Abstract
Background
Transcriptional regulation by alternative sigma (σ) factors represents an important mechanism that allows bacteria to rapidly regulate transcript and protein levels in response to changing environmental conditions. While the role of the alternative σ factor σB has been comparatively well characterized in L. monocytogenes, our understanding of the roles of the three other L. monocytogenes alternative σ factors is still limited. In this study, we employed a quantitative proteomics approach using Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) to characterize the L. monocytogenes σL, σH, and σC protein regulons. Proteomic comparisons used a quadruple alternative σ factor mutant strain (ΔBCHL) and strains expressing a single alternative σ factor (i.e., σL, σH, and σC; strains ΔBCH, ΔBCL, and ΔBHL) to eliminate potential redundancies between σ factors.
Results
Among the three alternative σ factors studied here, σH provides positive regulation for the largest number of proteins, consistent with previous transcriptomic studies, while σL appears to contribute to negative regulation of a number of proteins. σC was found to regulate a small number of proteins in L. monocytogenes grown to stationary phase at 37°C. Proteins identified as being regulated by multiple alternative σ factors include MptA, which is a component of a PTS system with a potential role in regulation of PrfA activity.
Conclusions
This study provides initial insights into global regulation of protein production by the L. monocytogenes alternative σ factors σL, σH, and σC. While, among these σ factors, σH appears to positively regulate the largest number of proteins, we also identified PTS systems that appear to be co-regulated by multiple alternative σ factors. Future studies should not only explore potential roles of alternative σ factors in activating a “cascade” of PTS systems that potentially regulate PrfA, but also may want to explore the σL and σC regulons under different environmental conditions to identify conditions where these σ factors may regulate larger numbers of proteins or genes.
Keywords: Listeria monocytogenes, Alternative sigma factors, Quantitative proteomics
Background
The foodborne pathogen Listeria monocytogenes uses complex regulatory mechanisms to adapt to a variety of environmental conditions and to cause listeriosis, a life-threatening infection, in humans and animals. A key mechanism used by L. monocytogenes to regulate transcript and protein levels in order to adapt to changing environmental conditions is through alternative sigma (σ) factors. Alternative σ factors reprogram the RNA polymerase holoenzyme to recognize specific promoters and hence allow for rapid induction of transcription of potentially large groups of genes under specific environmental conditions [1]. In L. monocytogenes, four alternative σ factors, σB, σC, σH, and σL, have been identified. However, σC has only been described in L. monocytogenes strains that group into lineage II, a well defined phylogenetic group that includes serotypes 1/2a and 1/2c [2-4]. A number of studies that have explored σB-mediated stress response as well as σB-mediated gene expression and protein production in L. monocytogenes[1,5-16] have shown that this alternative σ factor controls a large regulon and contributes to both stress response and virulence.
σH, σL, and σC have not been as extensively characterized as σB in L. monocytogenes, at least partially because studies to date have only identified limited phenotypic consequences of null mutations in these σ factors in L. monocytogenes. Among these three alternative σ factors, σH appears to control the largest regulon; Chaturongakul et al. (2011) identified 97 and 72 genes as positively and negatively regulated by σH, respectively, in L. monocytogenes strain 10403S [7]. While a L. monocytogenes EGD-e sigH mutant was reported to have significantly impaired growth in minimal medium and under alkaline stress conditions as well as slightly reduced virulence potential in a mouse model [17], phenotypic studies in a L. monocytogenes 10403S ΔsigH strain did not find evidence for an effect of this mutation on virulence in a guinea pig model, cell invasion and intracellular growth, or resistance to heat stress [7]. With regard to σL, 31 and 20 genes were identified as positively and negatively regulated, respectively, by this σ factor, in L. monocytogenes 10403S [7]. A more recent study in L. monocytogenes EGD-e identified 237 and 203 genes as positively regulated by σL when the parent and ΔsigL mutant strains were grown at 3°C and 37°C, respectively; most of the 47 genes that showed positive regulation by σL under both temperatures were located within prophage A118 [18]. Phenotypic and gene expression studies also support a potential contribution of σL to L. monocytogenes growth under different stress conditions, most notably osmotic and low temperature stress [19,20]. L. monocytogenes σL has also been reported to be involved in resistance to the antimicrobial peptide mesentericin Y105 [21]. Finally, studies conducted to date on the L. monocytogenes σC regulon typically identified few genes as σC-dependent. Chaturongakul et al. (2011) were only able to identify and confirm, by qRT-PCR, a single gene (lmo0422) as σC-dependent; lmo0422, which encodes LstR, a lineage II specific thermal regulator, is in the same operon as sigC and this finding is consistent with previous data suggesting that the sigC operon is auto-regulated [3,7]. Zhang et al. (2005) also found some evidence that σC may contribute to thermal resistance in the L. monocytogenes lineage II strain 10403S, when grown to log phase [3]; by contrast, Chaturongakul et al. (2011) did not find any evidence for reduced heat resistance when an independent L. monocytogenes 10403S ΔsigC strain was grown to stationary phase prior to heat exposure [7].
Previous studies [7] have suggested considerable overlap between different L. monocytogenes alternative σ factor regulons (e.g., between the σB and the σH regulon), suggesting the potential for redundancies as well as compensation for deletion of a single alternative σ factor by other σ factors. We thus hypothesized that an experimental approach that eliminates these potential redundancies is needed to gain a better understanding of the roles of σC, σH, and σL in regulating production of specific proteins in L. monocytogenes. As an experimental approach, we selected to create an L. monocytogenes 10403S quadruple mutant with a non-polar deletion of all four genes that encode alternative σ factors (i.e., strain ΔBCHL) as well as corresponding mutants with deletions of three alternative σ factors (ΔBCH, ΔBCL, and ΔBHL), which thus expressed only σL, σH, and σC, respectively. These strains were then used for proteomic comparisons between the quadruple mutant strain and the three different strains expressing only a single alternative σ factor. We particularly focused on exploring the contributions of these alternative σ factors to regulating protein production as, despite availability of a number of proteomics data sets on the σB regulon [15,16], only a single proteomics study on the σL regulon is available [22]. While alternative σ factors directly regulate transcription of genes, it is increasingly clear that alternative σ factors also make important indirect contributions to protein production via mechanisms other than transcriptional activation of a σ factor dependent promoter upstream of a protein encoding gene, including through regulation of non-coding RNAs or through direct transcriptional up-regulation of a protein that in turn, directly or indirectly, affects production of other proteins [23]. The goal of this proteomics study was thus to specifically identify additional post-transcriptional regulatory pathways that are linked to the action of alternative σ factors in L. monocytogenes.
Results and discussion
Proteomic comparisons between L. monocytogenes mutants expressing only σL, σH, and σC and a quadruple mutant that does not express any alternative σ factors, all grown to stationary phase at 37°C, showed that (i) σH provides, among these three alternative σ factors, positive regulation for the largest number of proteins, consistent with previous transcriptomic studies [7]; (ii) σL appears to contribute to negative regulation of a number of proteins; (iii) σC regulates a small number of proteins in L. monocytogenes grown to stationary phase at 37°C; and (iv) proteins regulated by multiple alternative σ factors include MptA, which has a potential role in regulation of PrfA.
