Summary
The first step in attachment of Chlamydia to host cells is thought to involve reversible binding to host heparan sulfate proteoglycans (HSPGs), polymers of variably sulfated repeating disaccharide units coupled to diverse protein backbones. However, the key determinants of HSPG structure that are involved in Chlamydia binding are incompletely defined. A previous genome-wide Drosophila RNAi screen suggested that the level of HSPG 6-O sulfation rather than the identity of the proteoglycan backbone maybe a critical determinant for binding (Elwell et al., 2008). Here, we tested in mammalian cells whether SULF1 or SULF2, human endosulfatases which remove 6-O sulfates from HSPGs, modulate Chlamydia infection. Ectopic expression of SULF1 or SULF2 in HeLa cells, which decreases cell surface HSPG sulfation, diminished C. muridarum binding and decreased vacuole formation. ShRNA depletion of endogenous SULF2 in a cell line that primarily expresses SULF2 augmented binding and increased vacuole formation. C. muridarum infection of diverse cell lines resulted in downregulation of SULF2 mRNA. In a murine model of acute pneumonia, mice genetically deficient in both endosulfatases or in SULF2 alone demonstrated increased susceptibility to C. muridarum lung infection. Collectively, these studies demonstrate that the level of HSPG 6-O sulfation is a critical determinant of C. muridarum infection in vivo and that 6-O endosulfatases are previously unappreciated modulators of microbial pathogenesis.
Introduction
Chlamydiae are obligate intracellular bacteria that are associated with a wide spectrum of diseases in humans. C. trachomatis is the most common bacterial cause of sexually transmitted diseases and non-congenital infertility in Western countries and the leading cause of acquired blindness in developing countries (Mandell et al., 2010). C. pneumoniae is an important cause of upper and lower respiratory tract infections and has been associated with a variety of chronic diseases including atherosclerosis (Campbell et al., 2003). The capacity of Chlamydia to cause sexually transmitted, ocular, and respiratory tract infections and the extraordinary prevalence and array of these infections make them public concerns of primary importance.
All Chlamydia species share a dimorphic life cycle in which they alternate between an extracellular, spore-like form, the elementary body (EB), and an intracellular, metabolically active but non-infectious form, the reticulate body (RB) (Moulder, 1991). EBs are rapidly internalized by most cultured cells, even nonprofessional phagocytes such as epithelial cells, suggesting that the receptor(s) is widespread and/or that there are multiple receptors (Dautry-Varsat et al., 2005). The initial step in infection, binding to host cells, is a two-step process involving reversible binding to host heparan sulfate proteoglycans (HSPGs) followed by higher affinity irreversible binding (Carabeo et al., 2001, Wuppermann et al., 2001, Su et al., 1996). Subsequent entry occurs through receptor-mediated endocytosis in clathrin-coated pits and/or pinocytosis in non-clathrin-coated pits (Hybiske et al., 2007, Dautry-Varsat et al., 2005).
The involvement of HSPGs in the initial interaction of Chlamydia species, including C. trachomatis, C. muridarum, and C. pneumoniae (Wuppermann et al., 2001, Su et al., 1996), with host cells is intriguing. HSPGs regulate a diverse array of biological processes, including the interaction of numerous growth factors and morphogens with their receptors (Esko et al., 2001). HSPGs are extraordinarily heterogeneous structures that are composed of a linear array of repeating dissacharide units that are variably sulfated and that are covalently linked to various core proteins. HSPGs can be sulfated at four different sites (N-, 3-O, and 6-O of glucosamine and 2-O of uronic acid) during biosynthesis at the Golgi, giving rise to almost limitless structural diversity. 6-O-sulfation is subject to regulation, both through the action of 6-O-sulfotransferases during biosynthesis in the Golgi and by post-synthetic editing of 6-O-sulfation by dedicated HSPG-specific endosulfatases at the cell surface.
Two extracellular HSPG-specific 6-O-endosulfatases, SULF1 and SULF2, have been characterized in humans, mice, quail and other species (Morimoto-Tomita et al., 2002, Dhoot et al., 2001). These neutral pH endosulfatases are secreted and act on cell surface or on extracellular HSPGs to remove 6-O sulfates from internal glucosamine residues present in highly sulfated subdomains of HSPGs. Studies in cell lines and in transgenic mice reveal important functions of these endosulfatases in development and in neoplasia. Expression of the Sulfs in different tissues or cell lines is variable and suggests in part overlapping functions; some tissues, such as lungs, express both SULF1 and SULF2, while others express SULF1 or SULF2 predominantly (Lum et al., 2007). Their functional redundancy in development has been revealed by detailed studies of transgenic mice. While individual SULF1- or SULF2- deficient mice exhibit a relatively mild phenotype affecting general growth and lung development, the double knockout mice exhibit severe developmental defects and early post-natal lethality (Holst et al., 2007, Ai et al., 2007, Lamanna et al., 2006, Lum et al., 2007, Lamanna et al., 2007, Ratzka et al., 2008). Many tumor or tumor-derived cells lines demonstrate enhanced expression of SULF2, and depletion of SULF2 in several cancer cell lines decreases cellular growth and cell migration (Nawroth et al., 2007, Lai et al., 2008b, Lemjabbar-Alaoui et al., 2010, Phillips et al., 2012) suggesting that SULF2 could be a novel therapeutic target. SULF1, on the other hand, has been suggested to be a tumor suppressor (Khurana et al., 2011, Lai et al., 2008a). The biological activity of the Sulfs relates to their ability to modulate growth factor/morphogen signaling pathways, including Wnts, Bone Morphogenic Proteins, glial cell-derived neurotrophic factor, transforming growth factor-β, FGF2, and PDGF-BB (Dhoot et al., 2001, Li et al., 2005, Lai et al., 2003, Otsuki et al., 2010, Yue et al., 2008, Lai et al., 2008b).
