Abstract
Background
A large number of insect chemosensory genes from different gene subfamilies have been identified and annotated, but their functional diversity and complexity are largely unknown. A systemic examination of expression patterns in chemosensory organs could provide important information.
Methodology/Principal Findings
We identified 92 putative chemosensory genes by analysing the transcriptome of the antennae and female sex pheromone gland of the purple stem borer Sesamia inferens, among them 87 are novel in this species, including 24 transcripts encoding for odorant binding proteins (OBPs), 24 for chemosensory proteins (CSPs), 2 for sensory neuron membrane proteins (SNMPs), 39 for odorant receptors (ORs) and 3 for ionotropic receptors (IRs). The transcriptome analyses were validated and quantified with a detailed global expression profiling by Reverse Transcription-PCR for all 92 transcripts and by Quantitative Real Time RT-PCR for selected 16 ones. Among the chemosensory gene subfamilies, CSP transcripts are most widely and evenly expressed in different tissues and stages, OBP transcripts showed a clear antenna bias and most of OR transcripts are only detected in adult antennae. Our results also revealed that some OR transcripts, such as the transcripts of SNMP2 and 2 IRs were expressed in non-chemosensory tissues, and some CSP transcripts were antenna-biased expression. Furthermore, no chemosensory transcript is specific to female sex pheromone gland and very few are found in the heads.
Conclusion
Our study revealed that there are a large number of chemosensory genes expressed in S. inferens, and some of them displayed unusual expression profile in non-chemosensory tissues. The identification of a large set of putative chemosensory genes of each subfamily from a single insect species, together with their different expression profiles provide further information in understanding the functions of these chemosensory genes in S. inferens as well as other insects.
Introduction
Olfaction plays an important role in various crucial behaviors of insects, such as locating food resources, plant and animal hosts and finding sexual partners. The periphery process of insect olfaction is generally thought to involve two main steps. Firstly, external chemical volatiles enter into the chemosensilla of insect antennae or other sensory tissues, and then are captured by odorant binding proteins (OBPs) [1], [2], [3] or chemosensory proteins (CSPs) [4], [5] which are highly abundant in the lymph of chemosensilla. Secondly, the OBP or CSP bound chemical volatiles are transported to the olfactory receptor proteins (ORs) [6], [7], [8] located on dendrite membranes, triggering the transduction of chemical signals to electric signals. In addition, some other chemosensory proteins have also been proposed to play a role in insect olfaction. Two important ones are sensory neuron membrane proteins (SNMPs) [9], [10] and ionotropic receptors (IRs) [1], [11], [12].
Identification and expression profiling of chemosensory genes are of primary importance for exploring their functions and the mechanisms of insect olfaction. In the early studies, the main method used to identify insect chemosensory genes was direct cloning [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], which normally involves designing degenerate primers, amplifying the fragment and obtaining the full length gene sequences by Rapid Amplification of cDNA Ends (RACE). This method is very time-consuming and inefficient, identifying only one gene each time. Later, the genome sequencing and annotation projects have allowed to find large-scale new chemosensory genes in B. mori [24], [25] and several other insect species [25], [26], [27], including first identification of insect ORs from Drosophila melanogaster [28]. Recently, with development of the next generation sequencing techniques, large scale chemosensory genes have also been identified from insects whose genomes have not been sequenced, as reported in Spodoptera littoralis [29], [30], Manduca sexta [31], Cydia pomonella [32] and Helicoverpa armigera [33].
Although great numbers of chemosensory genes have been molecularly identified from insects of almost all insect orders, their exact functions are mostly unknown, as these genes were identified mainly based on the sequence similarity to reported genes. The expression profiles, particularly the tissue distribution, could provide important information on the functions of the chemosensory genes [24], [34], [35], [36], [37], [38], [39], [40], [41], [42].
The purple stem borer (also called pink stem borer), Sesamia inferens (Lepidoptera: Noctuidae) is a polyphagous insect pest found in many Asian countries [43]. It damages a variety of crops including rice, corn, sugarcane, and has become one of the major rice pests in China since 1990s [44], [45]. In this study, we conducted a transcriptome analysis of adult antennae and female sex pheromone glands of S. inferens, and identified 92 putative chemosensory transcripts comprising of 24 OBPs, 24 CSPs, 2 SNMPs, 39 ORs and 3 IRs. We further conducted a comprehensive examination on the expression profile of these transcripts regarding to different tissues and life stages by Reverse Transcription-PCR (RT-PCR) for all transcripts and by Quantitative Real Time RT-PCR (qRT-PCR) for selected 16 genes. The results clearly depicted different expression profiles among different chemosensory genes families between chemosensory and non-chemosensory tissues, as well as between adults and larvae developmental stages.
Results
Transcriptome Sequencing and Sequence Assembly
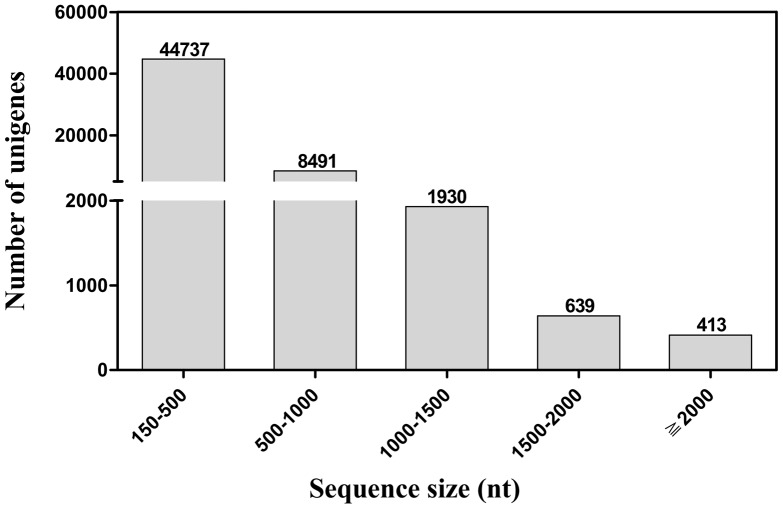
We carried out a next generation sequencing project on a cDNA library constructed from the mixture sample of antennae and female sex pheromone glands of S. inferens using Illuminna HiSeq™ 2000 platform. The transcriptome sequencing provided about 54 million reads (4.86 Gb), which were assembled into 175,059 contigs (≥75 bp) with a mean length of 195 bp and the N50 length of 234 bp. These contigs were further assembled into 126,081 scaffolds with a mean of 243 bp and the N50 length of 308 bp. After clustering and redundancy filtering, we finally acquired 56,210 unigenes (≥150 bp) with a mean length of 394 bp and the N50 length of 460 bp. We called these 56,210 ones unigenses according to some recently published papers [33], [46], although each of them may not necessarily represents a unique gene. Of the 56,210 unigenes, those with a sequence length more than 500 bp accounted for 20.41% of the transcriptome assembly (Figure 1). All the unigenes were referred to as transcripts here after and given a unique unigene id.
Figure 1. Distribution of unigene size in the S. inferens transcriptome assembly.
Homology Analysis and Gene Ontology (GO) Annotation
Among 56,210 transcripts, 21,796 were matched by the Blastx homology search to the entries in NCBI non-redundant (nr) protein database with a cut-off E-value of 10−5. The highest match percentage (16.20%) is to Tribolium castaneum sequences followed by the sequences of Bombyx mori (13.21%), Camponotus floridanus (5.96%), Harpegnathos saltator (5.88%) and Anopheles gambiae str. PEST (5.41%) (Figure 2).
Figure 2. Percentage of homologous hits of the S. inferens transcripts to other insect species.
The S. inferens transcripts were searched by BLASTx against the non-redundancy protein database with a cutoff E-value 10−5. Species which have more than 1% matching hits to the S. inferens transcripts are shown.
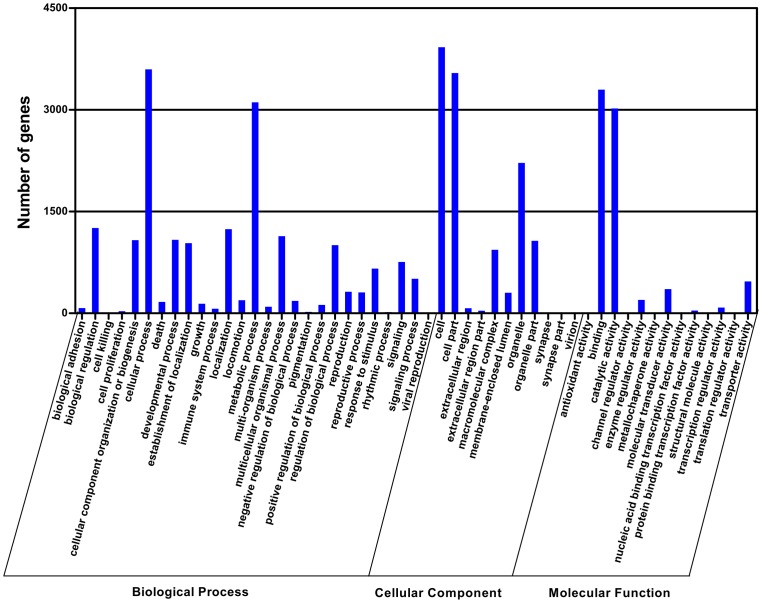
The Gene Ontology (GO) annotation was used to classify the transcripts into the functional groups according to the GO category. Of 56,210 transcripts, 7,195 ones (12.8%) could be annotated based on sequence homology. As one transcript could align to more than one biological processes, 7,195 transcripts resulted in 18,224 alignments in biological process category, 12119 in cellular component category and 7,509 in molecular function category. In these categories, there were a high percentage of transcripts in the subcategories such as cellular process (49.99%), metabolic process (43.25%), cell (54.54%), cell part (49.25%), binding (45.83%) and catalytic activity (41.97%) (Figure 3). In addition, some chemosensory transcripts were highly abundant in the transcriptome dataset, with 14 of 20 most abundant transcripts encoding for OBPs and CSPs (Figure 4).
Figure 3. Gene ontology (GO) classification of the S. inferens transcripts with Blast2GO program.
Figure 4. Top 50 most abundant transcripts in the S. inferens transcriptome dataset.
Odorant binding proteins (PBPs, GOBPs, OBPs and ABP) are indicated by red, chemosensory proteins (CSPs and SAPs) are indicated by green, and the other genes are indicated by blue. The genes expression abundance is indicated as the Reads Per Kilobase per Million mapped reads (RPKM) values. The transcript annotation by homologous comparisons with Blastx is indicated in Table 1 for chemosensory transcripts and Table S1 for the non-chemosensory transcripts.