σH positively regulates a large number of proteins and appears to directly and indirectly contribute to transport and metabolism of β-glucosides
Our proteomic comparison identified 15 proteins as positively regulated by σH, as supported by higher protein levels (Fold change (FC) ≥ 1.5; p-valuec (pc) < 0.05) in L. monocytogenes ΔBCL as compared to the ΔBCHL strain (Table 1); four of these 15 proteins also showed higher levels in the parent strain (which expresses all four alternative σ factors) as compared to the quadruple mutant. Overall, positive fold changes for these proteins (in ΔBCL versus ΔBCHL) ranged from 1.55 to 3.39. These 15 proteins represented nine role categories (e.g., “energy metabolism”; “amino acid biosynthesis”; “transport and binding proteins”, see Figure 1); a Monte Carlo simulation of Fisher’s exact test did not find a significant association between positively regulated genes and role categories (p = 0.06); however, individual Fisher’s exact tests did show overrepresentation of proteins in the role category “amino acid biosynthesis” among the 15 proteins that were found to be positively regulated by σH (with a significant p-value; p < 0.01; Odds Ratio = 6.26). Some of the 15 proteins positively regulated by σH have likely roles in stress adaptation and virulence, including Lmo1439 (superoxide dismutase, SodA) [24] and Lmo0096 (mannose-specific PTS system IIAB component, MptA), which has been linked to regulation of the virulence gene regulator PrfA [25]. Previously reported transcriptomic studies [7] only identified the coding gene for one of these 15 proteins (i.e., Lmo1454) as σH-dependent; lmo1454 (rpoD) was also identified as preceded by a σH consensus promoter, suggesting direct transcriptional regulation by σH. In addition, the coding gene for Lmo2487, one of these 15 proteins, is in an operon with lmo2485, which was previously reported to be positively regulated by σH, even though no upstream σH consensus promoter was identified, suggesting indirect regulation [7]. RNA-Seq data from our group (unpublished data) found clear evidence (FC > 5 and likelihood of being positively regulated by σH > 0.95) for σH-dependent transcript levels for only two of the genes encoding these 15 proteins, including lmo1454 and lmo0239; importantly, RNA-Seq data allow for quantification with similar sensitivity as qRT-PCR [14]. lmo1454 thus has been consistently identified as a gene that is directly up-regulated by σH, as supported by proteomics and transcriptomic studies and identification of an upstream σH-dependent promoter. Many of the other proteins identified here as showing σH-dependent production, on the other hand, appear to be regulated indirectly by σH, possibly at the post-transcriptional level. While future efforts will be needed to confirm σH-dependent production of these proteins (e.g., through Western blot or translational reporter fusions) and to explore the mechanisms of regulation, our data identified and further characterized a σH-dependent pathway that involves indirect effects of σH. Specifically, we found that both Lmo0027 (a component of a β-glucoside specific PTS system) and BglA (a β-glucosidase) showed higher protein levels in the presence of σH. As lmo0027 is preceded by a σH consensus promoter, these findings suggest a model where σH directly activates transcription of lmo0027, which facilitates PTS-based import of beta-glucosides into the cell. We hypothesize that these β-glucosides then lead to an increase in the levels of BglA (through a yet to be defined mechanism), facilitating the use of β-glucosides in downstream pathways involved in energy acquisition (e.g., glycolysis, the pentose phosphate pathway).
Table 1.
Proteins found to be differentially regulated by σH, as determined by a proteomic comparison between L. monocytogenes 10403S ΔBCL and ΔBCHL
| Proteina | Fold change Δ BCL /ΔBCHL | Description | Gene name | Role categoryb | Sub-Role categoryb | Promoterd | Sigma factor |
|---|---|---|---|---|---|---|---|
|
Proteins with positive fold change (>1.5) and p < 0.05 (indicating positive regulation by σH) | |||||||
| Lmo0027 |
1.55 |
beta-glucoside-specificPTS system IIABC component |
lmo0027 |
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
aggacacgtgtatgcgtggagtcctcgaatga |
SigmaH |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
|
|
| |
|
|
|
Energy metabolism |
Pyruvate dehydrogenase |
|
|
| Lmo0096 |
3.39 |
mannose-specific PTS system IIAB component ManL |
mptA |
Energy metabolism |
Pyruvate dehydrogenase |
tggcacagaacttgca |
SigmaL |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
|
|
| |
|
|
|
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
|
|
| Lmo0239 |
1.82 |
cysteinyl-tRNA synthetase |
cysS |
Protein synthesis |
tRNA aminoacylation |
ttgcaaggaattttattgctgttataatag |
SigmaA |
| Lmo0319 |
1.77 |
beta-glucosidase |
bglA |
Energy metabolism |
Sugars |
N/A |
N/A |
| Lmo0356 |
2.16 |
YhhX family oxidoreductase |
lmo0356 |
Energy metabolism |
Fermentation |
tggctaagtacagcgctagtgtagtactat |
SigmaA |
| |
|
|
|
Energy metabolism |
Electron transport |
|
|
| |
|
|
|
Central intermediary metabolism |
Other |
|
|
| Lmo1001 |
1.65 |
hypothetical protein |
lmo1001 |
Unclassified |
Role category not yet assigned |
N/A |
N/A |
| Lmo1070 |
2.18 |
similar to B. subtilis YlaN protein |
lmo1070 |
Hypothetical proteins |
Conserved |
ttgcgtggcaaataaattatgctatact |
SigmaA |
| Lmo1255 |
1.60 |
trehalose-specific PTS system IIBC component |
lmo1255 |
Energy metabolism |
Pyruvate dehydrogenase |
ttgcgctttcaactgatttatagtatagt |
SigmaA |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
|
|
| |
|
|
|
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
|
|
| Lmo1439 |
1.66 |
superoxide dismutase |
sodA |
Cellular processes |
Detoxification |
ttgcaagcatttagggagcatggtaggct |
SigmaA |
| |
|
|
|
|
|
gtttaacttttgagtttcagggaaa |
SigmaB |
| Lmo1454c |
1.85 |
RNA polymerase sigma factor RpoD |
rpoD |
Transcription |
Transcription factors |
gttttaaaaccgctaaatgatggtat |
SigmaB |
| |
|
|
|
|
|
aggacttttgctttttgtggcgaatat |
SigmaH |
| |
|
|
|
|
|
ttgactttttagcaaaaatacagtatctt |
SigmaA |
| Lmo2006 |
1.60 |
acetolactate synthase catabolic |
alsS |
Amino acid biosynthesis |
Aspartate family |
ttgcaataattcttttgagtagtataat |
SigmaA |
| |
|
|
|
Amino acid biosynthesis |
Pyruvate family |
|
|
| Lmo2064 |
2.01 |
large conductance mechanosensitive channel protein |
mscL |
Cellular processes |
Adaptations to atypical conditions |
tttcacatcgcagttagatgttttatact |
SigmaA |
| Lmo2487 |
1.65 |
hypothetical protein |
lmo2487 |
Hypothetical proteins |
Conserved |
N/A |
N/A |
| Lmo2614 |
2.05 |
50S ribosomal protein L30 |
rpmD |
Protein synthesis |
Ribosomal proteins: synthesis and modification |
ttgattactacccctaacccgtgtataat |
SigmaA |
| Lmo2621 |
1.63 |
50S ribosomal protein L24 |
rplX |
Protein synthesis |
Ribosomal proteins: synthesis and modification |
ttgattactacccctaacccgtgtataat |
SigmaA |
|
Proteins with negative fold change (< -1.5) and p < 0.05 (indicating negative regulation by σH) | |||||||
| Lmo1877 |
−1.61 |
formate-tetrahydrofolate ligase |
fhs |
Amino acid biosynthesis |
Aspartate family |
|
|
| |
|
|
|
Protein synthesis |
tRNA aminoacylation |
|
|
| |
|
|
|
Amino acid biosynthesis |
Histidine family |
|
|
| |
|
|
|
Purines, pyrimidines, nucleosides, and nucleotides |
Purine ribonucleotide biosynthesis |
|
|
| |
|
|
|
Biosynthesis of cofactors, prosthetic groups, and carriers |
Pantothenate and coenzyme A |
|
|
| Lmo2094 |
−7.35 |
hypothetical protein |
lmo2094 |
Energy metabolism |
Sugars |
|
|
| Lmo2097 |
−3.17 |
galactitol-specific PTS system IIB component |
lmo2097 |
Energy metabolism |
Pyruvate dehydrogenase |
|
|
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
|
|
| |
|
|
|
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
|
|
| Lmo2098 |
−2.33 |
galactitol-specific PTS system IIA component |
lmo2098 |
Energy metabolism |
Pyruvate dehydrogenase |
|
|
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
|
|
| Transport and binding proteins | Carbohydrates, organic alcohols, and acids | ||||||
aProtein names are based on the L. monocytogenes EGD-e locus.
