Abstract
Duchenne muscular dystrophy (DMD), one of the most common and lethal genetic disorders, and the mdx mouse myopathies are caused by a lack of dystrophin protein. These dystrophic muscles contain sporadic clusters of dystrophin-expressing revertant fibers (RFs), as detected by immunohistochemistry. RFs are known to arise from muscle precursor cells with spontaneous exon skipping (alternative splicing) and clonally expand in size with increasing age through the process of muscle degeneration/regeneration. The expansion of revertant clusters is thought to represent the cumulative history of muscle regeneration and proliferation of such precursor cells. However, the precise mechanisms by which RFs arise and expand are poorly understood. Here, to test the effects of mutation types and aging on RF expansion and muscle regeneration, we examined the number of RFs in mdx mice (containing a nonsense mutation in exon 23) and mdx52 mice (containing deletion mutation of exon 52) with the same C57BL/6 background at 2, 6, 12, and 18months of age. Mdx mice displayed a significantly higher number of RFs compared to mdx52 mice in all age groups, suggesting that revertant fiber expansion largely depends on the type of mutation and/or location in the gene. A significant increase in the expression and clustering levels of RFs was found beginning at 6months of age in mdx mice compared with mdx52 mice. In contrast to the significant expansion of RFs with increasing age, the number of centrally nucleated fibers and embryonic myosin heavy chain-positive fibers (indicative of cumulative and current muscle regeneration, respectively) decreased with age in both mouse strains. These results suggest that mutation types and aging differently affect revertant fiber expansion in mdx and mdx52 mice.
Introduction
Duchenne muscular dystrophy (DMD) is the most common genetic muscular disease and is characterized by progressive muscle degeneration. It occurs with a frequency of about 1 out of 3,500 boys, and is caused by mutations in the dystrophin (DMD) gene [1]–[3]. The mutation leads to progressive myopathy and muscle weakness coupled with cycles of muscle degeneration. Death eventually occurs due to severe respiratory and/or cardiac failure at approximately 20–30years of age [4], [5]. There is currently no effective cure for DMD [6]–[8]. The DMD gene is located on the X-chromosome and is characterized as one of the largest and more complex genes in humans, containing 79 exons and spanning more than 2.4million base pairs [9], [10]. The DMD gene encodes at least 18 protein isoform products of dystrophin with tissue specific alternative promoters [11]–[26]. The main skeletal muscle isoform is the largest known, consisting of 3,685-amino acids (427kDa). In skeletal muscles, dystrophin plays a central role in organizing a multi-protein complex at the sarcolemma, and linking cytoskeleton proteins to extracellular matrix proteins [27], [28]. The full-length dystrophin protein can be divided into actin-binding NH2- (N) terminal, rod, cysteine-rich, and COOH- (C) terminal domains [14], [20]. Interestingly, most of the known functions of the protein are assigned to the N- and C- terminals and the cysteine-rich domain [29]. In contrast, the central rod domain, consisting of 24 spectrin repeats with 4 hinges, and spanning about half the length of the protein, appears to be less essential to proper function [30].
Most DMD mutations, which cause disruption of the open reading frame, occur within the central rod domain, thus preventing translation of the crucial C-terminal domain [31]–[33]. In the case of Becker muscular dystrophy (BMD), a milder form of muscular dystrophy also caused by mutations in the DMD gene, most mutations occur in the same rod domain regions, but the mutated mRNA transcripts preserve the open reading-frame and are thus translated into a truncated yet partially functional protein [34]. The mdx mouse, an animal model of human DMD, harbors a natural nonsense point mutation in exon 23, while the mdx52 mouse has a deletion mutation of exon 52 owing to gene-targeting method [35]–[38]. Mdx mice were originally discovered with C57BL/10 background, but they were later backcrossed with C57BL/6 background for comparison with other mouse models including mdx52 mice with C57BL/6 background [39]. Both mutants exhibit a lack of dystrophin expression and cycles of muscle degeneration/regeneration [40], [41]. Thus, the comparison between mdx and mdx52 mice with the same genetic background should be quite useful in understanding pathogenic mechanisms dependent on different types of nonsense and exon-deletion mutations, which are found in approximately 10–15% and 50–60% of the DMD patients, respectively [42], [43].
Interestingly, skeletal muscles in most DMD patients and animal models including mouse and dog models display sporadic dystrophin-positive muscle fibers called “revertant fibers” (RFs) in an otherwise dystrophin-negative background [44]–[51]. Danko et al report distinct frequencies of RFs in different mdx mutants (mdx2cv, 3cv, 4cv, and 5cv mutants) [50]. These dystrophin-positive fibers arise from spontaneous exon skipping (alternative splicing) with a loss of up to 30 exons, leading to the production of in-frame truncated proteins [52]. In mdx mice, these RFs expand with age through cycles of muscle degeneration/regeneration and the activation of muscle precursor cells [40], [52]. In human DMD patients, the increase of RFs correlates significantly with their age up to early teens [53]. These revertant events are thought to arise within a subset of muscle precursor cells which proliferate in response to muscle degeneration and participate in regeneration of muscle fibers [40]. These fibers often produce clonal clusters that can expand to up to 100 fibers measuring more than 1mm in length by 18months of age [52]. To date, there is no report of either genomic deletion or secondary mutation as the mechanism facilitating the restoration of dystrophin in RFs. Our previous studies using various transgenic mouse models, including micro-dystrophin transgenic mdx mice and utrophin overexpressing mice, as well as irradiated mdx models, clearly demonstrated that expansion of RF clusters is dependent on muscle regeneration [40], [54]–[57]. The expansion of RF clusters reflects the cumulative history of skeletal muscle regeneration and is thought to provide a useful index for functional evaluation of therapies that diminish muscle degeneration [40].