Identification of Putative Chemosensory Genes
By homology analysis, we identified a total of 92 transcripts that belong to gene families putatively involved in insect chemoperception, including OBPs (24 transcripts), CSPs (24 transcripts), SNMPs (2 transcripts), ORs (39 transcripts) and IRs (3 transcripts) (Table 1 and Table 2). Of the 92 transcripts, 5 transcripts were the same as sequences deposited in the GenBank: 3 PBPs (GenBank accession number: JF927621.1, JN984058.1, JF927622.1), one GOBP (EU825760.1) and one OR (EU825763.1), while the other 87 transcripts found in the current study were new in S. inferens. Compared with insects in which the putative chemosensory genes had been identified by analyzing either genome or transcriptome, the number of the putative chemosensory genes identified by the current study in S. inferens (total: 92; OBP : CSP: SNMP : OR: IR = 24∶ 24 : 2∶ 39 : 3) was similar to the numbers found in M. sexta (total: 94; OBP : CSP : SNMP: OR : IR = 18∶ 21 : 2∶ 47 : 6) and H. armigera (total: 99; OBP : CSP : SNMP : OR : IR = 26∶ 12 : 2∶ 47 : 12), but less than that of S. littoralis (total: 127; OBP : CSP : SNMP : OR : IR = 36∶ 21 : 2∶ 47 : 17) and B. mori (total: 147; OBP: CSP: SNMP: OR: IR = 44∶18: 2∶72: 11) (Figure 5).
Table 1. The Blastx match of S. inferens putative OBPs, CSPs and SNMPs genes.
| Gene Name | Gene ID | Acc. number | ORF Length (bp) | Complete ORF | Signal Peptide | Best Blastx Match | ||||
| Name | Acc. number | Species | E value | Identity (%) | ||||||
| Pheromone Binding Protein (PBP) | ||||||||||
| PBP1 | 2820 | AEQ30019.1 | 312 | NO | NO | pheromone binding protein 1 | AEQ30019.1 | [Sesamia inferens] | 7.00E-71 | 99 |
| PBP2 | 5089 | AEX58642.1 | 177 | NO | NO | pheromone binding protein 2 | AEX58642.1 | [Sesamia inferens] | 3.00E-33 | 98 |
| PBP3 | 620 | AEQ30020.1 | 261 | NO | NO | pheromone binding protein 3 | AEQ30020.1 | [Sesamia inferens] | 4.00E-57 | 100 |
| General odorant binding protein (G0BP) | ||||||||||
| GOBP1 | 5080 | KC887506 | 495 | Yes | Yes | general odorant binding protein 1 | ABI24159.1 | [Agrotis segetum] | 3.00E-88 | 92 |
| GOBP2 | 586 | ACJ07121.1 | 414 | NO | NO | general odorant binding protein 2 | ACJ07121.1 | [Sesamia inferens] | 8.00E-96 | 100 |
| Odorant Binding Protein (OBP) | ||||||||||
| OBP1 | 52675 | KC887507 | 321 | Yes | NO | odorant-binding protein 3 precursor | NP_001140187.1 | [Bombyx mori] | 2.00E-56 | 65 |
| OBP2 | 38179 | KC887508 | 252 | NO | NO | odorant binding protein | ACX53743.1 | [Heliothis virescens] | 3.00E-43 | 82 |
| OBP3 | 46919 | KC887509 | 345 | NO | NO | odorant-binding protein | AEX07274.1 | [Helicoverpa assulta] | 2.00E-72 | 89 |
| OBP4 | 34399 | KC887510 | 237 | NO | NO | odorant binding protein | ACX53761.1 | [Heliothis virescens] | 4.00E-15 | 48 |
| OBP5 | 2896 | KC887511 | 429 | NO | Yes | OBP2 | B54586.1 | [Helicoverpa armigera] | 2.00E-83 | 85 |
| OBP6 | 50001 | KC887512 | 390 | Yes | Yes | odorant binding protein LOC100301496 precursor | NP_001153664.1 | [Bombyx mori] | 8.00E-36 | 41 |
| OBP7 | 39062 | KC887513 | 237 | NO | NO | odorant binding protein | EHJ67765.1 | [Danaus plexippus] | 3.00E-32 | 68 |
| OBP8 | 4911 | KC887514 | 303 | NO | NO | odorant-binding protein 4 | NP_001140188.1 | [Bombyx mori] | 1.00E-39 | 49 |
| OBP9 | 43124 | KC887515 | 300 | NO | NO | odorant-binding protein 5 precursor | NP_001140189.1 | [Bombyx mori] | 1.00E-36 | 60 |
| OBP10 | 5115 | KC887516 | 402 | NO | NO | odorant binding protein | ADY17882.1 | [Spodoptera exigua] | 6.00E-74 | 79 |
| OBP11 | 24721 | KC887517 | 138 | NO | NO | OBP4 | AEB54584.1 | [Helicoverpa armigera] | 1.00E-12 | 57 |
| OBP12 | 48911 | KC887518 | 348 | NO | NO | OBP5 | AEB54581.1 | [Helicoverpa armigera] | 1.00E-49 | 61 |
| OBP13 | 12194 | KC887519 | 522 | Yes | NO | OBP5 | AEB54581.1 | [Helicoverpa armigera] | 1.00E-30 | 48 |
| OBP14 | 24034 | 150 | NO | NO | antennal binding protein 8 | AAL60426.1 | [Manduca sexta] | 3.00E-31 | 83 | |
| OBP15 | 49422 | KC887521 | 438 | NO | NO | antennal binding protein 4 | EHJ65654.1 | [Danaus plexippus] | 1.00E-38 | 70 |
| OBP16 | 43398 | KC887522 | 222 | NO | NO | antennal binding protein 3 | AAL60413.1 | [Manduca sexta] | 5.00E-69 | 84 |
| OBP17 | 5096 | KC887523 | 213 | NO | NO | odorant-binding protein 1 | AFG72998.1 | [Cnaphalocrocis medinalis] | 6.00E-34 | 66 |
| OBP18 | 19425 | 169 | NO | NO | antennal binding protein 4 | AAL66739.1 | [Mamestra brassicae] | 3.00E-32 | 86 | |
| ABPX | 587 | KC887520 | 348 | NO | NO | antennal binding protein X-1 | AAP57463.1 | [Agrotis ipsilon] | 4.00E-55 | 87 |
| Chemosensory Protein (CSP) | ||||||||||
| CSP1 | 18859 | 150 | NO | NO | chemosensory protein 9 precursor | NP_001037069.1 | [Bombyx mori] | 2.00E-26 | 80 | |
| CSP2 | 21255 | 183 | NO | NO | chemosensory protein | AAF71290.2 | [Mamestra brassicae] | 6.00E-32 | 89 | |
| CSP3 | 21930 | 171 | NO | NO | chemosensory protein CSP2 | ABM67689.1 | [Spodoptera exigua] | 3.00E-25 | 77 | |
| CSP4 | 27050 | KC907741 | 207 | NO | NO | chemosensory protein 8 | ADV36661.1 | [Antheraea yamamai] | 2.00E-29 | 71 |
| CSP5 | 2822 | 120 | NO | NO | CSP4 | AEX07269.1 | [Helicoverpa armigera] | 1.00E-34 | 89 | |
| CSP6 | 2823 | KC907742 | 387 | Yes | Yes | chemosensory protein | AAF71290.2 | [Mamestra brassicae] | 8.00E-75 | 86 |
| CSP7 | 2855 | KC907743 | 123 | NO | NO | CSP2 | AEX07265.1 | [Helicoverpa armigera] | 2.00E-41 | 91 |
| CSP8 | 30460 | KC907744 | 219 | NO | NO | chemosensory protein | AAF19653.1 | [Mamestra brassicae] | 2.00E-42 | 83 |
| CSP9 | 32869 | KC907745 | 203 | NO | NO | chemosensory protein 13 | BAG71921.1 | [Papilio xuthus] | 1.00E-42 | 82 |
| CSP10 | 35445 | KC907746 | 187 | NO | NO | chemosensory protein 9 precursor | NP_001037069.1 | [Bombyx mori] | 6.00E-22 | 73 |
| CSP11 | 37159 | KC907747 | 207 | NO | NO | chemosensory protein | EHJ67380.1 | [Danaus plexippus] | 2.00E-36 | 87 |
| CSP12 | 604 | KC907748 | 336 | Yes | Yes | sensory appendage protein-like protein | AAK14793.1 | [Mamestra brassicae] | 2.00E-36 | 65 |
| CSP13 | 48349 | KC907749 | 375 | Yes | Yes | chemosensory protein | ACX53825.1 | [Heliothis virescens] | 9.00E-43 | 63 |
| CSP14 | 49098 | KC907750 | 336 | Yes | Yes | chemosensory protein | ACX53817.1 | [Heliothis virescens] | 8.00E-49 | 69 |
| CSP15 | 50431 | KC907751 | 324 | Yes | Yes | chemosensory protein | EHJ67380.1 | [Danaus plexippus] | 8.00E-57 | 86 |
| CSP16 | 5090 | KC907752 | 375 | Yes | Yes | chemosensory protein | ACX53727.1 | [Heliothis virescens] | 3.00E-46 | 67 |
| CSP17 | 5091 | KC907753 | 375 | Yes | Yes | chemosensory protein | ACX53727.1 | [Heliothis virescens] | 4.00E-47 | 66 |
| CSP18 | 5116 | KC907754 | 435 | Yes | Yes | chemosensory protein CSP1 | ABM67688.1 | [Spodoptera exigua] | 1.00E-66 | 76 |
| CSP19 | 5123 | KC907755 | 369 | Yes | Yes | CSP6 | AEX07267.1 | [Helicoverpa armigera] | 5.00E-57 | 86 |
| CSP20 | 5124 | KC907756 | 387 | Yes | Yes | chemosensory protein | AAF71289.1 | [Mamestra brassicae] | 8.00E-71 | 82 |
| CSP21 | 591 | KC907757 | 444 | Yes | Yes | chemosensory protein CSP1 | ABM67686.1 | [Plutella xylostella] | 1.00E-52 | 57 |
| CSP22 | 622 | KC907758 | 291 | NO | NO | chemosensory protein | ACX53806.1 | [Heliothis virescens] | 3.00E-57 | 81 |
| CSP23 | 650 | KC907759 | 384 | Yes | Yes | chemosensory protein 2 | AAM77040.1 | [Heliothis virescens] | 3.00E-68 | 83 |
| CSP24 | 717 | KC907760 | 271 | NO | NO | chemosensory protein | ACX53719.1 | [Heliothis virescens] | 4.00E-56 | 91 |
| Sensory Neuron Membrane Protein (SNMP) | ||||||||||
| SNMP1 | 43998 | KC907737 | 270 | NO | NO | Sensory neuron membrane protein1 | Q8I9S2.1 | [Mamestra brassicae] | 1.00E-69 | 92 |
| SNMP2 | 5122 | KC907738 | 1118 | NO | NO | Sensory neuron membrane protein2 | B2RFN2.1 | [Heliothis virescens] | 0 | 83 |
Note: PBP1, PBP2, PBP3 and GOBP2 were previously deposited by others. Genes without accession number represent that the gene fragments obtained in this study were less than 200 bp in length. Gene fragments less than 200 bp are unable to be deposited in the GenBank, and thus no accession numbers were provided for these genes.
