Abstract
Obesity is a major risk factor for a myriad of disorders such as insulin resistance and diabetes. The mechanisms underlying these chronic conditions are complex but low grade inflammation and alteration of the endogenous stress defense system are well established. Previous studies indicated that impairment of HSP-25 and HSP-72 was linked to obesity, insulin resistance and diabetes in humans and animals while their induction was associated with improved clinical outcomes. In an attempt to identify additional components of the heat shock response that may be dysregulated by obesity, we used the RT2-Profiler PCR heat shock array, complemented with RT-PCR and validated by Western blot and immunohistochemistry. Using adipose tissue biopsies and PBMC of non-diabetic lean and obese subjects, we report the downregulation of DNAJB3 cochaperone mRNA and protein in obese that negatively correlated with percent body fat (P = 0.0001), triglycerides (P = 0.035) and the inflammatory chemokines IP-10 and RANTES (P = 0.036 and P = 0.02, respectively). DNAJB positively correlated with maximum oxygen consumption (P = 0.031). Based on the beneficial effect of physical exercise, we investigated its possible impact on DNAJB3 expression and indeed, we found that exercise restored the expression of DNAJB3 in obese subjects with a concomitant decrease of phosphorylated JNK. Using cell lines, DNAJB3 protein was reduced following treatment with palmitate and tunicamycin which is suggestive of the link between the expression of DNAJB3 and the activation of the endoplasmic reticulum stress. DNAJB3 was also shown to coimmunoprecipiate with JNK and IKKβ stress kinases along with HSP-72 and thus, suggesting its potential role in modulating their activities. Taken together, these data suggest that DNAJB3 can potentially play a protective role against obesity.
Introduction
Obesity is a medical and social problem worldwide and is a major risk factor of a myriad of health complications, particularly insulin resistance, diabetes, hypertension and cardiovascular disorders that contribute substantially to a significant reduction of both life quality and expectancy [1], [2], [3], [4]. Sedentary lifestyle and increased energy intake are known as key contributing factors to this chronic condition.
The mechanisms underlying obesity are complex but chronic-low grade inflammation and impairment of the endogenous stress defence system in key metabolic sites are considered as the main determinants that govern obesity and its associated complications [5], [6], [7], [8], [9]. Recent investigations indicated that the inflammatory and stress responses pathways are highly integrated and they work most-likely in vicious cycles, which largely explain the myriad of disorders associated with obesity [6], [10], [11]. The perturbation of the inflammatory and stress responses is known to activate several stress kinases but the best characterized are the c-Jun NH2 terminal kinase (JNK) and the inhibitor of κB kinase-β (IKKβ), both of them can phosphorylate the insulin receptor substrate-1 (IRS-1) and rendering this crucial intermediate a poor substrate for the activated insulin receptor [12], [13], [14], [15], [16].
Heat shock response (HSR); a crucial host-defense system against various pathological, physiological and environmental stressors, is one of the key pathways that was shown to be impaired in obesity-induced insulin resistance. HSR involves the immediate activation of a set of highly conserved heat shock proteins (HSPs) referred to as molecular chaperones and their master regulators; the heat shock transcription factors (HSFs) [17], [18], [19]. To maintain normal protein homeostasis (proteostasis), HSPs act with other components of the proteostasis network as a sensor to detect misfolded and aggregated proteins and through non-covalent interactions, they assist in their proper folding or targeting them to degradation through the ubiquitin-dependent proteasome and lysosome-mediated autophagy [19], [20], [21], [22]. In addition to their chaperone activity, certain HSPs have the ability to mitigate damages resulting from metabolic disorders by physically interacting with key stress and apoptotic enzymes and suppressing the inflammatory response, alleviating various forms of metabolic stress and promoting cell survival by blocking caspase-dependent apoptosis [20], [23]. HSPs can also be released into the circulation, but in contrast to their intracellular roles, extracellular HSPs exert an immune-stimulatory effect by interacting with pattern recognition receptors, such as toll-like receptors, and thereby activate the host inflammatory response [24], [25].
HSPs are broadly classified, on the basis of their apparent molecular weight, amino acid sequences and functions into distinct families. In mammalian cells, six major families of HSPs with distinct functional classes have been described to date and they consist of HSP-110/HSPH, HSP-90/HSPC, HSP-70/HSPA, HSP-60/HSPD, HSP-40/DNAJ and the small HSPs/HSPB [26], [27]. Some members of these families are ubiquitously expressed, whereas others are expressed in response to a wide variety of stress conditions and thus, highlighting their critical role in maintaining cellular homeostasis and tissue integrity [28].
The status of the HSR and its role in the pathophysiology of obesity and its complications began to be unraveled. Recent studies suggested that HSR is impaired in obesity-induced insulin resistance both in humans and experimental animal models [29], [30], [31]. The initial studies were carried out on muscle biopsies from type 2 diabetic (T2D) patients and they showed a reduction of HSP-72 expression that correlated with the degree of insulin resistance [8], [9]. These observations were further supported in experimental animal models demonstrating impaired expression of HSP-72 in the rat model of streptozotocin-induced diabetes [32] and reduced expression of both HSP-25 and HSP-72 in the insulin-resistant aged rats [52], [53]. Consistent with these findings, therapies that induce specific HSPs such as heat therapy [33], electrical therapy [34], physical exercise [35] and pharmacological drugs [36], [37], [38] are associated with beneficial outcome as monitored by improved glucose homeostasis, enhanced insulin sensitivity, reduction of visceral adiposity and suppression of the chronic inflammatory state. Taken together, these data highlight the importance of the HSR in mitigating damages associated obesity-mediated insulin resistance. A deep understanding of the status of the HSR in obese subjects prone to insulin resistance and T2D will be therefore of extreme importance.
In an attempt to identify and characterize additional components of the HSR that may be aberrantly expressed in obese subjects, we used the human RT2-Profiler PCR Array targeting the HSR which allows simultaneous screening of the expression profile of 84 heat shock-related genes and compared their expression pattern to control normal-weight subjects. We hypothesize that this pathway-focused approach will lead to the identification of additional genes in the HSR that may be directly linked to obesity. Using this targeted approach, we report in this study the downregulation of DNAJB3, a member of the HSP-40 in obese subject both at the RNA and protein levels. Since physical exercise is known to modulate the stress response, to reduce inflammation and to improve insulin signaling, we investigated its possible effect on the expression of DNAJB3. We report here for the first time that physical exercise increased the expression of DNAJB3 in a manner that was concomitant with decreased phosphorylation of JNK in obese subjects. Using cell lines, the reduction of DNAJB3 protein was linked to the activation of the endoplasmic reticulum stress and in coimmunoprecipitation assays; DNAJB3 was part of a complex containing HSP-72 along with JNK and IKKβ stress kinases. Taken together, our data support the protective role that DNAJB3 may play against obesity.
Materials and Methods
Study population
The study was conducted on adult male and female subjects consisting of lean (BMI = 20–24.9 kg/m2) and obese (BMI = 30–40 kg/m2). Informed written consent was obtained from all subjects before their participation in the study which was approved by the Review Board of Dasman Diabetes Institute and carried out in line with the guideline ethical declaration of Helsinki. Participants that followed any physical exercise within the last 6 months prior to this study, morbid obese (i.e. BMI >40 kg/m2) and participants with prior major illness or intake of medications and/or supplements known to influence the body composition, bone mass were excluded from the study. The physical characteristics of the participating subjects are shown in Table 1.
Table 1. Physical characteristics of subjects at baseline.
| Lean (n = 54) | Obese (n = 66) | P-value | |
| Age (year) | 37.24±10.89 | 44.88±12.12 | 0.0004 |
| Gender (Males/ Females) | 18/36 | 37/29 | 0.013 |
| BMI (kg/m2) | 22.39±2.09 | 34.57±2.95 | <0.0001 |
| PBF (%) | 27.37±5.13 | 38.37±5.01 | <0.0001 |
| Waist (cm) | 80.74±15.84 | 108.53±12.16 | <0.0001 |
| Hip (cm) | 92.29±14.67 | 118.55±8.27 | <0.0001 |
| Waist/Hip | 0.86±0.10 | 0.92±0.10 | 0.003 |
Data are presented as mean ± SD. BMI (body mass index), PBF (percent body fat).
Exercise Protocol
All eligible subjects were enrolled to a supervised exercise program at the Fitness and Rehabilitation Center (FRC) of Dasman Diabetes Institute. Prior to exercise prescription, each individual underwent a symptom-limited maximal incremental cardiopulmonary exercise test “CPET” (COSMED Quark, Italy) using an electromagnetically braked cycle ergometer. The CPET was primarily used to determine the maximum heart rate (max HR) as well as the response to aerobic exercise as measured by the maximum oxygen consumption (VO2 Max) for each subject. Thereafter, a physical fitness assessment test was performed to determine muscle strength and endurance along with flexibility by performing grip strength (dynamometer), push-ups (upper body strength), sit-ups and forward bending test (both upper and lower body flexibility). The exercise training involves a combination of both moderate intensity of aerobic exercise and resistance training using either treadmill or cycling. Each exercise session includes 10 minutes warming-up and cooling down steps at 50–60% of max HR, along with 40 minutes of the prescribed exercise program at 65–80% of max HR. For the duration of the 3-months period, participants exercised 3 to 5 times per week and they were instructed to reach and maintain the recommended heart rate range. This was achieved by regular monitoring of the heart rate during the aerobic training. Strength training was performed 2 to 3 times a week according to the program plan. Exercise intensity, duration and blood pressures were recorded for each session. All trainings were supervised by qualified fitness professionals from FRC. To assess the effectiveness of the exercise, the same physical stress and fitness tests were performed for all subjects at the end of the exercise program.
