Abstract
Background
Considerable efforts have been devoted to evaluating the association of the receptor for advanced glycation end-products (gene AGER and protein: RAGE) genetic variants to coronary artery disease (CAD); the results, however, are often irreproducible. To generate more information, we sought to explore four common polymorphisms of AGER and its circulating forms associated with the risk of CAD via a meta-analysis.
Methodology/Principal Findings
Articles were identified by searching PubMed, EMBASE, Wanfang and CNKI databases before March 2013. Qualified articles had case-control designs and investigated AGER four polymorphisms (T-429C, T-374A, Gly82Ser, G1704A) or circulating soluble RAGE (sRAGE) or endogenous secretory RAGE (esRAGE) levels associated with CAD. Twenty-seven articles involving 39 independent groups fulfilled the predefined criteria. Overall, no significance was observed for all examined polymorphisms under allelic and dominant models. When restricting groups to CAD patients with diabetes mellitus or renal disease, deviations of risk estimates from the unity were stronger than overall estimates for all polymorphisms except for G1704A due to limited available studies. For example, under dominant model, having -429C allele increased the odds of developing CAD in diabetic patients by 1.22-fold (95% confidence interval (95% CI) 0.99–1.51; P = 0.06; I 2 = 6.7%) compared with that of overall estimate of 1.15-fold (95% CI: 0.97–1.36; P = 0.111; I 2 = 18.0%). Circulating sRAGE levels were non-significantly lower in CAD patients than in controls, whereas this reduction was totally and significantly reversed in CAD patients with diabetes mellitus (weighted mean difference: 185.71 pg/ml; 95% CI: 106.82 to 264.61 pg/ml). Circulating esRAGE levels were remarkably lower in CAD patients, as well as in subgroups with or without diabetes mellitus and without renal disease.
Conclusions
Our findings demonstrated that association of AGER genetic polymorphisms with CAD was potentiated in patients with diabetes mellitus or renal disease. Practically, circulating esRAGE might be a powerful negative predictor for the development of CAD.
Introduction
Coronary artery disease (CAD) is a leading cause of morbidity and mortality worldwide. Family studies suggest a strong genetic background: men with 2 or more affected parents or siblings relative to men without family history have a 3.4-fold increased risk of developing myocardial infarction [1]. One of the potential candidate genes that account for an inherited predisposition to CAD is the receptor for advanced glycation end-products (gene: AGER and protein: RAGE), which is a multiligand receptor, belonging to the immunoglobulin superfamily of cell surface molecules. The activation of AGER can evoke a wide range of signaling pathways that trigger inflammation, atherogenesis and vasoconstriction leading to coronary dysfunction, atherosclerosis and thrombosis [2]. Moreover, circulating soluble RAGE (sRAGE) levels, which were in dose-dependent association with angiographic observations, were observed to be significantly lower in angiographically-confirmed CAD patients than in healthy controls [3], [4]. By contrast, circulating sRAGE levels were significantly higher in patients with acute myocardial infarction, independent of the presence of diabetes mellitus [5]. Therefore association between circulating sRAGE and CAD must be confirmed in larger studies.
The gene encoding AGER is mapped on chromosome 6p21.3 and spans 3.27 kb with 11 exons. The genomic sequence of AGER gene is highly polymorphic with many alleles that exhibit different functional properties and heterogeneous distributions across populations [6]–[8]. Considerable efforts have been devoted to evaluating the contributory role of AGER genetic defects in the development of CAD; the results, however, are not often reproducible. As a caveat, failure to replicate might be attributable to the ethnicity-specific genetic profiles, the individual underpowered studies, and the lack of consideration for confounders. To generate more information, we sought to assess the association of four common polymorphisms (T-429C, T-374A, Gly82Ser, G1704A) of AGER and its circulating forms with CAD via a meta-analysis of individual participant data from qualified case-controls studies, while addressing between-study heterogeneity and publication bias. Selection of these four polymorphisms is relatively straightforward if three or more unduplicated studies are available for a certain polymorphism.
Methods
Meta-analysis of observational studies poses particular challenges due to the inherent biases and differences in study designs. In this context, we carried out this meta-analysis in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guideline [9] (see Supplementary Table S1).
Search Strategy
A literature search was conducted of PubMed, EMBASE, Wanfang (http://www.wanfangdata.com.cn) and China National Knowledge Infrastructure (CNKI, http://www.cnki.net) databases covering the period from the earliest possible year to March 15, 2013. The following subject terms were used in the search: “advanced glycation end products”, “RAGE”, “AGER”, “coronary heart disease”, “coronary syndrome” or “isch[a]emic heart disease” or “vascular disease” or “myocardial infarction” or “atherosclerosis” or “arteriosclerosis” or “coronary stenosis” or “coronary artery disease” or “coronary disease” or “CAD” or “CHD” or “ACS”, combined with “gene” or “allele” or “genotype” or “polymorphism” or “variant” or “mutation”. The research was also supplemented by reviews of reference lists, hand-searching of relevant journals, and correspondence with authors. Search results were limited to studies on a case-control design and articles published in English or Chinese language.
Study Selection
Two investigators (F.P. and W.N.) independently obtained the full texts of articles deemed as potentially eligible according to the titles and abstracts. If necessary, we emailed the contributing authors to avoid double counting of participants recruited in more than one study by the same group. Where more than one publication of the same study population existed, we abstracted data from the most recent or most complete publication.
Inclusion/Exclusion Criteria
Our analyses were restricted to studies that fulfilled the following inclusion criteria (all must be satisfied): (1) clinical endpoint (dependent variable): CAD or myocardial infarction (MI); (2) study design: either retrospective or nested case-control; (3) independent parameters: either genotypes/alleles of at least one examined polymorphism or circulating sRAGE or endogenous secretory RAGE (esRAGE). Studies were excluded (one was sufficient) if they investigated the progression, severity, phenotype modification, response to treatment or survival, as well as if they were meeting abstracts, case reports/series, editorials, review articles, or non-English and non-Chinese articles.
Data Extraction
Data were extracted from qualified articles independently by two investigators (F.P. and W.N.) according to a standardized Excel template (Microsoft Corp, Redmond, WA). Quality assessment was performed in duplicate with κ agreement rate of 0.98. Discrepancies were adjudicated by discussion and a consensus was reached. When three or more studies investigated the same polymorphism in AGER gene, published data were synthesized accordingly.
Collected data included the first author, publication year, ethnicity, CAD subtype (CAD or MI), study design, case-control status, genotypes/alleles of examined polymorphisms, circulating sRAGE and/or esRAGE levels, and the demographic records (if available), such as age, gender, matched information, percentage of diabetes and renal disease, body mass index, smoking, systolic and diastolic blood pressure.
Statistical Analysis
To maximize power to detect a true association, only allelic and dominant models were adopted to estimate risk effects of AGER genetic polymorphisms on CAD. The random-effects model using the DerSimonian & Laird method was employed to calculate weighted odds ratio (OR) and the corresponding 95% confidence interval (95% CI). Comparisons of circulating sRAGE and esRAGE levels between patients and controls were expressed as weighted mean difference (WMD) with 95% CI.
Between-study heterogeneity was assessed by χ2 test, and was quantified using the I 2 statistic (ranging from 0 to 100%), which is defined as the percentage of the observed between-study variability that is due to heterogeneity rather than chance. Cumulative analyses were performed for all polymorphisms according to the ascending date of publication in order to identify the impact of the first-published article on subsequent publications, and the evolution of the pooled estimates over time.
Predefined subgroup analyses were performed a priori according to the CAD endpoints (CAD and MI), descents of study population (Caucasian, East Asian, Middle Eastern, African), study design (retrospective and prospective), matched information on age and/or gender, and total sample sizes (<300 subjects: small study and ≥300 subjects: large study). Meta-regression analyses were performed to evaluate the extent to which different study-level variables, including age, gender, body mass index, smoking, systolic and diastolic blood pressure, explained the heterogeneity of pooled risk estimates of AGER genetic polymorphisms examined or circulating sRAGE and esRAGE levels on CAD.