Table 2. The Blastx match of S. inferens putative ORs and IRs genes.
| Gene Name | Gene ID | Acc. number | ORF Length (bp) | Complete ORF | TMD(NO) | Best Blastx Match | ||||
| Name | Acc. number | Species | E value | Identity (%) | ||||||
| Odorant Receptor (OR) | ||||||||||
| OR1 | 11700 | KC960453 | 561 | NO | 4 | odorant receptor | AEF32141.1 | [Spodoptera exigua] | 4.00E-94 | 73 |
| OR2 | 49820 | KC960454 | 1422 | Yes | 7 | olfactory receptor-2 | BAG71415.1 | [Mythimna separata] | 0 | 96 |
| OR3 | 11970 | KC960455 | 288 | NO | 0 | olfactory receptor 10 | ACC63238.1 | [Helicoverpa armigera] | 2.00E-61 | 96 |
| OR4 | 21368 | 126 | NO | 0 | putative chemosensory receptor 17 | CAG38118.1 | [Heliothis virescens] | 9.00E-07 | 83 | |
| OR5 | 44838 | KC960456 | 264 | NO | 0 | olfactory receptor 12 | ACF32963.1 | [Helicoverpa armigera] | 3.00E-61 | 84 |
| OR6 | 34021 | KC960457 | 243 | NO | 0 | olfactory receptor 63 | NP_001166620.1 | [Bombyx mori] | 3.00E-23 | 68 |
| OR7 | 16167 | 167 | NO | 0 | candidate odorant receptor 2 | ACS45308.1 | [Helicoverpa assulta] | 1.00E-24 | 79 | |
| OR8 | 27099 | KC960458 | 198 | NO | 0 | olfactory receptor-like receptor | BAG12809.1 | [Bombyx mori] | 1.00E-20 | 60 |
| OR9 | 52605 | KC960459 | 471 | NO | 2 | olfactory receptor 36 | NP_001166892.1 | [Bombyx mori] | 1.00E-65 | 67 |
| OR10 | 22505 | 177 | NO | 0 | putative chemosensory receptor 21 | CAG38122.1 | [Heliothis virescens] | 1.00E-15 | 58 | |
| OR11 | 11122 | KC960460 | 267 | NO | 1 | olfactory receptor 13 | NP_001166603.1 | [Bombyx mori] | 2.00E-35 | 64 |
| OR12 | 1887 | KC960461 | 354 | NO | 1 | odorant receptor 42 | ABK27852.1 | [Bombyx mori] | 2.00E-42 | 56 |
| OR13 | 26406 | KC960462 | 192 | NO | 0 | olfactory receptor 60 | NP_001155301.1 | [Bombyx mori] | 8.00E-26 | 75 |
| OR14 | 34752 | KC960463 | 207 | NO | 2 | putative chemosensory receptor 7 | CAD31853.1 | [Heliothis virescens] | 2.00E-24 | 58 |
| OR15 | 37297 | KC960464 | 257 | NO | 1 | putative odorant receptor OR12 | AFC91721.1 | [Cydia pomonella] | 9.00E-42 | 76 |
| OR16 | 39913 | KC960465 | 147 | NO | 0 | putative odorant receptor OR17 | AFC91725.1 | [Cydia pomonella] | 2.00E-17 | 54 |
| OR17 | 10394 | KF008005 | 246 | NO | 0 | olfactory receptor 33 | NP_001103623.1 | [Bombyx mori] | 1.00E-32 | 57 |
| OR18 | 10399 | KF008006 | 462 | NO | 1 | olfactory receptor 22 | NP_001166613.1 | [Bombyx mori] | 6.00E-87 | 74 |
| OR19 | 11474 | KC960466 | 1080 | NO | 6 | olfactory receptor-like | NP_001116817.1 | [Bombyx mori] | 2.00E-162 | 68 |
| OR20 | 1458 | KC960467 | 522 | NO | 3 | olfactory receptor 56 | NP_001166617.1 | [Bombyx mori] | 6.00E-110 | 76 |
| OR21 | 5112 | KC960468 | 1002 | NO | 2 | putative chemosensory receptor 16 | CAG38117.1 | [Heliothis virescens] | 9.00E-169 | 77 |
| OR22 | 43193 | KC960469 | 300 | NO | 1 | odorant receptor 23 | DAA05981.1 | [Bombyx mori] | 1.00E-18 | 41 |
| OR23 | 4444 | KC960470 | 732 | NO | 3 | olfactory receptor | EHJ63141.1 | [Danaus plexippus] | 6.00E-77 | 47 |
| OR24 | 54083 | KC960471 | 726 | NO | 3 | olfactory receptor 44 | NP_001166607.1 | [Bombyx mori] | 2.00E-83 | 80 |
| OR25 | 53466 | KC960472 | 647 | NO | 3 | olfactory receptor 49 | NP_001166614.1 | [Bombyx mori] | 9.00E-67 | 59 |
| OR26 | 53488 | KC960473 | 546 | NO | 0 | odorant receptor 30 | DAA05986.1 | [Bombyx mori] | 4.00E-90 | 68 |
| OR27 | 53951 | KC960474 | 616 | NO | 0 | olfactory receptor | BAG71423.2 | [Mythimna separata] | 7.00E-114 | 74 |
| OR28 | 54580 | KC960475 | 774 | NO | 4 | olfactory receptor 16 | NP_001104832.2 | [Bombyx mori] | 5.00E-137 | 71 |
| OR29 | 54690 | KC960476 | 624 | NO | 0 | olfactory receptor-1 | BAG71414.1 | [Mythimna separata] | 5.00E-143 | 81 |
| OR30 | 54930 | KC960477 | 714 | NO | 3 | olfactory receptor 64 | NP_001166621.1 | [Bombyx mori] | 4.00E-85 | 65 |
| OR31 | 54964 | KC960478 | 750 | NO | 4 | olfactory receptor-like receptor | EHJ72218.1 | [Danaus plexippus] | 8.00E-78 | 42 |
| OR32 | 55698 | KC960479 | 1080 | NO | 5 | olfactory receptor 29 | NP_001166894.1 | [Bombyx mori] | 6.00E-176 | 66 |
| OR33 | 5924 | KC960480 | 672 | NO | 3 | putative odorant receptor OR24 | AFC91732.1 | [Cydia pomonella] | 3.00E-83 | 61 |
| OR34 | 7341 | KF008007 | 921 | NO | 4 | putative chemosensory receptor 15 | CAG38116.1 | [Heliothis virescens] | 1.00E-108 | 69 |
| OR35 | 54102 | KC960481 | 435 | NO | 3 | putative chemosensory receptor 21 | CAG38122.1 | [Heliothis virescens] | 3.00E-89 | 75 |
| OR36 | 55898 | KC960482 | 1209 | Yes | 6 | putative chemosensory receptor 21 | CAG38122.1 | [Heliothis virescens] | 1.00E-91 | 40 |
| OR37 | 11050 | KF008008 | 336 | NO | 0 | odorant receptor OR24 | NP_001155300.1 | [Bombyx mori] | 1.00E-10 | 44 |
| OR38 | 2802 | KC960483 | 469 | NO | 2 | olfactory receptor 35 | NP_001103476.1 | [Bombyx mori] | 3.00E-36 | 51 |
| OR39 | 11752 | KC960484 | 450 | NO | 2 | odorant receptor 38 | ABK27851.1 | [Bombyx mori] | 7.00E-49 | 59 |
| Ionotropic Receptor (IR) | ||||||||||
| IR93a | 11522 | KC907739 | 384 | NO | 1 | ionotropic receptor 93a, isoform B | NP_732567.1 | [Drosophila melanogaster] | 2.00E-23 | 39 |
| IR75d | 14944 | 168 | NO | 0 | putative chemosensory ionotropic receptor IR75d | ADR64683.1 | [Spodoptera littoralis] | 2.00E-26 | 95 | |
| IR76b | 1261 | KC907740 | 1629 | Yes | 3 | putative chemosensory ionotropic receptor IR76b | ADR64687.1 | [Spodoptera littoralis] | 0 | 84 |
Note: Genes without accession number represent that the gene fragments obtained in this study were less than 200 bp in length. Gene fragments less than 200 bp are unable to be deposited in the GenBank, and thus no accession numbers were provided for these genes.
Figure 5. The number of chemosensory genes in different insect species, obtained from genome (*) or antenna transcriptome (#).
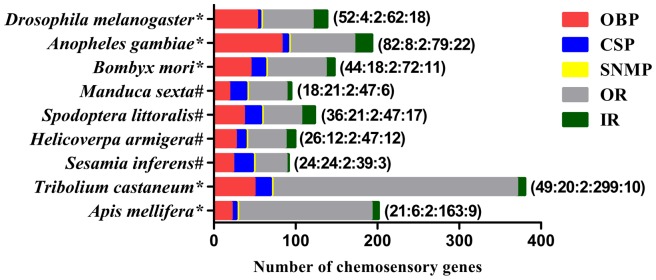
The digits by the histogram bars represent number of chemosensory genes in different subfamilies (OBP:CSP:SNMP:OR:IR). The data are obtained from the current study for S. inferens and from the references [6], [10], [70], [71] for Drosophila melanogaster, [6], [10], [70], [71] for Anopheles gambiae, [10], [24], [38], [65], [71], [72] for Bombyx mori, [6], [10], [70], [71] for Tribolium castaneum and Apis mellifera, [31] for Manduca sexta, [29], [30] for Spodoptera littoralis and [33] for Helicoverpa armigera.
Of the 92 chemosensory transcripts, we carried out the validation experiments for the transcripts encoding for 11 OBPs, 3CSPs, and 6 ORs by RT-PCR and confirmed their identity by sequencing the PCR products. The sequences obtained from positive clones were of ≧99% identical at the nucleic acid level with the corresponding sequences from the transcriptome, indicating that the assembly of the transcripts was adequate.