Blood and tissue sampling
Venous peripheral blood and subcutaneous adipose tissue biopsies were obtained before starting the exercise (baseline) and after 3-months of exercise. Peripheral blood mononuclear cells (PBMCs) were prepared from blood using Ficoll-Hypaque density gradient centrifugation method. Plasma and serum were prepared using vacutainer tubes and then aliquoted and stored at −80°C. Subcutaneous superficial adipose tissue biopsies (∼1 g) were obtained from the periumbilical area by surgical biopsy after a local anesthesia. Once removed, the biopsy was rinsed in cold PBS, divided into 4 pieces and stored appropriately until assayed.
Anthropometric measurements and blood biochemistry
Anthropometric measurements were taken at the baseline and after 3-months of exercise. Whole-body composition was determined by dual-energy radiographic absorptiometry device (Lunar DPX, Lunar radiation, Madison, WI). Glucose and lipid profiles were measured on the Siemens Dimension RXL chemistry analyzer (Diamond Diagnostics, Holliston, MA). HbA1c was determined using the VariantTM device (BioRad, Hercules, CA). Plasma levels of inflammatory and metabolic markers were measured using bead-based multiplexing technology. Median fluorescence intensities were collected on a Bioplex-200 system using Bio-plex Manager software version 6 (BioRad, Hercules, CA). Lipid peroxidation was assessed by measuring plasma levels of malonaldehyde, using TBARs Assay Kit (Cayman Chemical Company, Ann Arbor, MI). Serum levels of ROS were determined using the OxiSelect™ ROS Assay Kit (Cell Biolabs Inc, San Diego, CA). All the above assays were carried out according to the instructions of the manufacturers.
RNA extraction and Reverse Transcription
Total RNA was extracted from PBMC using AllPrep RNA/Protein Kit (Qiagen, Inc., Valencia, CA) and adipose tissue using The RNeasy Lipid Tissue Mini Kit (Qiagen, Inc., Valencia, CA). Quantity and quality of the RNA were determined using the Epoch spectrophotometer system (BioTek Instruments, Inc., Winooski, VT). 1 μg of each RNA sample was reverse-transcribed to cDNA using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA) or RT2 First Strand Kit (SABioscience/Qiagen, Valencia, CA). All the procedures were carried out according to the manufacturer's instructions.
Measurement of gene expression by Real-time Quantitative PCR
Human Heat Shock Protein RT2-profiler PCR Arrays (SABiosciences/Qiagen, Inc., Valencia, CA) is a quantitative SYBR Green-based real-time PCR for analyzing the expression of focused panel of genes simultaneously. It contains 84 specific cDNA fragments of heat shock related genes, plus five housekeeping genes for normalization consisting of beta-2-microglobulin, ribosomal protein L13a, hypoxanthine guanine phosphoribosyl transferase 1, glyceraldehyde-3-phosphate dehydrogenase and actin beta. For each array, 200 ng of the template cDNAs were mixed with RT2 Real time SYBR Green qPCR master mix. Equal aliquots of this mixture were loaded to each well containing pre-dispensed gene-specific primer sets as recommended by the manufacturer and run on the Rotor-Disc 100 system (Qiagen, Inc., Valencia, CA). Rotor-Gene Q version 2 software was used to calculate the cycle threshold (CT) values for all the genes on each PCR array (Qiagen, Inc., Valencia, CA). Each replicate CT was normalized to the average CT of 5 reference genes present in each run. The following formula was used to calculate the relative amount of transcripts in the obese and lean groups after normalization with the five internal controls: ΔΔCT = ΔCT (obese group)-ΔCT (control group) for RNA samples as described elsewhere [67]. Changes in gene expression between the two groups were determined using a 2-tailed t-test and the difference was presented as a fold increase/decrease. Only genes showing more than 1.5-fold change with a P-value less than 0.05 were retained. Genes showing differential expression were further validated by conventional quantitative real time PCR using primers corresponding to the genes of interest.
Cell Culture, plasmids and transfections
Human embryonic kidney (HEK-293), human acute monocytic leukemia (THP1) and L6 rat skeletal muscle cell lines were obtained from American Type Culture Collection (Rockville, Baltimore, MD). They were cultured in Eagle's Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Stimulation of cells with inflammatory cytokines (R & D Systems, Minneapolis, MN), H2O2, palmitate and tunicamycin (Sigma Aldrich, St. Louis, MO) was done for overnight. The plasmids used in this study consisted of pCMV-DNAJB3 (OriGene Technologies, Inc., Rockville, MD) which encodes for human DNAJB3 was cloned as an NH2-terminal fusion with Myc-FLAG tag. pCMV-ATF-6 (a kind gift from Dr. Ron Prywes, Dept. Biological Sciences, Columbia University, New York, USA) encodes for ATF-6 protein as an NH2-terminal fusion with FLAG tag. pcDNA3.1 (Invitrogen, Carlsbad, CA) was used as a control empty vector. For transient transfection assays, HEK-293T cells (at ∼80% of confluence) were transfected with 20 μg of DNA using Lipofectamine method as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Following transfection, cells were incubated in complete EMEM media for 48 hours and then, harvested for coimmunoprecipitation experiments.
Coimmunoprecipitations
Approximately 2×107 of HEK-293 transfected cells were washed twice with ice-cold PBS and lysed in 1 ml of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 1% Triton X-100) containing a cocktail of protease inhibitors Mini Complete (Roche Diagnostics, Laval, Quebec) for 30 min at 4°C. Extracts were centrifuged at 14,000 rpm for 10 minutes at 4°C to remove cell debris. 500 μg of total cell lysates were added to 100 μl of a 50% slurry of anti-FLAG M2 affinity agarose beads (Sigma Aldrich, St. Louis, MO), preequilibrated with cold washing buffer (50 mM Tris-HCl pH 7.4 and 150 mM NaCl) and incubated overnight at 4°C with continuous end-over-end rotation. Protein complex was collected by centrifugation and washed four times in washing buffer and bound proteins were eluted with 100 μl of 3xFLAG tag peptide at 150 μg/ml as recommended by the manufacturer (Sigma Aldrich, St. Louis, MO). To detect the endogenous formation of DNAJB3/JNK/HSP-72 complex, HEK293 cells were lysed as described above and 500 μg of total cell lysates were used in immunoprecipitation assays using either anti-DNAJB3 or anti-HSP-72 antibodies for pull down. Eluted samples were then fractionated on SDS-PAGE and transferred to PVDF membranes for immunoblotting as described below.
Western blot analysis
Western blots were carried out on whole PBMC extracts or cell extracts prepared in RIPA buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 1% Triton ×100, 1 mM EDTA, 0.5% Sodium deoxycholate and 0.1% SDS). Protein concentration was determined by Bradford method using globulin as a standard and 20 μg of proteins were resolved on 10% SDS-PAGE gels. Proteins were then transferred onto PVDF membranes, blocked with 5% non-fat dried milk in Tris-buffered saline containing 0.05% Tween 20 (TBST) for 1 h at room temperature (RT) and then probed with the primary antibody for overnight at 4°C. After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at RT and finally, protein bands were visualized by chemiluminescence and the images were captured by using the Versadoc 5000 system (BioRad, Hercules, CA). The primary antibodies used in this study are raised against DNAJB3 (Proteintech Group, Inc., Chicago, IL), DNAJC5B (Abcam, Cambridge, MA), DNAJB7 (Atlas Antibodies, Stockholm, Sweden). The remaining anti-HSPs antibodies were purchased from Enzo Life Sciences (Life Sciences International, Inc. Plymouth Meeting, PA) and they are raised against HSP-60, the constitutive HSP-70 (HSC-70), the inducible HSP-70 (HSP-72) and HSP-90. Antibodies against total JNK, Phospho-JNK and IKKβ were purchased from Cell Signaling (Cell Signaling Technology,Inc., Danvers, MA). Actin (Santa Cruz Biotechnology, Santa Cruz, CA) and GAPDH (Millipore, Temecula, CA) were used as internal controls. For densitometric analysis, the intensity of the bands was determined using Quantity One Software (BioRad, Hercules, CA).
Immunohistochemistry
Formalin fixed, paraffin embedded adipose tissue samples were prepared and used to make sections for immunohistochemical studies as described previously [68]. Briefly, sections were deparaffinized and the antigens were retrieved at high-temperature using antigen unmasking solution (Dako, Denmark). The endogenous peroxidase was quenched using 3% H2O2 (Merck Schuchardt, Gemany) for 60 min at RT. Sections were blocked with 5% fat-free milk for 60 min at RT followed by 1% BSA for another 60 min and then, incubated at 4°C for overnight with primary antibodies. After washing, sections were stained with horseradish conjugated secondary antibody (Dako, Denmark) for 60 minutes at RT. Colors were developed using DAB kit (Dako, Denmark) and sections were counterstained with hematoxylin (Sigma Aldrich, St. Louis, MO). Quantification of the immunohistochemical staining data was done using Aperio software version 6.3 (Molecular Devices, Downingtown, PA) with an established arbitrary threshold.