Publication bias was assessed by the Egger’s test and the trim-and-fill method. The latter was to estimate the number and outcomes of theoretically missing studies resulting from publication bias. P<0.05 was considered statistical significance, with the exception of I 2 and Egger’s statistics, for which a significance level was set at P<0.1 [10]. All statistical analyses were carried out with STATA software (StataCorp, TX, version 11.2 for Windows).
Results
Eligible Studies
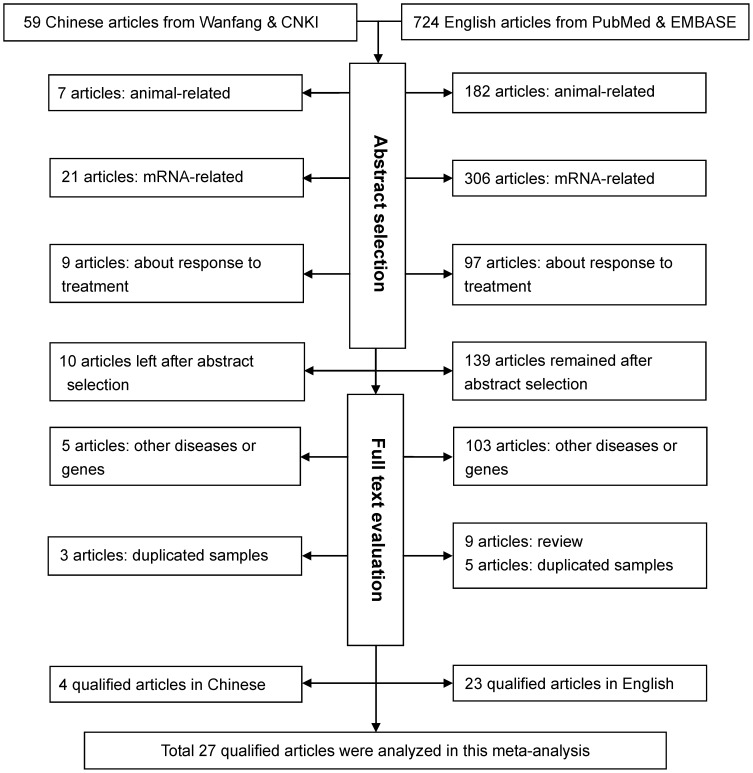
Characteristics of all qualified studies in this meta-analysis are summarized in Table 1 and Table 2. The initial search yielded 783 potentially relevant articles. Applying our inclusion/exclusion criteria left 27 qualified articles [3], [5], [8], [11]–[34]. A flow diagram schematized the process of excluding articles with specific reasons (Figure 1). These 27 articles were published between 2004 and 2012, with 4 articles written in Chinese [22], [27], [30], [31] and the others in English. One article was grouped by ethnicity [13] and hypertension [24] respectively, three by CAD subtypes (CAD, MI, CAD with and without restenosis) [14], [17], [25], and five by diabetes mellitus [5], [8], [19], [20], [23]. These independent subgroups were treated separately, and accordingly there were 39 groups in final analyses.
Table 1. Characteristics of qualified studies.
| Author (year) | Ethnicity | Matched | Disease | Diabetes (%) | Renal disease (%) | Study design |
| Kirbis (2004) | Caucasian | NA | CAD | 100.00 | NA | Retrospective |
| Falcone (2004) | Caucasian | NA | CAD | 0.00 | 0.00 | Retrospective |
| dos Santos (2005) (Caucasian) | Caucasian | NA | CAD | 100.00 | 97.20 | Retrospective |
| dos Santos (2005) (African) | African | NA | CAD | 100.00 | 97.20 | Retrospective |
| Falcone (2005) | Caucasian | NA | CAD | 0.00 | 0.00 | Retrospective |
| Hofmann (2005) (MI) | Caucasian | NA | MI | 24.00 | NA | Prospective |
| Hofmann (2005) (CAD-diabetes) | Caucasian | NA | CAD | 100.00 | NA | Prospective |
| Hofmann (2005) (CAD+diabetes) | Caucasian | NA | CAD | 0.00 | NA | Prospective |
| Zee (2006) | Caucasian | NA | MI | 8.90 | NA | Prospective |
| Yoon (2007) | Asian | NA | CAD | 26.00 | NA | Retrospective |
| Lu (2008) (−restenosis) | Asian | NA | CAD | 100.00 | 0.00 | Prospective |
| Lu (2008) (+restenosis) | Asian | NA | CAD | 100.00 | 0.00 | Prospective |
| Mulder (2008) | Caucasian | age, gender | CAD | 19.00 | 0.00 | Retrospective |
| Kucukhuseyin (2009) (+diabetes) | Middle Eastern | NA | CAD | 100.00 | NA | Retrospective |
| Kucukhuseyin (2009) (−diabetes) | Middle Eastern | NA | CAD | 0.00 | NA | Retrospective |
| Lu (2009) (−diabetes) | Asian | NA | CAD | 0.00 | NA | Retrospective |
| Lu (2009) (+diabetes) | Asian | NA | CAD | 100.00 | NA | Retrospective |
| Mahajan (2009) | Asian | NA | CAD | 0.00 | 0.00 | Retrospective |
| Peng (2009) (+diabetes) | Asian | NA | CAD | 100.00 | 0.00 | Retrospective |
| Peng (2009) (−diabetes) | Asian | NA | CAD | 100.00 | 0.00 | Retrospective |
| Yan (2009) (+diabetes) | Asian | NA | CAD | 100.00 | NA | Retrospective |
| Yan (2009) (−diabetes) | Asian | NA | CAD | 0.00 | NA | Retrospective |
| Pu (2009) | Asian | NA | CAD | 100.00 | NA | Retrospective |
| Gao (2010) (−hypertension) | Asian | age, gender | CAD | 0.00 | 0.00 | Retrospective |
| Gao (2010) (+hypertension) | Asian | age, gender | CAD | 0.00 | 0.00 | Retrospective |
| McNair (2010) | Caucasian | age, gender | MI | 0.00 | NA | Prospective |
| McNair (2010) (+restenosis) | Caucasian | NA | MI | 0.00 | NA | Retrospective |
| McNair (2010) (−restenosis) | Caucasian | NA | MI | 0.00 | NA | Retrospective |
| Xie (2010) | Asian | NA | CAD | 13.39 | 0.00 | Retrospective |
| Hou (2011) | Asian | NA | CAD | 0.00 | NA | Retrospective |
| Boiocchi (2011) | Caucasian | age, gender | MI | 27.00 | NA | Retrospective |
| Cai (2011) (CAD) | Asian | NA | CAD | 45.50 | 0.00 | Retrospective |
| Cai (2011) (MI) | Asian | NA | MI | 46.20 | 0.00 | Retrospective |
| Park (2011) (+diabetes) | Asian | age, gender | MI | 100.00 | 0.00 | Retrospective |
| Park (2011) (−diabetes) | Asian | age, gender | MI | 0.00 | 0.00 | Retrospective |
| Peng (2011) | Asian | NA | CAD | 100.00 | 0.00 | Retrospective |
| Lu (2011) | Asian | NA | CAD | 0.00 | NA | Retrospective |
| Aydogan (2012) | Middle Eastern | NA | CAD | 0.00 | NA | Retrospective |
| Selejan (2012) | Caucasian | NA | MI | 35.00 | 0.00 | Retrospective |
Abbreviations: NA, not available; CAD, coronary artery disease; MI, myocardial infarction.