Despite their emerging important roles in growth, development, and neoplasia, the role of HSPG sulfation in microbial pathogenesis is largely underexplored. We have recently shown that Chlamydia co-opts the FGF2 signaling pathway in an HSPG-dependent manner to enhance infection (Kim et al., 2011). FGF2 binds directly to EBs, where it may function as a bridging molecule to facilitate binding of EBs with HSPGs to activate FGFR. Since FGF2-FGFR interactions are stabilized by HSPGs and are affected by HSPG sulfation (Rusnati et al., 1994, Lundin et al., 2000), these results suggest that Sulfs may modulate Chlamydia binding. A study using chemically modified synthetic heparin molecules has shown that 6-O sulfation on the heparin is critical for Chlamydia attachment to mammalian cells (Yabushita et al., 2002). In a genome-wide Drosophila RNAi screen designed to identify host genes required for Chlamydia infection, we identified 3 genes involved in heparan sulfate biosynthesis and postsynthetic editing (Elwell et al., 2008): (i) Sulf1, whose depletion increased infection; (ii) 2-sulfotransferase, an enzyme required to add sulfate to the 2-O position of uronic acid in HS, which must occur before the addition of the sulfate at the 6-O position, and whose depletion decreased infection; and (iii) 6-sulfotransferase, an enzyme required to add sulfate to the 6-O position of glucosamine with HS after the 2-O sulfation step, and whose depletion decreased infection. These results suggested that sulfation is critical for Chlamydia infection. We further utilized the Drosophila S2 cell system to systematically deplete each of the 4 different core proteoglycans by RNAi (perlecan, syndecan, and 2 glypicans) and found that depletion of each of the known classes of proteoglycans partially decreases vacuole formation (unpublished data). These results suggested that 6-O-sulfation, rather than the identity of core proteoglycans, is an important determinant of Chlamydia binding. We were particularly interested in the Sulfs, as these enzymes have not previously been shown to modulation pathogenesis.
In this study, we tested the hypothesis that the Sulfs regulate Chlamydia infection in vitro and in vivo, using C. muridarim (also known as Mouse Pneumonitis), a murine strain that is closely related to C. trachomatis and that has served as a useful model for genital tract and pulmonary disease (Ramsey et al., 2009). Overexpression of SULF1 or SULF2 decreased Chlamydia binding and vacuole formation, whereas depletion of SULF2 enhanced infection. In a mouse model of acute pneumonia, mice deficient in SULF2 or in both SULF1 and SULF2 exhibited enhanced susceptibility to Chlamydia infection. Collectively, these studies suggest that HSPG 6-O sulfation is a critical determinant of Chlamydia infection in in vivo and implicate the HSPG-specific endosulfatases as novel modulators of pathogenesis.
Results
Ectopic expression of SULF1 or SULF2 decreases Chlamydia binding and vacuole formation
We first examined endogenous expression of SULF1 and SULF2 mRNA and protein in HeLa cells. qRT-PCR analysis revealed that SULF1 and SULF2 mRNA expression was low (relative to GAPDH mRNA) but detectable in HeLa cells, with steady state levels of SULF2 mRNA approximately ~3 fold greater than those of SULF1 (Fig S1). Endogenous SULF2 protein was too low to detect by immunoblot analysis (data not shown), and SULF1 proteins levels were not evaluated due to lack of commercially available antibodies. The low expression of Sulfs in HeLa cells makes this cell line useful for testing the effect of overexpression of Sulfs on Chlamydia infection.
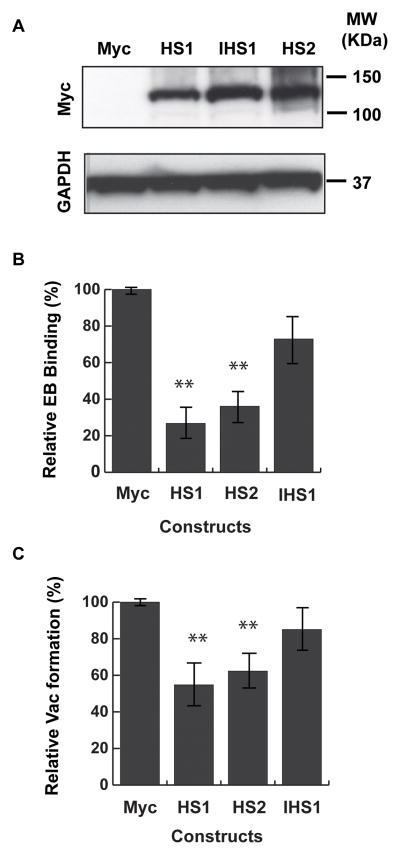
Transient transfection of Myc-tagged human SULF1 or SULF2 in HeLa cells resulted in similarly robust expression as assayed by immunoblot analysis with Myc antibodies (Fig 1A). Immunofluorescence (IF) microscopy of the Sulf-transfected cells with anti-Myc antibodies revealed intracellular expression as well as cell surface associated and extracellular protein, (Fig S2, white arrows) as SULF1 and SULF2 are known to be secreted onto the cell surface and extracellularly (Morimoto-Tomita et al., 2002, Tang et al., 2009) (Fig S2, see bottom panels). We compared C. muridarum binding and vacuole formation in HeLa cells transiently transfected with Myc-SULF1, Myc-SULF2, or vector control. For binding studies, we quantified the number of cell-surface associated EBs per cell at 1 hr post infection. For vacuole formation, we quantified the average number of vacuoles per cell at 20–24 hpi. We observed a 60–70% decrease in Chlamydia binding (Fig 1B) and, similarly, a 40% decrease in vacuole formation (Fig 1C and Fig S2) compared to vector-transfected cells. Under conditions that reduced vacuole formation, there was no decrease in vacuole size and no alteration in vacuole morphology, although there were fewer vacuoles per cell (Fig S2).