In this study, we employed two newly developed mouse models, mdx mice with C57BL/6 background, and mdx52 mice [37], [39]. We compared their RF generation and long-term expansion up to 18months of age. Interestingly, mdx mice exhibited a higher number of RFs compared to mdx52 mice, indicating the occurrence of revertant events largely depends on the type of mutation present in the DMD gene. To our surprise, although both mouse models showed an increase in RFs through 18months, the number of centrally nucleated fibers (CNFs) and embryonic myosin heavy chain (eMHC)-positive fibers (as cumulative and current indicators of muscle regeneration, respectively) decreased with age in both the strains except for an increase in CNFs between 2 to 6months of age in mdx mice. Overall, the dynamics of muscle regeneration associated with age was more markedly altered in mdx than mdx52 mice. These data reported here show that mutation types and aging differently affect RF expansion and muscle regeneration in mdx and mdx52 mice.
Materials and Methods
Ethics Statement
All animal works were conducted according to relevant national and international guidelines. Animal study was approved by the Ethics Committee for the Treatment of Laboratory Animals of the National Center of Neurology and Psychiatry, and the Animal Care and Use Committee (ACUC) of the University of Alberta. All animals were euthanized by cervical dislocation by trained personnel.
Animals
Mdx mice with C57BL/6 background, mdx52 mice, and C57BL/6 mice as controls were used in this study [37], [39], [58]. The genetic background of mdx mice was changed into C57BL/6 as previously described [39]. Male and female mdx and mdx52 mice at 2, 6, 12, and 18months of age were used in this study. Homozygous mutation of the DMD gene was confirmed in female mice by genotyping. Male C57BL/6 mice at 2months of age were used as controls. Mice were euthanized by cervical dislocation; then, tibialis anterior (TA) and gastrocnemius (GC) muscles were excised and immediately frozen in liquid nitrogen-cooled isopentane. Samples were stored at −80°C until used for immunohistochemistry and histochemistry.
Immunohistochemical analysisAntibodies
The rabbit polyclonal antibody against C-terminal domain (position at 3,661–3,677 amino acids) in human dystrophin was used to detect revertant fibers (Abcam, Bristol, UK). eMHC expressed in newly regenerated muscle fibers was detected with mouse monoclonal anti-rabbit developmental MHC antibody (Leica Biosystems, Newcastle upon Tyne, UK).
Immunohistochemistry
Dystrophin-positive RFs were detected by immunohistochemistry. Transverse frozen sections (7μm thick) from TA and GC muscles were cut using a Leica CM1900 cryostat (Leica Micro-systems, Wetzlar, Germany). Serial sections were picked up on poly-L-lysine-coated glass microscope slides and air-dried for 30min. Unfixed sections were then blocked in phosphate-buffered saline (PBS) with 20% goat serum, 0.1% TritonX-100 for one hour at room temperature. Dystrophin was detected with rabbit polyclonal primary antibody against human dystrophin C-terminal (1∶400) in the blocking solution by overnight incubation at 4°C. After washing 3 times with PBS, the primary antibody was detected with AlexaFluorTM 488-conjugated goat anti-rabbit IgG secondary antibody (1∶2,000) (Molecular Probes, OR, USA) with one-hour room temperature incubation. Nuclear counterstaining was performed with 4',6-diamidino-2-phenylindole (DAPI) in a mounting agent (Vectashield; Vector Laboratories, CA, USA). Expression of eMHC and its co-localization with RFs were assessed by triple staining using anti-developmental MHC antibody (1∶20), anti-dystrophin antibody (1∶400), and DAPI after blocking with Mouse on Mouse (M.O.M.) blocking reagent (Vector Laboratories).
Assessment of RFs and eMHC-positive fibers
The muscle RF assessment was performed according to our previous study [40]. RFs and eMHC-positive fibers in entire TA or GC muscle sections were observed using a fluorescence microscope (Nikon Eclipse TE 2000-U; Nikon, Tokyo, Japan) with a 20X objective lens. Muscle fibers were regarded as dystrophin-positive only when more than half the membrane circumference was stained in cross-sections. RFs immediately adjacent to each other were characterized as a single cluster. For closer comparison of RFs and eMHC-positive fibers in mice of different groups, at least 8 serial sections at every 100μm from the muscle belly were stained with antibodies to assess the following: the number of RFs in a cross-section, the number of revertant clusters, the maximum number of RFs in a cluster, and the number of eMHC-positive fibers. For data per mouse, the highest number among serial sections was averaged between left and right muscles for each of the aforementioned criteria. Large clusters of eMHC-positive fibers (clusters composed of more than one hundred eMHC-positive fibers, which arose due to severe degeneration and were independent of age) were excluded from counting.
Hematoxylin and Eosin staining
Hematoxylin and eosin (HE) staining was performed with Mayer's hematoxylin and eosin solutions as previously described [41]. A DMR microscope (Leica Micro-systems) was used for bright field microscopy with a 20x objective lens. The percentage of CNFs was calculated in 300 to 1,000 fibers in each left and right muscle, and was averaged between two muscles per mouse.
Statistical analysis
All data were reported as mean values ± standard deviation (S.D.). The statistical differences between the age groups or the strains were assessed by one-way ANOVA with a Tukey–Kramer multiple comparison test. Pearson correlation coefficient was performed between the number of RFs in a section, the number of RF clusters, and the maximum number of RFs in a single cluster. P<0.05 was considered statistically significant.