Statistical analysis
Statistical analyses were performed with SAS version 9.2 (SAS Institute Inc, Cary, NC). Unless otherwise stated, all descriptive statistics for the variables in the study were reported as means ± standard deviation (SD). Student's t-test was used to determine significance of difference in means between the two groups. Correlations between variables were calculated with the Spearman's rank correlation test. Differences were considered statistically significant at P-values less than 0.05.
Results
Baseline characteristics of study population
The physical characteristics of the two groups enrolled in this study are displayed in Table 1. As expected, body mass index (BMI), percent body fat (PBF), and waist and hip circumferences were significantly higher in obese group compared to lean group (P<0.0001). Given that age and gender were significantly different between the two groups, we adjusted the clinical and metabolic parameters displayed in Table 2 accordingly. Obese subjects had higher systolic blood pressure (SBP; P = 0.01) and lower capacity of maximal oxygen uptake (V02 Max; P = 0.03) as shown in Table 2. They also had decreased levels of HDL and increased levels of TG (P<0.0001 and P = 0.0002, respectively). Compared to lean group, the metabolic profile is dysregulated in obese subjects as indicated by increased levels of C-peptide (P = 0.03), glucagon (P = 0.048), leptin (P<0.0001) and PAI-1 (P = 0.013). Likewise, obese subjects displayed higher inflammatory response as measured by IP-10 and RANTES chemokines (P = 0.003 and P = 0.012, respectively), but they did not differ significantly in the rest of the inflammatory mediators as well as the oxidative stress markers (Table 2 and data not shown).
Table 2. Clinical and biochemical characteristics of subjects at baseline.
| Lean (n = 54) | Obese (n = 66) | P-value | |
| Resting HR (beat/min) | 80.71±14.51 | 77.43±8.15 | 0.73 |
| SBP (mmHg) | 113.00±10.81 | 127.50±11.89 | 0.01 |
| DBP (mmHg) | 76.43±6.33 | 82.00±10.14 | 0.13 |
| VO2, max (ml/kg/min) | 21.63±3.76 | 17.48±4.83 | 0.03 |
| Cholesterol (mmol/l) | 5.03±0.92 | 5.26±1.02 | 0.31 |
| HDL (mmol/l) | 1.43±0.49 | 1.13±0.24 | <0.0001 |
| LDL (mmol/l) | 3.13±0.85 | 3.43±0.96 | 0.14 |
| TG (mmol/l) | 0.91±0.43 | 1.52±0.91 | 0.0002 |
| Glucose (mmol/l) | 5.02±0.64 | 5.45±0.92 | 0.17 |
| HBA1C (%) | 5.47±0.40 | 5.93±1.03 | 0.056 |
| C-peptide (ng/ml) | 2.44±0.71 | 3.68±2.31 | 0.03 |
| Glucagon (ng/ml) | 0.65±0.11 | 0.71±0.15 | 0.048 |
| GLP-1 (ng/ml) | 2.62±0.83 | 2.68±1.54 | 0.68 |
| Insulin (ng/ml) | 2.35±1.12 | 4.51±2.11 | 0.12 |
| Leptin (ng/ml) | 4.82±3.03 | 9.72±6.91 | <0.0001 |
| PAI-1 (ng/ml) | 3.19±1.55 | 4.14±1.53 | 0.013 |
| TNF-α (pg/ml) | 23.70±9.30 | 25.70±13.20 | 0.53 |
| IL-1β (pg/ml) | 1.20±0.47 | 1.35±0.85 | 0.83 |
| IL-6 (pg/ml) | 4.99±2.18 | 4.68±1.87 | 0.54 |
| IL-10 (pg/ml) | 1.97±1.39 | 2.33±2.24 | 0.43 |
| IP-10 (ng/ml) | 0.39±0.14 | 0.60±0.32 | 0.003 |
| RANTES (ng/ml) | 1.29±0.60 | 1.78±0.86 | 0.012 |
| ROS (mM) | 1.38±0.37 | 1.58±0.63 | 0.24 |
| TBARS (μM) | 1.29±0.48 | 1.52±0.43 | 0.11 |
Data were adjusted for age and gender and presented as mean ± SD. HR (heart rate), SBP (systolic blood pressure), DBP (diastolic blood pressure), VO2 Max (maximum oxygen consumption), HDL (high density lipoprotein), LDL (low density lipoprotein) and TG (triglycerides).
RT2 Profiler PCR Array for Hsp-related genes
Despite the central role that HSPs play against a variety of diseases, only a few studies that documented their role in obesity, insulin resistance and diabetes. In addition, these studies were primarily focused on HSP-25, HSP-72 and to a lesser extent; HSP-60 [8], [9], [33], [39]. Therefore, we decided to perform an expression profiling analysis on a selected set of Hsp-related genes using RT2-Profiler PCR heat shock array. RNA samples prepared from PBMC of 6 non-diabetic subjects divided into lean and obese (n = 3 for each group). Among the 84 Hsp-related genes screened by this approach and after normalization with the reference genes, only seven genes showed differential expression between the two groups in which, five genes were up-regulated and two genes were down-regulated in obese subjects (Data not shown). Members of the Hsp-40 family, showing a downregulation in obese participants are the subject of the current investigation.
Reduced expression of members of the Hsp-40 family in PBMC and adipose tissue of obese subjects
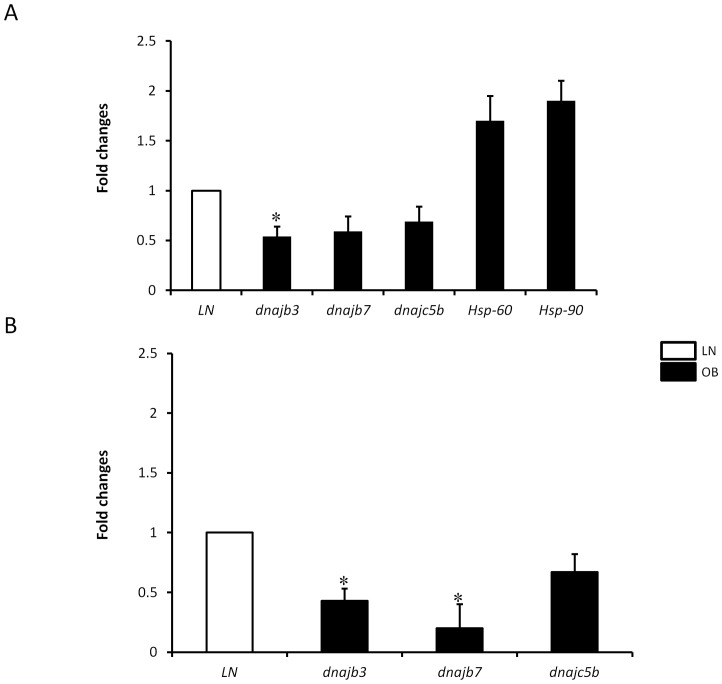
To validate the RT2-Profiler PCR array data, we performed real-time PCR analysis on individual genes using RNA from PBMC of 10 additional non-diabetic obese and lean subjects (n = 5 for each group). We investigated the expression profile dnajc5b and dnajb7 that were identified in the initial RT2-Profiler screening array, but subsequently, we included other heat shock-related genes, including dnajb3 gene that were not represented in the RT2-profiler heat shock array (Table 3). The inclusion of dnajb3 was based on a recent descriptive gene expression profiling study showing a downregulation of dnajb3 mRNA in obese mice compared to lean mice [40]. Consistent with the RT2-Profiler PCR array data, dnajc5b and dnajb7 showed more than 1.5-fold decrease in obese compared to lean group (Fig. 1A). Likewise, the expression of dnjab3 was also significantly reduced in obese subjects (P = 0.037). In contrast, Hsp-60 and Hsp-90 expression was increased by more than 1.5-fold in obese subjects, albeit this increase was not statistically significant (Fig. 1A). No change has been found in the expression of other Hsp-related genes (data not shown). Given the central role played by the adipose tissue in the pathophysiology of obesity, we next investigated whether obesity triggers a reduction in the expression levels of dnaj genes in this dynamic organ. Using real time PCR analysis, we found a more pronounced reduction in the expression of dnajb3 (2.3-fold; P = 0.026), dnajb7 (4-fold; P = 0.04) and dnajc5b (1.7-fold) in obese subjects (Fig. 1B). Taken together, these results indicate that obesity is associated with a significant reduction in the expression of the 3 members of Hsp-40 both in PBMC and adipose tissue.