Fig. 1. Overexpression of SULF1 or SULF2 in HeLa cells decreases Chlamydia binding and vacuole formation.
HeLa cells were transiently transfected with the vector expressing Myc-tag alone (Myc) or Myc-tagged SULF1 (HS1), Myc-tagged SULF2 (HS2), or a catalytically inactive version of Myc-tagged SULF1 (IHS1). (A) Levels of Myc-tagged transfected protein were determined by immunoblotting with an antibody to Myc. GAPDH serves as a loading control. (B) For quantitation of EB binding by immunofluorescence microscopy, HeLa cells were transfected for 48 hrs and then infected with C. muridarum for 1 hr. The number of EBs bound per cell was normalized to vector-transfected controls. For vector-transfected controls, the average number of bound EBs/cell was 10.5. Shown is the mean EB binding (± SEM) relative to vector-transfected controls for at least 5 independent experiments. At least 150 cells were counted in each experiment (C) HeLa cells were transfected for 48 hrs and then infected with C. muridarum for 24 hrs. The average number of vacuoles per cell was normalized to vector-transfected controls. For vector-transfected controls, the average number of vacuoles per cell was 0.54. Shown is the mean (± SEM) number of vacuoles per cells normalized to vector-transfected controls for at least 3 experiments. At least 500 cells were counted in each experiment **p < 0.01 ANOVA.
To confirm that the catalytic activity of Sulfs was required for binding and vacuole formation, we ectopically expressed a catalytically inactive version of SULF1, in which the critical cysteines are changed to alanine (IHS1) (Tang et al., 2009). When Myc-IHS1 was overexpressed at levels similar to Myc-SULF1 or Myc-SULF2, there was no decrease in Chlamydia binding or vacuole formation compared to vector-transfected cells (Figs 1B, 1C, and S2). Together, these results suggest that increased expression of either SULF1 or SULF2 can negatively regulate Chlamydia infection in a manner dependent on their enzymatic activity.
SULF2 limits Chlamydia infection
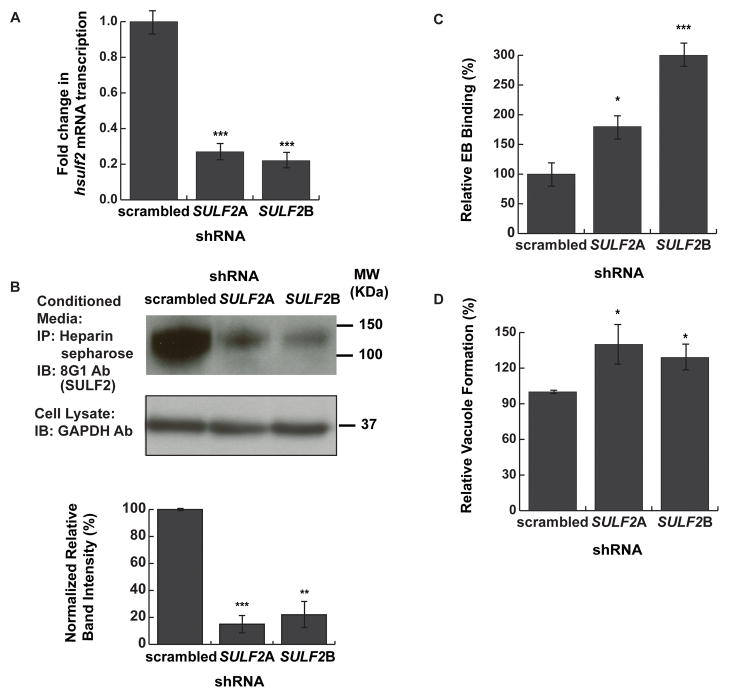
We next tested whether depletion of endogenous SULF1 or SULF2 would enhance Chlamydia infection, as the resultant increased cell surface HSPG 6-O sulfation would be predicted to increase binding and vacuole formation. To avoid issues caused by functional redundancy of the Sulfs, we utilized HaCaT cells, a human keratinocyte cell line which expresses high levels of SULF2 mRNA but barely detectable levels of SULF1 mRNA (Fig S3A). HaCaT cells were transduced with a lentivirus containing either one of two specific human SULF2 shRNAs or a scrambled control shRNA. Puromycin-selected HaCaT cells were examined for depletion of SULF2 mRNA by qRT-PCR (Fig. 2A). Transduction of two different SULF2 shRNAs resulted in at least a 5-fold reduction in SULF2 mRNA levels (Fig 2A). Consistent with previous reports (Lamanna et al., 2006), SULF1 expression was increased by 2~3 fold upon depletion of SULF2 but the total SULF1 mRNA levels were still negligible compared to SULF2 (Fig S3B). To verify that transduction of SULF2 shRNA decreased SULF2 protein levels, we immunoprecipitated secreted SULF2 using heparin sepharose beads followed by immunoblotting with an antibody to SULF2 (Fig 2B, upper panel). As quantified by densitometry, depletion of SULF2 by either shRNA decreased secreted SULF2 levels by over 5-fold (Fig 2B, lower panel).
Fig 2. Depletion of SULF2 increased Chlamydia binding.