Among the 87 new putative chemosensory genes, 4 OBPs, 12 CSPs and 3 ORs contained complete open reading frame (ORF); 9 CSPs and one OR (OR2) were of full-length (Table 1 and Table 2). These genes were obtained by transcriptome analysis and RACE.
Expression Profile of the Putative Chemosensory Transcripts
To investigate the general expression profiles, RT-PCR measurements for all 92 transcripts were conducted (Table 3, Figure S1 and Table S2), and 16 selected transcripts were further quantified by qRT-PCR (Figure 6) to validate the RT-PCR results. As a result, the overall relative expression profiles of these transcripts in different tissues and stages obtained by the two methods were similar. In addition, there was a clear agreement between transcript abundance estimated by transcritptome analysis and the expression level measured by RT-PCR. Fourteen of top 20 highly abundant transcripts (Unigene586, Unigene2823, Unigene2855, Unigene2820, Unigene5096, Unigene5080, Unigene587, Unigene5089, Unigene2821, Unigene2896, Unigene5091, Unigene5090, Unigene591 and Unigene5115) (Figure 4) were highly expressed in the antennae (GOBP2, CSP6, CSP7, PBP1, OBP16, GOBP1, ABPX, PBP2, PBP3, OBP5, CSP17, CSP16, CSP21 and OBP10) (Table 3). This suggested that the RT-PCR could be used as an effective mean to investigate the general expression profiles and the relative levels of the putative chemosensory genes among different tissues and developmental stages.
Table 3. Expression of putative chemosensory genes in larvae and different adult tissues of S. inferens.
| Gene | Tissue | |||||||||||||
| La | PG | A♀ | A♂ | H♀ | H♂ | T♀ | T♂ | Ab♀ | Ab♂ | L♀ | L♂ | W♀ | W♂ | |
| Pheromone Binding Protein (PBP) | ||||||||||||||
| PBP1 | * | *** | *** | * | * | * | * | * | ||||||
| PBP2 | *** | *** | * | * | * | * | ** | ** | * | * | ||||
| PBP3 | * | *** | *** | * | * | * | * | * | * | * | ||||
| General odorant binding protein (GOBP) | ||||||||||||||
| GOBP1 | * | *** | *** | * | * | * | * | * | * | * | * | |||
| GOBP2 | *** | *** | * | * | * | * | ||||||||
| Oorant binding protein (OBP) | ||||||||||||||
| OBP1a | *** | ** | ||||||||||||
| OBP2a | ** | ** | * | * | ** | * | *** | *** | ** | ** | ** | ** | ||
| OBP3 | * | *** | *** | * | * | * | *** | * | * | |||||
| OBP4 | *** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ||
| OBP5 | *** | *** | * | * | * | ** | * | |||||||
| OBP6 | * | *** | *** | * | * | |||||||||
| OBP7a | ** | ** | ** | * | ||||||||||
| OBP8 | *** | *** | * | * | ||||||||||
| OBP9 | * | * | * | * | * | * | * | * | ||||||
| OBP10a | * | *** | *** | * | * | * | * | * | ||||||
| OBP11 a | * | *** | *** | ** | * | *** | ** | ** | ** | |||||
| OBP12 | * | ** | * | ** | * | |||||||||
| OBP13 | ** | ** | *** | ** | * | * | * | ** | ** | * | ||||
| OBP14 | *** | ** | *** | *** | * | * | *** | *** | ** | ** | *** | *** | ** | ** |
| OBP15a | *** | ** | * | |||||||||||
| OBP16a | *** | *** | * | * | ||||||||||
| OBP17 | *** | *** | * | * | ||||||||||
| OBP18 | * | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | ||
| ABPXa | *** | *** | * | * | ||||||||||
| Chemosensory Protein (CSP) | ||||||||||||||
| CSP1 | ** | *** | *** | *** | *** | *** | ** | ** | *** | *** | *** | *** | ||
| CSP2 | ** | ** | ** | ** | * | * | *** | ** | ** | ** | *** | *** | *** | *** |
| CSP3 | ** | ** | *** | *** | ** | ** | *** | ** | ** | ** | *** | *** | *** | |
| CSP4 | *** | *** | *** | *** | ** | ** | * | ** | *** | *** | *** | *** | ||
| CSP5 | *** | ** | *** | *** | * | * | ** | *** | * | ** | *** | *** | *** | *** |
| CSP6 | *** | *** | *** | *** | * | * | *** | *** | *** | *** | *** | *** | *** | *** |
| CSP7 | * | *** | *** | *** | * | * | *** | *** | *** | *** | *** | *** | *** | *** |
| CSP8a | * | * | *** | *** | ** | ** | * | ** | *** | *** | *** | *** | ||
| CSP9a | * | * | * | ** | ** | ** | ||||||||
| CSP10 | ** | * | *** | *** | *** | *** | * | ** | ** | *** | *** | *** | ||
| CSP11a | ** | * | ** | * | * | *** | ** | |||||||
| CSP12 | * | *** | *** | * | ** | * | *** | *** | *** | *** | ||||
| CSP13a | *** | *** | *** | *** | ** | ** | ** | ** | *** | *** | *** | *** | ||
| CSP14a | ** | ** | ** | ** | ** | ** | ** | *** | *** | ** | ** | |||
| CSP15 | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | ||
| CSP16 | *** | *** | *** | *** | * | * | *** | *** | *** | *** | *** | *** | *** | *** |
| CSP17 | ** | ** | *** | *** | *** | *** | ** | * | ** | *** | *** | *** | ||
| CSP18 | *** | *** | *** | *** | *** | ** | ** | ** | *** | *** | *** | *** | ||
| CSP19a | * | *** | *** | ** | * | * | * | ** | ** | ** | ** | |||
| CSP20 | *** | ** | *** | *** | * | * | *** | *** | ** | * | *** | *** | *** | *** |
| CSP21a | * | *** | *** | * | * | * | ** | ** | * | * | ||||
| CSP22a | *** | ** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | ||
| CSP23 | *** | *** | *** | *** | * | * | *** | *** | *** | *** | *** | *** | *** | *** |
| CSP24 | *** | * | *** | *** | ** | ** | ** | ** | *** | *** | *** | *** | ||
| Sensory Neuron Membrane Protein (SNMP) | ||||||||||||||
| SNMP1 | *** | *** | * | * | * | * | * | |||||||
| SNMP2 | * | *** | *** | *** | ? | ? | ** | ** | ** | ** | ** | *** | *** | *** |
| Odorant Receptor (OR) | ||||||||||||||
| OR1 | *** | *** | ||||||||||||
| OR2(OR83b)a | *** | *** | ||||||||||||
| OR3a | * | * | ||||||||||||
| OR4a | ** | ** | ||||||||||||
| OR5a | *** | ** | ||||||||||||
| OR6a | ** | ** | ** | |||||||||||
| OR7 | *** | |||||||||||||
| OR8 | ** | ** | ||||||||||||
| OR9 | *** | *** | ||||||||||||
| OR10 | ** | ** | ||||||||||||
| OR11 | *** | *** | ||||||||||||
| OR12 | *** | *** | ||||||||||||
| OR13 | *** | *** | ||||||||||||
| OR14 | ** | ** | ||||||||||||
| OR15 | *** | *** | ||||||||||||
| OR16a | *** | |||||||||||||
| OR17 | ** | ** | ||||||||||||
| OR18 | * | * | ||||||||||||
| OR19a | *** | *** | ||||||||||||
| OR20 | *** | ** | ||||||||||||
| OR21a | * | *** | ||||||||||||
| OR22a | * | * | ||||||||||||
| OR23 | *** | * | ||||||||||||
| OR24 | *** | ** | ||||||||||||
| OR25 | * | ** | *** | *** | * | ** | ** | *** | ** | ** | ** | ** | ** | |
| OR26a | *** | ** | ||||||||||||
| OR27a | ** | *** | ||||||||||||
| OR28 | * | * | ||||||||||||
| OR29 | *** | |||||||||||||
| OR30 | *** | *** | ||||||||||||
| OR31a | *** | *** | ||||||||||||
| OR32 | * | ** | *** | *** | ** | * | * | |||||||
| OR33a | ** | * | *** | *** | * | * | * | * | ||||||
| OR34 | ** | * | *** | *** | * | * | ** | * | ** | * | * | * | ||
| OR35 | ** | ** | ||||||||||||
| OR36a | ** | ** | ||||||||||||
| OR37a | ** | * | ||||||||||||
| OR38a | *** | ** | ||||||||||||
| OR39 | * | ** | ** | * | * | * | * | * | ||||||
| Ionotropic Receptor (IR) | ||||||||||||||
| IR93a | * | * | ** | ** | * | * | * | |||||||
| IR75d | * | ** | ** | * | * | |||||||||
| IR76b | ** | ** | ** | |||||||||||
The relative expression levels of genes in the same tissue were calculated by the ratio of the RT-PCR bands intensity between target gene and internal reference gene SinfGAPDH [73](Figure S1). *, **and *** indicate the intensity ratio of 0.20-0.59, 0.60-0.99, 1.00-1.39, respectively; the blank indicates no signal. The band intensity was calculated by Bio-Rad-Quantity one 4.6.2 software). La, larvae (third instar); Adult tissues include PG, pheromone glands; A, antennae; H, heads (without antennae); T, thoraxes; Ab, abdomens (female without PG); L, legs and W, wings. ♀: female, ♂: male. Superscript “a” followed the gene name represents that the expression level of the gene was obtained by two biological replications.