Table 3. Primer sequences used for real time PCR to analyze gene expression status of human heat shock-related genes.
| Genes | Primers |
| dnajb3 | Forward: 5'-ATCCGAGGCCATCAAGAAG-3' Reverse: 5'-CCACCTGCTTGAATCTCCTC-3' |
| dnajb7 | Forward: 5'-CGGAGGTGGAAGTCATTTTG-3' Reverse: 5'-AGGAGCTTCCTGGACGATTT-3' |
| dnajc5b | Forward: 5'-ACGGTGGAACAGTTTTGCAGC-3' Reverse: 5'-TTTCTTCATTTGATGCTCCCTTA-3' |
| Hsp-60 | Forward: 5'-GATGTCCTGGGCTGTTTCAT-3' Reverse: 5'-GCCTCGATCAAACTTCATGC-3' |
| Hsp-90 | Forward: 5'-ACTTAGCCAAGATGCCTGAGG-3' Reverse: 5'-CACCCCCAAGAAGTTCACAC-3' |
| hspe1 | Forward: 5'-GTGCAGTGGAGGGAAAAGAA-3' Reverse: 5'-CGGCCTATTGAGGACAATTT-3' |
| hspa14 | Forward: 5'-GTGCAGTGGAGGGAAAAGAA-3' Reverse: 5'-CGGCCTATTGAGGACAATTT-3' |
| GAPDH | Forward: 5'-AGGGCTGCTTTTAACTCTGGT-3' Reverse: 5'-CCCCACTTGATTTTGGAGGGA-3' |
Figure 1. Downregulation of members of Hsp-40 in obese subjects.
Total RNA was isolated from PBMC (A) and adipose tissue biopsies (B) of lean (n = 14) and obese (n = 17) non-diabetic participants and subjected to quantitative analysis using real-time PCR. The data are presented as fold changes in obese compared to lean subjects. * P<0.05 as determined using student's t-test.
Validation of the transcriptomic data by Western Blotting and Immunohistochemistry
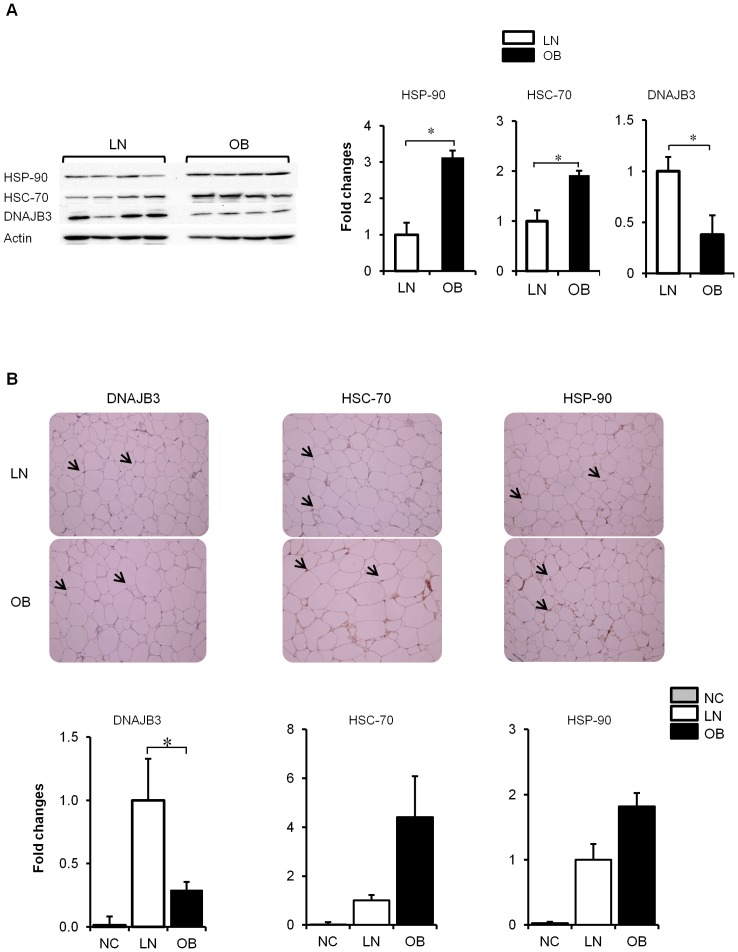
In order to validate the RT-PCR data on these members of the Hsp-40 at the protein level, we performed Western blot analysis on PBMCs and immunohistochemical (IHC) analysis on adipose tissue from selected subjects. As shown in Figure 2A, Western blot analysis performed on lean and obese subjects (n = 4 for each group) showed a major reduction in the expression of DNAJB3 protein in obese subjects (P<0.05). Under the same conditions, we were unable to validate the expression of DNAJC5B and DNAJB7 (data not shown). In contrast to DNAJB3, the expression levels of HSC-70 and HSP-90 were increased in obese subjects (Fig. 2A). Consistent with Western blot analysis, IHC studies on adipose tissue biopsies isolated from lean (n = 4) and obese (n = 11) subjects indicated a significant reduction of DNAJB3 (P<0.05) and an increase in the expression of HSP-70 and HSP-90 (Fig. 2B). Hence both Western blot and IHC data were in complete agreement with each other and they fully support the gene expression data obtained on dnajb3, Hsp-70 and Hsp-90.
Figure 2. Obesity triggers a downregulation of DNAJB3 protein.
(A) Total proteins were extracted from PBMC of lean (n = 4) and obese (n = 4) non-diabetic participants and subjected to western blot using the indicated antibodies. The bands were quantified as described in materials and methods and the relative intensity was determined after correction with actin that was used as internal control to monitor loading efficiency. The data are presented at the bottom as fold changes compared to lean group. The blots shown are representative of at least three independent experiments with consistent results. (B) Immunohistochemical staining using subcutaneous adipose biopsies from lean (n = 4) and obese (n = 11) non-diabetic participants. Aperio software was used to quantify positive staining (indicated by arrows) and the values are illustrated at the bottom as fold changes compared to lean. As negative control (NC) for the experiment, the primary antibodies were omitted. * P<0.05 as determined using student's t-test.
Effect of physical exercise on DNAJB3 expression
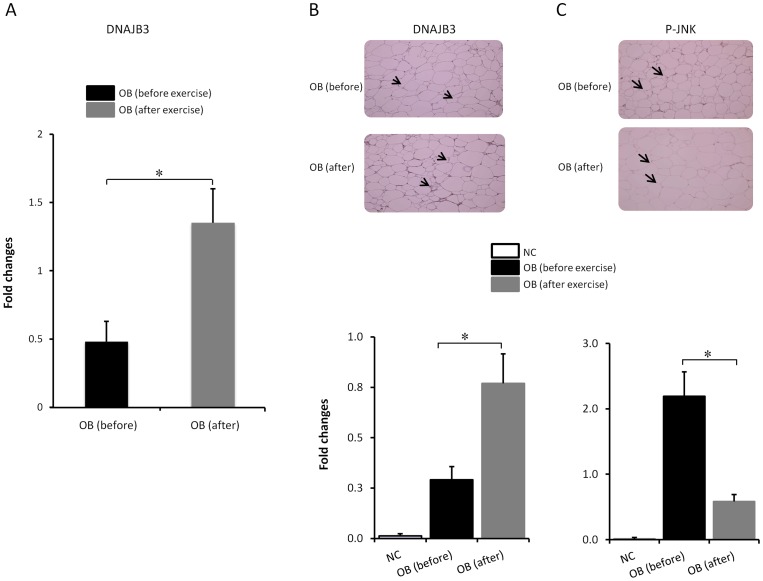
The beneficial effect of physical exercise on weight loss and improving clinical manifestations associated with obesity prompted us to test if it has an effect on the expression of DNAJB3 and if so, whether these levels correlated with the post-exercise training data on physical and metabolic parameters. To investigate the effectiveness of the exercise protocol assigned for participants, we performed a pairwise comparison of physical, clinical and metabolic parameters in obese subjects (n = 24) before and after physical exercise. Table 4 shows that although there was no significant change in the BMI, waist and hip after 3 months of exercise, there was a significant reduction of PBF and SBP (P = 0.023 and P = 0.01, respectively) and increase in VO2 Max (P = 0.01) along with reduced inflammatory response as indicated by reduced levels of TNF-α and IL-6 (P = 0.004 and P = 0.012, respectively). Exercise triggered also a decrease in TBARS levels (P = 0.004). In order to assess the effect of exercise on the expression of DNAJB3, fat adipose tissue biopsies were collected from obese subjects that completed the exercise program and used to monitor the expression of DNAJB3 (n = 10). As shown in Figure 3A, there was a significant increase in the levels of DNAJB3 mRNA in obese subjects after exercise (P = 0.005). Consistent with the RT-PCR data, IHC analysis confirmed that DNAJB3 protein is significantly increased in the adipose tissue of obese subjects after physical exercise (Fig. 3B; P = 0.003). Taken together, these data suggest that exercise can interfere with obesity-mediated repression of DNAJB3. Since obesity is known to trigger the activation of the stress kinase JNK; particularly its phosphorylation, and with the perspective to establish a correlation between the levels of DNAJB3 and activated JNK, we measured the levels of phosphorylated JNK in adipose tissue by IHC before and after exercise. In contrast to the pattern observed for DNAJB3, there was a significant increase in the levels phosphorylated JNK in obese subjects compared to lean subjects (P = 0.016, data not shown). Furthermore, the expected increase in phosphorylated JNK in obese was significantly reduced by physical exercise (Fig. 3C; P = 0.0013). No effect was observed for total JNK before and after exercise (data not shown). From this experiment we concluded that there was an inverse correlation between DNAJB3 and activated JNK.