HaCaT cells were transduced with lentivirus expressing scrambled shRNA or two different shRNAs targeting SULF2 (SULF2A and SULF2B) and selected for puromycin resistance. (A) Total mRNA was isolated from shRNA-transduced cells, and SULF2 levels were quantified by qRT-PCR. mRNA expression was compared to GAPDH and then further normalized to scrambled shRNA-transduced cells. (B) Secreted SULF2 from conditioned media from the puromycin-selected shRNA transduced cells was affinity purified by binding to heparin-sepharose beads and immunoblotted with a monoclonal antibody to SULF2 (8G1). As a control for cell number, lysates of the transduced cells were immunoblotted with GAPDH. The intensity of SULF2 bands was quantitated and compared to GAPDH by densitometry using ImageJ and then further normalized to scrambled shRNA-treated cells. The lower panel shows the average from 3 blots each for scrambled or SULF2 shRNA-depleted cells. (C) EB binding was quantified from Puromycin-selected shRNA-transduced HaCaT cells by immunofluorescence microscopy. The number of EBs bound per cell was normalized to scrambled shRNA-transduced controls. For scrambled shRNA-transduced controls, the average number of bound EBs/cell was 2.4. Shown is the mean EB binding (± SEM) relative to scrambled shRNA transduced controls from 3 independent experiments. At least 150 cells were counted in each experiment. (D) Vacuole formation was quantified from Puromycin-selected shRNA-transduced HaCaT cells by immunofluorescence microscopy. The number of vacuoles per cell was normalized to scrambled shRNA-transduced controls. For scrambled shRNA-transduced controls, the average number of vacuoles per cell was 0.29. Shown is the mean number of vacuoles/cell (± SEM) relative to scrambled shRNA transduced controls from 3 independent experiments. At least 500 cells were counted in each experiment. * p<0.05, *** p<0.001 ANOVA
The puromycin selected shRNA-transduced cells were examined for Chlamydia binding and vacuole formation. Depletion of SULF2 by either of the two SULF2-directed shRNAs resulted in a 2 to 3 fold increase in Chlamydia binding (Fig 2C, p<0.05) and 30–40% increase in vacuole formation (Fig 2D, p<0.05). While the effect of SULF2 protein depletion on Chlamydia infection was modest compared to the efficiency of depletion, the fact that it was observed with two different shRNAs makes off-target effects unlikely. Together, our results indicate that endogenous endosulfatase activity limits Chlamydia infection
Chlamydia infection decreases SULF2 expression
In previous work, we have established that FGF2 enhances Chlamydia binding, while Chlamydia infection stimulates FGF2 transcription and enhances production and release of FGF2, thus establishing a positive feedback loop that increases secondary infection (Kim et al., 2011). We therefore tested the notion that Chlamydia infection might regulate SULF expression. We assayed three different cell lines that primarily express SULF2: HeLa cells, HaCaT cells, and H292 cells, a human lung mucoepidermoid cell line (Lemjabbar-Alaoui et al., 2010). Each cell line was infected with Chlamydia, and mRNA levels were examined by RT-PCR at 24 hrs post-infection (hpi). Although we could not detect significant changes in SULF1 mRNA expression at this time point, SULF2 mRNA expression was decreased by 70% in HeLa cells, by 40% HaCaT cells, and by 60% in H292 cells (Fig 3). Together, these results indicate that Chlamydia infection results in decreased SULF2 expression in multiple cell lines. Decreased SULF2 protein, particularly in cells that express little SULF1, might cause local increases cell associated levels of 6-O sulfation and thus could lead to increased susceptibility to Chlamydia infection.
Fig 3. Chlamydia infection decreases SULF2 expression.
HeLa, HaCaT, and H292 cells were infected with C. muridarum. At 24 hpi, total RNA was isolated and SULF1 and SULF2 mRNA levels were quantified by qRT-PCR. SULF1 or SULF2 mRNA was normalized to GAPDH mRNA and compared to uninfected cells. * p<0.05, ** p<0.01 ANOVA
SULF1 and SULF2 limit Chlamydia infection in vivo
Our results thus far suggest that overexpression of SULF1 or SULF2 decreases Chlamydia binding while depletion of SULF2 (in the absence of significant SULF1 expression) enhances Chlamydia infection. We next tested whether Sulf activity would modulate Chlamydia infection in vivo. For these experiments, we assessed the susceptibility to infection of wild type compared to SULF knockout mice, utilizing a well-described murine model of acute pneumonia in which mice are intranasally infected with C. muridarum. This mouse model faithfully recapitulates important aspects of C. trachomatis and C. pneumoniae pneumonia (Rank, 2006, He et al., 2011, Coalson et al., 1987, Murthy et al., 2004).
We confirmed that Chlamydia replicates in the lung upon intranasal inoculation and determined the optimal duration of infection for our experiments. Whole lungs were harvested at 5 and 48 hpi, frozen, thawed, and homogenized to release infectious progeny. HeLa cells were infected with these lysates, and progeny quantified by vacuole formation from this second round of infection. While no infectious progeny could be isolated at 5 hpi, EB production was robust at 48 hpi (Supplemental Fig 4A), indicating that replication had occurred in the lungs. Consistent with this finding, lung inflammation, as assessed by H&E staining, was clearly evident at 48 hpi (Supplemental Fig 4B).