Table 2. Characteristics of study populations in qualified studies.
| Author (year) | Age, yr | Males, % | BMI, kg/m2 | Smoking | SBP, mmHg | DBP, mmHg | sRAGE, pg/ml | esRAGE, pg/ml |
| Kirbis (2004) | 59.3/66.9 | 64.9/43.2 | 28.7/27.8 | 44/14.1 | 146/145 | 83/85 | NA | NA |
| Falcone (2004) | 61.8/59.6 | 79.4/77.4 | 26.1/24.9 | 73.7/45.2 | NA | NA | NA | NA |
| dos Santos (2005) (Caucasian) | 61.8/62.4 | 55.93/40.23 | 27.9/28.3 | NA | NA | NA | NA | NA |
| dos Santos (2005) (African) | 59.5/58.7 | 41.33/25 | 28.7/28.6 | NA | NA | NA | NA | NA |
| Falcone (2005) | 64.1/63.2 | NA | 25.7/25.6 | 49.09/31.1 | NA | NA | 966/1335 | NA |
| Hofmann (2005) (MI) | NA | NA | NA | NA | NA | NA | NA | NA |
| Hofmann (2005) (CAD-diabetes) | NA | NA | NA | NA | NA | NA | NA | NA |
| Hofmann (2005) (CAD+diabetes) | NA | NA | NA | NA | NA | NA | NA | NA |
| Zee (2006) | NA | NA | NA | NA | NA | NA | NA | NA |
| Yoon (2007) | 55.73/53.18 | NA | 25.01/23.41 | 84.9/77.3 | 119.86/114.16 | 77.13/75.84 | NA | NA |
| Lu (2008) (−restenosis) | 67/61 | NA | NA | 29.6/ | 140/ | 83/ | NA | 220/480 |
| Lu (2008) (+restenosis) | 65/61 | NA | NA | 33.7/14.7 | 137/137 | 81/80 | NA | 160/300 |
| Mulder (2008) | 64.7/63.4 | 78/72 | 27.7/25.5 | 68/72 | NA | NA | 1373/1299 | NA |
| Kucukhuseyin (2009) (+diabetes) | 61.42/57.96 | 42/49 | 27.48/25.52 | 40.7/49 | 135.59/123.6 | 85.13/76.2 | NA | NA |
| Kucukhuseyin (2009) (−diabetes) | 58.42/57.96 | 21/49 | 25.81/25.81 | 77.1/49 | 127.3/123.6 | 79.8/123.6 | NA | NA |
| Lu (2009) (−diabetes) | 64.8/56.3 | 74.16/49.57 | NA | 30.3/16.8 | 130/127 | 79/79 | NA | NA |
| Lu (2009) (+diabetes) | 66.5/62.8 | 68.90/36.09 | NA | 26.9/12.3 | 137/135 | 79/79 | NA | NA |
| Mahajan (2009) | 44.4/41.6 | 81/67.5 | 22.68/22.55 | 47/38 | 125.47/121.95 | 81.65/77.5 | 892.39/1611.9 | NA |
| Peng (2009) (+diabetes) | 64/63 | 63/58 | 25.41/25.65 | 41.6/25.1 | 136/135 | NA | NA | 270/290 |
| Peng (2009) (−diabetes) | 64/63 | 63/58 | 25.41/25.65 | 41.6/25.1 | 136/135 | NA | NA | 270/290 |
| Yan (2009) (+diabetes) | 66.2/62.3 | 70.19/45 | NA | 30.5/20 | 137/136 | 80/81 | 673.6/473.6 | 230/290 |
| Yan (2009) (−diabetes) | 64.1/58.6 | 72.48/53.03 | NA | 35.6/13.6 | 128/123 | 79/75 | 669.8/759.6 | 390/470 |
| Pu (2009) | 67.06/63.41 | 68.91/36.09 | NA | 26.89/12.25 | 137.19/135.22 | 79.16/79.25 | NA | 220/310 |
| Gao (2010) (−hypertension) | 60.8/61 | 74.3/42.4 | NA | NA | 120/118.3 | 72.7/71.7 | NA | NA |
| Gao (2010) (+hypertension) | 63.5/60.6 | 69/49.5 | NA | NA | 146.2/156 | 82.4/88.2 | NA | NA |
| McNair (2010) | NA | NA | NA | NA | NA | NA | 910.5/1302.5 | NA |
| McNair (2010) (+restenosis) | 61.5/60 | NA | 25/25 | NA | 148/125 | 74/78 | 610.6/1287 | NA |
| McNair (2010) (−restenosis) | 66.1/60 | NA | 29/25 | NA | 153/125 | 70/78 | 1143.8/1287 | NA |
| Xie (2010) | NA | NA | NA | NA | NA | NA | NA | NA |
| Hou (2011) | 57.7/58.2 | 78.15/77.34 | NA | 61.34/33.2 | NA | NA | NA | NA |
| Boiocchi (2011) | 59/62 | 83/64 | 26.1/25.6 | NA | NA | NA | NA | NA |
| Cai (2011) (CAD) | 65.5/61 | 58.3/53.8 | 25.4/25.5 | 25.1/17.1 | 132.5/130.2 | 78.9/78.6 | 691.53/652.55 | NA |
| Cai (2011) (MI) | 65.9/61 | 75.7/53.8 | 25/25.5 | 43.1/17.1 | 128.9/130.2 | 76.3/78.6 | 724.01/652.55 | NA |
| Park (2011) (+diabetes) | 64.2/62.2 | 50/50 | 23.8/24.9 | 44.4/27.8 | NA | NA | 610/450 | NA |
| Park (2011) (−diabetes) | 64.2/62.2 | 50/50 | 23.8/24.9 | 44.4/27.8 | NA | NA | 600/370 | NA |
| Peng (2011) | 68/64 | 63/53 | 25/25.7 | 24.9/25.9 | 139/137 | 78/79 | NA | 260/310 |
| Lu (2011) | 63.7/61.8 | 65.18/58.03 | 24.3/24.2 | 48.1/38.4 | NA | NA | NA | NA |
| Aydogan (2012) | 60.02/58.1 | NA | 25.92/25.52 | NA | 131.01/122.34 | 82.08/75.74 | NA | NA |
| Selejan (2012) | NA | NA | NA | NA | NA | NA | 122.15/125.68 | NA |
Abbreviations: NA, not available; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Figure 1. Flow diagram of search strategy and study selection.
Study Characteristics
Out of 39 qualified groups, 21 included East Asian, 14 included Caucasian, 4 included Middle Eastern, and 1 included African. Seven groups were reportedly matched in age and/or gender between patients and controls. There were 32 groups designed retrospectively and 7 groups prospectively. The patients of 10 groups were clinically diagnosed as MI.
Four polymorphisms of AGER gene were examined, including T-429C (rs1800625 in the promoter), T-374A (rs1800624 in the promoter), Gly82Ser (rs2070600 in exon 3) and G1704A (rs184003 in intron 7), and their results were extracted and synthesized in this meta-analysis. In detail, there were 10 (patients/controls: 1945/2013), 14 (2796/2209), 14 (2145/4966), and 3 (1075/1173) groups evaluating the association of these four polymorphisms with CAD. With regard to circulating sRAGE and esRAGE, there were respectively 13 (patients/controls: 1578/1275) and 8 (1752/1860) groups.