Table 4. Physical and clinical characteristics of 24 obese subjects before and after exercise.
| Before exercise | After exercise | P-value | |
| Age (year) | 47.78±13.02 | – | – |
| Gender (M/F) | 17/7 | – | – |
| BMI (kg/m2) | 34.60±2.95 | 33.93±2.44 | 0.18 |
| PBF (%) | 37.48±5.35 | 36.63±5.71 | 0.022 |
| Waist (cm) | 108.93±11.32 | 107.33±9.10 | 0.102 |
| Hip (cm) | 117.51±9.42 | 117.88±6.59 | 0.19 |
| Resting HR (beat/min) | 77.43±8.15 | 77.00±10.30 | 0.89 |
| SBP (mmHg) | 127.50±11.89 | 119.29±8.29 | 0.01 |
| DBP (mmHg) | 82.00±10.14 | 77.86±4.26 | 0.11 |
| VO2 Max (ml/kg/min) | 17.48±4.83 | 19.66±5.17 | 0.01 |
| Cholesterol (mmol/l) | 5.26±1.03 | 5.18±0.99 | 0.28 |
| HDL (mmol/l) | 1.16±0.25 | 1.05±0.25 | 0.073 |
| LDL (mmol/l) | 3.35±0.95 | 3.40±0.94 | 0.72 |
| TG (mmol/l) | 1.71±0.93 | 1.63±0.71 | 0.76 |
| Glucose (mmol/l) | 5.76±1.08 | 5.91±1.03 | 0.84 |
| HbA1C (%) | 6.17±1.44 | 5.99±0.56 | 0.46 |
| C-peptide (ng/ml) | 2.89±1.48 | 3.40±1.16 | 0.11 |
| Glucagon (ng/ml) | 0.63±0.16 | 0.54±0.10 | 0.056 |
| GLP-1 (ng/ml) | 2.31±1.66 | 1.83±0.45 | 0.052 |
| Insulin (ng/ml) | 3.04±1.96 | 1.52±1.15 | 0.155 |
| Leptin (ng/ml) | 7.01±3.60 | 5.03±2.88 | 0.705 |
| PAI-1 (ng/ml) | 3.33±1.30 | 3.06±1.17 | 0.32 |
| TNF-α (pg/ml) | 21.60±14.45 | 13.62±10.56 | 0.004 |
| IL-6 (pg/ml) | 3.85±1.35 | 2.43±1.21 | 0.012 |
| IP-10 (ng/ml) | 0.47±0.18 | 0.30±0.19 | 0.097 |
| RANTES (ng/ml) | 1.75±0.67 | 1.62±0.63 | 0.98 |
| TBARS (μM) | 1.46±0.30 | 0.97±0.23 | 0.004 |
Data are presented as mean ± SD. Paired t-test was used to compare differences in obese before and after physical exercise.
Figure 3. Physical exercise restores the expression of DNAJB3.
(A) Quantitative analysis of DNAJB3 mRNA levels in the adipose tissue from obese before exercise (n = 10) and after 3 months of exercise (n = 10) using real-time PCR. (B and C) Immunohistochemical staining using subcutaneous adipose biopsies from obese subjects before exercise (n = 11) and after 3 months of exercise (n = 7) using DNAJB3 (B) and Phopsho-JNK (C) antibodies. Arrows indicate the positive staining. Aperio software was used for quantification and the values are illustrated at the bottom as fold changes after exercise. Student's t-test for two group analysis was done to compare the expression of DNAJB3 (B) and JNK (C) in obese before and after exercise. *: P<0.05.
Correlation analysis of DNAJB3 with physical and clinical profiles, inflammatory and metabolic stress markers
To understand the physiological consequence of the reduction of DNAJB3 in obese subjects on the clinical profile as well as the inflammatory and metabolic stress responses, we investigated their correlation with DNAJB3 mRNA levels using Spearman's rank test before and after exercise. Table 5 shows that before exercise, there was a negative correlation between levels of DNAJB3 and the indicators of obesity such as BMI (r2 = −0.71; P<0.0001) and PBF (r2 = −0.66; P = 0.0001). There was also a negative correlation with TG (r2 = −0.36; P<0.035) as well as with the pro-inflammatory chemokines IP-10 and RANTES (r2 = −0.37; P<0.036 and r2 = −0.40; P = 0.02, respectively). No correlation was found for the other parameters (Table 5 and data not shown). After exercise, the increased expression of DNAJB3 mRNA in obese correlated negatively with the PBF (r2 = −0.53; P = 0.044) and positively with RANTES (r2 = 0.75; P<0.008) (Table 5).
Table 5. Correlation between DNAJB3 mRNA expression and physical, clinical and biochemical parameters.
| Before exercise | After exercise | |||
| R2 | P-value | R2 | P-value | |
| BMI | −0.709 | <0.0001 | −0.128 | 0.650 |
| PBF | −0.656 | 0.0001 | −0.526 | 0.044 |
| SLM | −0.296 | 0.12 | 0.378 | 0.165 |
| HR | −0.02 | 0.945 | −0.119 | 0.727 |
| SBP | 0.038 | 0.894 | 0.136 | 0.690 |
| DBP | 0.136 | 0.629 | 0.391 | 0.235 |
| VO2 Max | 0.556 | 0.031 | 0.220 | 0.515 |
| HDL | 0.301 | 0.07 | −0.193 | 0.594 |
| TG | −0.358 | 0.035 | −0.407 | 0.214 |
| Leptin | −0.322 | 0.064 | −0.014 | 0.965 |
| PAI-1 | −0.162 | 0.362 | 0.277 | 0.383 |
| IP10 | −0.367 | 0.036 | 0.259 | 0.416 |
| RANTES | −0.404 | 0.020 | 0.747 | 0.008 |
The correlation was based on ΔΔCT method and it was done on non-diabetic participants before exercise consisting of lean (n = 14), obese before exercise (n = 21) and obese after exercise (n = 17). Correlation was assessed by using Spearman's rank correlation coefficient.
DNAJB3 binds to JNK and IKKβ stress kinases
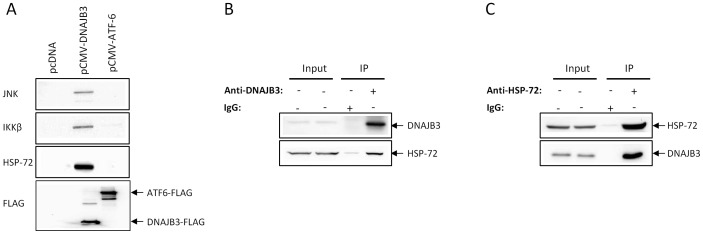
In order to complement the in vivo data shown above, we undertook a series of in vitro experiments using cell lines. Based on the inverse correlation between the levels of DNAJB3 and activated JNK (Fig. 3B and 3C) and given the importance of stress kinases such as JNK, IKKβ in obesity and insulin resistance, we initially sought to determine if there is an interaction between DNAJB3 and these stress kinases. For this purpose, HEK-293 cells were transfected with pCMV-DNAJB3 and investigated the partners of interaction that might bind to DNAJB3 by coimmunoprecipitation as described in materials and methods. As negative controls, we transfected cells with pCMV-ATF-6 and pcDNA3.1 mock vector. As shown in Figure 4, we were able to detect the presence of JNK and IKKβ bands in the immunocomplex prepared from cells transfected with DNAJB3 clone. Under the same conditions, these bands were not detected in lysates prepared from cells transfected with either ATF-6 clone or with the empty vector and thus, demonstrating the specificity of the interactions. To rule out the possibility of differences in transfection efficiency between clones and/or binding affinity of the recombinant proteins to the anti-FLAG conjugated beads, we probed the membranes with anti-FLAG antibody and found that both DNAJB3 and ATF-6 clones are adequately expressed in transfected cells and they bind equally to the anti-FLAG beads (Fig. 4). Given that HSP-72 was shown in previous studies to bind and inactivate JNK and IKKβ and taking into consideration the cochaperone role of DNAJB3, we postulated that HSP-72 might be part of the coimmunoprecipated complex. Probing the membranes with anti-HSP-72 antibody revealed indeed the presence of HSP-72 in complex obtained from cell transfected with DNAJB3 clone but not from ATF-6 clone or the control vector (Fig. 4A). Our findings prompted us to investigate whether endogenous DNAJB3 could form a complex with JNK/HSP-72 by immunoprecipitation using untransfected cells using either anti-DNAJB3 or anti-HSP-72 antibody. While the interaction of JNK with either DNAJB3 or HSP-72 was inconclusive (data not shown), we were able to confirm the interaction between DNAJB3 and HSP-72 using either anti-DNAJB3 (Fig. 4B) or anti-HSP-72 (Fig. 4C) to pull down the immunocomplex.
Figure 4. DNAJB3 forms a complex with HSP-72 and stress kinases in vitro by coimmunoprecipitation.