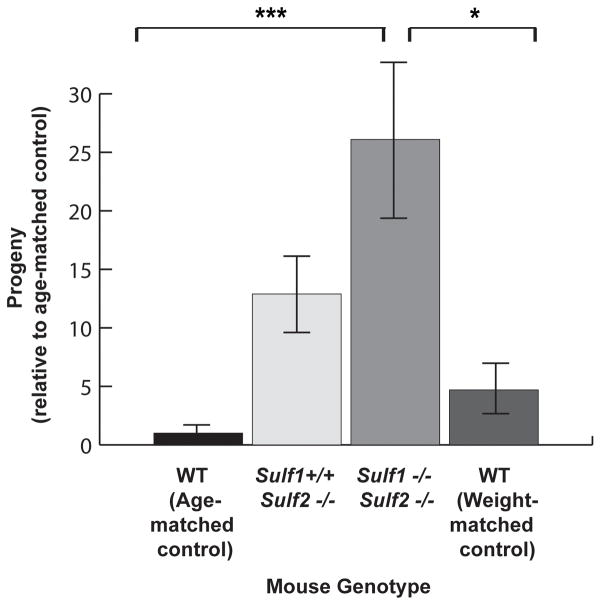
Sulf2−/− single knockout mice and Sulf1−/−Sulf2−/− double knockout C57BL/6 mice (Lum et al., 2007) were derived by breeding. We were unable to generate sufficient numbers of Sulf1−/− single knockout mice and thus could not assess their susceptibility to infection. Because Sulf1−/−Sulf2−/− mice demonstrated decreased body size and weight compared to wild-type mice (data not shown and (Lamanna et al., 2006, Lum et al., 2007, Ai et al., 2007)), we utilized two different control groups: wild type littermates that were age-matched (10–12 week old mice that weighed more than the Sulf1−/−Sulf2−/− mice) or that were weight-matched (younger mice that weighed the same as the Sulf1−/−Sulf2−/− mice). Control littermates, Sulf2−/− mice, and Sulf1−/−Sulf2−/− mice were infected intranasally with C. muridarum (5 × 106 IFU), lungs harvested at 48 hrs, and infectious progeny quantified. Consistent with our cell culture based results, we observed a statistically significant enhanced susceptibility to Chlamydia infection in the Sulf1−/−Sulf2−/− mice, with a mean increase of 26-fold in IFU compared to the age-matched controls (P <0.001) and a 5-fold increase in IFU compared to the weight-matched controls (P <0.05). A trend towards increased susceptibility was observed in the Sulf2−/− knockout mice, with a 13-fold increase in mean production of infectious progeny compared to the age-matched controls. However, this result but did not reach statistical significance as determined by ANOVA, consistent with redundant roles of SULF1 and SULF2. We conclude that by controlling HSPG 6-O sulfation, Sulfs regulate susceptibility to Chlamydia infection both in vitro and in vivo and thus function as novel modulators of microbial pathogenesis.
Discussion
The variability in HPSG sulfation accounts in part for the enormous complexity and diversity in the glycan code, allowing subtle regulation of important cellular processes ranging from development to cell adhesion and growth factor signaling (Bernfield et al., 1999, Lamanna et al., 2007, Esko et al., 2001). Pathogens can also subvert cell surface gycosaminoglycans for binding (Chen et al., 2008), but the precise role of the sulfation of specific residues has only been elucidated in a few instances. For example, HSPG 3-O sulfation is critical for Herpes Simplex Virus binding (Shukla et al., 1999) to host cells, while 6-O- and N-sulfation of GlcNAc of HS is crucial for Coxsackie B virus binding through secondary receptors (Zautner et al., 2006). While 6-O-sulfated heparin mimics have been reported to inhibit Chlamydia entry (Yabushita et al., 2002), we provide the first evidence that the endosulfatases SULF1 and SULF2, which control the level of 6-O-sulfation on cell surface HSPGs, are important modulators of Chlamydia infection. We demonstrate that overexpression of either SULF1 or SULF2, which has been shown to decrease surface HSPG 6-O sulfation (Hossain et al., 2010), results in diminished Chlamydia binding and vacuole formation, while depletion of SULF2 in cells that predominantly express SULF2 results in increased Chlamydia binding and vacuole formation. Most importantly, we use genetically modified mice to show that mice deficient in SULF2 or in both SULF1 and SULF2 show increased susceptibility to Chlamydia-induced pneumonia after intranasal inoculation. Our study is amongst the few that extends findings from cell culture to an in vivo animal model of the pathogenesis of Chlamydia infections. Whether Sulfs play a role in modulating genital tract infection will be of interest to determine in the future.
There is a long history of the involvement of HSPGs in Chlamydia attachment. Despite some reports suggesting that Chlamydia might synthesize an HSPG-like molecule (Zhang et al., 1992, Chen et al., 1996, Chen et al., 1997, Rasmussen-Lathrop et al., 2000), the general consensus in the field is that Chlamydia co-opt host cell HSPGs for the initial low affinity reversible binding step (Dautry-Varsat et al., 2005). Various glycosaminoglycans have been shown to competitively inhibit binding (Yabushita et al., 2002). Studies utilizing chemically derived host cell mutants deficient in one or more biosynthetic steps have pointed to a role in host cell HSPGs (Zhang et al., 1992, Rasmussen-Lathrop et al., 2000). However, these studies have been limited by conflicting results (Stephens et al., 2006), possibly reflecting the presence of multiple or compensatory mutations in these chemically mutagenized genomes. More recently, we have shown in a genome-wide RNAi screen performed in Drosophila S2 cells that host enzymes involved in HSPG biosynthesis were required for intracellular infection (Elwell et al., 2008). While this paper was in preparation, Rosmarin et al performed a loss-of-function genetic screen in human haploid cells to identify host factors important in Chlamydia infection and found that inactivation of three host genes involved in HSPG biosynthesis decreased the susceptibility of these cells to C. trachomatis infection (Rosmarin et al., 2012). Our work, in conjunction with our previous RNAi screen (Elwell et al., 2008), provides strong evidence that the HSPG 6-O sulfation plays a critical role in the initial binding of Chlamydia to host cells. Our recent finding of a role for FGF2 in mediating HSPG-dependent binding (Kim et al., 2011) suggests the intriguing possibility that 6-O sulfation may have dual roles in Chlamydia infection: it may contribute to initial interactions between Chlamydia and the host cell as well as stabilize a receptor complex, comprised of FGF2, HSPG, and FGFR (Rusnati et al., 1994), to which Chlamydia binds and utilizes for entry.
Our studies do not rule out an additional role for HSPG 6-O-sulfation in post-binding steps during Chlamydia infection. For example, HSPGs have also been reported to play roles in viral entry, in addition to serving as a binding receptor or co-receptor for viruses (Chen et al., 2008, Zautner et al., 2006). Our finding that the modulation of 6-O sulfation affected vacuole formation in parallel with binding without affecting vacuole morphology suggests that if there is a separate role in entry, it is likely to be minimal.