bRole Categories and Sub-Role categories are based on JCVI classification [26].
cReported as positively and directly regulated by σH in Chaturongakul et al., 2011 [7].
dPromoters were identified based on RNA-Seq data (Orsi et al., unpublished) or previously published data. -10 and -35 (σA, σB, σH) and -12 and -24 (σL) regions are underlined. N/A indicates that a promoter was not identified.
Figure 1.
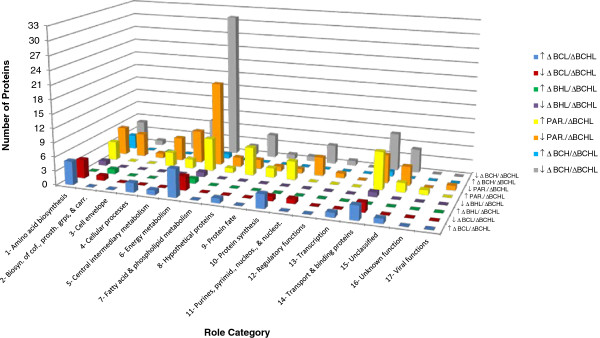
Functional role category classification of alternative σ factor dependent proteins. Functional role category classification of σH positively-regulated (blue), σH negatively-regulated (red), σC positively-regulated (green), σC negatively-regulated (purple), σL positively-regulated (turquoise), and σL negatively-regulated (gray) proteins; and proteins with higher levels in L. monocytogenes parent strain 10403S (PAR.) compared to ΔBCHL (yellow) and lower levels in PAR. compared to ΔBCHL (orange). Role category numbers correspond to: (1) Amino acid biosynthesis; (2) Biosynthesis of cofactors, prosthetic groups, and carriers; (3) Cell envelope; (4) Cellular processes; (5) Central intermediary metabolism; (6) Energy metabolism; (7) Fatty acid and phospholipid metabolism; (8) Hypothetical proteins; (9) Protein fate; (10) Protein synthesis; (11) Purines, pyrimidines, nucleosides, and nucleotides; (12) Regulatory functions; (13) Transcription; (14) Transport and binding proteins; (15) Unclassified; (16) Unknown function; (17) Viral functions. One protein may be classified into more than one role category. Statistical analysis of contingency tables for regulons with > 10 proteins (i.e., proteins positively regulated by σH; proteins negatively regulated by σL; proteins with higher or lower levels in the parent strain) found that role categories were not randomly distributed among proteins negatively regulated by σL and proteins with lower levels in the parent strain.
Our proteomic comparison also identified four proteins that showed lower levels in the strain expressing σH, suggesting (indirect) negative regulation by σH; three of these four proteins also showed lower levels in the parent strain (which expresses all four alternative σ factors) as compared to the quadruple mutant. None of the genes encoding these proteins showed significantly higher transcript levels in a ΔsigH strain in a transcriptomic study [7]. However, the coding gene for Lmo1877, one of these four proteins, is in an operon with lmo1876, which was previously reported to be negatively regulated by σH[7]. Overall, global indirect down-regulation of proteins by σH does not seem to play an important role in stationary phase L. monocytogenes 10403S.
σL appears to contribute to negative regulation of a number of proteins
Our proteomic comparison identified only two proteins (Lmo0096 and Lmo2006) as positively regulated by σL, as supported by higher protein levels (FC ≥ 1.5; pc < 0.05) in L. monocytogenes ΔBCH as compared to the ΔBCHL strain (Table 2). Both of these proteins also showed higher levels in the parent strain (which expresses all four alternative σ factors) as compared to the quadruple mutant. Lmo0096 (MptA) is annotated as the mannose-specific PTS system IIAB component, while Lmo2006 (AlsS) is annotated as an acetolactate synthase. Both lmo0096 and lmo2006 have previously been reported to be positively regulated by σL at the transcriptomic level [18]. Lmo0096 was also reported as showing lower levels in an L. monocytogenes EGD-e rpoN (σL) mutant in a 2-DE based proteomic analysis [22] and the lmo0096 gene was found to be preceded by a putative σL consensus promoter in the same study, further supporting positive regulation of the gene encoding this protein by σL.
Table 2.