(A) HEK-293 cells transfected Flag-tagged DNAJB3 and proteins lysates were coimmunprecipitated with anti-Flag antibody. Eluted proteins were subjected to western blot analysis using JNK, IKKβ and HSP-72 antibodies. Flag-tagged ATF-6 vector and pcDNA empty vector were run in parallel and used as controls. Anti-Flag antibody was also used to monitor for transfection efficiency and binding of the recombinant proteins to the anti-Flag antibody. To investigate endogenous formation of DNAJB3/HSP-72/JNK complex, whole cell lysate proteins were prepared from HEK293 cells and used to pull down the immunocomplex using either anti-DNAJB3 (B) or anti-HSP-72 (C) antibody. Eluted proteins were separated on SDS-PAGE and subjected to Western blotting using the appropriate antibodies as indicated.
DNAJB3 expression is reduced in vitro upon activation of the ER stress
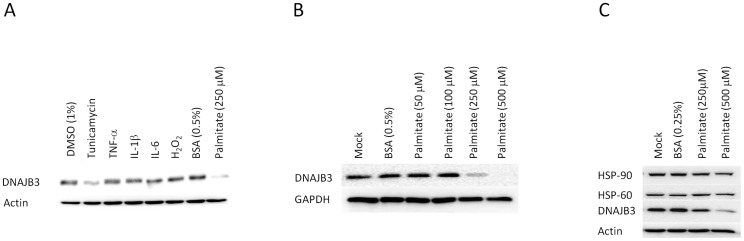
Low grade chronic metabolic inflammation, hyperlipidemia, and enhanced oxidative and endoplasmic reticulum (ER) stress responses are cardinal features that lead to obesity and its further progression to insulin resistance and T2D. In the context of obesity, no previous study reported the existence of mediators that could positively or negatively modulate the expression of DNAJB3. To gain new insight into the molecular mechanisms involved in regulating the expression of DNAJB3 in vitro using cell lines, we stimulated THP-1 and L6 cells with an array of mediators that elicit inflammation, oxidative stress and ER stress. To this end, cells were stimulated with classic inflammatory cytokines such as IL-1β, IL-6 and TNF-α, H2O2 to elicit oxidative stress, and palmitate and tunicamycin; both of them are potent inducers of the ER stress [41], [42]. As shown in Figure 5A, neither inflammatory cytokines nor H2O2 had an effect on the expression of DNAJB3 in THP-1 cells. By contrast, treatment of cells with palmitate resulted in a reduction of DNAJB3 protein in both THP-1 (Fig. 5A) and L6 (Fig. 5B) cells, while it had no effect on HSP-60 and HSP-90 proteins (Fig. 5C). In an attempt to elucidate whether the effect of palmitate on the expression of DNAJB3 was due to the activation of the ER stress or through a different pathway, we stimulated cells with tunicamycin; a chemical drug that specifically activate the ER stress. As shown in Figure 5A, tunicamycin triggers also a reduction of DNAJB3 protein. The observed downregulation of DNAJB3 following activation of ER stress by tunicamycin and palmitate is consistent with the previous study that showed a link between activation of ER stress and downregulation of DNAJB3 gene expression upon stimulation of cardiomyocytes with doxazosin [43].
Figure 5. Downregulation of DNAJB protein in vitro following activation of ER stress by palmitate and tunicamycin.
Western blot analysis using protein lysates from THP-1 cells (A) and L6 cells (B) after stimulation with 25 ng/ml of TNF-α, IL-1β and IL-6, 30 μM of H2O2, 1 μg/ml of tunicamycin and from 50 to 500 μM of palimate for overnight. DMSO at 1% and 0.5% BSA were used as controls for vehicles. Actin and GAPDH were used as internal controls to monitor for loading efficiency. (C) Effect of plamitate on the expression of HSP-60 and HSP-90 along with DNAJB3. The blots shown are representatives of at least three independent experiments with consistent results.
Discussion
The current study was designed to identify unexplored components of the heat shock response that might be aberrantly expressed in obese subjects and playing a possible role in the pathophysiology of obesity and insulin resistance. By complementing the in vivo works with the in vitro studies, our main findings are: 1) the expression of DNAJB3 was significantly reduced in obese subjects and physical exercise restored its normal expression; 2) DNAJB3 formed a complex with JNK, IKKβ and HSP-72 and; 3) a clear association between activation of the ER stress and reduction of DNAJB3 protein in vitro. The observed decrease in the expression of DNAJB3 in obese subjects was independent of the gender effect as both males and females showed similar reduction of DNAJB3 mRNA levels (data not shown). Our data demonstrating for the first time that obesity triggers a reduction in the expression of a co-chaperone at both mRNA and protein levels in human subjects add further evidence that impairment of the heat shock response is one of the key steps that lead to the development of various metabolic disorders associated with obesity.
Human DNAJB3; also known as MSJ-1 in mouse [44], is one of the 49 members of the HSP-40 family of proteins and it acts as a co-chaperone to regulate the activity and specificity of HSP-70; a major component of the proteostasis network [27], [45], [46]. DNAJB members exert their role by stimulating the ATPase activity of HSP-70 through their J-domain, thereby keeping the bound substrates in successive refolding cycles [47], [48]. They are also known for their ability to deliver a diverse set of substrates to HSP-70 and thus, determining substrate specificity [46], [49]. Given their dual mode of action, DNAJB-type proteins are classified among the strongest protectors against protein toxicity associated with protein aggregation [50], [51]. This makes DNAJB family of proteins interesting targets for therapy against protein folding diseases either through functional modulation of their activity or by increasing their expression.
There is a widespread clinical interest in the protective role of HSPs against a variety of diseases, including obesity, insulin resistance and diabetes. Attenuation of this important host defense system is associated with various clinical manifestations and pathological disorders. The findings of our current investigation confirm the previous gene expression profiling study in which DNAJB3 was among the list of genes downregulated in obese mice compared to lean mice [40]. In our model, the significant decrease of DNAJB3 in obese subjects, its correlation with inflammatory markers and fat levels and the restoration of its normal expression after a defined exercise protocol suggest that DNAJB3 might potentially play a protective role in obesity, insulin resistance and T2D. In support of this, the decrease of DNAJB3 was more pronounced in diabetic than in non-diabetics subjects (data not shown). Recently, it was proposed that T2D is the result of a metabolic paradigm in which metabolic inflammation, insulin resistance and impairment of the HSR work in a vicious cycle [11]. Obesity, sedentary lifestyle and high fat calorie perpetuate this cycle by lowering HSPs and thus, leading to metabolic inflammation and impairment of insulin signaling [5], [16], [29]. The downregulation of DNAJB3 in clinically relevant tissue organ can be added to the list of component of the HSR that are attenuated by obesity in human subjects. Our data illustrate also the complexity of the HSR to protect from metabolic disorders associated with obesity. The previous studies that investigated the status of the HSR in the context of obesity, insulin resistance and diabetes were carried out on the skeletal muscle of T2D patients and they showed a reduced expression of HSP-72 that correlates with the degree of insulin resistance [8], [9], [33]. The findings in human subjects were further supported in experimental animal models demonstrating impaired expression of HSP-72 in the rat model of streptozotocin-induced diabetes [32] and reduced expression of both HSP-25 and HSP-72 in the insulin-resistant aged rats [52], [53]. As a note of caution, our data did not explain the exact significance of this reduction to obesity and this may represent a limitation of this study, nonetheless, further studies that are beyond the scope of this work such as using DNAJB3 knockout mouse animal models as well as treatment with pharmacological modulators of DNAJB3 are warranted.
Beside their chaperone activity, HSPs are well known for their anti-inflammatory and anti-stress properties by binding to JNK and IKKβ stress kinases and concomitantly suppressing their activities [11], [54]. In order to gain new insights into the role that DNAJB3 may play in the context of obesity and the functional consequences associated with its reduction in obese subjects, we sought to investigate the partners of interaction that associate with it using coimmunoprecipiation assays. Under our experimental conditions, we identified HSP-72 and two stress kinases namely JNK and IKKβ as part of the complex that specifically copurified with DNAJB3 protein (Fig. 4). Interestingly, all these partners have been linked to obesity, insulin resistance and T2D [5], [10], [16]. Our current findings raise a series of fundamental questions for future follow-up studies to elucidate the role of this understudied co-chaperone protein on metabolic diseases. One of the eminent questions is the functional consequences of these interactions on the activity of these stress kinases and if so, does DNAJB3 acts alone or in cooperation with HSP-72? Additionally, does DNAJB3 interact directly with these proteins or not? Does heat therapy induce the expression of DNAJB3 such as it is the case for HSP-72? For instance, overexpression of HSP-72 by prior heat conditioning or by ectopic expression can markedly block the activation of JNK both in vitro and in vivo [33], [55], [56] and prevent NF-κB [57]. Together, these observations illustrate the detrimental consequences associated with the activation of JNK and IKKβ stress kinases in key metabolic sites when the HSR is blunted.