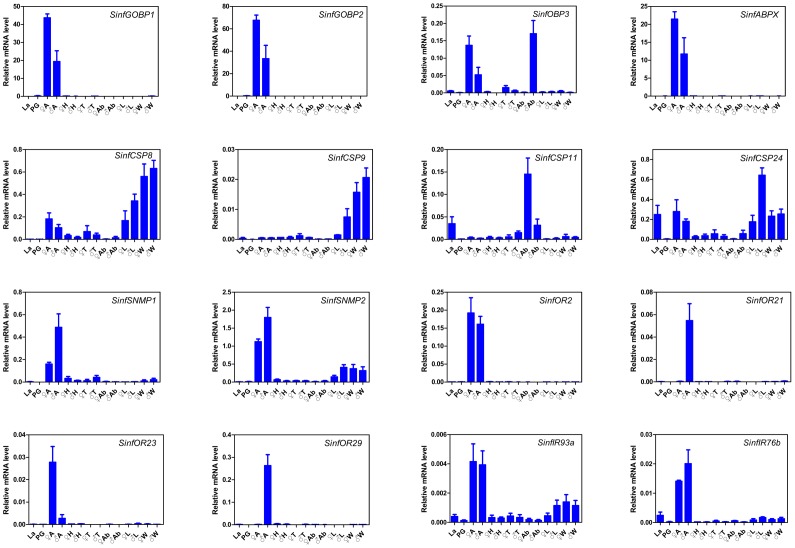
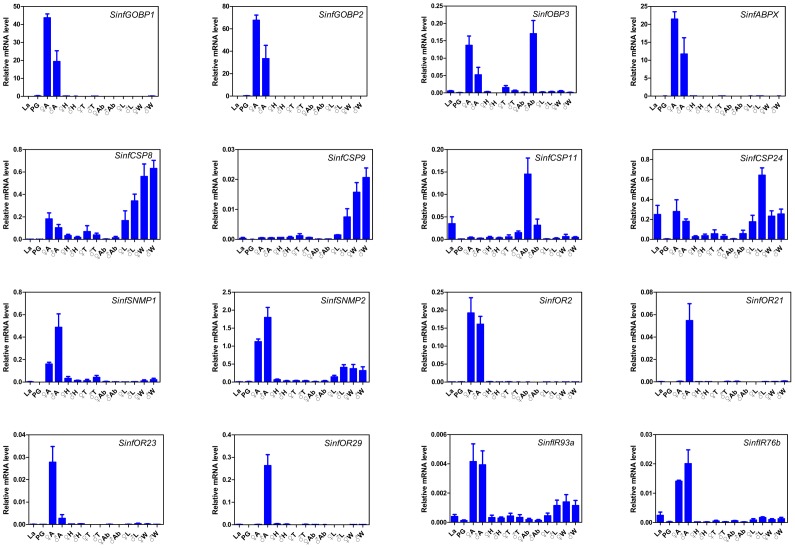
Figure 6. Relative expression levels of 16 putative chemosensory transcripts using qRT-PCR.
La, larvae whole body; PG, female pheromone glands; A, antennae; H, heads; T, thoraxes; Ab, abdomens (female without PG); L, legs; W, wings; ♀, female, ♂, male.
The investigation showed that almost all the transcripts were expressed in the antennae, 40–50% expressed in other tested tissues and only <15% expressed in heads. In addition, the numbers of detected transcripts were similar in male and female moth antennae (91 and 90, respectively), showing no sex bias in chemosensory gene expression (Figure 7A). Thirty nine chemosensory transcripts were detected in female pheromone glands and larvae (Figure 7B).
Figure 7. Tissue distribution of the 92 S. inferens chemosensory transcripts.
A: The proportion of chemosensory genes expressed in larvae, female pheromone gland and other tissues of male and female adults. B: The number of chemosensory gene in each subfamily expressed in larvae, female pheromone glands, and female and male antennae. The digits by the histogram represent number of genes in each subfamily (OBP:CSP:SNMP:OR:IR).
OBP Transcript Expression
The tissue expression profiles are shown in Table 3 and Figure 6. Interestingly, OBP1 was the only antenna-specific OBP transcript. The 3 PBP transcripts and 2 GOBP transcripts displayed highly antenna biased expression, and other antenna highly expressed transcripts included OBP5, OBP6, OBP8, OBP10, OBP15, OBP16, OBP17 and ABPX. The transcripts OBP4 and OBP18 had a similar expression level between antennae and non-antenna tissues. OBP14 was the only OBP transcripts found in all tissues.
Interestingly, the transcripts of PBP1, PBP3 and others (OBP2, OBP3, OBP4, OBP6, OBP7, OBP9, OBP12, OBP13, and OBP14) were also detected in the larvae. Three PBP transcripts were not detected in the pheromone glands, while GOBP1, OBP2, OBP4, OBP9, OBP10, OBP11, OBP13 and OBP14 were detected in the pheromone glands (Table 3, Figure 6 and Figure S1).
CSP Transcript Expression
Compared to OBP transcripts, CSP transcripts were highly expressed in non-olfactory tissues as well as olfactory tissues. Among the 24 newly identified CSP transcripts, 21 displayed a wide range of tissue distribution, and 7 CSP transcripts (CSP2, CSP5-7, CSP16, CSP20 and CSP23) were expressed in all 14 tissues. Most of CSP transcripts were highly expressed in larvae and in pheromone glands (Table 3, Figure 6 and Figure S1).
SNMP Transcript Expression
Two SNMPs homologs were also obtained from S. inferens transcriptome. In comparison, SNMP1 encoding a protein with 78% identity to SNMP1 of B. mori (GenBank accession number: NP_001037186) was highly expressed in the antennae, whilst SNMP2 encoding a protein with 83% identity to SNMP2 of Heliothis virescens (GenBank accession number: B2RFN2.1) was also expressed in remarkable levels in other tissues such as legs and wings (Table 3, Figure 6 and Figure S1).
OR Transcript Expression
Of the 39OR transcripts identified in S. inferens, 34 were expressed only in antennae of both sexes at lower level, relative to the expression level of the OBP and CSP transcripts. OR16 was female-specific while OR7 and OR29 were male-specific. In addition, two ORs, OR23 and OR26 were expressed at much higher levels in female antennae than in male antennae, while OR27 and OR21 were more highly expressed in male antennae than in female antennae. Only 5 OR transcripts, (OR6, OR25, and OR32-34) were expressed broadly in several tissues, including the female sex pheromone glands and the larvae (Table 3, Figure 6 and Figure S1).
IR Transcript Expression
All 3 IR transcripts of S. inferens were expressed at a high level in the antennae, and also at low levels in other tissues. In comparison, IR76b was more specifically detected in the antennae than the other two IRs (Table 3, Figure 6 and Figure S1).
Phylogenetic Analyses
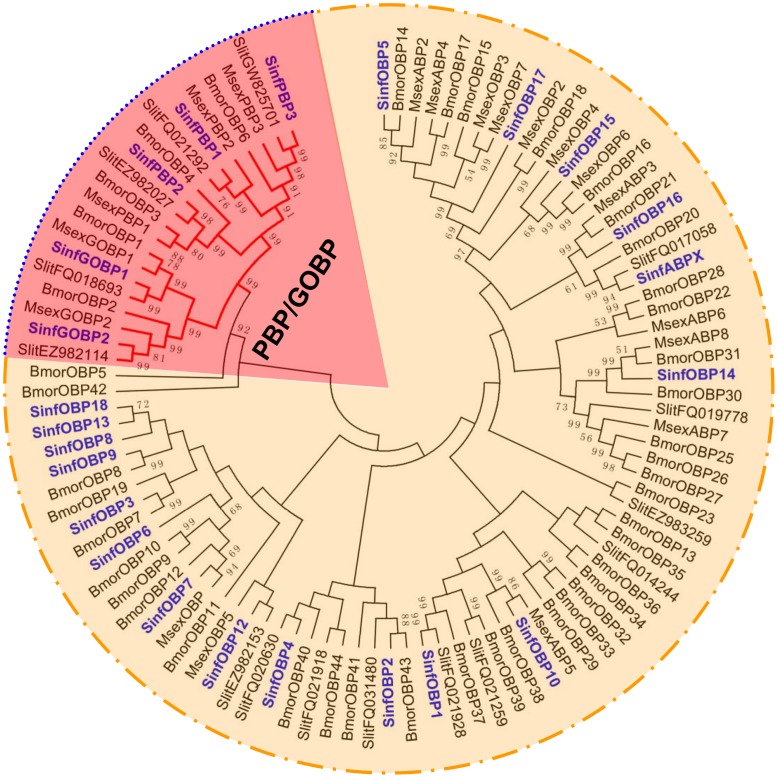
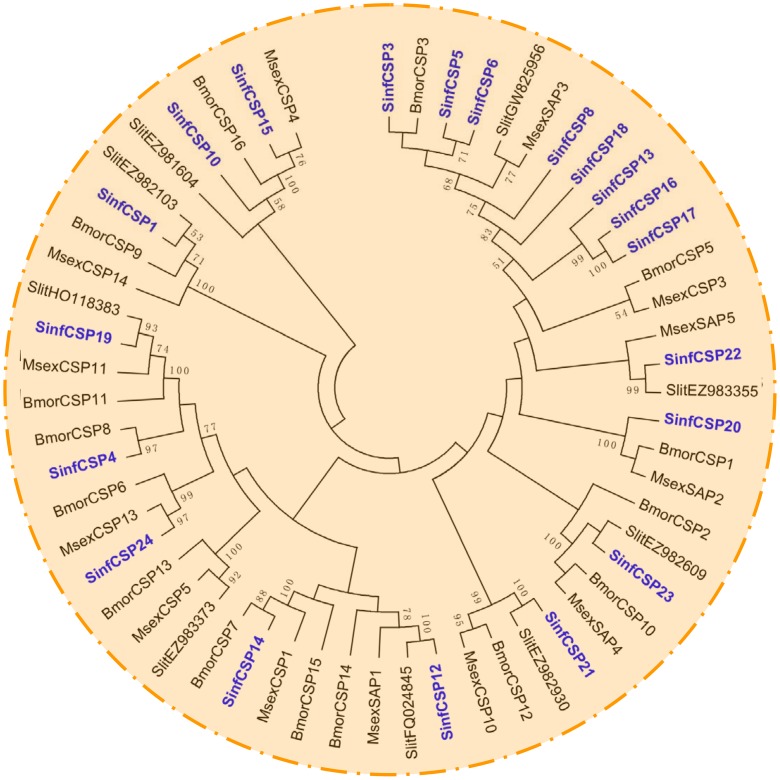
A phylogenetic tree of OBPs was constructed using protein sequences of the OBPs from S. inferens, M. sexta, S. littoralis and B. mori (Figure 8). It was shown that all PBP and GOBP sequences were clustered into distinct clades from other OBPs. More interestingly, the identified SinOBP sequences were clustered in each subclass (PBP1, PBP2, PBP3, GOBP1 and GOBP2) with at least one lepidopteran orthologue (Figure 8). Among the 24 putative CSPs, 20 sequences were clustered with at least one lepidopteran orthologous gene (Figure 9).
Figure 8. Phylogenetic tree of putative OBPs from S. inferens, M. sexta, S. littoralis and B. mori.
PBP/GOBP clade is marked in red. The S. inferens translated unigenes are shown in blue. Accession numbers are given in Table S3. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values indicated at the nodes are bootstrap values based on 1000 replicates, and the bootstrap values <50% are not shown. Sinf, Sesamia inferens; Msex, Manduca sexta; Slit, Spodoptera littoralis; Bmor, Bombyx mori.
Figure 9. Phylogenetic tree of putative CSPs from S. inferens, M. sexta, S. littoralis and B. mori.