Overall Analyses of AGER Genetic Polymorphisms
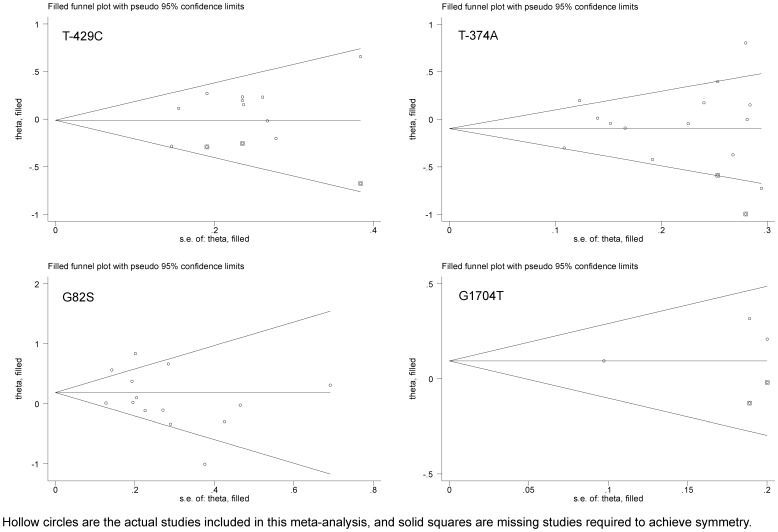
The fact that only three groups were available for G1704A precluded further subgroup analyses. Pooling all qualified groups detected no statistical significance for AGER gene four polymorphisms in association with CAD under allelic and dominant models (Figure 2 and Tables 3–5). Further restricting groups to CAD patients with diabetes mellitus found that deviations of risk estimates from the unity were stronger than the overall estimates, except for Gly82Ser under allelic model. For example, under dominant model, having -429C allele increased the odds of developing CAD in diabetic patients by 1.22-fold (95% CI: 0.99–1.51) compared with that of overall estimate of 1.15-fold (95% CI: 0.97–1.36). Contrastingly, the risk magnitude was alleviated for T-429C, T-374A, and Gly82Ser in CAD patients without diabetes mellitus. Moreover, in CAD patients without renal disease, deviations of risk estimates from the unity was enhanced, albeit non-significant, than the overall estimates, especially for Gly82Ser (allelic model: OR = 1.26; 95% CI: 0.92–1.74 and dominant model: OR = 1.28; 95% CI: 0.83–1.96). Significant heterogeneity was observed for T-374A (allelic model only) and Gly82Ser. There was high probability of publication bias for T-374A and Gly82Ser in CAD patients without diabetes mellitus. Further evidence of selective publication indicated that there were respectively three, two, and two missing groups required to make the funnel plot symmetrical for T-429C, T-374A and G1704A (Figure 3).
Figure 2. Overall estimates of AGER gene four polymorphisms examined for CAD under allelic model.
The summary odds ratio (OR) is shown by the middle of a solid diamond whose left and right extremes represent the corresponding 95% confidence interval (95% CI). Horizontal axis represents ORs, which were calculated against controls for each study.
Table 3. Overall and subgroup analyses of AGER gene T-429C polymorphism with the risk of developing CAD, and exploration of between-study heterogeneity and publication bias.
| Groups and subgroups | Studies (cases/controls), n (n/n) | Allele model | Dominant model | ||
| OR; 95% CI; P | I 2 (P χ2); P Egger | OR; 95% CI; P | I 2 (P χ2); P Egger | ||
| Total studies | 10 (1945/2013) | 1.09; 0.93–1.38; 0.301 | 23.7% (0.225); 0.118 | 1.15; 0.97–1.36; 0.111 | 18.0% (0.277); 0.097 |
| Total studies in DM | 6 (1004/1191) | 1.15; 0.96–1.38; 0.118 | 0.0% (0.515); 0.792 | 1.22; 0.99–1.51; 0.06 | 6.7% (0.373); 0.554 |
| Total studies in non-DM | 4 (941/822) | 1.04; 0.77–1.39; 0.815 | 49.3% (0.115); 0.013 | 1.07; 0.81–1.4; 0.644 | 30.8% (0.228); 0.026 |
| Total studies in non-RD | 5 (914/1094) | 1.1; 0.9–1.33; 0.349 | 0.0% (0.74); 0.61 | 1.17; 0.94–1.45; 0.159 | 0.0% (0.787); 0.553 |
| CAD endpoint | |||||
| CAD | 9 (1604/1672) | 1.17; 1.01–1.36; 0.073 | 0.0% (0.814); 0.642 | 1.24; 1.05–1.46; 0.012 | 0.0% (0.693); 0.411 |
| MI | 1 (341/341) | 0.75; 0.56–1.0; 0.05 | NA | 0.8; 0.58–1.11; 0.181 | NA |
| Descent of populations | |||||
| Caucasian | 3 (686/751) | 1.04; 0.71–1.52; 0.855 | 69.1% (0.039); 0.356 | 1.06; 0.75–1.5; 0.742 | 51.9% (0.125); 0.335 |
| East Asian | 6 (1184/1206) | 1.11; 0.93–1.33; 0.266 | 0.0% (0.841); 0.901 | 1.17; 0.96–1.42; 0.13 | 0.0% (0.886); 0.465 |
| African | 1 (75/56) | 1.93; 0.91–4.09; 0.088 | NA | 2.77; 1.18–6.53; 0.02 | NA |
| Study design | |||||
| Retrospective | 7 (1390/1315) | 1.23; 1.05–1.45; 0.011 | 0.0% (0.928); 0.046 | 1.29; 1.08–1.55; 0.013 | 0.0% (0.737); 0.035 |
| Prospective | 3 (555/698) | 0.82; 0.64–1.01; 0.052 | 0.0% (0.677); 0.403 | 0.85; 0.66–1.12; 0.232 | 0.0% (0.678); 0.531 |
| Age and/or gender | |||||
| Matched | 2 (330/369) | 1.26; 0.9–1.78; 0.184 | 0.0% (0.944); 1.0 | 1.33; 0.92–1.91; 0.13 | 0.0% (0.933); 1.0 |
| Unclear | 8 (1615/1644) | 1.06; 0.88–1.28; 0.538 | 27.2% (0.202); 0.18 | 1.12; 0.92–1.91; 0.275 | 30.6% (0.183); 0.184 |
| Total samples | |||||
| <300 subjects | 8 (1234/1304) | 1.19; 1.0–1.42; 0.045 | 71.6% (0.06); 0.83 | 0.97; 0.66–1.43; 0.884 | 62.9% (0.101); 0.425 |
| ≥300 subjects | 2 (711/709) | 0.91; 0.62–1.35; 0.654 | 0.0% (0.741); 0.779 | 1.25; 1.04–1.52; 0.021 | 0.0% (0.598); 1.0 |
Abbreviations: DM, diabetes mellitus; RD, renal disease; CAD, coronary artery disease; MI, myocardial infarction; OR, odds ratio; 95% CI: 95% confidence interval; NA, not available.
Table 5. Overall and subgroup analyses of AGER gene Gly82Ser and G1704A polymorphisms with the risk of developing CAD, and exploration of between-study heterogeneity and publication bias.