Proteins found to be differentially regulated by σL, as determined by a proteomic comparison between L. monocytogenes 10403S ΔBCH and ΔBCHL
| Proteina | Fold change Δ BCH /ΔBCHL | Description | Gene name | Role categoryb | Sub-Role categoryb |
|---|---|---|---|---|---|
|
Proteins with positive fold change (>1.5) and p < 0.05 (indicating positive regulation by σL) | |||||
| Lmo0096d,f |
64.16 |
mannose-specific PTS system IIAB component ManL |
mptA |
Energy metabolism |
Pyruvate dehydrogenase |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
| |
|
|
|
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
| Lmo2006g |
3.41 |
acetolactate synthase catabolic |
alsS |
Amino acid biosynthesis |
Aspartate family |
| |
|
|
|
Amino acid biosynthesis |
Pyruvate family |
|
Proteins with negative fold change (< -1.5) and p < 0.05 (indicating negative regulation by σL) | |||||
| Lmo0027c,e |
−3.62 |
beta-glucoside-specific PTS system IIABC component |
lmo0027 |
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
| |
|
|
|
Energy metabolism |
Pyruvate dehydrogenase |
| Lmo0130 |
−3.64 |
hypothetical protein |
lmo0130 |
Unclassified |
Role category not yet assigned |
| Lmo0178 |
−2.07 |
hypothetical protein |
lmo0178 |
Regulatory functions |
Other |
| Lmo0181 |
−3.25 |
multiple sugar transport system substrate-binding protein |
lmo0181 |
Transport and binding proteins |
Unknown substrate |
| Lmo0260 |
−1.68 |
hydrolase |
lmo0260 |
Hypothetical proteins |
Conserved |
| Lmo0278 |
−1.67 |
maltose/maltodextrin transport system ATP-binding protein |
lmo0278 |
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
| Lmo0319c,e |
−2.96 |
beta-glucosidase |
bglA |
Energy metabolism |
Sugars |
| Lmo0343 |
−3.94 |
transaldolase |
tal2 |
Energy metabolism |
Pentose phosphate pathway |
| Lmo0344 |
−4.69 |
short chain dehydrogenase |
lmo0344 |
Energy metabolism |
Biosynthesis and degradation of polysaccharides |
| Lmo0345 |
−6.04 |
ribose 5-phosphate isomerase B |
lmo0345 |
Energy metabolism |
Pentose phosphate pathway |
| Lmo0346 |
−2.74 |
triosephosphate isomerase |
tpiA2 |
Energy metabolism |
Glycolysis/gluconeogenesis |
| Lmo0348 |
−2.41 |
dihydroxyacetone kinase |
lmo0348 |
Fatty acid and phospholipid metabolism |
Biosynthesis |
| |
|
|
|
Energy metabolism |
Sugars |
| Lmo0391 |
−1.67 |
hypothetical protein |
lmo0391 |
|
|
| Lmo0401 |
−2.16 |
alpha-mannosidase |
lmo0401 |
Unclassified |
Role category not yet assigned |
| Lmo0517e |
−3.21 |
phosphoglycerate mutase |
lmo0517 |
Energy metabolism |
Glycolysis/gluconeogenesis |
| Lmo0521 |
−2.23 |
6-phospho-beta-glucosidase |
lmo0521 |
Energy metabolism |
Sugars |
| Lmo0536 |
−1.97 |
6-phospho-beta-glucosidase |
lmo0536 |
Central intermediary metabolism |
Other |
| Lmo0574 |
−1.65 |
6-phospho-beta-glucosidase GmuD |
lmo0574 |
Central intermediary metabolism |
Other |
| Lmo0640 |
−1.78 |
oxidoreductase |
lmo0640 |
Energy metabolism |
Fermentation |
| |
|
|
|
Central intermediary metabolism |
Other |
| |
|
|
|
Energy metabolism |
Electron transport |
| Lmo0643 |
−2.61 |
transaldolase |
lmo0643 |
Energy metabolism |
Pentose phosphate pathway |
| Lmo0689 |
−1.71 |
chemotaxis protein CheV |
lmo0689 |
Cellular processes |
Chemotaxis and motility |
| Lmo0690 |
−2.44 |
flagellin |
flaA |
Cellular processes |
Chemotaxis and motility |
| Lmo0692 |
−1.66 |
chemotaxis protein CheA |
cheA |
Cellular processes |
Chemotaxis and motility |
| Lmo0813 |
−2.04 |
fructokinase |
lmo0813 |
Energy metabolism |
Sugars |
| Lmo0930 |
−1.88 |
hypothetical protein |
lmo0930 |
Unclassified |
Role category not yet assigned |
| Lmo1242 |
−1.59 |
hypothetical protein |
lmo1242 |
Hypothetical proteins |
Conserved |
| Lmo1254 |
−2.10 |
alpha-phosphotrehalase |
lmo1254 |
Energy metabolism |
Biosynthesis and degradation of polysaccharides |
| Lmo1348 |
−2.42 |
glycine cleavage system T protein |
gcvT |
Energy metabolism |
Amino acids and amines |
| Lmo1349 |
−2.68 |
glycine cleavage system P-protein |
gcvPA |
Energy metabolism |
Amino acids and amines |
| |
|
|
|
Central intermediary metabolism |
Other |
| Lmo1350e |
−2.11 |
glycine dehydrogenase subunit 2 |
gcvPB |
Central intermediary metabolism |
Other |
| |
|
|
|
Energy metabolism |
Amino acids and amines |
| Lmo1388e |
−2.02 |
ABC transport system |
tcsA |
Unclassified |
Role category not yet assigned |
| Lmo1389 |
−2.32 |
simple sugar transport system ATP-binding protein |
lmo1389 |
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
| Lmo1538e |
−1.89 |
glycerol kinase |
glpK |
Energy metabolism |
Other |
| Lmo1699 |
−1.92 |
Methyl-accepting chemotaxis protein |
lmo1699 |
Cellular processes |
Chemotaxis and motility |
| Lmo1730 |
−2.55 |
lactose/L-arabinose transport system substrate-binding protein |
lmo1730 |
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
| Lmo1791 |
−1.75 |
hypothetical protein |
lmo1791 |
|
|
| Lmo1812 |
−1.70 |
L-serine dehydratase iron-sulfur-dependent alpha subunit |
lmo1812 |
Energy metabolism |
Amino acids and amines |
| |
|
|
|
Energy metabolism |
Glycolysis/gluconeogenesis |
| Lmo1856 |
−1.65 |
purine nucleoside phosphorylase |
deoD |
Purines, pyrimidines, nucleosides, and nucleotides |
Salvage of nucleosides and nucleotides |
| Lmo1860 |
−1.64 |
peptide-methionine (S)-S-oxide reductase |
msrA |
Protein fate |
Protein modification and repair |
| Lmo1877 |
−2.14 |
formate-tetrahydrofolate ligase |
fhs |
Amino acid biosynthesis |
Aspartate family |
| |
|
|
|
Protein synthesis |
tRNA aminoacylation |
| |
|
|
|
Amino acid biosynthesis |
Histidine family |
| |
|
|
|
Purines, pyrimidines, nucleosides, and nucleotides |
Purine ribonucleotide biosynthesis |
| |
|
|
|
Biosynthesis of cofactors, prosthetic groups, and carriers |
Pantothenate and coenzyme A |
| Lmo1954e |
−1.97 |
phosphopentomutase |
deoB |
Purines, pyrimidines, nucleosides, and nucleotides |
Salvage of nucleosides and nucleotides |
| Lmo1993 |
−1.81 |
pyrimidine-nucleoside phosphorylase |
pdp |
Purines, pyrimidines, nucleosides, and nucleotides |
Salvage of nucleosides and nucleotides |
| Lmo2094 |
−28.99 |
hypothetical protein |
lmo2094 |
Energy metabolism |
Sugars |
| Lmo2097 |
−12.12 |
galactitol-specific PTS system IIB component |
lmo2097 |
Energy metabolism |
Pyruvate dehydrogenase |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
| |
|
|
|
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
| Lmo2098 |
−3.96 |
galactitol-specific PTS system IIA component |
lmo2098 |
Energy metabolism |
Pyruvate dehydrogenase |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
| |
|
|
|
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
| Lmo2160 |
−2.37 |
sugar phosphate isomerase/epimerase |
lmo2160 |
Hypothetical proteins |
Conserved |
| Lmo2161 |
−2.58 |
hypothetical protein |
lmo2161 |
Hypothetical proteins |
Conserved |
| Lmo2362 |
−1.87 |
glutamate/gamma-aminobutyrate antiporter |
lmo2362 |
Transport and binding proteins |
Amino acids, peptides and amines |
| Lmo2425 |
−1.59 |
glycine cleavage system H protein |
gcvH |
Energy metabolism |
Amino acids and amines |
| Lmo2481 |
−1.52 |
pyrophosphatase PpaX |
ppaX |
Central intermediary metabolism |
Other |
| Lmo2529 |
−1.72 |
ATP synthase F1 beta subunit |
atpD2 |
Energy metabolism |
ATP-proton motive force interconversion |
| Lmo2648 |
−2.50 |
hypothetical protein |
lmo2648 |
Unclassified |
Role category not yet assigned |
| Lmo2664 |
−1.72 |
L-iditol 2-dehydrogenase |
lmo2664 |
Central intermediary metabolism |
Other |
| |
|
|
|
Energy metabolism |
Glycolysis/gluconeogenesis |
| |
|
|
|
Energy metabolism |
Electron transport |
| |
|
|
|
Energy metabolism |
TCA cycle |
| |
|
|
|
Energy metabolism |
Fermentation |
| Lmo2696 |
−2.68 |
dihydroxyacetone kinase L subunit |
lmo2696 |
Energy metabolism |
Sugars |
| |
|
|
|
Fatty acid and phospholipid metabolism |
Biosynthesis |
| Lmo2697 |
−3.10 |
dihydroxyacetone kinase |
lmo2697 |
Hypothetical proteins |
Conserved |
| Lmo2743 | −2.71 | transaldolase | tal1 | Energy metabolism | Pentose phosphate pathway |
aProtein names are based on the L. monocytogenes EGD-e locus.