Results
Distinct patterns of revertant fiber expression and clustering in mdx and mdx52 mice
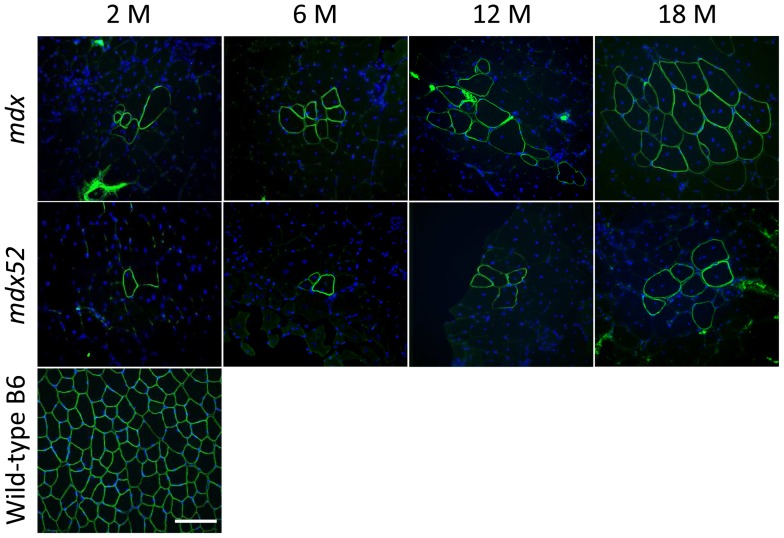
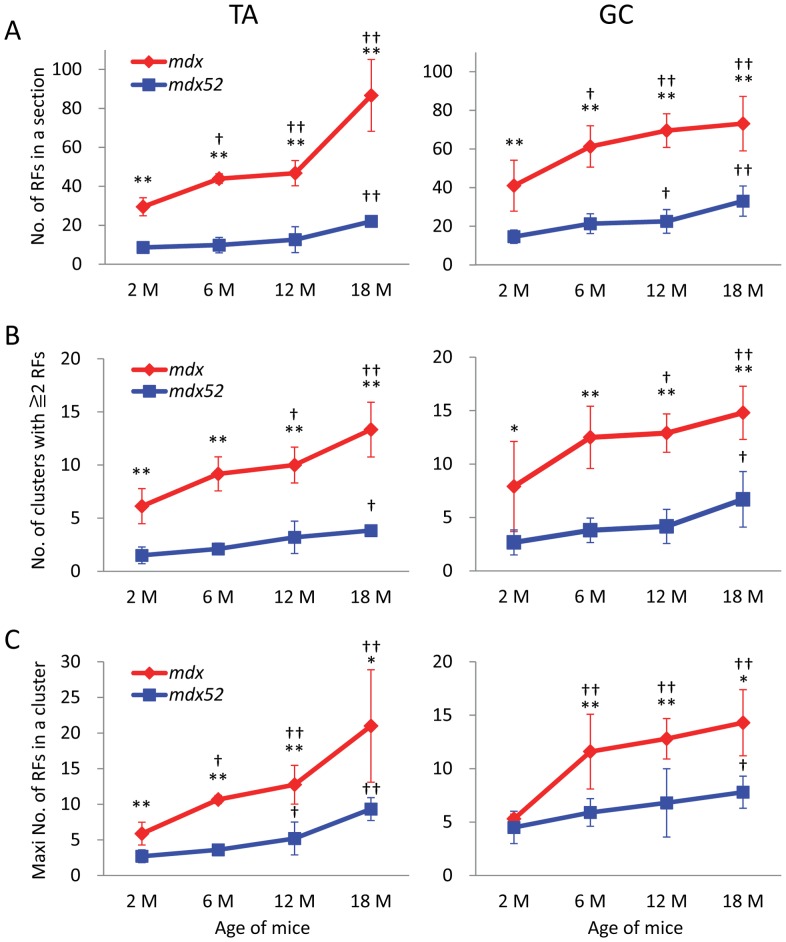
To investigate the effect of different mutation types of the DMD gene on the generation and expansion of RF clusters, we examined RFs in TA and GC muscles of mdx mice with C57BL/6 background and mdx52 mice (also with C57BL/6 background). Mdx mice contain a nonsense mutation in exon 23, and mdx52 mice contain a deletion mutation in exon 52 [36], [37]. RFs were observed in all age groups in both mdx and mdx52 mice and centrally-located nuclei were found in most RFs ( Figure 1 ). A rabbit polyclonal antibody against the C-terminal domain of dystrophin was used to detect revertant dystrophin expression because this amino acid-region is reported to be retained in most of the truncated dystrophin or RF proteins in mdx and DMD patients [49], [52], [59]. Mdx mice clearly showed a higher number of RFs and RF clusters in TA and GC muscles across all age groups in comparison to mdx52 mice ( Figures 1 , 2A, and B ). There were significantly fewer RFs and RF clusters in mdx52 mice when compared to mdx mice. Mdx mice also showed a significantly higher number of RFs per cluster compared with mdx52 mice, except at 2months of age in GC muscle ( Figure 2C ).
Figure 1. Dystrophin-positive revertant fibers with central nuclei at ages of 2, 6, 12, and 18 months in mdx and mdx52 mice.
Representative immunohistochemical images of maximum clusters of RFs in TA muscles are shown in each group. Mdx shows a higher maximum number of RFs than mdx52 in all age groups. Wild-type C57BL/6 muscle at 2months old is displayed as a control. An anti-dystrophin C-terminal antibody (green) and DAPI staining (blue) were used. M: months. 20x objective lens, scale bar = 100μm.
Figure 2. Mutation- and age-related expression of dystrophin-positive revertant fibers in TA and GC muscles from mdx and mdx52 mice.
(A) The number of RFs in one TA or GC section. (B) The number of RF clusters containing 2 or more positive fibers. (C) The maximum number of RFs in a single cluster. Mdx mice have a significantly higher number of RFs in all criteria than mdx52 mice except for 2months of age in maximum number of RFs per cluster. The number of RFs in all criteria increases with age. Values are mean ± S.D. (n = 3–6 mice per each group). *P<0.05, **P<0.01 between mdx and mdx52 mice; †P<0.05, ††P<0.05 compared to 2months old. M: months.