The S. inferens translated unigenes are shown in blue. Accession numbers are given in Table S3. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values indicated at the nodes are bootstrap values based on 1000 replicates, and the bootstrap values <50% are not shown. Sinf, Sesamia inferens; Msex, Manduca sexta; Slit, Spodoptera littoralis; Bmor, Bombyx mori.
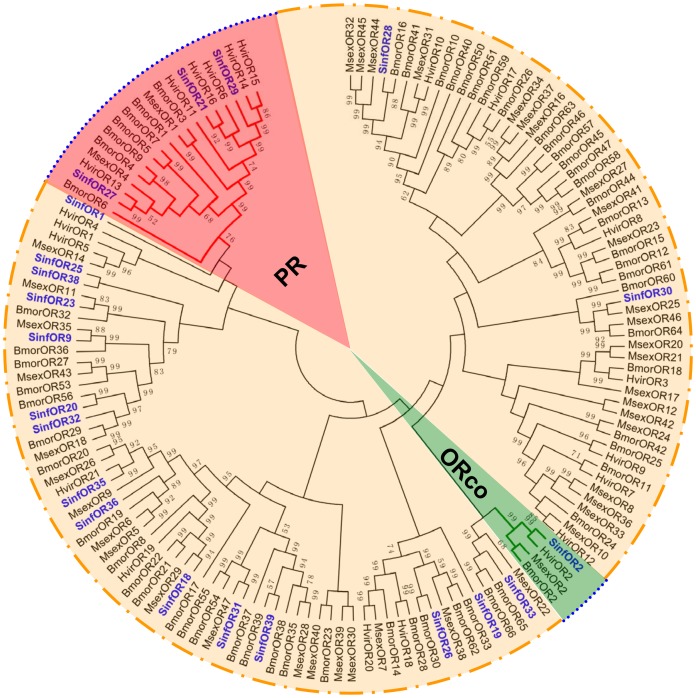
In the OR phylogenetic tree, SinfOR2 was clustered with other lepidopteran OR2 (ORco) sequences, and three SinfORs (OR21, OR27 and OR29) were clustered in the lepidopteran pheromone receptor (PR) clade (Figure 10). The majority of the identified SinfORs had at least one lepidopteran orthologue, with only two (SinfOR1 and SinfOR19) having no counterpart.
Figure 10. Phylogenetic tree of putative ORs from S. inferens, M. sexta, H. virescens and B. mori.
PR clade is marked in red and ORco in green. The S. inferens translated unigenes are shown in blue. Accession numbers are given in Table S3. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values indicated at the nodes are bootstrap values based on 1000 replicates, and the bootstrap values <50% are not shown. Sinf, Sesamia inferens; Msex, Manduca sexta; Hvir, Heliothis virescenss; Bmor, Bombyx mori.
Discussion
In the S. inferens transcriptome data of this study, only 38.8% of 56,210 transcripts have homologous matches to the entries of GenBank with the cutoff value of 10−5, and only 12.8% can be annotated to one or more GO term by the GO analyses, which is similar to M. sexta [31] and S. littoralis [30], indicating that a large number of S. inferens transcripts are either non-coding or homologous with genes that do not have any GO term. In addition, 87 chemosensory transcripts are first reported in S. inferens. Further studies using this transcriptome data could provide insights into insect physiology and pest control strategy [47].
The total number (92) of chemosensory transcripts identified in the current study is similar with those reported in M. sexta (94) and H. armigera (99), but much lower than those of 5 species whose genome has been sequenced, D. melanogaster, A. gambiae, B. mori, T. castaneum and A. mellifera. The chemosensory gene numbers in B. mori (147) and S. littorallis (127) is 1.6 and 1.4 times, respectively of that in S. inferens (Figure 5), suggesting there is a high chance to identify more S. inferens chemosensory genes. On the other hand, CSP transcripts found in S. inferens (24) are more than the CSP genes identified in B. mori genome (18) and in D. melanogaster genome (4). Therefore, it is more likely that we have identified all the CSPs, while missed out some larvae-biased OBPs and lower expressed ORs. These also imply the plant host adaptation and species-specific sex pheromone perception of lepidopteran insects during evolution.
The phylogenetic analysis of SinfOBPs, SinfCSPs and SinfORs suggest that the identified chemosensory transcripts in S. inferens covered main repertoires of the chemosensory genes of the insect. It is worth noting that two ORs (SinfOR1 and SinfOR19) had no counterpart in other species, indicating that the two ORs may represent new types of OR. However, as SinfOR1 was a fragment with only 187 amino acids, it is possible that counterparts might be found, when the full length sequence is available and used in the analysis.
The tissue distribution profiles of all 92 S. inferens chemosensory genes were investigated by RT-PCR, which were confirmed by an additional qRT-PCR measurementusing16 selected genes. Among three subfamilies (CSPs, OBPs and ORs) of the chemosensory gene, CSPs are highly expressed and most widely distributed in chemosensory tissues as well as in non-chemosensory tissues, suggesting CSPs in insects may also involve in other functions apart from chemosensation [48], [49], [50], such as female survival and reproduction in Spodoptera exigua [51], limb regeneration in Periplaneta americana [52] and embryo development in Apis mellifera [53]. In our present study, OBPs are usually highly expressed in the antennae relative to other chemosensory tissues (legs, wings, female sex pheromone glands). However, about half the OBP transcripts are also weakly expressed in non-chemosensory tissues (thorax and abdomen) (Figure 7A), indicating that these OBPs may also have other functions. On the other hand, OBP transcripts that are exclusively expressed in antennae and legs (such as PBPs, GOBPs, OBP8, OBP15-17 and ABPX) may play important role in chemosensory. Interestingly, both PBP1 and PBP3 were detected with weak signals in larvae, similar to that reported in S. littoralis larvae [54]. Poivet et al (2012) suggested that the S. littoralis PBPs in larvae were used to perceive the sex pheromone adsorbed on or deposited on the eggs when female moths ovipositing on the leaves of the host plants, and this perception thus could promote the food search. The larva-expressed PBPs may play similar roles in S. inferens.
In contrast to CSPs and OBPs, OR transcripts are highly restricted in the antennae and expressed at lower levels. This olfactory tissue specific expression profile is well consistent with the specific functional role that OR gene family plays in the moth olfaction [7], [55], [56], [57]. Our study also revealed some OR transcripts (OR25, OR33 and OR34) have a very high expression level in non-chemosensory tissues (thoraxes and abdomens). It is interesting that two SNMP transcripts displayed very different expression profiles, with SNMP1 being highly antennal biased, while SNMP2 was ubiquitously expressed in most tested tissues and larvae. This may suggest that SNMP1 is important in chemosensory, while SNMP2 have other functions in addition to (if any) chemosensation.
In conclusion, we identified members within each subfamily of chemosensory gene family by analysing the trancriptomic sequencing data of antennae and female sex pheromone glands from S. inferens. This provides a rich resource for investigation and elucidation of the chemosensation in S. inferens. As the first step towards understanding their functions, we conducted a comprehensive and comparative examination of the chemosensory gene expression patterns, and demonstrated a wide distribution of these chemosensory proteins. In particular, the expression of SNMPs, IRs and some ORs in non-chemosensory tissues indicate new insights on their roles in insect physiology.
Materials and Methods
Insects Rearing and Collection
The purple stem borer S. inferens was originally collected from a rice field in the Jiangsu Provincial Academy of Agricultural Sciences, Nanjing, China. To collect the insect naturally occurred in the above mentioned field, ethical approval was not required, because the purple stem borer is a common insect pest in South China including Nanjing city, and the insects in the above mentioned field was naturally occurred without any special property. The collected larvae were reared on fresh wild rice stem in glass bottles (d = 7cm, h = 11cm) until pupation and sexed as pupae [58]. Rearing conditions were 28±1°C, 70–80% RH and a 14 h light:10 h dark photoperiod. Adults were provided with a cotton swab dipped in 10% honey solution and renewed daily. Antennae of both sexes and female pheromone glands of 1–5 day-old adults were collected for transcriptome sequencing. Antennae from 3-day-old adults of both sexes were collected for PCR validation of the chemosensory gene sequences obtained from transcriptomic analysis. Antennae, heads (without antennae), thoraxes, abdomens (female without pheromone glands), legs and wings from 3-day-old virginal male and female, female sex pheromone glands of same adult age, and larvae of third instar were dissected and collected in two replications for detection of the tissue expression by RT-PCR. All samples were collected during the first hour of the photoperiod and stored at -70°C until use.
cDNA Library Construction and Illumina Sequencing
Total RNA was extracted using TRIzol reagent (Invitrogen), cDNA library construction and Illumina sequencing of the sample were performed at Beijing Genomics Institute (BGI)-Shenzhen, Shenzhen, China [59]. The mRNA was purified from 20 µg of total RNA (a mixture of RNAs from antennae and pheromone glands at 5∶1 ratio) using oligo (dT) magnetic beads and fragmented into short sequences in the presence of divalent cations at 94°C for 5 min. Then, the first-strand cDNA was generated using random hexamer-primed reverse transcription, followed by synthesis of the second-strand cDNA using RNaseH and DNA polymerase I. After the end repair and ligation of adaptors, the products were amplified by PCR and purified using the QIAquick PCR Purification Kit to create a cDNA library, and sequenced on the HisSeq™ 2000 platform.
De novo Assembly of Short Reads and Gene Annotation
Transcriptome de novo assembly is carried out with short reads assembling program SOAPdenovo [60]. SOAPdenovo first combines reads with a certain length of overlap, to form longer fragments without N (N represent unknown sequence) to produce contigs. The reads are then mapped back to contigs, by using paired-end reads that enable identification of contigs from the same transcript and the distances between these contigs. Next, SOAPdenovo connects the contigs based on the paired-end reads for gap filling between each two contigs to build scaffold sequences with the least Ns. Such sequences are defined as unigenes. In this study, all the clean reads were submitted and available from the NCBI/SRA data base (SRA experiment accession number: SRX286371, BioProject accession number: PRJNA205103).
The Unigenes larger than 150 bp were first aligned by BlASTX to protein databases, including Nr, Swiss-Prot, KEGG and COG (e-value<10−5), retrieving proteins with the highest sequence similarity with the given unigenes along with their protein functional annotations. Then, we used Blast2GO program [61] to get GO annotation of the unigenes, and got GO functional classification by using WEGO software [62].
Expression Abundance Analysis of the Unigenes
The expression abundance of these unigenes were calculated by the RPKM (Reads Per Kilobase per Million mapped reads) method [63], using the formula: RPKM (A) = (10,00,000×C×1,000)/(N×L). In the formula, RPKM (A) is the expression abundance of gene A; C is the number of reads that uniquely aligned to gene A; N is total number of reads that uniquely aligned to all genes; and L is the number of bases on gene A. The RPKM method is able to eliminate the influence of different gene lengths and sequencing discrepancy on the calculation of expression abundance.