bRole Categories and Sub-Role categories are based on JCVI classification [26].
cReported as negatively regulated by σL in Chaturongakul et al., 2011 [7].
dReported as downregulated in a rpoN (σL) mutant compared to wildtype L. monocytogenes EGD-e in Arous et al., 2004 [22].
eReported as upregulated in a rpoN (σL) mutant compared to wildtype L. monocytogenes EGD-e in Arous et al., 2004 [22].
fPreceded by a putative σL promoter; tggcacagaacttgca; -12 and -24 regions are underlined.
gPreceded by a putative σA promoter; ttgcaataattcttttgagtagtataat; -10 and -35 regions are underlined.
A total of 56 proteins showed lower levels in the presence of σL (in the comparison between the ΔBCH and the ΔBCHL strain), suggesting indirect negative regulation of these proteins by σL (Table 2); two of the genes encoding these proteins had previously been shown to have higher transcript levels in a ΔsigL null mutant as compared to a parent strain, further supporting negative regulation by σL[7]. Twenty-one of the proteins with evidence for negative regulation by σL also showed lower protein levels in the parent strain as compared to the ΔBCHL strain (Additional file 1: Table S1), further supporting their negative regulation. Four of these 21 proteins as well as three other proteins found to be negatively regulated by σL in this study were also reported as showing higher transcript levels in an L. monocytogenes EGD-e rpoN (σL) mutant [22] (Table 2), supporting their negative regulation by σL. Overall, the 56 proteins identified here as negatively regulated by σL represented 13 role categories (e.g., energy metabolism, transport and binding proteins, central intermediary metabolism), including 31 proteins in the energy metabolism role category; statistical analyses showed overrepresentation of the role category “energy metabolism” (p < 0.01; Odds Ratio = 5.6) among these 56 proteins. Specific proteins identified as negatively regulated by σL included flagellin (FlaA), chemotaxis protein CheA, and a glutamate-γ-aminobutyric acid (GABA) antiporter (Lmo2362, GadC, GadT2), which have known roles in stress adaptation or virulence in L. monocytogenes[1,27].
σC regulates a small number of proteins
Previous studies indicated a role for σC in L. monocytogenes thermal adaptive response as well as in cold adaptation [3,13], however only a few genes have been identified as part of the σC regulon [7]. Similarly, we were only able to identify one protein, Lmo0096, that showed higher protein levels (FC ≥ 1.5; pc < 0.05) in the presence of σC (i.e., the comparison between the ΔBHL and the ΔBCHL strain; Table 3). Lmo0096 has been previously reported to be induced under cold stress in L. monocytogenes[28], supporting a role of σC in response to temperature stress in the bacterium. By comparison, the transcriptomic study by Chaturongakul et al., 2011 only identified lmo0422, which is in the same operon as sigC (lmo0423), as positively regulated by σC[7].
Table 3.
Proteins found to be differentially regulated by σC, as determined by a proteomic comparison between L. monocytogenes 10403S ΔBHL and ΔBCHL
| Proteina | Fold change ΔBHL/ΔBCHL | Description | Gene name | Role categoryb | Sub-Role categoryb |
|---|---|---|---|---|---|
|
Proteins with positive fold change (>1.5) and p < 0.05 (indicating positive regulation by σC) | |||||
| Lmo0096c |
3.19 |
mannose-specific PTS system IIAB component ManL |
mptA |
Energy metabolism |
Pyruvate dehydrogenase |
| |
|
|
|
Amino acid biosynthesis |
Aromatic amino acid family |
| |
|
|
|
Transport and binding proteins |
Carbohydrates, organic alcohols, and acids |
|
Proteins with negative fold change (< -1.5) and p < 0.05 (indicating negative regulation by σC) | |||||
| Lmo2094 |
−1.82 |
hypothetical protein |
lmo2094 |
Energy metabolism |
Sugars |
| Lmo1902 | −1.61 | 3-methyl-2-oxobutanoate hydroxymethyltransferase | panB | Biosynthesis of cofactors, prosthetic groups, and carriers | Pantothenate and coenzyme A |
aProtein names are based on the L. monocytogenes EGD-e locus.
bRole Categories and Sub-Role categories are based on JCVI classification [26].
cPreceded by a putative σL promoter; tggcacagaacttgca; -12 and -24 regions are underlined.
We also identified two proteins, Lmo2094 and Lmo1902, that showed higher protein levels in the absence of σC, suggesting negative regulation of these proteins by σC (Table 3). By comparison, the transcriptomic study by Chaturongakul et al. (2011) identified three different genes, representing two operons (lmo1854; lmo2185 and lmo2186), that showed lower transcript levels in the parent strain compared to the ΔsigC mutant, suggesting negative regulation by σC[7]. While our data are consistent with previous findings of a limited σC regulon in L. monocytogenes 10403S, it is possible that σC- dependent gene regulation only occurs under specific conditions (e.g., heat stress [3]) and that more complete identification of the σC regulon requires transcriptomic and proteomic studies under specific conditions that remain to be defined. In addition, future experiments using an L. monocytogenes strain that expresses sigC from an inducible promoter may also allow for identification of additional proteins that show σC-dependent production; this strategy applied to other alternative σ factors may also allow for identification of additional proteins that show σH- or σL-dependent production.