We then compared the expression and clustering levels of RFs at 2, 6, 12, and 18months of age for mdx and mdx52 strains individually. In TA and GC muscles from mdx and mdx52 mice, the number of RF, RF clusters, and the maximum number of RFs per cluster significantly increased compared to 2months of age ( Figures 2A, B and C ). The Pearson correlation coefficient showed a strong correlation among these three criteria regarding expression and clustering levels of RFs in each strain (squared correlation coefficient; R2 = 0.27–0.84, P<0.05). A significant increase in the number of RFs and the maximum number of RFs per cluster at 6months of age was found only in mdx mice. Mdx mice at 12months old also showed significant increase in the number of RF clusters compared to 2months old. In contrast to the dramatic RF expansion with age in mdx mice, mdx52 mice showed only a moderate increase in RFs with age across all measured criteria. A significant increase in RF expression and clustering was found at 18months in mdx mice compared with mdx52 mice. Levels of RF expression/clustering in mdx52 mice never reached levels seen in mdx mice. This result is not attributable to genetic background, since both mouse strains have a C57BL/6 background. The mdx52 mouse, which has fewer RFs and shows little change in RF expansion with age, may be a good model for testing of new therapies aimed at restoring dystrophin expression, because pre-existing RFs can disturb accurate assessments of the restored dystrophin expression owing to therapeutic treatment.
Dynamics of muscle regeneration with age is different between mdx and mdx52 mice
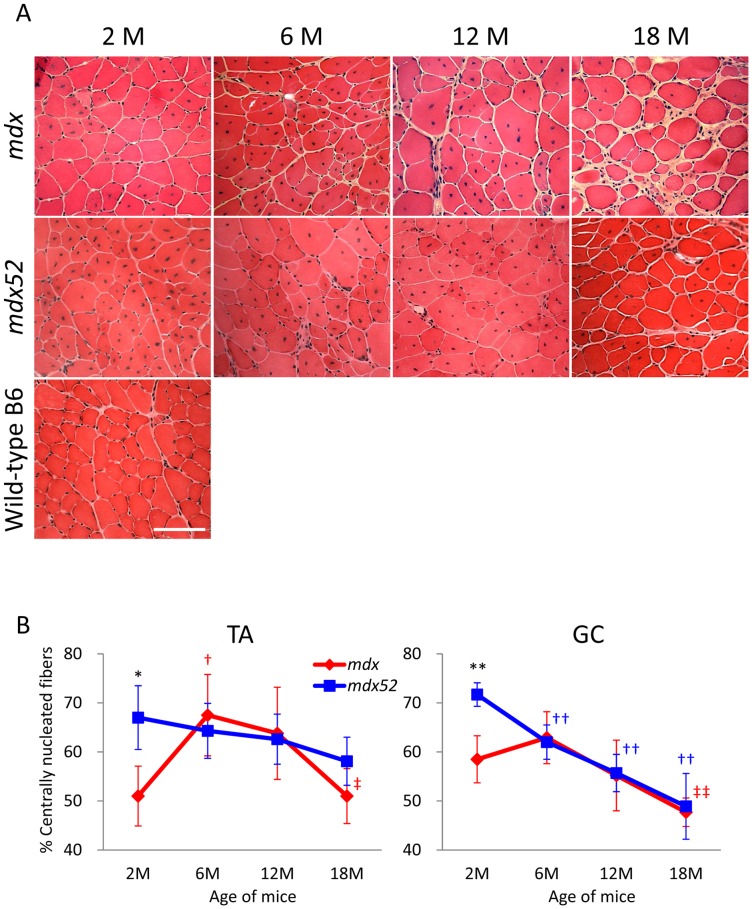
To examine whether muscle regeneration is physiologically correlated with RF expansion, we assessed CNFs and eMHC-positive fibers, which are cumulative and current indicators of muscle regeneration, respectively, in mdx and mdx52 mice. During muscle regeneration, regenerated skeletal muscle fibers are centrally nucleated as opposed to mature muscle fibers which are peripherally nucleated [60], [61]. Also, centrally-located nuclei in mdx mice are reported to persist long-term, indicating that the percentage of CNFs represents the accumulated history of regeneration up to the present [62], [63]. Following HE staining, mdx mice showed a significantly lower percentage of CNFs than mdx52 mice at 2months, followed by an increase in percent CNFs resembling observations in mdx52 mice at 6months, and a subsequent decrease in CNFs at 12 and 18months ( Figures 3A and B ). At 18months old in mdx mice, a significant decrease in CNFs was found in both TA and GC muscles when compared to 6months (the age of the peak CNF percentage). In contrast to the dynamics of CNFs in mdx mice, the percentage of CNFs in mdx52 mice consistently and significantly decreased over time from the peak of 2months old. This finding indicates that there is a different peak in muscle regeneration between mdx and mdx52 mice, which is before 6months of age and before 2months of age, respectively.
Figure 3. Distinct changes in the percentage of centrally nucleated fibers by mutations and age in mdx and mdx52 mice.
(A) Representative images of TA muscles from mdx and mdx52 mice at ages 2, 6, 12 and 18months with hematoxylin and eosin staining. Wild-type C57BL/6 muscle at 2months of age is displayed as a control. M: months. Scale bar = 100μm. (B) The percentage of centrally nucleated fibers in TA and GC muscles from mdx and mdx52 mice. Three hundred to one thousand myofibers were counted in left and right muscles and the percentage of CNFs was averaged between the two muscles per mouse. Values are mean ± S.D. (n = 3–6 mice per group). *P<0.05, **P<0.01 between mdx and mdx52 mice; †P<0.05, ††P<0.01 compared to 2months old; ‡P<0.05, ‡‡P<0.01 compared to 6months old. Symbol colors are accordant with the color of mice (red; mdx, blue; mdx52).