| Groups and subgroups | Studies (cases/controls), n (n/n) | Allele model | Dominant model | ||
| OR; 95% CI; P | I 2 (P χ2); P Egger | OR; 95% CI; P | I 2 (P χ2); P Egger | ||
| Gly82Ser polymorphism | |||||
| Total studies | 14 (2145/4966) | 1.12; 0.9–1.41; 0.316 | 69.0% (<0.001); 0.259 | 1.12; 0.82–1.52; 0.477 | 75.8% (<0.001); 0.707 |
| Total studies in DM | 5 (678/900) | 1.11; 0.88–1.39; 0.381 | 26.0% (0.248); 0.415 | 1.2; 0.77–1.87; 0.423 | 66.8% (0.017); 0.235 |
| Total studies in non–DM | 9 (1467/4066) | 1.08; 0.77–1.5; 0.673 | 76.6% (<0.001); 0.028 | 1.04; 0.68–1.59; 0.851 | 79.6% (<0.001); 0.023 |
| Total studies in non–RD | 5 (914/1097) | 1.26; 0.92–1.74; 0.148 | 73.8% (0.004); 0.562 | 1.28; 0.83–1.96; 0.262 | 80.1% (<0.001); 0.484 |
| CAD endpoint | |||||
| CAD | 12 (1739/3058) | 1.15; 0.9–1.47; 0.278 | 72.8% (<0.001); 0.348 | 1.15; 0.81–1.62; 0.435 | 79.1% (<0.001); 0.789 |
| MI | 2 (406/1908) | 0.91; 0.58–1.45; 0.702 | 0.0% (0.881); 1.0 | 0.91; 57–1.46; 0.696 | 0.0% (0.878); 1.0 |
| Descent of populations | |||||
| Caucasian | 4 (538/3408) | 0.9; 0.61–1.33; 0.601 | 0.0% (0.898); 0.48 | 0.9; 0.61–1.33; 0.592 | 0.0% (0.893); 0.473 |
| East Asian | 7 (1407/1396) | 1.29; 1.01–1.66; 0.046 | 72.5% (0.001); 0.968 | 1.32; 0.95–1.83; 0.099 | 75.6% (<0.001); 0.404 |
| Middle Eastern | 3 (200/162) | 0.81; 0.32–2.06; 0.662 | 85.5% (0.003); 0.425 | 0.83; 0.18–3.89; 0.807 | 91.3% (<0.001); 0.872 |
| Study design | |||||
| Retrospective | 8 (1393/1201) | 1.21; 0.88–1.67; 0.239 | 80.7% (<0.001); 0.496 | 1.24; 0.78–1.97; 0.372 | 85.7% (<0.001); 0.951 |
| Prospective | 6 (752/3765) | 0.96; 0.76–1.22; 0.75 | 0.0% (0.939); 0.911 | 0.93; 0.72–1.21; 0.605 | 0.0% (0.958); 0.81 |
| Age and/or gender | |||||
| Matched | 2 (330/370) | 1.82; 1.16–2.87; 0.01 | 63.9% (0.096); 1.0 | 2.08; 1.23–3.51; 0.006 | 63.4% (0.098); 1.0 |
| Unclear | 12 (1815/4596) | 1.02; 0.81–1.28; 0.864 | 61.8% (0.002); 0.225 | 0.99; 0.73–1.34; 0.926 | 68.5% (<0.001); 0.84 |
| Total samples | |||||
| <300 subjects | 12 (1434/4255) | 0.99; 0.79–1.24; 0.909 | 0.0% (0.693); 0.105 | 0.88; 0.68–1.15; 0.353 | 0.0% (0.972); 0.275 |
| ≥300 subjects | 2 (711/711) | 1.15; 0.88–1.51; 0.309 | 70.9% (<0.001); 1.0 | 1.17; 0.81–1.68; 0.414 | 77.0% (<0.001); 1.0 |
| G1704A polymorphism | |||||
| Total studies | 3 (1075/1173) | 1.16; 1.0–1.36; 0.057 | 0.0% (0.552); 0.28 | 1.1; 0.92–1.31; 0.307 | 0.0% (0.907); 0.41 |
Abbreviations: DM, diabetes mellitus; RD, renal disease; CAD, coronary artery disease; MI, myocardial infarction; OR, odds ratio; 95% CI: 95% confidence interval; NA, not available.
Figure 3. Trim-and-fill funnel plots for studies evaluating the effect of AGER gene four examined polymorphisms on CAD.
Hollow circles are the actual studies included in this meta-analysis, and solid squares are missing studies required to achieve symmetry.
Overall Analyses of Circulating RAGE Forms
Circulating sRAGE levels were lower in CAD patients than in controls with the difference being non-significant (Table 6). However, this reduction was totally and significantly reversed in CAD patients with diabetes mellitus (WMD: 185.71 pg/ml; 95% CI: 106.82 to 264.61 pg/ml), without evidence of heterogeneity or publication bias. Relative to controls, circulating esRAGE levels were consistently and significantly lower in CAD patients, as well as in subgroups with or without diabetes mellitus, and without renal disease. However, significant heterogeneity obsessed these comparisons, and the probability of publication bias was low.
Table 6. Overall and subgroup analyses of circulating sRAGE and esRAGE levels with CAD, and exploration of between-study heterogeneity and publication bias.
| sRAGE and esRAGE levels | Studies (cases/controls),n (n/n) | WMD; 95% CI; P | I 2 (P χ2); P Egger |
| sRAGE (pg/ml) | |||
| Total studies | 13 (1578/1275) | −123.12; −294.63 to 48.39; 0.159 | 99.7% (<0.001); 0.664 |
| Total studies in DM | 3 (232/161) | 185.71; 106.82 to 264.61; <0.001 | 0.0% (0.634); 1.0 |
| Total studies in non-DM | 11 (1400/1168) | −177.9; −363.33 to 7.54; 0.06 | 99.7% (<0.001); 0.861 |
| Total studies in non-RD | 9 (1250/1031) | −54.46; −216.05 to 107.13; 0.509 | 98.9% (<0.001); 0.966 |
| CAD endpoint | |||
| CAD | 6 (1002/864) | −136.54; −370.88 to 97.8; 0.253 | 98.2% (<0.001); 0.261 |
| MI | 7 (576/411) | −110.61; −362.23 to 141.01; 0.389 | 99.8% (<0.001); 0.863 |
| Descent of populations | |||
| Caucasian | 6 (493/467) | −257.72; −509.63 to −5.8; 0.045 | 99.9% (<0.001); 0.721 |
| East Asian | 7 (1085/808) | 15.26; −104.78 to 135.29; 0.803 | 91.8% (<0.001); 0.512 |
| Study design | |||
| Retrospective | 12 (1542/1245) | −100.25; −291.8 to 91.31; 0.305 | 99.7% (<0.001); 0.662 |
| Prospective | 1 (36/30) | −392.0; −471.43 to −366.57; <0.001 | NA |
| Age and/or gender | |||
| Matched | 4 (153/117) | 14.92; −381.82 to 411.66; 0.941 | 98.3% (<0.001); 0.079 |
| Unclear | 9 (1425/1158) | −182.65; −402.44 to 37.14; 0.103 | 99.8% (<0.001); 0.877 |
| Total samples | |||
| <300 subjects | 3 (929/830) | −86.94; −411.07 to 237.19; 0.599 | 99.3% (<0.001); 0.857 |
| ≥300 subjects | 10 (619/445) | −134.55; −347.88 to 78.78; 0.216 | 99.7% (<0.001); 0.136 |
| esRAGE (pg/ml) | |||
| Total studies | 9 (1752/1860) | −84.27; −133.94 to −34.61; 0.001 | 96.4% (<0.001); 0.675 |
| Total studies in DM | 8 (1603/1728) | −84.6; −140.64 to −28.56; 0.003 | 96.8% (<0.001); 0.711 |
| Total studies in non-DM | 1 (149/132) | −80.0; −109.38 to −50.62; <0.001 | NA |
| Total studies in non-RD | 6 (1095/1346) | −117.82; −222.58 to −13.05; 0.028 | 98.2% (<0.001); 0.681 |
| CAD endpoint | |||
| CAD | 8 (1698/1806) | −90.25; −142.34 to −38.17; 0.001 | 96.8% (<0.001); 0.764 |
| MI | 1 (54/54) | −20.0; −110.61 to 70.61; 0.665 | NA |
| Descent of populations | |||
| East Asian | 9 (1752/1860) | −84.27; −133.94 to −34.61; 0.001 | 96.4% (<0.001); 0.675 |
| Study design | |||
| Retrospective | 7 (1538/1503) | −52.12; −74.48 to −26.76; <0.001 | 79.1% (<0.001); 0.332 |
| Prospective | 2 (214/357) | −199.7; −317.3 to −82.1; 0.001 | 97.6% (<0.001); 1.0 |
| Age and/or gender | |||
| Matched | 1 (54/54) | −20.0; −110.61 to 70.61; 0.665 | NA |
| Unclear | 8 (1698/1806) | −90.25; −142.34 to −38.17; 0.001 | 96.8% (<0.001); 0.764 |
| Total samples | |||
| <300 subjects | 4 (1184/1237) | −45.62; −85.61 to −5.62; 0.025 | 88.4% (<0.001); 0.724 |
| ≥300 subjects | 5 (568/623) | −116.06; −192.92 to −39.12; 0.003 | 96.8% (<0.001); 0.366 |
Abbreviations: WMD, weighted mean difference; 95% CI, 95% confidence interval; DM, diabetes mellitus; RD, renal disease; CAD, coronary artery disease; MI, myocardial infarction; NA, not available.