Proteins regulated by multiple alternative σ factors include MptA, which has a potential role in regulation of PrfA
Our data reported here also provided an opportunity to gather further insight into genes and proteins that are co-regulated by multiple σ factors and, consequently, into regulatory networks among different alternative σ factors. To facilitate these analyses, we also compared the protein levels between the L. monocytogenes parent strain and the ΔBCHL strain (which does not express any alternative σ factors). This analysis identified (i) 33 proteins that showed significantly higher levels (FC ≥ 1.5; pc < 0.05) in the parent strain as compared to the ΔBCHL strain (Additional file 1: Table S1) and (ii) 44 proteins that show lower levels in the parent as compared to the ΔBCHL mutant (Additional file 1: Table S1). Approximately 40% of the proteins that showed differential production (either up or down) are involved in energy metabolism and transport and binding functions (Figure 1). Among the 33 proteins that showed higher levels in the parent strain, (i) two were also found to be positively regulated by σH; (ii) one was also positively regulated by σH and σL, and (iii) one was also positively regulated by σH, σL and σC (Figure 2; Table 4). In addition, 12 of the 29 proteins that were found to be positively regulated in the parent strain, were also found to be positively regulated by σB in a recent proteomics study, which compared L. monocytogenes parent strain 10403S and ΔsigB mutant grown to stationary phase under the same conditions as used here [23]. While these 12 proteins likely represent proteins that are positively regulated by σB, the other 17 proteins that showed higher levels in the parent strain as compared to the ΔBCHL strain, but were not identified as positively regulated by any of the alternative σ factors, represent candidate proteins for redundant co-regulation by multiple alternative σ factors. Future experiments using an L. monocytogenes strain that only expresses σB (i.e., a ΔCHL strain) may help to not only further define the σB regulon, but also allow for further refinement of genes and proteins co-regulated by multiple alternative σ factors. Regulatory redundancy among multiple alternative σ factors has also previously been demonstrated through analyses of Bacillus subtilis alternative σ factor mutants; in particular, certain phenotypes displayed by a B. subtilis triple alternative σ factor deletion mutant were not found among single or double mutants of each of the three alternative σ factors, suggesting regulatory overlaps [29].
Figure 2.
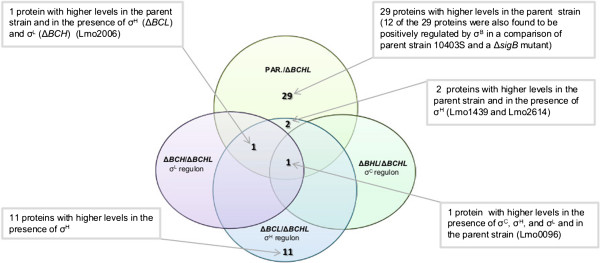
Venn diagram of proteins identified as showing higher protein levels in comparisons of (i) L. monocytogenes parent strain 10403S (PAR.) and Δ BCHL ; (ii) ΔBCH and ΔBCHL (identifying genes positively regulated by σL); ΔBCL and ΔBCHL (identifying genes positively regulated by σH); and ΔBHL and ΔBCHL (identifying genes positively regulated by σC). Twelve of the 29 proteins that were found to be positively regulated in the parent strain were also found to be positively regulated by σB in a recent proteomics study, which compared L. monocytogenes parent strain 10403S and a ΔsigB mutant [23]; these proteins include Lmo2748, Lmo2213, Lmo2158, Lmo2047, Lmo1830, Lmo0913, Lmo0796, Lmo0794, Lmo0722, Lmo0654, Lmo0539, and Lmo0265. The 17 proteins that show higher levels in the parent strain as compared to the ΔBCHL strain, but were not identified as positively regulated by any of the alternative σ factors include Lmo1540, Lmo2610, Lmo1422, Lmo1421, Lmo1602, Lmo1426, Lmo1428, Lmo2205, Lmo2398, Lmo1601, Lmo0554, Lmo1634, Lmo0110, Lmo2558, Lmo0783, Lmo0134, and Lmo0098.
Table 4.
Proteins found to be differentially regulated by at least two of the three alternative sigma factors studied here
| |
Regulation byb |
Regulation by σBc | Differential levels in comparison between parent and ΔBCHL | ||
|---|---|---|---|---|---|
| Proteina | σH | σL | σC | ||
| Lmo0027 |
+ |
- |
NDR |
NDR |
- |
| Lmo0096 (MptA) |
+ |
+ |
+ |
NDR |
+ |
| Lmo0319 (BglA) |
+ |
- |
NDR |
NDR |
- |
| Lmo1877 (Fhs) |
- |
- |
NDR |
NDR |
- |
| Lmo2006 (AlsS) |
+ |
+ |
NDR |
NDR |
+ |
| Lmo2094 |
- |
- |
- |
NDR |
- |
| Lmo2097 |
- |
- |
NDR |
NDR |
- |
| Lmo2098 | - | - | NDR | NDR | NDR |
aWhere available, protein name is shown in parenthesis.
bProteins that were identified here as positively (+) or negatively (−) regulated (absolute FC > 1.5; p < 0.05) by a given σ factor are shown; NDR (“not differentially regulated”) indicates that a protein was not found to be differentially regulated between strains with and without a given alternative σ factor.
cData for proteins differentially regulated by σB were obtained from Mujahid et al. [23]; this study compared protein levels between the 10403S parent strain and an isogenic ΔsigB strain.
Among the 44 proteins that showed lower levels in the parent strain as compared to the ΔBCHL mutant (Additional file 1: Table S1), (i) two also showed evidence for negative regulation by σH and σL (Lmo2097 and Lmo1877); (ii) one also showed evidence for negative regulation by σH, σL, and σC (Lmo2094; located in the same operon as lmo2097). Among these 44 proteins, statistical analyses showed overrepresentation of three role categories, including (i) “energy metabolism” (p < 0.01; Odds Ratio = 3.02), (ii) “biosynthesis of cofactors, prosthetic groups, and carriers” (p = 0.04; Odds Ratio = 2.72), and (iii) “purines, pyrimidines, nucleosides, and nucleotides” (p = 0.04; Odds Ratio = 3.29), as well as underrepresentation of the role category “hypothetical proteins” (p = 0.01; Odds Ratio = 0.208).
Overall, our data provide additional evidence that a number of genes and proteins are co-regulated by more than one σ factor. This is consistent with previous microarray studies [7] that have reported considerable overlaps between σ factor regulons in L. monocytogenes, in particular between the σH and the σB regulon. We also identified some proteins with particularly striking patterns of co-regulation, including (i) members of the lmo2093-lmo2099 operon, specifically Lmo2094, which was found to be negatively regulated by σH, σL, and σC and Lmo2097 and Lmo2098, which were found to be negatively regulated by σH and σL (Table 4) and (ii) MptA (Lmo0096), which was found to be positively regulated by σH, σL, and σC (Table 4). Lmo2094 shows particularly striking negative regulation of protein production by σH, σL, and σC with respective fold changes of −7.35, -28.99, and −1.82. Although Lmo2094 is annotated as a fuculose-phosphate aldolase, it is part of an operon in which most of the other genes with assigned functions are annotated as being involved in the galactitol degradation pathway. Specifically, the lmo2093 to lmo2099 operon encodes components of a putative PTS galactitol family permease [30], including the PTS system galactitol-specific enzyme IIC (Lmo2096), IIB (Lmo2097), and IIA (Lmo2098) components, as well as a transcription antiterminator (Lmo2099), a tagatose-6-phosphate kinase/1-phosphofructokinase (Lmo2095), an L-fuculose-phosphate aldolase (Lmo2094), and a hypothetical protein (Lmo2093). Therefore, it is possible that Lmo2094 is also involved in this pathway functioning as a tagatose-1,6-biphosphate aldolase. This enzyme converts tagatose-1,6,-biphosphate into glyceraldehyde 3-phosphate and dihydroxyacetone phosphate, which allows both tagatose and galactitol to be used as energy sources for glycolysis [31].