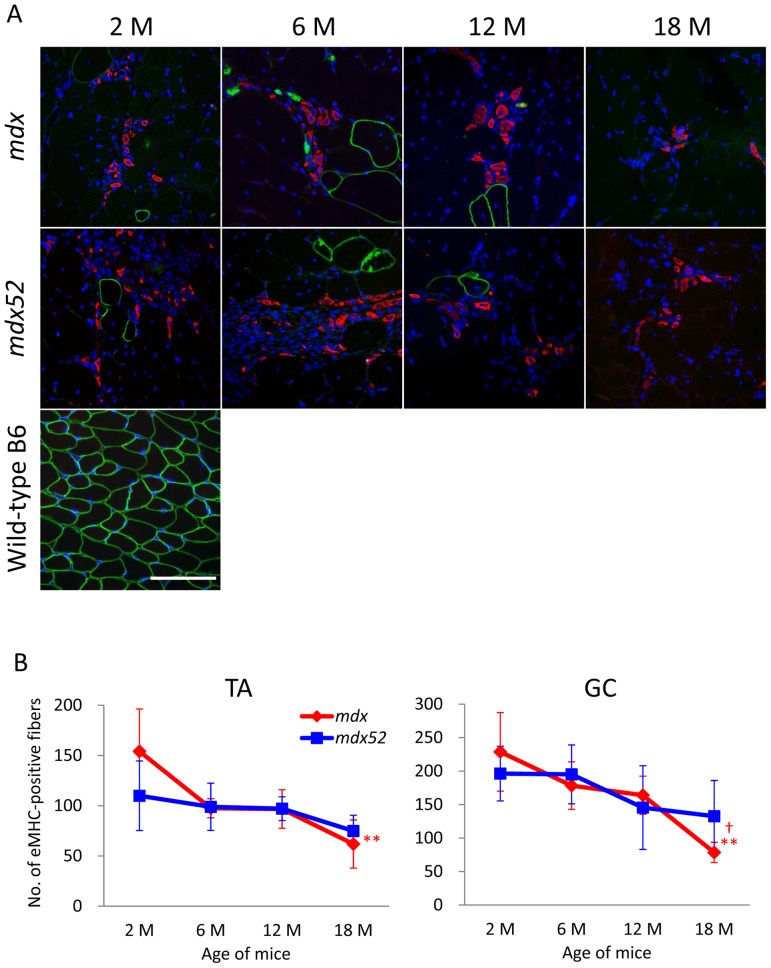
To examine the possible correlation between ongoing muscle regeneration and RF expansion, we analyzed the co-localization of eMHC with revertant dystrophin and the number of eMHC-positive fibers, which are found short-term during muscle regeneration. Interestingly, eMHC was not expressed in RFs at any age in mdx and mdx52 mice ( Figure 4A ). The number of eMHC-positive fibers was similar between mdx and mdx52 mice at each age ( Figure 4A and B ). However, expression dynamics of eMHC with age was different between the strains. In mdx mice, the number of eMHC-positive fibers was the highest at 2months of age and significantly decreased at 18months of age in TA and GC muscles, while in mdx52 mice, the number was slightly but not significantly decreased through 18months.”. These data from CNFs and eMHC-positive fibers indicate that in mdx and mdx52 mice the dystrophic skeletal muscles actively regenerate throughout their life span but the muscle regeneration activity with age is different between the strains.
Figure 4. No expression of eMHC in RFs and attenuation of ongoing muscle regeneration in aged mdx and mdx52 mice.
(A) Triple staining of mdx and mdx52 mice for RF (green), eMHC (red), and nucleus (blue). Revertant dystrophin is not co-localized with newly regenerated eMHC-positive fibers in TA and GC muscles from mdx and mdx52 mice at any age. The pictures are representative GC muscles from mdx and mdx52 mice at each age. 20x objective lens, scale bar = 100μm. (B) The number of eMHC-positive fibers. Values are mean ± S.D. (n = 3–6 mice per group). A significant decrease in the number of eMHC-positive fibers is found only at 18months old in mdx mice (**P<0.01 compared to 2months old, †P<0.05 compared to 6 and 12months old). Symbol colors are accordant with the color of mice (red; mdx, blue; mdx52).
Discussion
Dystrophin RFs were first described more than 20years ago by Hoffman et al [44], however, the mechanisms behind both the generation and expansion of RFs remain poorly understood. Lu et al previously reported that revertant events arise in individual muscle satellite cells at around birth [52]. RFs appear at around birth as short segments, approximately 10µm in length, of sporadic single muscle fibers. One of the most interesting characteristics of the RFs is their substantial expansion with age. In mdx mice, RFs continuously expand at least up to 18months, resulting in clusters containing more than 100 fibers and traversing 1mm or more of muscle fiber length. However, RFs never accumulate to a sufficient number to significantly ameliorate dystrophic muscle pathology [40].
Our previous study with irradiated mdx mice aged up to 2months supported the hypothesis that muscle regeneration is essential for RF expansion in mdx mice [40]. The expansion of RFs has been attributed to the combined effects of cycles of muscle degeneration/regeneration and increased survival of the fibers containing truncated dystrophin [40], [64]. However, in this paper, we demonstrated that mutation types and aging differently affect generation and expansion of dystrophin-positive RFs in mdx and mdx52 mice.