Subgroup Analyses
As for T-429C, significance was reached in patients with CAD (compared with MI) under both models, in populations of African descent under dominant model, in studies under retrospective design and involving more than 300 subjects (Table 3). As for T-374A, significance was reached in populations of Middle Eastern (under allelic model) and African (under both models) descents, and in studies with age and/or gender-matched controls under allelic model (Table 4). As for Gly82Ser, significance was reached in populations of East Asian descent under both models, and in studies with age and/or gender-matched controls under dominant model (Table 5). As expected, heterogeneity was greatly improved in subgroups except for Gly82Ser in East Asians. Likewise, publication bias was also greatly improved in subgroups except for T-429C in retrospectively-designed studies.
Table 4. Overall and subgroup analyses of AGER gene T-374A polymorphism with the risk of developing CAD, and exploration of between-study heterogeneity and publication bias.
| Groups and subgroups | Studies (cases/controls), n (n/n) | Allele model | Dominant model | ||
| OR; 95% CI; P | I 2 (P χ2); P Egger | OR; 95% CI; P | I 2 (P χ2); P Egger | ||
| Total studies | 14 (2796/2209) | 0.97; 0.82–1.14; 0.713 | 61.7% (0.001); 0.567 | 0.97; 0.82–1.13; 0.658 | 31.7% (0.122); 0.876 |
| Total studies in DM | 7 (1058/1246) | 0.94; 0.74–1.21; 0.644 | 63.2% (0.012); 0.822 | 0.91; 0.72–1.17; 0.465 | 41.1% (0.117); 0.717 |
| Total studies in non-DM | 7 (1738/963) | 1.0; 0.78–1.27; 0.987 | 66.0% (0.007); 0.417 | 1.02; 0.82–1.26; 0.89 | 28.2% (0.213); 0.838 |
| Total studies in non-RD | 5 (886/899) | 0.87; 0.72–1.05; 0.137 | 11.3% (0.341); 0.57 | 0.91; 0.73–1.12; 0.363 | 0.0% (0.91); 0.577 |
| CAD endpoint | |||||
| CAD | 12 (1764/1634) | 0.98; 0.81–1.18; 0.801 | 55.2% (0.011); 0.71 | 0.96; 0.81–1.13; 0.595 | 15.3% (0.295); 0.972 |
| MI | 2 (1032/575) | 0.95; 0.58–1.54; 0.821 | 89.1% (0.002); 1.0 | 0.99; 0.58–1.69; 0.976 | 83.2% (0.015); 1.0 |
| Descent of populations | |||||
| Caucasian | 5 (1552/1069) | 0.89; 0.71–1.1; 0.279 | 67.1% (0.016); 0.817 | 0.95; 0.75–1.2; 0.662 | 46.2% (0.115); 0.516 |
| East Asian | 5 (976/926) | 0.98; 0.81–1.17; 0.806 | 0.0% (0.649); 0.627 | 0.99; 0.8–1.22; 0.893 | 0.0% (0.872); 0.86 |
| Middle Eastern | 3 (193/158) | 1.57; 1.09–2.24; 0.014 | 26.8% (0.255); 0.969 | 1.37; 0.88–2.13; 0.168 | 0.0% (0.659); 0.596 |
| African | 1 (75/56) | 0.48; 0.27–0.86; 0.013 | NA | 0.36; 0.18–0.74; 0.006 | NA |
| Study design | |||||
| Retrospective | 11 (2241/1511) | 0.97; 0.8–1.18; 0.751 | 64.3% (0.002); 0.198 | 0.93; 0.77–1.11; 0.425 | 31.3% (0.149); 0.573 |
| Prospective | 3 (555/698) | 0.99; 0.72–1.37; 0.956 | 50.9% (0.131); 0.176 | 1.12; 0.86–1.46; 0.408 | 11.1% (0.325); 0.095 |
| Age and/or gender | |||||
| Matched | 1 (691/234) | 0.74; 0.6–0.92; 0.005 | NA | 0.76; 0.55–1.03; 0.079 | NA |
| Unclear | 13 (2105/1975) | 1.01; 0.85–1.19; 0.697 | 56.1% (0.007); 0.805 | 1.0; 0.85–1.18; 0.991 | 25.6% (0.185); 0.493 |
| Total samples | |||||
| <300 subjects | 11 (1394/1264) | 0.97; 0.79–1.2; 0.808 | 59.0% (0.007); 0.579 | 0.96; 0.79–1.17; 0.659 | 23.0% (0.225); 0.992 |
| ≥300 subjects | 3 (1402/945) | 0.96; 0.71–1.31; 0.811 | 79.0% (0.008); 0.552 | 0.98; 0.72–1.33; 0.892 | 66.8% (0.049); 0.331 |
Abbreviations: DM, diabetes mellitus; RD, renal disease; CAD, coronary artery disease; MI, myocardial infarction; OR, odds ratio; 95% CI: 95% confidence interval; NA, not available.
Circulating sRAGE levels were significantly lower in CAD patients of Caucasian descent and in prospectively-designed studies than controls (Table 6). With regard to circulating esRAGE, significant lower levels were observed in studies under retrospective or prospective design, and in small or large studies than controls, especially for the prospective and large studies (Table 6).
Cumulative and Met-regression Analyses
Regarding four examined polymorphisms of AGER and its circulating forms, cumulative risk estimates tended to be stable with accumulating data over time under both models (data not shown).
To explore the extent to which study-level variables explain heterogeneity among individual estimates, a set of meta-regression analyses were undertaken. Unfortunately, none of the confounders including age, gender, body mass index, smoking, systolic and diastolic blood pressure could explain large part of heterogeneity for all examined polymorphisms and circulating sRAGE and esRAGE levels (data not shown). Because meta-regression analyses involved studies of limited sample sizes, it might be underpowered to detect a small or moderate effect.
Discussion
On the basis of 27 studies involving 7585 CAD patients and 9240 controls, we evaluated the association of AGER genetic polymorphisms and circulating sRAGE and esRAGE levels with the risk of developing CAD. The two noteworthy findings of this study were that (1) despite the overall null association, there was a contributory role of common variants in AGER gene to CAD in patients with diabetes mellitus or renal disease; (2) circulating esRAGE might be a powerful negative predictor for the development of CAD. Moreover, our findings demonstrated that the existence of diversity of ethnicity, study design, case-control matched information and sample size across studies might result to the presence of heterogeneity.
More recently, Wang and colleagues have synthesized data from 17 studies on AGER three genetic polymorphisms (T-429C, T-374A, Gly82Ser) and the risk of CAD, and they failed to observe any suggestive association [6], consistent with the pooled results of this meta-analysis. Extending beyond overall comparisons, we noticed that risk effects of AGER genetic variants on CAD were strikingly potentiated in patients with diabetes mellitus or renal disease. As indicated by clinical investigations, over-expression of AGER gene can enhance inflammatory reaction and matrix metalloprotease expression in plaque macrophages of diabetic patients [35]. Moreover, AGER expression was found to be closely associated with the worsening of chronic kidney disease [36]. Furthermore, circulating esRAGE levels were remarkably lower in type 2 diabetic subjects without chronic kidney disease than in nondiabetic controls, but gradually increased in accordance with progression of chronic kidney disease [37]. On the basis of previous work and the findings of this study, it is reasonable to hypothesize that diabetes mellitus and/or renal disease might precipitate the occurrence of CAD via the inheritance of genetic defects leading to the transcriptional activation of AGER.