The regulation of SULF1 and SULF2 transcription is incompletely understood. TGF-β upregulates SULF1 expression while P53 increases SULF2 expression (Chau et al., 2009, Yue et al., 2008). The mechanism and pathways by which Chlamydia modulate SULF2 expression remain to be explored. In cell types that predominantly express SULF2, we found that SULF2 expression was consistently reduced 40–70% upon Chlamydia infection. The work presented here suggests the intriguing possibility that Chlamydia-induced down regulation of Sulf transcription could function in a feedforward loop that enhances secondary infection and increases susceptibility to Chlamydia infection. Through the predicted subsequent decrease in SULF2 production and secretion, cells in the local environment could have higher levels of surface 6-O sulfation. They may thus be more susceptible to secondary infection with Chlamydia and/or with other pathogens, including including Human Immunodeficiency virus, Herpes Simplex Virus, Human Papilloma virus, Syphilis, and Neisseria gonorrhea, which are common co-infecting pathogens (Mandell et al., 2010). Together with our recent findings that FGF2 expression and secretion is also enhanced upon Chlamydia infection (Kim et al., 2011), it is clear that this organism may utilize multiple mechanisms to facilitate local spread of infection as well as to enhance susceptibility to other sexually transmitted diseases.
In summary, our studies reveal a novel role for 6-O endosulfatases in modulating Chlamydia infection. Furthermore, these studies underscore the potential for using therapies directed against HSPGs as multivalent antimicrobials (Tiwari et al., 2012). It will be of interest to determine whether modulating HSPG sulfation is a common theme in microbial pathogenesis. If so, combination therapy involving antagonists of the Sulfs, in conjunction with HSPG mimetics, may be a potent and broad strategy to prevent infection with multiple STDs.
Materials and Methods
Reagents
8G1, a SULF2 monoclonal mAb, was developed by immunizing a SULF2 null mouse (Sulf-1−/−Sulf2+/+) with human SULF2 protein as previously described (Lemjabbar-Alaoui et al., 2010). Other antibodies were obtained from the following sources: goat anti-Chlamydia MOMP was purchased from Fitzgerald; mouse anti-GAPDH from Chemicon; HRP-rabbit anti-goat IgG from Zymed; goat anti-mouse IgG HRP from Amersham Biosciences; all fluorescently labeled secondary antibodies and phalloidin from Molecular Probes; Effectene, QIAshredder, RNeasy kit, RNase-free DNase, and cDNA synthesis kit from Qiagen; SYBR GreenER qPCR SuperMix from Invitrogen; Heparin sepharose beads from GE health care; Puromycin from Millipore.
Cell culture and Chlamydia propagation
HeLa 229, L929, and H292 cells were obtained from ATCC and passaged as previously described (van Ooij et al., 1997, Lemjabbar-Alaoui et al., 2010). HaCaT cells were obtained from Rosen lab (UCSF) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum. C. muridarim was propagated in L929 cells grown in suspension culture and purified by discontinuous density gradient centrifugation with Renografin (Mallinckrodt, Inc) as previously described (Caldwell et al., 1981).
Immunofluorecent quantitation of C. trachomatis binding and vacuole formation
Cells were grown on glass coverslips in 24-well plates in DMEM containing 10% FBS overnight, and were infected with C. trachomatis for 1 hr at a multiplicity of infection (MOI) of 2–10 for binding studies and an MOI of 0.5–1 for vacuole formation studies. For quantitation of binding, non-adherent bacteria were removed by washing three times with PBS, and the infected cells were fixed with 4% PFA for 30 min. For quantitation of vacuole formation, PBS-washed cells were incubated in fresh DMEM containing 10% FBS for 20–24 hr. The cells were fixed and were permeabilized with 0.2% Triton X-100 for 15 min, followed by blocking in 2% FBS/1% Fish Skin Gelatin in PBS for 30 min. Bound EBs or vacuoles were visualized by staining with anti-Chlamydia MOMP antibody followed by staining with Alexa-488 conjugated secondary antibody. The host cell was visualized by staining the actin cytoskeleton with phalloidin-Alexa 594. Immunofluorescence images were acquired under identical exposure time for each set of experiments on a Nikon TE2000 perfect focus microscope using Nikon Elements. Metamorph (Molecular devices) or Adobe Photoshop CS4 was used to quantify nuclei, EBs, or vacuoles.
For binding studies, the number of bound EBs per cell was quantified and then normalized to the average number of bound EBs in untreated samples. For vacuole formation studies, the number of vacuoles and the number of nuclei per field was quantified to yield an average number of vacuoles per cell (vacuole formation). We chose this metric rather than measuring the fraction of cells infected because multiple vacuoles per cell are often observed at high multiplicities of C. muridarum infection. We then normalized to untreated samples (relative vacuole formation). Under the conditions of our experiments, increased binding efficiency (ie number of EBs bound per cell) is also reflected in increased vacuole formation (ie number of vacuoles/cell).
For all experiments utilizing image analysis, a minimum of 10 fields was analyzed per treatment. Data were compiled from at least 3 independent experiments unless otherwise specified. Data is presented as relative EB binding (normalized to untreated samples) or relative vacuoles formation (number of vacuoles/cell normalized to untreated samples).
Transfection studies
HeLa cells were seeded on 12 mm glass coverslips in 6 well plates and transfected with Effectene according to the manufacturer’s instructions. 48 hrs after transfection, cells were infected with Chlamydia at 37 °C. Transfected protein levels were determined by immunoblotting as previously described (Kim et al., 2011).