Our paradoxical result reported here strongly challenges our previous hypothesis, since rapid expansion of RFs and a decrease in regenerated CNFs and eMHC-positive fibers occur concurrently at older ages ( Figures 1 – 4 ). This is in sharp contrast to previous experiments with transgenic mdx muscles [40], [65]. No expansion of RFs was observed in any transgenic mdx mice expressing mini- or micro-dystrophin in spite of the significant muscle growth before the age of 5 weeks. Instead, the number of RFs decreased with age in these transgenic mdx mice. Our observations here lead us to propose two possibilities to explain RF expansion with age: first, mechanisms other than muscle regeneration, such as secondary DNA mutation, up-regulation of short dystrophin isoforms, or age-related changes affecting splicing machinery may be involved in RF expansion in older muscle fibers. The expansion of RFs independent of muscle regeneration supports the existence of a mechanism in aged muscles that Lu et al described previously; i.e. RF expansion may represent the progressive increase in a territory of factors, each of which determines a specific pattern of exon skipping (alternative splicing), and which spread by diffusion within each fiber and between neighboring fibers [52]. This hypothesis predicts that RF clusters grow within the existing stable (mature) muscle fibers. A second possibility is that the increase of RFs with age may be related to RF stability combined with the expression of partially functional dystrophin proteins; thus, more stable RFs would accumulate with age and prevent degeneration/regeneration cycles in themselves and in closely surrounding fibers. This could lead to a decrease in muscle regeneration with age in both mouse strains. Nevertheless, muscle regeneration appears to be fundamentally required for RF expansion because central nucleation was found in most RFs through 18months in this study, and our previous study demonstrated that RF expansion does not occur in the absence of regeneration, even when degeneration continues after irradiation [40]. Another hypothesis to possibly explain these conflicting observations is that regenerating muscle fibers with central nucleation and/or eMHC in older dystrophic mice might change to mature fibers without them more rapidly than in younger dystrophic mice; thus, a reduced number of CNFs and eMHC-positive fibers might not reflect the frequency of regeneration in older muscles, although there is currently no evidence to support this hypothesis.
As no such phenomenon like RFs has been described in other genetic disorders, the mechanism of their genesis and expansion is still open to speculation, although it seems modification of splicing is involved. Thus, RFs could be an interesting model with which to investigate the mechanisms of spontaneous exon skipping (alternative splicing). Future molecular analysis of RFs will also provide invaluable information toward the development of molecular therapies, such as antisense-mediated exon skipping, which are aimed at inducing the production of revertant-like dystrophin-positive fibers for the treatment of DMD [8], [66], [67].
Acknowledgments
We thank Dr. Shin'ichi Takeda (Department of Molecular Therapy, National Institute of Neuroscience, National Center of Neurology and Psychiatry) for providing the mdx52 mouse samples for this study and for useful discussions.
Funding Statement
This work was supported by the University of Alberta Faculty of Medicine and Dentistry, Parent Project Muscular Dystrophy (USA), The Friends of Garrett Cumming Research Fund, HM Toupin Neurological Science Research Fund, Muscular Dystrophy Canada, Canada Foundation for Innovation, Alberta Advanced Education and Technology, and the Women and Children's Health Research Institute (WCHRI). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lovering RM, Porter NC, Bloch RJ (2005) The muscular dystrophies: from genes to therapies. Phys Ther 85: 1372–1388. [PMC free article] [PubMed] [Google Scholar]
- 2.Biggar WD, Klamut HJ, Demacio PC, Stevens DJ, Ray PN (2002) Duchenne muscular dystrophy: current knowledge, treatment, and future prospects. Clin Orthop Relat Res: 88–106. [DOI] [PubMed]
- 3. Duchenne (1867) The pathology of paralysis with muscular degeneration (paralysie myosclerotique), or paralysis with apparent hypertrophy. Br Med J 2: 541–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McNally EM, Pytel P (2007) Muscle diseases: the muscular dystrophies. Annu Rev Pathol 2: 87–109. [DOI] [PubMed] [Google Scholar]
- 5. Hyser CL, Mendell JR (1988) Recent advances in Duchenne and Becker muscular dystrophy. Neurol Clin 6: 429–453. [PubMed] [Google Scholar]
- 6. Lu QL, Yokota T, Takeda S, Garcia L, Muntoni F, et al. (2011) The status of exon skipping as a therapeutic approach to duchenne muscular dystrophy. Mol Ther 19: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pichavant C, Aartsma-Rus A, Clemens PR, Davies KE, Dickson G, et al. (2011) Current status of pharmaceutical and genetic therapeutic approaches to treat DMD. Mol Ther 19: 830–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffman EP, Bronson A, Levin AA, Takeda S, Yokota T, et al. (2011) Restoring dystrophin expression in Duchenne muscular dystrophy muscle progress in exon skipping and stop codon read through. Am J Pathol 179: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, et al. (1987) Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50: 509–517. [DOI] [PubMed] [Google Scholar]
- 10. Hoffman EP Brown RH Jr., Kunkel LM (1987 ) Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51: 919–928. [DOI] [PubMed] [Google Scholar]
- 11. Holder E, Maeda M, Bies RD (1996) Expression and regulation of the dystrophin Purkinje promoter in human skeletal muscle, heart, and brain. Hum Genet 97: 232–239. [DOI] [PubMed] [Google Scholar]
- 12. Feener CA, Koenig M, Kunkel LM (1989) Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus. Nature 338: 509–511. [DOI] [PubMed] [Google Scholar]