To shed some light on this hypothesis, we further evaluated the relation between circulating RAGE forms, which can serve as RAGE blockers and might be applicable to human diseases, and CAD, and it is worth noting that circulating esRAGE might be a powerful negative predictor of CAD, even with the presence of diabetes mellitus or renal disease. Specifically, esRAGE is an alternative splicing product of AGER mRNA, constituting approximately 20% of sRAGE levels in humans [38]. There is growing evidence that circulating esRAGE was decreased in both types 1 and 2 diabetic patients, and its low levels were associated with the severity of cardiac dysfunction in patients with heart failure [2], [32], [39], [40], a finding which is also mirrored in this meta-analysis. In fact, therapies targeting RAGE have been undertaken in experimental models and are proven to be effective in reducing atherosclerosis in diabetic mice [41], [42]. Also, the soundness of our results on esRAGE was bolstered by further restricting subgroup analyses to the prospectively-designed and large studies, which are deemed to be less prone to selective publication and change results. This study, to our knowledge, represents the first meta-analysis to date evaluating the association of circulating sRAGE and esRAGE with CAD.
Heterogeneity in a meta-analysis is mostly produced by differences in study-level characteristics. Our subgroup analyses indicated that ethnicity, study design, matched information and sample size might be potential sources of heterogeneity. For example, risk estimate of 82Ser allele on CAD was 1.29 in East Asians, but was 0.9 in Caucasians. A possible explanation may be due to divergent genetic backgrounds or linkage patterns, and usually a variant is in close linkage with another nearby causal variant in one ethnic group but not in another [43], [44]. As a consequence, there is a need to construct a database of CAD-susceptibility genes or variants in each racial/ethnic group.
Meta-analysis is a powerful tool to summarize results of individual studies; however, it is important to recognize certain limitations. First, most qualified studies were retrospective in design, precluding further comments on causality. Second, albeit low probability of publication bias in this meta-analysis, potential selection bias cannot be ruled out, because we only retrieved articles published in English or Chinese. Third, heterogeneity persisted in some subgroups, limiting the interpretation of our pooled estimates. Fourth, as most studies in this meta-analysis enrolled subjects aged more than 50 years, large studies in a younger population of CAD patients are of special interest, because genetic factors may exert great contribution to those in whom CAD develops at a younger age and in the absence of strong environmental risk factors [45]. Fifth, we selected only four polymorphisms from AGER gene, and did not cover other CAD-susceptibility genes, such as angiotensin II receptor, type 1 [44] and matrix metalloproteinase family genes [46]. Therefore, the jury must refrain from drawing a final conclusion until large, well-designed, prospective studies confirm or refute our findings.
Taken together, our findings collectively demonstrated that association of AGER genetic polymorphisms with CAD was potentiated in patients with diabetes mellitus or renal disease. From a practical standpoint, circulating esRAGE might be a powerful negative predictor for the development of CAD. Nevertheless, we hope that this study will establish background data for further investigations into the mechanisms of AGER gene and relevant pathway genes in the development of CAD.
Supporting Information
Checklist of items to include when reporting a systematic review or meta-analysis.
(DOC)
Funding Statement
This work was supported by the Shanghai Rising Star Program (11QA1405500) and the National Natural Science Foundation of China (30900808). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Niu W, Liu Y, Qi Y, Wu Z, Zhu D, et al. (2012) Association of interleukin-6 circulating levels with coronary artery disease: a meta-analysis implementing mendelian randomization approach. Int J Cardiol 157: 243–252. [DOI] [PubMed] [Google Scholar]
- 2. Hegab Z, Gibbons S, Neyses L, Mamas MA (2012) Role of advanced glycation end products in cardiovascular disease. World J Cardiol 4: 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Falcone C, Emanuele E, D’Angelo A, Buzzi MP, Belvito C, et al. (2005) Plasma levels of soluble receptor for advanced glycation end products and coronary artery disease in nondiabetic men. Arterioscler Thromb Vasc Biol 25: 1032–1037. [DOI] [PubMed] [Google Scholar]
- 4. Koyama H, Yamamoto H, Nishizawa Y (2007) RAGE and soluble RAGE: potential therapeutic targets for cardiovascular diseases. Mol Med 13: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park HJ, Baek JY, Shin WS, Kim DB, Jang SW, et al. (2011) Soluble receptor of advanced glycated endproducts is associated with plaque vulnerability in patients with acute myocardial infarction. Circ J 75: 1685–1690. [DOI] [PubMed] [Google Scholar]
- 6. Wang J, Zou L, Song Z, Lang X, Huang S, et al. (2012) Meta-analysis of RAGE gene polymorphism and coronary heart disease risk. PLoS One 7: e50790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hudson BI, Stickland MH, Futers TS, Grant PJ (2001) Effects of novel polymorphisms in the RAGE gene on transcriptional regulation and their association with diabetic retinopathy. Diabetes 50: 1505–1511. [DOI] [PubMed] [Google Scholar]
- 8. Peng WH, Lu L, Wang LJ, Yan XX, Chen QJ, et al. (2009) RAGE gene polymorphisms are associated with circulating levels of endogenous secretory RAGE but not with coronary artery disease in Chinese patients with type 2 diabetes mellitus. Arch Med Res 40: 393–398. [DOI] [PubMed] [Google Scholar]
- 9.Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151: 264–269, W264. [DOI] [PubMed]
- 10. Bowden J, Tierney JF, Copas AJ, Burdett S (2011) Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med Res Methodol 11: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falcone C, Campo I, Emanuele E, Buzzi MP, Zorzetto M, et al. (2004) Relationship between the -374T/A RAGE gene polymorphism and angiographic coronary artery disease. Int J Mol Med 14: 1061–1064. [PubMed] [Google Scholar]
- 12. Kirbis J, Milutinovic A, Steblovnik K, Teran N, Terzic R, et al. (2004) The -429 T/C and -374 T/A gene polymorphisms of the receptor of advanced glycation end products gene (RAGE) are not risk factors for coronary artery disease in Slovene population with type 2 diabetes. Coll Antropol 28: 611–616. [PubMed] [Google Scholar]
- 13. dos Santos KG, Canani LH, Gross JL, Tschiedel B, Pires Souto KE, et al. (2005) The -374A allele of the receptor for advanced glycation end products gene is associated with a decreased risk of ischemic heart disease in African-Brazilians with type 2 diabetes. Mol Genet Metab 85: 149–156. [DOI] [PubMed] [Google Scholar]
- 14. Hofmann MA, Yang Q, Harja E, Kedia P, Gregersen PK, et al. (2005) The RAGE Gly82Ser polymorphism is not associated with cardiovascular disease in the Framingham offspring study. Atherosclerosis 182: 301–305. [DOI] [PubMed] [Google Scholar]
- 15. Zee RY, Romero JR, Gould JL, Ricupero DA, Ridker PM (2006) Polymorphisms in the advanced glycosylation end product-specific receptor gene and risk of incident myocardial infarction or ischemic stroke. Stroke 37: 1686–1690. [DOI] [PubMed] [Google Scholar]
- 16. Yoon SJ, Park S, Shim CY, Park CM, Ko YG, et al. (2007) Association of RAGE gene polymorphisms with coronary artery disease in the Korean population. Coron Artery Dis 18: 1–8. [DOI] [PubMed] [Google Scholar]
- 17. Lu L, Jin Pu L, Chen QJ, Wang L, Peng W, et al. (2008) Increased glycated albumin and decreased esRAGE concentrations are associated with in-stent restenosis in Chinese diabetic patients. Clin Chim Acta 396: 33–37. [DOI] [PubMed] [Google Scholar]