Immunoblot analysis
Cells were lysed for 15 min on ice in Lysis Buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 50mM NaF, 1% sodium deoxycholate, 0.1% SDS, 1 mM sodium orthovanadate, 0.1 mM okadaic acid, and Complete protease inhibitors (Roche Diagnostics)). Cell lysates were collected, centrifuged at 20,800 g for 15 min to remove cell debris, and the supernatant was boiled in NuPage 4X LDS sample buffer (Invitrogen) with 100 mM DTT for 10 min. Proteins in the supernatant were separated on 10% NuPAGE Novex Bis-Tris gels (Invitrogen) and transferred to 0.45 mm Trans-blot nitrocellulose membranes using iBlot (Invitrogen). Membranes were rinsed in water, followed by blocking with 3% milk (Upstate) in Tris-buffered saline (TBS) for 1 hr. Each membrane was incubated with the indicated antibody in 3% milk in TBS with 0.02% Tween-20 (TBST) overnight at 4 °C, followed by an incubation with the appropriate HRP-conjugated antibodies for 1 hr. HRP-conjugated antibodies were detected by ECL (Amersham Biosciences) according to the manufacturer’s protocol. For quantification, band intensity was analyzed using ImageJ analysis software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA,).
Isolation of RNA and DNA, cDNA synthesis, and Real Time PCR
RNA was isolated using the QIAshredder and the RNeasy kit according to the manufacturer’s instructions (Quiagen). RNA was treated with RNase-free DNase according to the manufacturer’s instruction. RNA concentrations were measured using Nanodrop Spectrophotometer (Thermo-scientific). One μg of RNA was reverse transcribed using the cDNA synthesis (Qiagen) kit in a 20 μL reaction. Quantitative PCR (qPCR) was performed with 2 μL of the cDNA preparation using SYBR GreenER qPCR SuperMix (Invitrogen) in a 25 μL reaction using DNA Engine Opticon-2 Real-Time PCR Detection System in the Opticon-2 Real-Time Cycler (BioRad). Primers for human SULF1 were 5′-cgtgctatgaaggaagatgga3′ forward and 5′-tgcccagttcgtttcagt-3 ′ reverse, for human sulf2 5 ′-cgtgctatgaaggaagatgga3 ′ forward and 5 ′-tgcccagttcgtttcagt-3 ′ reverse, and for human gapdh were 5 ′-CTTCTCTGATGAGGCCCAAG-3′ forward and 5′-GCAGCAAACTGGAAAGGAAG-3′ reverse. qPCR included initial denaturation at 94 °C for 10 min, followed by 35 cycles of 94 °C for 10 s, 53 °C for 15 s, 72 °C for 20 s, 72 °C for 1 s and then 72 °C for 10 min followed by a dissociation curve every 0.5 °C from 55 °C to 95 °C. In a dissociation curve, a single peak was confirmed in each of the amplified sequences. For the quantification of SULF1 or SULF2 expression relative to gapdh in different samples, the threshold cycle (Ct) values of targets were expressed as 2-ΔΔ Ct (fold) as described previously (Winer et al., 1999). Each sample was additionally amplified without reverse transcription reaction to confirm the absence of contaminating DNA in the RNA sample.
Lentiviral shRNA depletion
pLKO-puro vectors containing human SULF2 specific shRNAs (SULF2A: ccgggggcgaaagtcattggaatttctcgagaaattccaatgactttcgccctttttg, SULF2B: ccggcatcaatgagactcacaatttctcgagaaattgtgagtctcattgatgtttttga) or scrambled shRNA were purchased from Sigma. Human embryonic kidney 293 FT cells with transfected with Virapower lentiviral packaging mix (Invitrogen) according to manufacturer’s instructions. The next day, transfection complexes were removed and cells were allowed to produce virus for 48 hrs. Media containing virus were collected and used to directly transduce HaCaT cells. The cells were allowed to recover for 24 h and subject to puromycin selection (5 μg/ml). Puromycin resistant cells were harvested, RNA extracted, and qRT-PCTR performed to assess the depletion of SULF2 mRNA.
To assess efficiency of SULF2 protein depletion, culture media was collected from Lentiviral shRNA-transduced HACaT cells grown in 10 cm-dishes for 48 hr. Following centrifugation for 10 min at 240 × g, the clarified supernatants were incubated overnight with Heparin coated Sepharose beads (GE HealthCare) at 4 °C. The beads were washed, and bound proteins were eluted by boiling in NuPage 4X LDS sample buffer (Invitrogen) containing 100mM dithiothreitol followed by immunoblotting with anti-Sulf2 antibodies.
Animal experiments
Sulf1+/+Sulf2+/+, Sulf-1−/−Sulf2+/+, and Sulf1+/+Sulf2−/− C57BL/6 mice (Ratzka et al., 2008) were bred to generate Sulf1+/−Sulf2+/− and Sulf1+/−Sulf2−/− heterozygoes. The heterozygotes were then bred to generate Sulf1+/+Sulf2−/− and Sulf1−/−Sulf2−/−. double knockout mice. Genotyping was performed by PCR of tail vein DNA. Mice were fed a chow diet and water ad libitum throughout the study. Mice were anesthesized by isofurane inhalation and infected intranasally with 50 ul of PBS containing 5×106 IFU of C. muridarum. Mice were euthanized by CO2 inhalation and cervical dislocation at 48 hours post infection. Lungs were dissected intact and immediately placed on ice prior to freezing at −80 °C. All animal protocols were approved by the University of California, San Francisco Institutional Animal Care and Use Committee.