- 13. D'Souza VN, Nguyen TM, Morris GE, Karges W, Pillers DA, et al. (1995) A novel dystrophin isoform is required for normal retinal electrophysiology. Hum Mol Genet 4: 837–842. [DOI] [PubMed] [Google Scholar]
- 14. Nishio H, Takeshima Y, Narita N, Yanagawa H, Suzuki Y, et al. (1994) Identification of a novel first exon in the human dystrophin gene and of a new promoter located more than 500 kb upstream of the nearest known promoter. J Clin Invest 94: 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blake DJ, Weir A, Newey SE, Davies KE (2002) Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev 82: 291–329. [DOI] [PubMed] [Google Scholar]
- 16. Muntoni F, Cau M, Congiu R, Congia M, Cao A, et al. (1993) Identification of a novel T-insertion polymorphism at the DMD locus. Hum Genet 92: 103. [DOI] [PubMed] [Google Scholar]
- 17. Muntoni F, Cau M, Ganau A, Congiu R, Arvedi G, et al. (1993) Brief report: deletion of the dystrophin muscle-promoter region associated with X-linked dilated cardiomyopathy. N Engl J Med 329: 921–925. [DOI] [PubMed] [Google Scholar]
- 18. Wheway JM, Roberts RG (2003) The dystrophin lymphocyte promoter revisited: 4.5-megabase intron, or artifact? Neuromuscul Disord 13: 17–20. [DOI] [PubMed] [Google Scholar]
- 19. Nudel U, Zuk D, Einat P, Zeelon E, Levy Z, et al. (1989) Duchenne muscular dystrophy gene product is not identical in muscle and brain. Nature 337: 76–78. [DOI] [PubMed] [Google Scholar]
- 20. Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, et al. (1986) Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature 323: 646–650. [DOI] [PubMed] [Google Scholar]
- 21. Gorecki DC, Monaco AP, Derry JM, Walker AP, Barnard EA, et al. (1992) Expression of four alternative dystrophin transcripts in brain regions regulated by different promoters. Hum Mol Genet 1: 505–510. [DOI] [PubMed] [Google Scholar]
- 22. Lidov HG, Selig S, Kunkel LM (1995) Dp140: a novel 140 kDa CNS transcript from the dystrophin locus. Hum Mol Genet 4: 329–335. [DOI] [PubMed] [Google Scholar]
- 23. Byers TJ, Lidov HG, Kunkel LM (1993) An alternative dystrophin transcript specific to peripheral nerve. Nat Genet 4: 77–81. [DOI] [PubMed] [Google Scholar]
- 24. Lederfein D, Levy Z, Augier N, Mornet D, Morris G, et al. (1992) A 71-kilodalton protein is a major product of the Duchenne muscular dystrophy gene in brain and other nonmuscle tissues. Proc Natl Acad Sci U S A 89: 5346–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Austin RC, Howard PL, D'Souza VN, Klamut HJ, Ray PN (1995) Cloning and characterization of alternatively spliced isoforms of Dp71. Hum Mol Genet 4: 1475–1483. [DOI] [PubMed] [Google Scholar]
- 26. Tinsley JM, Blake DJ, Davies KE (1993) Apo-dystrophin-3: a 2.2kb transcript from the DMD locus encoding the dystrophin glycoprotein binding site. Hum Mol Genet 2: 521–524. [DOI] [PubMed] [Google Scholar]
- 27. Rybakova IN, Patel JR, Ervasti JM (2000) The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol 150: 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watkins SC, Cullen MJ, Hoffman EP, Billington L (2000) Plasma membrane cytoskeleton of muscle: a fine structural analysis. Microsc Res Tech 48: 131–141. [DOI] [PubMed] [Google Scholar]
- 29. Bies RD, Caskey CT, Fenwick R (1992) An intact cysteine-rich domain is required for dystrophin function. J Clin Invest 90: 666–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhasin N, Law R, Liao G, Safer D, Ellmer J, et al. (2005) Molecular extensibility of mini-dystrophins and a dystrophin rod construct. J Mol Biol 352: 795–806. [DOI] [PubMed] [Google Scholar]
- 31. Yokota T, Takeda S, Lu QL, Partridge TA, Nakamura A, et al. (2009) A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground. Arch Neurol 66: 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, Burke MM, Nagel N, et al. (2010) A duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient cavalier king charles spaniels is amenable to exon 51 skipping. PLoS One 5: e8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gualandi F, Rimessi P, Trabanelli C, Spitali P, Neri M, et al. (2006) Intronic breakpoint definition and transcription analysis in DMD/BMD patients with deletion/duplication at the 5' mutation hot spot of the dystrophin gene. Gene 370: 26–33. [DOI] [PubMed] [Google Scholar]
- 34. Covone AE, Lerone M, Romeo G (1991) Genotype-phenotype correlation and germline mosaicism in DMD/BMD patients with deletions of the dystrophin gene. Hum Genet 87: 353–360. [DOI] [PubMed] [Google Scholar]
- 35. Bulfield G, Siller WG, Wight PA, Moore KJ (1984) X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A 81: 1189–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, et al. (1989) The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science 244: 1578–1580. [DOI] [PubMed] [Google Scholar]
- 37. Araki E, Nakamura K, Nakao K, Kameya S, Kobayashi O, et al. (1997) Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in Duchenne muscular dystrophy. Biochem Biophys Res Commun 238: 492–497. [DOI] [PubMed] [Google Scholar]
- 38. Aoki Y, Nakamura A, Yokota T, Saito T, Okazawa H, et al. (2010) In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol Ther 18: 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang B, Miyagoe-Suzuki Y, Yada E, Ito N, Nishiyama T, et al. (2011) Reprogramming efficiency and quality of induced Pluripotent Stem Cells (iPSCs) generated from muscle-derived fibroblasts of mdx mice at different ages. PLoS Curr 3: RRN1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yokota T, Lu QL, Morgan JE, Davies KE, Fisher R, et al. (2006) Expansion of revertant fibers in dystrophic mdx muscles reflects activity of muscle precursor cells and serves as an index of muscle regeneration. J Cell Sci 119: 2679–2687. [DOI] [PubMed] [Google Scholar]