MptA, a component of a permease of the PTS mannose–fructose–sorbose family, which is another one of the seven PTS families of L. monocytogenes[30], showed the highest fold change in the ΔBCH strain as compared to the ΔBCHL strain, supporting σL dependent protein levels (FC = 64.16); fold changes supporting σH and σC dependent protein levels were 3.39 and 3.19, respectively. MptA is encoded by a gene that is part of a three-gene operon (mptACD[32], which also has been designated as manLMN[25]); these three genes encode a mannose-specific PTS system IIAB component, a mannose-specific PTS system IIC component, and a mannose-specific PTS system IID component, respectively [25,32]. Recently, it was suggested that during glucose uptake, MptA dephosphorylates, which directly, or indirectly, inhibits PrfA, the major positive regulator of L. monocytogenes virulence genes [25]. These findings thus provide for a hypothesis that redundant upregulation of MptA, through multiple alternative σ factors, may provide a critical initial step towards inactivation of PrfA.
Conclusions
Transcriptional regulation through the interplay between alternative σ factors represents an important component of L. monocytogenes stress response systems and the ability of this pathogen to regulate gene expression during infection. In addition to transcriptional regulation, alternative σ factors may also regulate protein production post-transcriptionally and/or post-translationally. To allow for further insights into the roles of different alternative σ factors in L. monocytogenes, we thus completed a global evaluation of alternative σ factor-dependent protein production patterns in L. monocytogenes stationary phase cells. In concert with previous transcriptomic studies, our data not only provide a further refinement of our understanding of the alternative σ factor regulons in this important pathogen, but also provide clear evidence for co-regulation, by multiple σ factors, of different PTS systems, including one PTS system that has been suggested to be linked to regulation of PrfA. Co-regulation by multiple σ factors can provide sensitive means for fine-tuning of gene expression and protein production under different environmental conditions, as well as redundancy that can ensure gene expression and protein production under different conditions. Consistent with the goals of this study, many of the proteins that were identified as showing production dependent on the presence of alternative σ factors appear to represent indirect regulation by a given σ factor, which will require future confirmation by protein based methods (e.g., Western blots, translational fusions).
Methods
Bacterial strains, mutant construction, and growth conditions
Splicing by overlap extension (SOE) PCR and allelic exchange mutagenesis was used to construct ΔBCL, ΔBHL, ΔBCH, and ΔBCHL mutant strains in an L. monocytogenes 10403S background as described previously [13] (Additional file 2: Table S2). All mutations were confirmed by PCR amplification and sequencing of the PCR product. Strains were grown to stationary phase in BHI at 37°C as described previously [33].
Protein isolation, iTRAQ labeling, and Nano-scale reverse phase chromatography and tandem mass spectrometry (nanoLC-MS/MS)
Protein isolation, digestion, and iTRAQ labeling were performed as previously described [33]. Briefly, proteins were isolated from a 25 ml culture of L. monocytogenes stationary phase cells. A noninterfering protein assay kit (Calbiochem) and 1D SDS-PAGE were used to verify protein concentration and quality. A total of 100 μg protein of each sample was denatured, reduced, and the cysteine residues were blocked. Protein samples were then digested with sequence-grade-modified trypsin at 37°C for 16 h, and protein digestion efficiency was assessed by SDS-PAGE. Tryptic peptides from L. monocytogenes parent strain 10403S and ΔBCL, ΔBHL, ΔBCH, and ΔBCHL mutant strains were each labeled with iTRAQ reagents, according to the manufacturer’s protocols.
Four labeled protein samples were combined for a single run and fractionated via Isoelectric focusing OffGel electrophoresis (OGE) using an Agilent 3100 OFFGEL Fractionator (Agilent, G3100AA), and subsequent nanoLC-MS/MS was carried out using a LTQ-Orbitrap Velos (Thermo-Fisher Scientific) mass spectrometer as previously described [33]. Two separate biological replicates of the entire proteomics experiment were run for each strain.
Protein identification and data analysis
All MS and MS/MS raw spectra from iTRAQ experiments were processed using Proteome Discoverer 1.1 for subsequent database search using in-house licensed Mascot Daemon; quantitative processing, protein identification, and data analysis were conducted as previously described [33].
The biological replicates of each experiment were analyzed independently. As described in [33], the Wilcoxon signed rank test was applied to peptide ratios for each identified protein to determine significant changes between strains. The Fisher’s Combined Probability Test was then used to combine FDR adjusted Wilcoxon p-values from each replicate into one test statistic for every protein to obtain a combined p-value (p-valuec). Proteins with peptide ratios exhibiting a Fisher’s Combined Probability Test p-valuec < 0.05 and an iTRAQ protein ratio ≥ 1.5 in both replicates were considered significantly differentially expressed. Statistical analyses were conducted using R statistical software.
A Monte Carlo simulation of Fisher’s exact test was used to determine whether the distribution of role categories among proteins identified as differentially regulated by a given σ factor was different from the role category distribution that would be expected by chance (based on the role category primary annotation for all L. monocytogenes EGD-e genes [26]). Individual Fisher’s exact tests were then used to determine whether individual role categories were over- or under- represented; uncorrected p-values were reported, allowing readers to apply corrections if deemed appropriate. Analyses were performed using all role categories assigned to a given gene in the JCVI-CMR L. monocytogenes EGD-e database. Analyses were only performed for regulons that contained 10 or more proteins (i.e., proteins positively regulated by σH; proteins negatively regulated by σL; proteins with higher or lower levels in the parent strain).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SM performed the experimental work and most of the data analysis that was not carried out at the Cornell Proteomics and Mass Spectrometry Core Facility and drafted the manuscript. RHO contributed to analysis of the data and helped to revise the manuscript. MW and KJB conceived of the study, and participated in its design and coordination and helped to draft or revise the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Proteins found to be differentially produced between L. monocytogenes parent strain 10403S and ΔBCHL.
Strains used in this study.
Contributor Information
Sana Mujahid, Email: sm832@cornell.edu.
Renato H Orsi, Email: rho2@cornell.edu.
Kathryn J Boor, Email: kjb4@cornell.edu.
Martin Wiedmann, Email: mw16@cornell.edu.
Acknowledgements
This work was funded by NIH-NIAID R01 AI052151 (K.J.B.). S. M. was partially supported by a New York Sea Grant Scholar Fellowship (RSHH-15). We thank the Proteomics and Mass Spectrometry Core Facility at Cornell University for their services and Dr. James Booth for assistance with statistical analyses.