RNA Isolation and cDNA Synthesis for Reverse Transcription-PCR
Total RNA was extracted by SV 96 Total RNA Isolation System (Promega, Madison, WI, USA) following the manufacturer’s instructions, in which a DNase digestion was included to avoid the genomic DNA contamination. RNA quality was checked with a spectrophotometer (NanoDropTM 1000, Thermo Fisher Scientific, USA). The single-stranded cDNA templates were synthesized using 1.2 µg total RNAs from various samples with oligo (dT) 18 primer as the anchor primers. The M-MLV Reverse Transcriptase (M-MLV) (TaKaRa, Dalian, Liaoning, China) was used for the cDNA synthesis, with the reaction conducted at 42°C for 1 h, and then stopped by heating at 70°C for 15 min.
RACE Amplification and Sequence Analysis
The SMART™ RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA) was used to amplify the 5′ and 3′ regions of target genes following the manufacturer’s instructions. The RACE PCR products were subcloned into pEASY-T3 cloning vector system (TransGene, Beijing, China) and positive clones were sequenced by GenScript (Nanjing, China). Full-length sequences were determined by assembling the cDNA fragments and the sequences obtained from the 5′ and 3′ RACE PCR. The RACE primers (Table S4) were designed using Primer Premier 5.0 (PREMIER Biosoft International, CA, USA).
The open reading frames (ORFs) of the putative chemosensory genes were predicted by using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The similarity searches were performed by using the NCBI-BLAST network server (http://blast.ncbi.nlm.nih.gov/). Putative N-terminal signal peptides of SinfOBPs and SinfCSPs were predicted by Signal IP 4.0 (http://www.cbs.dtu.dk/services/SignalP/) [64]. The TMDs (Transmembrane Domain) of SinfORs and SinfIRs were predicted using TMHMM Server Version2.0 (http://www.cbs.dtu.dk/services/TMHMM).
Phylogenetic Analyses
The phylogenetic trees were reconstructed for phylogenetic analyses of SinfOBPs, SinfCSPs and SinfORs, based on the amino sequences (the signal peptides of sequences had been removed) of the putative chemosensory genes and the sequences of other Lepidoptera insects. The OBP data set contained 23 sequences from S. inferens (amino acids >45 aa), 19 from M. sexta [29], [31] and 43 from B. mori. The CSP data set contained the 20 sequences from S. inferens (amino acids >40 aa), 13 from M. sexta [31], 9from S. littoralis [29] and 15 from B. mori [26]. The OR data set contained 21 OR sequences from S. inferens (amino acids >144 aa), 43 from M. sexta [31], 21 from H. virescens [41], [42] and 60 from B. mori [65]. The protein name and accession number of the genes used for phylogenetic tree building are listed in Table S3. Amino acid sequences were aligned with ClustalX 2.0 [66] and unrooted trees were constructed by MEGA5.0 [67] using the Neighbor-joining method, with Poisson correction of distances. Node support was assessed using a bootstrap procedure base on 1000 replicates.
Reverse Transcription-PCR Analysis
Gene specific primers across ORF of predicted chemosensory genes were designed using Beacon Designer 7.6 and Primer Premier 5.0 (PREMIER Biosoft International, CA, USA). The sequences of these primers were listed in Table S4. PCR experiments including negative controls (no cDNA template) were carried out in a MyCycler™ (Bio-Rad, USA) under the following conditions: 94°C for 4 min; 30 (35 for OBP13) cycles at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 40 sec, and final incubation for 10 min at 72°C. The reactions were performed in 12.5 µl with 0.5 µl of single-stranded cDNA, 2.0 mM MgCl2, 0.2 mM dNTP, 0.4 µM for each primer and 1.25 U rTaq DNA polymerase (TaKaRa, Dalian, Liaoning, China). PCR products were analyzed by electrophoresis on 2.0% w/v agrose gel in TAE buffer (40 mmol/L Tris-acetate, 2 mmol/L Na2EDTA·H2O) and the resulting bands were visualized with ethidium bromide and digitized using a GelDoc 2000 (Bio-Rad, USA). The control gene encoding for the S. inferens glyceraldehyde-3-phosphate dehydrogenase (SinfGAPDH) was used for quantification.
To detect the relative expression levels of the predicted chemosensory genes, the gels loaded with PCR products of different tissues were scanned for quantification of the band intensity, by using Bio-Rad-Quantity one 4.6.2 software. In addition, 32 transcripts were randomly chosen to perform a second biological replication in order to check the repeatability of the tissue expression. To validate the predicted sequences of chemosensory genes, the PCR products obtained from cDNA sample of adult antennae were purified using the AxyPrep™ PCR Cleanup Kit (Axygen), and then sub-cloned into a T/A plasmid using the pEASY-T3 cloning vector system (TransGene, China) following manufacturer's instructions. The plasmid DNA was used to transform into Trans1-T1 competent cells. Positive clones were checked by PCR and were sequenced by GenScript (Nanjing, China).
Quantitative Real Time-PCR Validation
The expression profiling of a total of 16 putative chemosensory genes was carried out to validate the accuracy of the RT-PCR results using quantitative real time-PCR (qRT-PCR) experiments. The qRT-PCR was performed on an ABI 7500 (Applied Biosystems, Foster City, CA, USA) using a mixture of 10 µl 2× SYBR Green PCR Master Mix, 0.4 µl each primer (10 µM), 2.5 ng of sample cDNA, and 6.8 µl sterilized ultrapure H2O. The reaction programs were 30s at 95°C, 40 cycles of 95°C for 5s and 60°C for 34s. The results were analyzed using the ABI 7500 analysis software SDS 1.4. The qRT-PCR primers (Table S4) were designed using Beacon Designer 7.7 (PREMIER Biosoft International, CA, USA). The mRNA levels were measured by qRT-PCR using the SYBR Premix ExTaq™ (TaKaRa, Dalian, Liaoning, China). This was followed by the measurement of fluorescence during a 55 to 95°C melting curve in order to detect a single gene-specific peak and to check the absence of primer dimer peaks, and a single and discrete peak was detected for all primers tested. Negative controls were non-template reactions (replacing cDNA with H2O).
Expression levels of 16 genes were calculated relative to the reference gene SinfGAPDH using the Q-Gene method in Microsoft Excel-based software of Visual Basic [68], [69] For each sample, three biological replications were performed with each biological replication measured in three technique replications.
Supporting Information
Expression of S. inferens chemosensory transcripts in whole larvae body and different adult tissues. GAPDH gene was used as a positive control and NC (no cDNA template) as a negative control. La, larvae whole body; PG, female pheromone glands; A, antennae; H, heads; T, thoraxes; Ab, abdomens (female without PG); L, legs; W, wings;♀, female, ♂, male. A, Expression of all chemosensory genes by using the first cDNA sample; B, Expression of 32 randomly chosen genes for checking the repeatability of the RT-PCR method by using the second cDNA sample.
(TIF)
The Blastx match of top 50 most abundant unigenes. Except for the putative chemosensory genes in S. inferens.
(DOC)
Data of band intensity of RT-PCR products. It is showing the repeatability of two biological replicates of 32 genes randomly chosen from the 92 ones. #: The band intensity was not calculated because of the irregular images, and were estimated by comparison with the normal bands.
(DOC)
Accession numbers for amino acid sequences of OBPs, CSPs and ORs used in phylogenetic analyses.
(DOC)
Primers used for RT-PCR, qRT-PCR and RACE.
(DOC)
Acknowledgments
We thank Master students Qing Guo, Zhao-Qun Li, Jin Zhang, Ke Yang and Zhan-Feng Ye (Nanjing Agricultural University, China) for help in collecting purple stem borer.
Funding Statement
This work was supported by a Special Fund for Agro-scientific Research in the Public Interest (201203036), and grants from the National Natural Science Foundation (31071978) and the Zhejiang Provincial Natural Foundation (Y3100384) of China.
References
- 1.Vogt RG, editor (2003) Biochemical diversity of odor detection:OBPs, ODEs and SNMPs. Insect pheromone biochemistry and molecular biology. London. Elsevier Academic Press. 397–451p.