- 18. Mulder DJ, van Haelst PL, Gross S, de Leeuw K, Bijzet J, et al. (2008) Skin autofluorescence is elevated in patients with stable coronary artery disease and is associated with serum levels of neopterin and the soluble receptor for advanced glycation end products. Atherosclerosis 197: 217–223. [DOI] [PubMed] [Google Scholar]
- 19. Kucukhuseyin O, Aydogan HY, Isbir CS, Isbir T (2009) Associations of -374T/A polymorphism of receptor for advanced glycation end products (RAGE) gene in Turkish diabetic and non-diabetic patients with coronary artery disease. In Vivo 23: 949–954. [PubMed] [Google Scholar]
- 20. Lu L, Pu LJ, Zhang Q, Wang LJ, Kang S, et al. (2009) Increased glycated albumin and decreased esRAGE levels are related to angiographic severity and extent of coronary artery disease in patients with type 2 diabetes. Atherosclerosis 206: 540–545. [DOI] [PubMed] [Google Scholar]
- 21. Mahajan N, Malik N, Bahl A, Dhawan V (2009) Receptor for advanced glycation end products (RAGE) and its inflammatory ligand EN-RAGE in non-diabetic subjects with pre-mature coronary artery disease. Atherosclerosis 207: 597–602. [DOI] [PubMed] [Google Scholar]
- 22. Pu L, Lu L, Shen W, Zhang Q, Fang D, et al. (2009) Relation of serum levels of endogeneous secretory receptor for advanced glycation end products and glycation albumin to coronary artery disease in type 2 diabetes mellitus. Chinese Journal of Microcriculation 19(4): 24–26. [Google Scholar]
- 23. Yan XX, Lu L, Peng WH, Wang LJ, Zhang Q, et al. (2009) Increased serum HMGB1 level is associated with coronary artery disease in nondiabetic and type 2 diabetic patients. Atherosclerosis 205: 544–548. [DOI] [PubMed] [Google Scholar]
- 24. Gao J, Shao Y, Lai W, Ren H, Xu D (2010) Association of polymorphisms in the RAGE gene with serum CRP levels and coronary artery disease in the Chinese Han population. J Hum Genet 55: 668–675. [DOI] [PubMed] [Google Scholar]
- 25. McNair ED, Wells CR, Mabood Qureshi A, Basran R, Pearce C, et al. (2010) Soluble receptors for advanced glycation end products (sRAGE) as a predictor of restenosis following percutaneous coronary intervention. Clin Cardiol 33: 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McNair ED, Wells CR, Mabood Qureshi A, Basran R, Pearce C, et al. (2010) Modulation of high sensitivity C-reactive protein by soluble receptor for advanced glycation end products. Mol Cell Biochem 341: 135–138. [DOI] [PubMed] [Google Scholar]
- 27. Xie G, Chen Y, Xiao H, Han C, Chen J (2010) Association of -374T/A polymorphism of RAGE gene with coronary heart disease in Chinese Han population. South China Journal of Cardiovascular Diseases 16: 468–470. [Google Scholar]
- 28. Boiocchi C, Bozzini S, Buzzi MP, Schirinzi S, Zorzetto M, et al. (2011) Age of onset of myocardial infarction: is promoter polymorphism of the RAGE gene implicated? Rejuvenation Res 14: 67–73. [DOI] [PubMed] [Google Scholar]
- 29. Cai XY, Lu L, Wang YN, Jin C, Zhang RY, et al. (2011) Association of increased S100B, S100A6 and S100P in serum levels with acute coronary syndrome and also with the severity of myocardial infarction in cardiac tissue of rat models with ischemia-reperfusion injury. Atherosclerosis 217: 536–542. [DOI] [PubMed] [Google Scholar]
- 30. Hou X, Jin J, Han S, Song Y, He X (2011) The study of the relationship between single-nucleotide polymorphism of receptor for advanced glycation end products and coronary angiography in a Positive. Prevention and Treatment of Cardio-Carebral-Vascular Disease 11: 272–274. [Google Scholar]
- 31. Lu W, Xie G, Feng B, Liu F (2011) Association of RAGE gene polymorphisms with the presence and the severity of coronary artery disease in non-diabetic Han populations. Journal of Soochow University Medical Science Edition 31: 454–458. [Google Scholar]
- 32. Peng WH, Jian WX, Li HL, Hou L, Wei YD, et al. (2011) Increased serum myeloid-related protein 8/14 level is associated with atherosclerosis in type 2 diabetic patients. Cardiovasc Diabetol 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aydogan HY, Kucukhuseyin O, Tekeli A, Isbir T (2012) Associations of receptor for advanced glycation end products -374 T/A and Gly82 Ser and peroxisome proliferator-activated receptor gamma Pro12Ala polymorphisms in Turkish coronary artery disease patients. Genet Test Mol Biomarkers 16: 134–137. [DOI] [PubMed] [Google Scholar]
- 34. Selejan SR, Poss J, Hewera L, Kazakov A, Bohm M, et al. (2012) Role of receptor for advanced glycation end products in cardiogenic shock. Crit Care Med 40: 1513–1522. [DOI] [PubMed] [Google Scholar]
- 35. Yamagishi S, Nakamura K, Matsui T, Ueda S, Fukami K, et al. (2008) Agents that block advanced glycation end product (AGE)-RAGE (receptor for AGEs)-oxidative stress system: a novel therapeutic strategy for diabetic vascular complications. Expert Opin Investig Drugs 17: 983–996. [DOI] [PubMed] [Google Scholar]
- 36. Hou FF, Ren H, Owen WF Jr, Guo ZJ, Chen PY, et al. (2004) Enhanced expression of receptor for advanced glycation end products in chronic kidney disease. J Am Soc Nephrol 15: 1889–1896. [DOI] [PubMed] [Google Scholar]
- 37. Kalousova M, Hodkova M, Kazderova M, Fialova J, Tesar V, et al. (2006) Soluble receptor for advanced glycation end products in patients with decreased renal function. Am J Kidney Dis 47: 406–411. [DOI] [PubMed] [Google Scholar]
- 38. Nakamura K, Yamagishi S, Adachi H, Matsui T, Kurita-Nakamura Y, et al. (2008) Serum levels of soluble form of receptor for advanced glycation end products (sRAGE) are positively associated with circulating AGEs and soluble form of VCAM-1 in patients with type 2 diabetes. Microvasc Res 76: 52–56. [DOI] [PubMed] [Google Scholar]
- 39. Chen G, Wu Y, Wang T, Liang J, Lin W, et al. (2012) Association between serum endogenous secretory receptor for advanced glycation end products and risk of type 2 diabetes mellitus with combined depression in the Chinese population. Diabetes Technol Ther 14: 936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katakami N, Matsuhisa M, Kaneto H, Matsuoka TA, Sakamoto K, et al. (2009) Serum endogenous secretory RAGE level is an independent risk factor for the progression of carotid atherosclerosis in type 1 diabetes. Atherosclerosis 204: 288–292. [DOI] [PubMed] [Google Scholar]
- 41. Bucciarelli LG, Wendt T, Qu W, Lu Y, Lalla E, et al. (2002) RAGE blockade stabilizes established atherosclerosis in diabetic apolipoprotein E-null mice. Circulation 106: 2827–2835. [DOI] [PubMed] [Google Scholar]
- 42. Yan SF, D’Agati V, Schmidt AM, Ramasamy R (2007) Receptor for Advanced Glycation Endproducts (RAGE): a formidable force in the pathogenesis of the cardiovascular complications of diabetes & aging. Curr Mol Med 7: 699–710. [PubMed] [Google Scholar]
- 43. Niu W, Qi Y, Wu Z, Liu Y, Zhu D, et al. (2012) A meta-analysis of receptor for advanced glycation end products gene: four well-evaluated polymorphisms with diabetes mellitus. Mol Cell Endocrinol 358: 9–17. [DOI] [PubMed] [Google Scholar]
- 44. Li Y, Li X, Jia N, Guo S, Chu S, et al. (2013) Meta-analysis of the association between angiotensin II receptor, type 1 gene A1166C polymorphism and coronary artery disease in Chinese populations. J Renin Angiotensin Aldosterone Syst 14: 82–90. [DOI] [PubMed] [Google Scholar]
- 45. Zintzaras E, Raman G, Kitsios G, Lau J (2008) Angiotensin-converting enzyme insertion/deletion gene polymorphic variant as a marker of coronary artery disease: a meta-analysis. Arch Intern Med 168: 1077–1089. [DOI] [PubMed] [Google Scholar]
- 46. Niu W, Qi Y (2012) Matrix metalloproteinase family gene polymorphisms and risk for coronary artery disease: systematic review and meta-analysis. Heart 98: 1483–1491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Checklist of items to include when reporting a systematic review or meta-analysis.
(DOC)