References
- Chaturongakul S, Raengpradub S, Wiedmann M, Boor KJ. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 2008;16(8):388–396. doi: 10.1016/j.tim.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, Scott VN, Gombas DE, Boor KJ, Wiedmann M. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl Environ Microbiol. 2004;70(10):5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Nietfeldt J, Zhang M, Benson AK. Functional consequences of genome evolution in Listeria monocytogenes: the lmo0423 and lmo0422 genes encode SigmaC and LstR, a lineage II-specific heat shock system. J Bacteriol. 2005;187(21):7243–7253. doi: 10.1128/JB.187.21.7243-7253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi RH, den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol. 2011;301(2):79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- O’Byrne CP, Karatzas KA. The role of Sigma B (Sigma B) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv Appl Microbiol. 2008;65:115–140. doi: 10.1016/S0065-2164(08)00605-9. [DOI] [PubMed] [Google Scholar]
- Oliver HF, Orsi RH, Wiedmann M, Boor KJ. Listeria monocytogenes {Sigma}B has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl Environ Microbiol. 2010;76(13):4216–4232. doi: 10.1128/AEM.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S, Raengpradub S, Palmer ME, Bergholz TM, Orsi RH, Hu Y, Ollinger J, Wiedmann M, Boor KJ. Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors SigmaB, SigmaC, SigmaH, and SigmaL in Listeria monocytogenes. Appl Environ Microbiol. 2011;77(1):187–200. doi: 10.1128/AEM.00952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S, Boor KJ. RsbT and RsbV contribute to SigmaB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl Environ Microbiol. 2004;70(9):5349–5356. doi: 10.1128/AEM.70.9.5349-5356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemekamp-Kamphuis HH, Wouters JA, de Leeuw PP, Hain T, Chakraborty T, Abee T. Identification of sigma factor Sigma B-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl Environ Microbiol. 2004;70(6):3457–3466. doi: 10.1128/AEM.70.6.3457-3466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser KR, Sue D, Wiedmann M, Boor K, O’Byrne CP. Role of SigmaB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is SigmaB dependent. Appl Environ Microbiol. 2003;69(4):2015–2022. doi: 10.1128/AEM.69.4.2015-2022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker LA, Evans SN, Hutkins RW, Benson AK. Role of Sigma(B) in adaptation of Listeria monocytogenes to growth at low temperature. J Bacteriol. 2000;182(24):7083–7087. doi: 10.1128/jb.182.24.7083-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead SM, Dykes GA. Influence of the sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int J Food Microbiol. 2004;91(1):63–72. doi: 10.1016/S0168-1605(03)00332-5. [DOI] [PubMed] [Google Scholar]
- Chan YC, Hu Y, Chaturongakul S, Files KD, Bowen BM, Boor KJ, Wiedmann M. Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J Food Prot. 2008;71(2):420–425. doi: 10.4315/0362-028x-71.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour SW, Filiatrault MJ, Wiedmann M, Boor KJ. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and Sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics. 2009;10:641-2164-10-641. doi: 10.1186/1471-2164-10-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abram F, Starr E, Karatzas KA, Matlawska-Wasowska K, Boyd A, Wiedmann M, Boor KJ, Connally D, O’Byrne CP. Identification of components of the Sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl Environ Microbiol. 2008;74(22):6848–6858. doi: 10.1128/AEM.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abram F, Su WL, Wiedmann M, Boor KJ, Coote P, Botting C, Karatzas KA, O’Byrne CP. Proteomic analyses of a Listeria monocytogenes mutant lacking SigmaB identify new components of the SigmaB regulon and highlight a role for SigmaB in the utilization of glycerol. Appl Environ Microbiol. 2008;74(3):594–604. doi: 10.1128/AEM.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea RB, Gahan CG, Hill C. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect Immun. 2004;72(2):717–727. doi: 10.1128/IAI.72.2.717-727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila M, Somervuo P, Rattei T, Korkeala H, Stephan R, Tasara T. Phenotypic and transcriptomic analyses of Sigma L-dependent characteristics in Listeria monocytogenes EGD-e. Food Microbiol. 2012;32(1):152–164. doi: 10.1016/j.fm.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Okada Y, Okada N, Makino S, Asakura H, Yamamoto S, Igimi S. The sigma factor RpoN (sigma54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol Lett. 2006;263(1):54–60. doi: 10.1111/j.1574-6968.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- Raimann E, Schmid B, Stephan R, Tasara T. The alternative sigma factor Sigma(L) of L. monocytogenes promotes growth under diverse environmental stresses. Foodborne Pathog Dis. 2009;6(5):583–591. doi: 10.1089/fpd.2008.0248. [DOI] [PubMed] [Google Scholar]
- Robichon D, Gouin E, Debarbouille M, Cossart P, Cenatiempo Y, Hechard Y. The rpoN (sigma54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J Bacteriol. 1997;179(23):7591–7594. doi: 10.1128/jb.179.23.7591-7594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arous S, Buchrieser C, Folio P, Glaser P, Namane A, Hebraud M, Hechard Y. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology. 2004;150(Pt 5):1581–1590. doi: 10.1099/mic.0.26860-0. [DOI] [PubMed] [Google Scholar]
- Mujahid S, Orsi RH, Vangay P, Boor KJ, Wiedmann M. Refinement of the Listeria monocytogenes σB regulon through quantitative proteomic analysis. Microbiology. 2013. in press. [DOI] [PMC free article] [PubMed]
- Archambaud C, Nahori MA, Pizarro-Cerda J, Cossart P, Dussurget O. Control of Listeria Superoxide Dismutase by phosphorylation. J Biol Chem. 2006;281(42):31812–31822. doi: 10.1074/jbc.M606249200. [DOI] [PubMed] [Google Scholar]
- Ake FM, Joyet P, Deutscher J, Milohanic E. Mutational analysis of glucose transport regulation and glucose-mediated virulence gene repression in Listeria monocytogenes. Mol Microbiol. 2011;81(1):274–293. doi: 10.1111/j.1365-2958.2011.07692.x. [DOI] [PubMed] [Google Scholar]
- Craig J, Venter Institute Comprehensive Microbial Resource. http://cmr.jcvi.org.
- Dons L, Eriksson E, Jin Y, Rottenberg ME, Kristensson K, Larsen CN, Bresciani J, Olsen JE. Role of Flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect Immun. 2004;72(6):3237–3244. doi: 10.1128/IAI.72.6.3237-3244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace G, Mazzeo MF, Sorrentino A, Spada V, Malorni A, Siciliano RA. Proteomics for the elucidation of cold adaptation mechanisms in Listeria monocytogenes. J Proteomics. 2010;73(10):2021–2030. doi: 10.1016/j.jprot.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Mascher T, Hachmann AB, Helmann JD. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J Bacteriol. 2007;189(19):6919–6927. doi: 10.1128/JB.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll R, Goebel W. The major PEP-phosphotransferase systems (PTSs) for glucose, mannose and cellobiose of Listeria monocytogenes, and their significance for extra- and intracellular growth. Microbiology. 2010;156(Pt 4):1069–1083. doi: 10.1099/mic.0.034934-0. [DOI] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39(Database issue):D583–D590. doi: 10.1093/nar/gkq1143. http://biocyc.org/ECOLI/new-image?object=GALACTITOLCAT-PWY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalet K, Cenatiempo Y, Cossart P, Hechard Y. European Listeria Genome Consortium. A Sigma(54)-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology. 2001;147(Pt 12):3263–3269. doi: 10.1099/00221287-147-12-3263. [DOI] [PubMed] [Google Scholar]
- Mujahid S, Bergholz TM, Oliver HF, Boor KJ, Wiedmann M. Exploration of the role of the non-Coding RNA SbrE in L. monocytogenes stress response. Int J Mol Sci. 2012;14(1):378–393. doi: 10.3390/ijms14010378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins found to be differentially produced between L. monocytogenes parent strain 10403S and ΔBCHL.
Strains used in this study.