- 2. Zhou JJ (2010) Odorant-binding proteins in insects. Vitam Horm 83: 241–272. [DOI] [PubMed] [Google Scholar]
- 3. Xu YL, He P, Zhang L, Fang SQ, Dong SL, et al. (2009) Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC genomics 10: 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pelosi P, Zhou JJ, Ban LP, Calvello M (2006) Soluble proteins in insect chemical communication. Cell Mol Life Sci 63: 1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pelosi P, Calvello M, Ban L (2005) Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem Senses 30 Suppl 1i291–292. [DOI] [PubMed] [Google Scholar]
- 6. Robertson HM, Warr CG, Carlson JR (2003) Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A 100 Suppl 214537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakagawa T, Sakurai T, Nishioka T, Touhara K (2005) Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307: 1638–1642. [DOI] [PubMed] [Google Scholar]
- 8. Krieger J, Klink O, Mohl C, Raming K, Breer H (2003) A candidate olfactory receptor subtype highly conserved across different insect orders. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 189: 519–526. [DOI] [PubMed] [Google Scholar]
- 9. Rogers ME, Sun M, Lerner MR, Vogt RG (1997) Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J Biol Chem 272: 14792–14799. [DOI] [PubMed] [Google Scholar]
- 10. Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, et al. (2009) The insect SNMP gene family. Insect Biochem Mol Biol 39: 448–456. [DOI] [PubMed] [Google Scholar]
- 11. Tunstall NE, Warr CG (2012) Chemical communication in insects: the peripheral odour coding system of Drosophila melanogaster. Adv Exp Med Biol 739: 59–77. [DOI] [PubMed] [Google Scholar]
- 12. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB (2009) Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McKenna MP, Hekmat-Scafe DS, Gaines P, Carlson JR (1994) Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J Biol Chem 269: 16340–16347. [PubMed] [Google Scholar]
- 14. Maleszka R, Stange G (1997) Molecular cloning, by a novel approach, of a cDNA encoding a putative olfactory protein in the labial palps of the moth Cactoblastis cactorum . Gene 202: 39–43. [DOI] [PubMed] [Google Scholar]
- 15. Picimbon JF, Gadenne C (2002) Evolution of noctuid pheromone binding proteins: identification of PBP in the black cutworm moth, Agrotis ipsilon. Insect Biochem Mol Biol 32: 839–846. [DOI] [PubMed] [Google Scholar]
- 16. Abraham D, Lofstedt C, Picimbon JF (2005) Molecular characterization and evolution of pheromone binding protein genes in Agrotis moths. Insect Biochem Mol Biol 35: 1100–1111. [DOI] [PubMed] [Google Scholar]
- 17. Xiu WM, Zhou YZ, Dong SL (2008) Molecular characterization and expression pattern of two pheromone-binding proteins from Spodoptera litura (Fabricius). J Chem Ecol 34: 487–498. [DOI] [PubMed] [Google Scholar]
- 18. Gong ZJ, Zhou WW, Yu HZ, Mao CG, Zhang CX, et al. (2009) Cloning, expression and functional analysis of a general odorant-binding protein 2 gene of the rice striped stem borer, Chilo suppressalis (Walker) (Lepidoptera: Pyralidae). Insect Mol Biol 18: 405–417. [DOI] [PubMed] [Google Scholar]
- 19. Calvello M, Brandazza A, Navarrini A, Dani FR, Turillazzi S, et al. (2005) Expression of odorant-binding proteins and chemosensory proteins in some Hymenoptera. Insect Biochem Mol Biol 35: 297–307. [DOI] [PubMed] [Google Scholar]
- 20. Nagnan-Le Meillour P, Cain AH, Jacquin-Joly E, Francois MC, Ramachandran S, et al. (2000) Chemosensory proteins from the proboscis of mamestra brassicae. Chem Senses 25: 541–553. [DOI] [PubMed] [Google Scholar]
- 21. Picimbon JF, Dietrich K, Angeli S, Scaloni A, Krieger J, et al. (2000) Purification and molecular cloning of chemosensory proteins from Bombyx mori. Arch Insect Biochem Physiol 44: 120–129. [DOI] [PubMed] [Google Scholar]
- 22. Ban L, Scaloni A, Brandazza A, Angeli S, Zhang L, et al. (2003) Chemosensory proteins of Locusta migratoria. Insect Mol Biol 12: 125–134. [DOI] [PubMed] [Google Scholar]
- 23. Liu SJ, Liu NY, He P, Li ZQ, Dong SL, et al. (2012) Molecular characterization, expression patterns, and ligand-binding properties of two odorant-binding protein genes from Orthaga achatina (Butler) (Lepidoptera: Pyralidae). Arch Insect Biochem Physiol 80: 123–139. [DOI] [PubMed] [Google Scholar]
- 24. Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH (2009) The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC genomics 10: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou JJ, Kan Y, Antoniw J, Pickett JA, Field LM (2006) Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem Senses 31: 453–465. [DOI] [PubMed] [Google Scholar]
- 26. Foret S, Wanner KW, Maleszka R (2007) Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol 37: 19–28. [DOI] [PubMed] [Google Scholar]
- 27. Zhou JJ, He XL, Pickett JA, Field LM (2008) Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol Biol 17: 147–163. [DOI] [PubMed] [Google Scholar]
- 28. Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, et al. (1999) A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22: 327–338. [DOI] [PubMed] [Google Scholar]
- 29. Legeai F, Malpel S, Montagne N, Monsempes C, Cousserans F, et al. (2011) An Expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC genomics 12: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poivet E, Gallot A, Montagne N, Glaser N, Legeai F, et al. (2013) A Comparison of the Olfactory Gene Repertoires of Adults and Larvae in the Noctuid Moth Spodoptera littoralis. PLoS One 8: e60263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, et al. (2011) Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci U S A 108: 7449–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bengtsson JM, Trona F, Montagne N, Anfora G, Ignell R, et al. (2012) Putative Chemosensory Receptors of the Codling Moth, Cydia pomonella, Identified by Antennal Transcriptome Analysis. PLoS One 7: e31620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y, Gu S, Zhang Y, Guo Y, Wang G (2012) Candidate Olfaction Genes Identified within the Helicoverpa armigera Antennal Transcriptome. PLoS One 7: e48260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pelletier J, Leal WS (2011) Characterization of olfactory genes in the antennae of the Southern house mosquito, Culex quinquefasciatus. J Insect Physiol 57: 915–929. [DOI] [PubMed] [Google Scholar]
- 35. Pelletier J, Leal WS (2009) Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PLoS One 4: e6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu SH, Wang SP, Zhang XY, Wu KM, Guo YY, et al. (2011) Identification and tissue distribution of odorant binding protein genes in the lucerne plant bug Adelphocoris lineolatus (Goeze). Insect Biochem Mol Biol 41: 254–263. [DOI] [PubMed] [Google Scholar]
- 37. Foret S, Maleszka R (2006) Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res 16: 1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gong DP, Zhang HJ, Zhao P, Lin Y, Xia QY, et al. (2007) Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem Mol Biol 37: 266–277. [DOI] [PubMed] [Google Scholar]
- 39. Robertson HM, Wanner KW (2006) The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res 16: 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E (2011) Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol 20: 189–199. [DOI] [PubMed] [Google Scholar]
- 41. Krieger J, Raming K, Dewer YM, Bette S, Conzelmann S, et al. (2002) A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci 16: 619–628. [DOI] [PubMed] [Google Scholar]
- 42. Krieger J, Grosse-Wilde E, Gohl T, Dewer YM, Raming K, et al. (2004) Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc Natl Acad Sci U S A 101: 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chai HN, Du YZ (2012) The Complete Mitochondrial Genome of the Pink Stem Borer, Sesamia inferens, in Comparison with Four Other Noctuid Moths. Int J Mol Sci 13: 10236–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu LN, Li CC, Hu BJ, Zhou ZY, Li XX (2011) Review of History, Present Situation and Prospect of Pink Stem Borer in China. Chinese Agricultural Science Bulletin 27: 244–248. [Google Scholar]
- 45. Gao Y, Hu Y, Fu Q, Zhang J, Oppert B, et al. (2010) Screen of Bacillus thuringiensis toxins for transgenic rice to control Sesamia inferens and Chilo suppressalis. J Invertebr Pathol 105: 11–15. [DOI] [PubMed] [Google Scholar]
- 46. Li SW, Yang H, Liu YF, Liao QR, Du J, et al. (2012) Transcriptome and Gene Expression Analysis of the Rice Leaf Folder, Cnaphalocrosis medinalis. PLoS One 7: e47401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou JJ, Field LM, He XL (2010) Insect odorant-binding proteins: Do they offer an alternative pest control strategy?. Outlooks Pest Manag 21: 31–34. [Google Scholar]
- 48. Gu SH, Wang SY, Zhang XY, Ji P, Liu JT, et al. (2012) Functional Characterizations of Chemosensory Proteins of the Alfalfa Plant Bug Adelphocoris lineolatus Indicate Their Involvement in Host Recognition. PLoS One 7: e42871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y, Dong X, Liu J, Hu M, Zhong G, et al. (2012) Molecular cloning, expression and molecular modeling of chemosensory protein from Spodoptera litura and its binding properties with Rhodojaponin III. PLoS One 7: e47611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo W, Wang X, Ma Z, Xue L, Han J, et al. (2011) CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet 7: e1001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong L, Luo Q, Rizwan-Ul-Haq M, Hu MY (2012) Cloning and characterization of three chemosensory proteins from Spodoptera exigua and effects of gene silencing on female survival and reproduction. Bull Entomol Res: 1–10. [DOI] [PubMed]
- 52. Nomura A, Kawasaki K, Kubo T, Natori S (1992) Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int J Dev Biol 36: 391–398. [PubMed] [Google Scholar]
- 53. Maleszka J, Foret S, Saint R, Maleszka R (2007) RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev Genes Evol 217: 189–196. [DOI] [PubMed] [Google Scholar]
- 54.Poivet E, Rharrabe K, Monsempes C, Glaser N, Rochat D, et al.. (2012) The use of the sex pheromone as an evolutionary solution to food source selection in caterpillars. Nat Commun: 1–7. [DOI] [PubMed]
- 55. Xu P, Garczynski SF, Atungulu E, Syed Z, Choo YM, et al. (2012) Moth sex pheromone receptors and deceitful parapheromones. PLoS One 7: e41653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wanner KW, Nichols AS, Allen JE, Bunger PL, Garczynski SF, et al. (2010) Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One 5: e8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, et al. (2004) Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci U S A 101: 16653–16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang HY, Li HD, Hang ZJ (2012) Difference in Susceptibility of Field Populations of Sesamia inferens (Walker) to Various Insecticides. China Rice 18: 29–33. [Google Scholar]
- 59.BGI (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Research. [DOI] [PMC free article] [PubMed]
- 60. Li R, Zhu H, Ruan J, Qian W, Fang X, et al. (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20: 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 62. Ye J, Fang L, Zheng H, Zhang Y, Chen J, et al. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34: W293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 64. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 65. Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, et al. (2009) Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol 19: 881–890. [DOI] [PubMed] [Google Scholar]
- 66. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 67.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al.. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. [DOI] [PMC free article] [PubMed]
- 68. Simon P (2003) Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19: 1439–1440. [DOI] [PubMed] [Google Scholar]
- 69.Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32: 1372–1374, 1376, 1378–1379. [PubMed]
- 70. Sanchez-Gracia A, Vieira FG, Rozas J (2009) Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb) 103: 208–216. [DOI] [PubMed] [Google Scholar]
- 71. Croset V, Rytz R, Cummins SF, Budd A, Brawand D, et al. (2010) Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6: e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Consortium THG (2012) Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang XM, Dai W, Guo YJ, Chen CX, Li N, et al. (2011) Semi-quantitative Analysis of Growth Hormone (GH) mRNA Expression of Clarias gariepinus in High Density Pond Culture. Acta Agriculturae Boreali-Sinica 26: 94–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of S. inferens chemosensory transcripts in whole larvae body and different adult tissues. GAPDH gene was used as a positive control and NC (no cDNA template) as a negative control. La, larvae whole body; PG, female pheromone glands; A, antennae; H, heads; T, thoraxes; Ab, abdomens (female without PG); L, legs; W, wings;♀, female, ♂, male. A, Expression of all chemosensory genes by using the first cDNA sample; B, Expression of 32 randomly chosen genes for checking the repeatability of the RT-PCR method by using the second cDNA sample.
(TIF)
The Blastx match of top 50 most abundant unigenes. Except for the putative chemosensory genes in S. inferens.
(DOC)
Data of band intensity of RT-PCR products. It is showing the repeatability of two biological replicates of 32 genes randomly chosen from the 92 ones. #: The band intensity was not calculated because of the irregular images, and were estimated by comparison with the normal bands.
(DOC)
Accession numbers for amino acid sequences of OBPs, CSPs and ORs used in phylogenetic analyses.
(DOC)
Primers used for RT-PCR, qRT-PCR and RACE.
(DOC)