either Tripol III* (20 nM) or DiPol III* (80 nM); DnaG primase concentration was 200 nM. Right, plot of DNA length versus time. (c) Left, leading- and lagging-strand replication products from bead-based reactions, resolved on denaturing agarose gels, using 320 nM DnaG primase. Right, quantitation of leading- and lagging-strand synthesis, normalized to the products of the TriPol replisome.
Our work shows that bead-based assays require four-fold more DiPol III* than Tripol III*, as we consistently observed less overall DNA synthesis using DiPol versus TriPol replisomes (Supplementary Fig. 1). This is probably due to the lower stability of DiPol III* at the replication fork, resulting in dissociation from the DNA during the extensive washing steps. Lower stability of DiPol III* is consistent with the lower processivity of DiPol compared to TriPol replisomes (see Fig. 1).
Next, we examined the two replisomes for differences in lagging-strand synthesis. In these experiments, we added a DNA trap after the wash steps to ensure that any polymerase that dissociated did not rebind the rolling circle substrate. We were surprised to find that the differences in DNA synthesis between the DiPol and TriPol replisome were even greater on the lagging strand than on the leading strand (Fig. 2c). This result implies that DiPol replisomes leave single-stranded DNA (ssDNA) gaps on the lagging strand. The DiPol replisome may not complete the extension of Okazaki fragments, or it may be deficient in primer utilization. To test the hypothesis that DiPol replisomes leave gaps on the lagging strand, we closely examined DNA products under the microscope. The YO-PRO-1 intercalator binds only to double-stranded DNA (dsDNA), so ssDNA gaps appear as dark regions between fluorescent segments of dsDNA. The pixel resolution of our experimental setup corresponded to about 600 bp. Therefore, ssDNA gaps less than 600 bp would not be observed as dark pixels, requiring indirect methods to analyze their pixel intensity. Nevertheless, we observed some ssDNA gaps that were large enough to extend over one or more pixels (Fig. 3a). These directly observed ssDNA gaps were five-fold more prevalent using the DiPol replisome than using the TriPol replisome (Supplementary Figs. 2 and 3). Because these gaps were small and infrequent when we used the TriPol replisome, we changed the conditions to favor longer Okazaki fragments and thus more frequent gaps, making it possible to take more accurate measurements to compare DiPol and TriPol replisomes.
Figure 3.
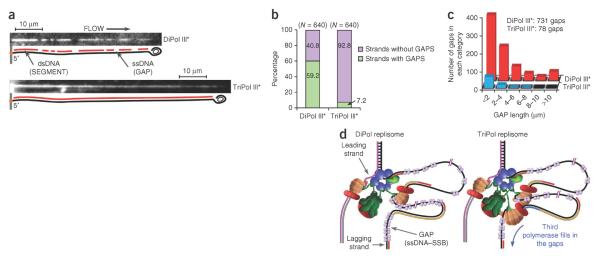
Analysis of ssDNA gaps in lagging strand products. (a) Magnified view of DNA products generated by DiPol and TriPol replisomes; the light and dark regions correspond to dsDNA segments and ssDNA gaps. (b) Comparative histogram showing the percentage of DNA strands with gaps (green) and without gaps (purple). (c) Histograms showing the distribution of gap length (in mm) using DiPol (red) and TriPol (blue) replisomes. (d) Model of TriPol and DiPol replisome action. Pol III cores are represented as right hands; with the β-clamp (red), clamp loader (dark green), DnaB helicase (blue hexamer), primase (light green) and SSB (purple). The τ-subunit C-terminal domains (IV and V) are illustrated as jointed lines that mediate connections to DnaB helicase and Pol III cores. The χψ subunits of the clamp loader are omitted for clarity. The TriPol replisome depicts two Pol III cores extending two Okazaki fragments simultaneously, although there are other ways a TriPol replisome can be used (see text). The left illustration depicts one lagging Pol III extending an RNA primer (red) to produce a DNA strand (yellow), and the other lagging Pol III core extends the DNA (blue) to fill a ssDNA gap.
The experiments described thus far were conducted using 250 nM primase, which generates 1–2 kb Okazaki fragments, whereas lowering primase concentration to 4 nM gives >20-kb Okazaki fragments9. Examination of DNA products using 3 nM primase showed numerous ssDNA gaps produced by the DiPol replisome (59.2% of the DNA molecules) compared to the TriPol replisome (only 7.2% of molecules) (Fig. 3a,b and Supplementary Videos 1 and 2). We also analyzed the gap-size distribution (Fig. 3c). This showed an even more pronounced difference between DiPol and TriPol replisomes. Of the 640 DNA strands analyzed (Fig. 3b), DiPol replisomes left 731 gaps, whereas TriPol replisomes left only 78 gaps. In addition, there were no long gaps in the TriPol III*-replicated material. The nucleotide size of ssDNA gaps is difficult to determine because ssDNA is bound to SSB and its contour length is ill-defined and different from dsDNA and ssDNA. Therefore, we report the gap length in microns (Fig. 3c).
The marked decrease in the length and number of ssDNA gaps demonstrates that the TriPol replisome, with its extra polymerase, conducts more efficient lagging-strand synthesis than the DiPol replisome does. One may question whether primase is more active with the TriPol replisome. In this case, primed sites may be further apart for DiPol replisomes and thus produce longer Okazaki fragments and/or ssDNA gaps. Our earlier study noted slightly longer Okazaki fragments produced by the DiPol versus TriPol replisomes, suggesting that primase is somewhat more active with the TriPol replisome3. However, the difference is too small to explain the large difference in ssDNA gaps between the DiPol and TriPol replisomes observed in this report.