Since physical exercise was shown to exert favorable effects on obesity, insulin resistance and diabetes; at least in part due to the induction of heat shock response [58], we speculated whether it can restore the expression of DNAJB3 in obese subjects with concomitant improvement of metabolic stress and clinical outcomes. As expected, our regular exercise protocol upregulated the expression of DNAJB3 and also reduced the expression of phosphoryated JNK (Fig. 3). While the negative effect of exercise on JNK phosphorylation is well established both in human and animals models [59], the upregulation of DNAJB3 by physical exercise is novel. In our case, the effect of exercise on the increase of DNAJB3 expression was at the mRNA and protein levels and it was observed in both PBMC and subcutaneous adipose tissue. Our results are similar to those reported for HSP-72 in which they showed that all the interventions that lead to the induction of HSP-72 expression; including exercise are associated with impairment of JNK phosphorylation with concomitant improvement of clinical outcomes in humans and animal models of obesity, insulin resistance and T2D [52], [58], [59], [60], [61]. Likewise, activated HSP-25 was shown to bind to IKKβ and inhibits its activity and thereby, improving insulin signaling in skeletal muscle from high fat diet-fed rats [36], [37], [56], [62]. In the present study, it is unclear whether there is a direct role of DNAJB3 on JNK and IKKβ activities or not, but from our immunopreciptation studies, the fact that DNAJB3 is part of a complex that contains JNK, IKKβ along with HSP-72 suggests that DNAJB3 might reside in the pathway modulating the activity of these stress kinases.
The chronic conditions associated with obesity such as low grade metabolic inflammation, hyperlipidemia, and enhanced oxidative and ER stress responses promoted us to explore the possible mechanisms involved in modulating the expression of DNAJB3. From our in vitro studies, only palmitate and tunicamycin were shown to trigger a significant reduction in the expression of DNAJB3 protein (Fig. 5). Under the same conditions, the expression of HSP-60 and HSP-90 were not affected by any of these treatments. Palmitate is a saturated free fatty acid well known for its cytotoxic effect. In addition to its ability to induce ER stress, palmitate acts also by increasing the levels of ceramide, reactive oxygen and nitric oxygen species, alteration of mitochondrial function [41], [42]. The observed inhibition of DNAJB3 expression with palmitate and its confirmation with tunicamycin suggest that the ER stress is involved in the downregulation of DNAJB3. Our observation is in agreement with a previous study showing more than 5-fold decrease in the expression of DNAJB3 mRNA following stimulation of HL-1 cardiomyocytes with doxazosin, a potent inducer of the ER stress [43]. In their study, the authors also detected the increased expression of two ER stress related transcription factors, namely CAAT enhancer binding protein β (C/EBPβ) and growth arrest and DNA-damage inducible protein 153 (GADD153) also known as C/EBP-homologous protein (CHOP) [43]. Interestingly, a recent study showed that stress-induced expression of CHOP was associated with repression MyoD, a gene involved in muscle differentiation [63]. The murine dnajb3 promoter has two C/EBP binding sites [64] and their role in the regulation of DNAJB3 following activation of the ER stress, if any, remains to be demonstrated. Further clarifications are also needed to confirm whether or not the observed inhibition of DNAJB3 in vitro following activation of the ER stress with palmitate occurs also in vivo in obese subjects. In our study population, we detected high levels of CHOP as well as the spliced form of the ER stress response protein, X-box-binding protein 1 (XBP1) in obese subjects (data not shown) which indicates that the ER stress is induced in obese subjects. On the other hand, our data did not rule out the effect of other inflammatory mediators since we only selected in our study a few representatives of inflammatory cytokines.
Another limitation of the study is the fact that obese subjects that participated in this study are relatively older than lean subjects (Table 1). In this regard, aging is well known to decrease the expression of HSPs such as HSP-72 [65], [66]. However, the fact that, in one hand, the expression of HSC-70 and HSP-90 were increased in obese subjects and on the other hand, the positive effect of exercise in restoring the normal expression of DNAJB3 suggest that aging is unlikely to affect the expression of DNAJB3.
In conclusion, we provided compelling evidence that the expression of the co-chaperone DNAJB3 is markedly reduced at the mRNA and protein levels in both PBMC and adipose tissue of obese subjects. We further demonstrated that physical exercise restores the normal expression of DNAJ3 to the levels comparable to lean subjects. Although we did not demonstrate the causal relationship between reduced expression of DNAJB3 and obesity, we demonstrated that DNAJB3 is part of a complex that contains key proteins involved in obesity, insulin resistance and T2D such as HSP-72, JNK and IKKβ. All together, our data support the suggestion that DNAJB3 can potentially play a protective role against obesity, and thus targeting DNAJB3 may have a potential therapeutic benefit for the control and management of obesity and insulin resistance.
Acknowledgments
We would like to thank Dr. Ron Prywes (Columbia University, New York, USA) for providing us with pCMV-ATF-6 clone. We are also indebted to staff at the Tissue Bank and Clinical Laboratory for their assistance throughout this study.
Funding Statement
This work was supported by the Kuwait Foundation for the Advancement of Sciences (KFAS) under project (RA-2010-003). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Eckel RH, Grundy SM, Zimmet PZ (2005) The metabolic syndrome. Lancet 365: 1415–1428. [DOI] [PubMed] [Google Scholar]
- 2. Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, et al. (2003) Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA 289: 76–79. [DOI] [PubMed] [Google Scholar]
- 3. Landsberg L, Aronne LJ, Beilin LJ, Burke V, Igel LI, et al. (2013) Obesity-related hypertension: pathogenesis, cardiovascular risk, and treatment: a position paper of the obesity society and the american society of hypertension. J Clin Hypertens (Greenwich) 15: 14–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flegal KM, Kit BK, Orpana H, Graubard BI (2013) Association of all-cause mortality with overweight and obesity using standard body mass index categories: a systematic review and meta-analysis. JAMA 309: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gregor MF, Hotamisligil GS (2011) Inflammatory mechanisms in obesity. Annu Rev Immunol 29: 415–445. [DOI] [PubMed] [Google Scholar]
- 6. Hummasti S, Hotamisligil GS (2010) Endoplasmic reticulum stress and inflammation in obesity and diabetes. Circ Res 107: 579–591. [DOI] [PubMed] [Google Scholar]
- 7. Furuhashi M, Ishimura S, Ota H, Miura T (2011) Lipid chaperones and metabolic inflammation. Int J Inflam 2011: 642612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruce CR, Carey AL, Hawley JA, Febbraio MA (2003) Intramuscular heat shock protein 72 and heme oxygenase-1 mRNA are reduced in patients with type 2 diabetes: evidence that insulin resistance is associated with a disturbed antioxidant defense mechanism. Diabetes 52: 2338–2345. [DOI] [PubMed] [Google Scholar]
- 9. Kurucz I, Morva A, Vaag A, Eriksson KF, Huang X, et al. (2002) Decreased expression of heat shock protein 72 in skeletal muscle of patients with type 2 diabetes correlates with insulin resistance. Diabetes 51: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 10. Hotamisligil GS (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140: 900–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hooper PL (2009) Inflammation, heat shock proteins, and type 2 diabetes. Cell Stress Chaperones 14: 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boura-Halfon S, Zick Y (2009) Phosphorylation of IRS proteins, insulin action, and insulin resistance. Am J Physiol Endocrinol Metab 296: E581–591. [DOI] [PubMed] [Google Scholar]
- 13. Neels JG, Olefsky JM (2006) Cell signaling. A new way to burn fat. Science 312: 1756–1758. [DOI] [PubMed] [Google Scholar]
- 14. Schenk S, Saberi M, Olefsky JM (2008) Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest 118: 2992–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gual P, Le Marchand-Brustel Y, Tanti JF (2005) Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie 87: 99–109. [DOI] [PubMed] [Google Scholar]
- 16. Vallerie SN, Hotamisligil GS (2010) The role of JNK proteins in metabolism. Sci Transl Med 2: 60rv65. [DOI] [PubMed] [Google Scholar]
- 17.Voellmy R (2006) Feedback regulation of the heat shock response. Handb Exp Pharmacol: 43–68. [DOI] [PubMed]
- 18. Voellmy R, Boellmann F (2007) Chaperone regulation of the heat shock protein response. Adv Exp Med Biol 594: 89–99. [DOI] [PubMed] [Google Scholar]
- 19. Morimoto RI (2011) The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol 76: 91–99. [DOI] [PubMed] [Google Scholar]
- 20. Westerheide SD, Morimoto RI (2005) Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem 280: 33097–33100. [DOI] [PubMed] [Google Scholar]
- 21. Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, et al. (2012) Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol 8: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benbrook DM, Long A (2012) Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol 34: 286–297. [PubMed] [Google Scholar]