- 41. Aoki Y, Yokota T, Nagata T, Nakamura A, Tanihata J, et al. (2012) Bodywide skipping of exons 45–55 in dystrophic mdx52 mice by systemic antisense delivery. Proc Natl Acad Sci U S A 109: 13763–13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokota T, Duddy W, Echigoya Y, Kolski H (2012) Exon skipping for nonsense mutations in Duchenne muscular dystrophy: too many mutations, too few patients? Expert Opin Biol Ther. [DOI] [PubMed]
- 43. Den Dunnen JT, Grootscholten PM, Bakker E, Blonden LA, Ginjaar HB, et al. (1989) Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. American Journal of Human Genetics 45: 835–847. [PMC free article] [PubMed] [Google Scholar]
- 44. Hoffman EP, Morgan JE, Watkins SC, Partridge TA (1990) Somatic reversion/suppression of the mouse mdx phenotype in vivo. J Neurol Sci 99: 9–25. [DOI] [PubMed] [Google Scholar]
- 45. Nicholson LV, Bushby KM, Johnson MA, Gardner-Medwin D, Ginjaar IB (1993) Dystrophin expression in Duchenne patients with “in-frame” gene deletions. Neuropediatrics 24: 93–97. [DOI] [PubMed] [Google Scholar]
- 46. Sherratt TG, Vulliamy T, Dubowitz V, Sewry CA, Strong PN (1993) Exon skipping and translation in patients with frameshift deletions in the dystrophin gene. American Journal of Human Genetics 53: 1007–1015. [PMC free article] [PubMed] [Google Scholar]
- 47. Yokota T, Hoffman E, Takeda S (2011) Antisense oligo-mediated multiple exon skipping in a dog model of Duchenne muscular dystrophy. Methods Mol Biol 709: 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arechavala-Gomeza V, Kinali M, Feng L, Guglieri M, Edge G, et al. (2010) Revertant fibres and dystrophin traces in Duchenne muscular dystrophy: implication for clinical trials. Neuromuscul Disord 20: 295–301. [DOI] [PubMed] [Google Scholar]
- 49. Winnard AV, Mendell JR, Prior TW, Florence J, Burghes AH (1995) Frameshift deletions of exons 3–7 and revertant fibers in Duchenne muscular dystrophy: mechanisms of dystrophin production. American Journal of Human Genetics 56: 158–166. [PMC free article] [PubMed] [Google Scholar]
- 50. Danko I, Chapman V, Wolff JA (1992) The frequency of revertants in mdx mouse genetic models for Duchenne muscular dystrophy. Pediatr Res 32: 128–131. [DOI] [PubMed] [Google Scholar]
- 51. Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, et al. (2009) Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol 65: 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lu QL, Morris GE, Wilton SD, Ly T, Artem'yeva OV, et al. (2000) Massive idiosyncratic exon skipping corrects the nonsense mutation in dystrophic mouse muscle and produces functional revertant fibers by clonal expansion. J Cell Biol 148: 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fanin M, Danieli GA, Cadaldini M, Miorin M, Vitiello L, et al. (1995) Dystrophin-positive fibers in Duchenne dystrophy: origin and correlation to clinical course. Muscle Nerve 18: 1115–1120. [DOI] [PubMed] [Google Scholar]
- 54. Wakeford S, Watt DJ, Partridge TA (1991) X-irradiation improves mdx mouse muscle as a model of myofiber loss in DMD. Muscle Nerve 14: 42–50. [DOI] [PubMed] [Google Scholar]
- 55. Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, et al. (1996) Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature 384: 349–353. [DOI] [PubMed] [Google Scholar]
- 56. Sakamoto M, Yuasa K, Yoshimura M, Yokota T, Ikemoto T, et al. (2002) Micro-dystrophin cDNA ameliorates dystrophic phenotypes when introduced into mdx mice as a transgene. Biochem Biophys Res Commun 293: 1265–1272. [DOI] [PubMed] [Google Scholar]
- 57. Pagel CN, Partridge TA (1999) Covert persistence of mdx mouse myopathy is revealed by acute and chronic effects of irradiation. Journal of the Neurological Sciences 164: 103–116. [DOI] [PubMed] [Google Scholar]
- 58. Fraley SM, Springer AD (1981) Memory of simple learning in young, middle-aged, and aged C57/BL6 mice. Behav Neural Biol 31: 1–7. [DOI] [PubMed] [Google Scholar]
- 59. Klein CJ, Coovert DD, Bulman DE, Ray PN, Mendell JR, et al. (1992) Somatic reversion/suppression in Duchenne muscular dystrophy (DMD): evidence supporting a frame-restoring mechanism in rare dystrophin-positive fibers. American Journal of Human Genetics 50: 950–959. [PMC free article] [PubMed] [Google Scholar]
- 60. Narita S, Yorifuji H (1999) Centrally nucleated fibers (CNFs) compensate the fragility of myofibers in mdx mouse. Neuroreport 10: 3233–3235. [DOI] [PubMed] [Google Scholar]
- 61. Wroblewski R, Edstrom L, Mair WG (1982) Five different types of centrally nucleated muscle fibres in man: elemental composition and morphological criteria. A study of myopathies, fetal tissue and muscle spindle. J Submicrosc Cytol 14: 377–387. [PubMed] [Google Scholar]
- 62. McGeachie JK, Grounds MD, Partridge TA, Morgan JE (1993) Age-related changes in replication of myogenic cells in mdx mice: quantitative autoradiographic studies. Journal of the Neurological Sciences 119: 169–179. [DOI] [PubMed] [Google Scholar]
- 63. Shavlakadze T, Davies M, White JD, Grounds MD (2004) Early regeneration of whole skeletal muscle grafts is unaffected by overexpression of IGF-1 in MLC/mIGF-1 transgenic mice. J Histochem Cytochem 52: 873–883. [DOI] [PubMed] [Google Scholar]
- 64. Garcia L, Peltekian E, Pastoret C, Israeli D, Armande N, et al. (1999) Computerised dystrophic muscle simulator: prospecting potential therapeutic strategies for muscle dystrophies using a virtual experimental model. J Gene Med 1: 43–55. [DOI] [PubMed] [Google Scholar]
- 65. Crawford GE, Lu QL, Partridge TA, Chamberlain JS (2001) Suppression of revertant fibers in mdx mice by expression of a functional dystrophin. Hum Mol Genet 10: 2745–2750. [DOI] [PubMed] [Google Scholar]
- 66. Fall AM, Johnsen R, Honeyman K, Iversen P, Fletcher S, et al. (2006) Induction of revertant fibres in the mdx mouse using antisense oligonucleotides. Genet Vaccines Ther 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yokota T, Duddy W, Partridge T (2007) Optimizing exon skipping therapies for DMD. Acta Myol 26: 179–184. [PMC free article] [PubMed] [Google Scholar]