To ensure that the gaps we observed were indeed ssDNA, we conducted several control experiments. In one control experiment, we stopped the buffer flow, which results in DNA recoiling, then restarted the flow to stretch the DNA again (Supplementary Video 3). Both dsDNA regions (fluorescent) and gaps (nonfluorescent) recoiled and re-extended together, supporting the notion that the gaps and dsDNA are on one continuous DNA molecule. Second, we used fluorescent SSB in place of YO-PRO-1. If the gaps were ssDNA, we should observe fluorescent SSB associated with the DiPol replisome products. This was indeed the case (Supplementary Video 4). Because some SSB associates with the lipid bilayer nonspecifically, we stopped the buffer flow, and SSB on DNA recoiled as expected. Additionally, we used excess Pol III* in the buffer flow, which should fill and remove any ssDNA gaps, and, indeed, the gaps disappeared (Supplementary Fig. 3). To ensure that our results were not influenced by the 1.5-pN force exerted on DNA by the hydrodynamic flow (100 μL min−1), we repeated the experiments at two lower flow rates, 30 μL min−1 and 10 μL min−1, corresponding to forces of ~0.4 pN and ~0.1 pN, respectively. These lower forces did not alter the prevalence of ssDNA gaps (Supplementary Fig. 4).
One explanation for ssDNA gaps is occasional premature termination of Okazaki fragments, termed `signal release'11–15. The factors contributing to the signal release mechanism are not well understood. Bacteriophage T4 and T7 replisomes, which are thought to contain only two polymerases, often use the signal release mechanism during replication13,15. Perhaps the E. coli cellular replisome permits two out of three polymerases to extend lagging-strand fragments and reduces the incidence of signal release. Notably, an EM study of the T4 system showed three polymerases at the fork in about 6% of DNA molecules16. Some of the gaps we observed were quite long and may be attributable to inefficient use of RNA primers by the DiPol replisome than to signal release. A TriPol replisome, with its additional DNA polymerase, may more efficiently capture RNA primers. Indeed, all three Pol III cores in TriPol can be active simultaneously3. However, the in vivo study indicated that only two polymerases function at the same time4, so the frequency with which all three polymerases are used simultaneously is not certain.
It may seem unnecessary to produce a replisome with a third Pol III to fill ssDNA gaps left by a DiPol replisome (Fig. 3d), considering that these gaps could simply be filled in the wake of the fork by other polymerases that are abundant in E. coli, including the high-fidelity Pol I (400 copies per cell)1. On the other hand, E. coli bacteria also contain high levels of lower fidelity Pol II (40 copies per cell17) and of the very low-fidelity Y-family Pol IV translesion polymerase (250 copies per cell18). Thus, a TriPol replisome containing three high-fidelity Pol III cores will increase the effective concentration of this sparse enzyme (10–20 molecules per cell). In summary, our observations point toward the conclusion that a third polymerase at the fork could be the norm rather than the exception. We think it will be exciting to determine if this finding can be generalized to eukaryotic systems as well.
The current study demonstrates that the TriPol replisome has two distinct advantages over the DiPol replisome: increased processivity and increased efficiency in lagging-strand synthesis. A more processive and efficient replisome is particularly important to a cell that must be capable of rapid cell duplication in order to outnumber other organisms under the intense selective pressure of competition and survival while replicating its genome from a single origin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health grant GM38839 to M.O.D. We thank A. van Oijen and D. Fenyo for helpful suggestions. We also thank L. Langston for suggestions and for critical reading of the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
R.E.G. and I.K. carried out experiments; R.E.G., I.K. and M.E.O. designed the experiments. R.E.G., I.K. and M.E.O. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Kornberg A, Baker TA. DNA replication. xiv. W.H. Freeman; New York: 1992. p. 931. [Google Scholar]
- 2.Sinha NK, Morris CF, Alberts BM. J. Biol. Chem. 1980;255:4290–4293. [PubMed] [Google Scholar]
- 3.McInerney P, Johnson A, Katz F, O'Donnell M. Mol. Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 4.Reyes-Lamothe R, Sherratt DJ, Leake MC. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson A, O'Donnell M. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 6.Blinkova A, et al. J. Bacteriol. 1993;175:6018–6027. doi: 10.1128/jb.175.18.6018-6027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao NY, Georgescu RE, Finkelstein J, O'Donnell ME. Proc. Natl. Acad. Sci. USA. 2009;106:13236–13241. doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granéli A, Yeykal CC, Robertson RB, Greene EC. Proc. Natl. Acad. Sci. USA. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zechner EL, Wu CA, Marians KJ. J. Biol. Chem. 1992;267:4045–4053. [PubMed] [Google Scholar]
- 10.Kim S, Dallmann HG, McHenry CS, Marians KJ. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 11.Tanner NA, et al. EMBO J. 2011;30:1830–1840. doi: 10.1038/emboj.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Marians KJ. J. Biol. Chem. 2000;275:34757–34765. doi: 10.1074/jbc.M006556200. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Nelson SW, Benkovic SJ. Mol. Cell. 2006;21:153–164. doi: 10.1016/j.molcel.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 14.McInerney P, O'Donnell M. J. Biol. Chem. 2007;282:25903–25916. doi: 10.1074/jbc.M703777200. [DOI] [PubMed] [Google Scholar]
- 15.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Nature. 2009;457:336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nossal NG, Makhov AM, Chastain PD, II, Jones CE, Griffith JD. J. Biol. Chem. 2007;282:1098–1108. doi: 10.1074/jbc.M606772200. [DOI] [PubMed] [Google Scholar]
- 17.Bonner CA, Hays S, McEntee K, Goodman MF. Proc. Natl. Acad. Sci. USA. 1990;87:7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.