- 23. Dremina ES, Sharov VS, Schoneich C (2012) Heat-shock proteins attenuate SERCA inactivation by the anti-apoptotic protein Bcl-2: possible implications for the ER Ca2+-mediated apoptosis. Biochem J 444: 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Asea A, Rehli M, Kabingu E, Boch JA, Bare O, et al. (2002) Novel signal transduction pathway utilized by extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. J Biol Chem 277: 15028–15034. [DOI] [PubMed] [Google Scholar]
- 25. Johnson JD, Fleshner M (2006) Releasing signals, secretory pathways, and immune function of endogenous extracellular heat shock protein 72. J Leukoc Biol 79: 425–434. [DOI] [PubMed] [Google Scholar]
- 26. Noble EG, Milne KJ, Melling CW (2008) Heat shock proteins and exercise: a primer. Appl Physiol Nutr Metab 33: 1050–1065. [DOI] [PubMed] [Google Scholar]
- 27. Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, et al. (2009) Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Calderwood SK, Murshid A, Prince T (2009) The shock of aging: molecular chaperones and the heat shock response in longevity and aging – a mini-review. Gerontology 55: 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCarty MF (2006) Induction of heat shock proteins may combat insulin resistance. Med Hypotheses 66: 527–534. [DOI] [PubMed] [Google Scholar]
- 30. Kondo T, Koga S, Matsuyama R, Miyagawa K, Goto R, et al. (2011) Heat shock response regulates insulin sensitivity and glucose homeostasis: pathophysiological impact and therapeutic potential. Curr Diabetes Rev 7: 264–269. [DOI] [PubMed] [Google Scholar]
- 31. Gupte AA, Bomhoff GL, Touchberry CD, Geiger PC (2011) Acute heat treatment improves insulin-stimulated glucose uptake in aged skeletal muscle. J Appl Physiol 110: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Najemnikova E, Rodgers CD, Locke M (2007) Altered heat stress response following streptozotocin-induced diabetes. Cell Stress Chaperones 12: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chung J, Nguyen AK, Henstridge DC, Holmes AG, Chan MH, et al. (2008) HSP72 protects against obesity-induced insulin resistance. Proc Natl Acad Sci U S A 105: 1739–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morino S, Kondo T, Sasaki K, Adachi H, Suico MA, et al. (2008) Mild electrical stimulation with heat shock ameliorates insulin resistance via enhanced insulin signaling. PLoS One 3: e4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Whitham M, Fortes MB (2008) Heat shock protein 72: release and biological significance during exercise. Front Biosci 13: 1328–1339. [DOI] [PubMed] [Google Scholar]
- 36. Gupte AA, Bomhoff GL, Morris JK, Gorres BK, Geiger PC (2009) Lipoic acid increases heat shock protein expression and inhibits stress kinase activation to improve insulin signaling in skeletal muscle from high-fat-fed rats. J Appl Physiol 106: 1425–1434. [DOI] [PubMed] [Google Scholar]
- 37. Gupte AA, Bomhoff GL, Swerdlow RH, Geiger PC (2009) Heat treatment improves glucose tolerance and prevents skeletal muscle insulin resistance in rats fed a high-fat diet. Diabetes 58: 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Literati-Nagy B, Kulcsar E, Literati-Nagy Z, Buday B, Peterfai E, et al. (2009) Improvement of insulin sensitivity by a novel drug, BGP-15, in insulin-resistant patients: a proof of concept randomized double-blind clinical trial. Horm Metab Res 41: 374–380. [DOI] [PubMed] [Google Scholar]
- 39. Marker T, Sell H, Zillessen P, Glode A, Kriebel J, et al. (2012) Heat shock protein 60 as a mediator of adipose tissue inflammation and insulin resistance. Diabetes 61: 615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aksu S, Koczan D, Renne U, Thiesen HJ, Brockmann GA (2007) Differentially expressed genes in adipose tissues of high body weight-selected (obese) and unselected (lean) mouse lines. J Appl Genet 48: 133–143. [DOI] [PubMed] [Google Scholar]
- 41. Lai E, Bikopoulos G, Wheeler MB, Rozakis-Adcock M, Volchuk A (2008) Differential activation of ER stress and apoptosis in response to chronically elevated free fatty acids in pancreatic beta-cells. Am J Physiol Endocrinol Metab 294: E540–550. [DOI] [PubMed] [Google Scholar]
- 42. Karaskov E, Scott C, Zhang L, Teodoro T, Ravazzola M, et al. (2006) Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology 147: 3398–3407. [DOI] [PubMed] [Google Scholar]
- 43. Eiras S, Fernandez P, Pineiro R, Iglesias MJ, Gonzalez-Juanatey JR, et al. (2006) Doxazosin induces activation of GADD153 and cleavage of focal adhesion kinase in cardiomyocytes en route to apoptosis. Cardiovasc Res 71: 118–128. [DOI] [PubMed] [Google Scholar]
- 44. Berruti G, Perego L, Borgonovo B, Martegani E (1998) MSJ-1, a new member of the DNAJ family of proteins, is a male germ cell-specific gene product. Exp Cell Res 239: 430–441. [DOI] [PubMed] [Google Scholar]
- 45. Hageman J, van Waarde MA, Zylicz A, Walerych D, Kampinga HH (2011) The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem J 435: 127–142. [DOI] [PubMed] [Google Scholar]
- 46. Kampinga HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buchberger A, Bukau B, Sommer T (2010) Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 40: 238–252. [DOI] [PubMed] [Google Scholar]
- 48. Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858. [DOI] [PubMed] [Google Scholar]
- 49. Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU (2002) Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci U S A 99 Suppl 416412–16418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hageman J, Rujano MA, van Waarde MA, Kakkar V, Dirks RP, et al. (2010) A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell 37: 355–369. [DOI] [PubMed] [Google Scholar]
- 51. Ye CF, Li H (2009) HSP40 ameliorates impairment of insulin secretion by inhibiting huntingtin aggregation in a HD pancreatic beta cell model. Biosci Biotechnol Biochem 73: 1787–1792. [DOI] [PubMed] [Google Scholar]
- 52. Atalay M, Oksala NK, Laaksonen DE, Khanna S, Nakao C, et al. (2004) Exercise training modulates heat shock protein response in diabetic rats. J Appl Physiol 97: 605–611. [DOI] [PubMed] [Google Scholar]
- 53. Gupte AA, Bomhoff GL, Geiger PC (2008) Age-related differences in skeletal muscle insulin signaling: the role of stress kinases and heat shock proteins. J Appl Physiol 105: 839–848. [DOI] [PubMed] [Google Scholar]
- 54. Simar D, Jacques A, Caillaud C (2012) Heat shock proteins induction reduces stress kinases activation, potentially improving insulin signalling in monocytes from obese subjects. Cell Stress Chaperones 17: 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gabai VL, Meriin AB, Mosser DD, Caron AW, Rits S, et al. (1997) Hsp70 prevents activation of stress kinases. A novel pathway of cellular thermotolerance. J Biol Chem 272: 18033–18037. [DOI] [PubMed] [Google Scholar]
- 56. Park KJ, Gaynor RB, Kwak YT (2003) Heat shock protein 27 association with the I kappa B kinase complex regulates tumor necrosis factor alpha-induced NF-kappa B activation. J Biol Chem 278: 35272–35278. [DOI] [PubMed] [Google Scholar]
- 57. Meldrum KK, Burnett AL, Meng X, Misseri R, Shaw MB, et al. (2003) Liposomal delivery of heat shock protein 72 into renal tubular cells blocks nuclear factor-kappaB activation, tumor necrosis factor-alpha production, and subsequent ischemia-induced apoptosis. Circ Res 92: 293–299. [DOI] [PubMed] [Google Scholar]
- 58. Gleeson M, Bishop NC, Stensel DJ, Lindley MR, Mastana SS, et al. (2011) The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat Rev Immunol 11: 607–615. [DOI] [PubMed] [Google Scholar]
- 59. Golbidi S, Mesdaghinia A, Laher I (2012) Exercise in the metabolic syndrome. Oxid Med Cell Longev 2012: 349710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ogawa K, Seta R, Shimizu T, Shinkai S, Calderwood SK, et al. (2011) Plasma adenosine triphosphate and heat shock protein 72 concentrations after aerobic and eccentric exercise. Exerc Immunol Rev 17: 136–149. [PubMed] [Google Scholar]
- 61. Teixeira-Lemos E, Nunes S, Teixeira F, Reis F (2011) Regular physical exercise training assists in preventing type 2 diabetes development: focus on its antioxidant and anti-inflammatory properties. Cardiovasc Diabetol 10: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alford KA, Glennie S, Turrell BR, Rawlinson L, Saklatvala J, et al. (2007) Heat shock protein 27 functions in inflammatory gene expression and transforming growth factor-beta-activated kinase-1 (TAK1)-mediated signaling. J Biol Chem 282: 6232–6241. [DOI] [PubMed] [Google Scholar]
- 63. Alter J, Bengal E (2011) Stress-induced C/EBP homology protein (CHOP) represses MyoD transcription to delay myoblast differentiation. PLoS One 6: e29498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meccariello R, Berruti G, Chianese R, De Santis R, Di Cunto F, et al. (2008) Structure of msj-1 gene in mice and humans: a possible role in the regulation of male reproduction. Gen Comp Endocrinol 156: 91–103. [DOI] [PubMed] [Google Scholar]
- 65. Gutsmann-Conrad A, Pahlavani MA, Heydari AR, Richardson A (1999) Expression of heat shock protein 70 decreases with age in hepatocytes and splenocytes from female rats. Mech Ageing Dev 107: 255–270. [DOI] [PubMed] [Google Scholar]
- 66. Kim G, Meriin AB, Gabai VL, Christians E, Benjamin I, et al. (2012) The heat shock transcription factor Hsf1 is downregulated in DNA damage-associated senescence, contributing to the maintenance of senescence phenotype. Aging Cell 11: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 68. Ghebeh H, Tulbah A, Mohammed S, Elkum N, Bin Amer SM, et al. (2007) Expression of B7-H1 in breast cancer patients is strongly associated with high proliferative Ki-67-expressing tumor cells. Int J Cancer 121: 751–758. [DOI] [PubMed] [Google Scholar]