Quantitation of lung bacterial burden
Lungs were thawed in SPG buffer to make a 10% (wt/vol) solution and then pulverised through 70 μm cell strainers using the rubber plunger from a sterile plastic syringe. Tissue homogenates were centrifuged at 500 ×g for 10 min at 4 °C to remove coarse tissue debris. 10-fold dilutions of the clarified supernatants were then added for 1 hr to 24 hr HeLa cell monolayers grown on coverslips. Unbound EBs were removed by washing 3x with media followed by incubation at 37 C in 5% CO2 for 24 hours. Vacuoles/cell were quantified by staining with a MOMP-specific monoclonal antibody conjugated to fluorescein isothiocyanate (1:3 dilution, Fitzgerald). Vacuole quantitation was normalized to age-matched wild type mice
Statistical analysis
Data are expressed as mean ± SE. Data were analyzed by ANOVA, followed by the Bonferroni multiple comparisons. All statistical calculations were performed using InStat software and GraphPad Prism 4.0. Differences were considered statistically significant at p < 0.05, compared with control.
Gene ID
Human SULF1 (GC08P070428) and SULF2 (GC20M046285)
Supplementary Material
Total RNA was isolated from HeLa cells. SULF1 and SULF2 mRNA levels were quantified by qRT-PCR analysis and normalized to GAPDH mRNA.
HeLa cells were transiently transfected with the vector expressing Myc-tag alone (Vector) or Myc-tagged SULF1 (HS1), Myc-tagged SULF2 (HS2), or a catalytically inactive version of Myc-tagged SULF1 (IHS1) and then infected with C. muridarum. At 24 hpi, cells were fixed and stained with antibodies to Myc (red), to visualize intracellular and extracellular SULFs, and to the Chlamydial Major Outer Membrane Protein (MOMP; Green) to visualize vacuoles. Bacterial and host DNA were detected by DAPI staining (blue). White arrows point to cells expressing Myc-tagged HS1, HS2, or IHS1. Note the presence of multiple vacuoles in the vector or IHS1-transfected cells (yellow arrows) and the decreased number of vacuoles per cell in SULF1- and SULF2-transfected cells compared to vector-transfected and IHS-transfected cells (MOMP staining; also see Fig 1C for quantitation). Scale bar = 20 microns
(A) Total RNA was isolated from HaCaT cells. SULF1 and SULF2 mRNA levels were quantified by qRT-PCR and normalized to GAPDH mRNA levels. (B) HaCaT cells were transduced with lentivirus encoding scrambled shRNA or two different shRNAs targeting SULF2 (SULF2A and SULF2B) and selected for puromycin resistance. Total RNA was isolated from shRNA-transduced cells and hsulf1 mRNA level was quantified by qRT-PCR. mRNA expression was normalized to GAPDH and then expressed as a fold change compared to scrambled shRNA-transduced cells.
Wild type mice were intranasally infected with 5×106 IFU of C. muridarum. (A) Infectious progeny were recovered from whole lungs at 5 and 48 hpi, stored frozen, and titered by quantitation of vacuole formation upon secondary infection of HeLa cells for 24 hrs. Whereas very few infectious progeny were recovered at 5 hpi, abundant viable progeny were detectable at 48 hpi. (B) H&E staining of representative lung section reveals abundant inflammatory infiltrates in Chlamydia-infected lungs at 48 hpi.
Fig. 4. Sulf-deficient mice are more susceptible to lung infection by Chlamydia.
Wild type (age or weight matched), Sulf1+/+Sulf2−/−, and Sulf1−/−Sulf2−/− mice were intranasally infected with 5×106 IFU of C. muridarum. At 48 hpi, lungs were harvested from mice and infectious progeny determined by plating serial dilutions onto HeLa cells followed by quantitation of vacuoles/cell at 24 hpi. Data was normalized to age-matched mice and were pooled from 4 experiments. Data were expressed as means ± SEM; ***p<0.001 compared to age-matched control; *p<0.05 compared to weight control. Data were analyzed by one-way ANOVA with post hoc Bonferroni correction.
Acknowledgments
We thank Drs. Wong Rong and Jay Debnath for use of their instruments, Dr. Inna Maltseva for sharing mice and providing advice for the mouse experiments, Dr. Megumi Morimoto for providing Sulf expression constructs, Dr. Kathleen Averette for helpful discussion and comments on the manuscript, and members of the Engel lab for advice and encouragement. This work was supported by grants from NIH RO1-AI073770 (JNE), NIH PO1-AI053194 (SR), the American Cancer Society grant 73087 (HLA), and the Deutsche Forschungsgemeinschaft grant DI 575/6 (TD).
Footnotes
The authors declare no conflicts of interest.
References
- Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP., Jr SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134:3327–3338. doi: 10.1242/dev.007674. [DOI] [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LA, Kuo CC. Chlamydia pneumoniae and atherosclerosis. Semin Respir Infect. 2003;18:48–54. doi: 10.1053/srin.2003.50006. [DOI] [PubMed] [Google Scholar]
- Carabeo RA, Hackstadt T. Isolation and characterization of a mutant Chinese hamster ovary cell line that is resistant to Chlamydia trachomatis infection at a novel step in the attachment process. Infect Immun. 2001;69:5899–5904. doi: 10.1128/IAI.69.9.5899-5904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau BN, Diaz RL, Saunders MA, Cheng C, Chang AN, Warrener P, et al. Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Res. 2009;69:1368–1374. doi: 10.1158/0008-5472.CAN-08-2742. [DOI] [PubMed] [Google Scholar]
- Chen JC, Stephens RS. Chlamydia trachomatis glycosaminoglycan-dependent and independent attachment to eukaryotic cells. Microb Pathog. 1997;22:23–30. doi: 10.1006/mpat.1996.0087. [DOI] [PubMed] [Google Scholar]
- Chen JC, Zhang JP, Stephens RS. Structural requirements of heparin binding to Chlamydia trachomatis. J Biol Chem. 1996;271:11134–11140. [PubMed] [Google Scholar]
- Chen Y, Gotte M, Liu J, Park PW. Microbial subversion of heparan sulfate proteoglycans. Mol Cells. 2008;26:415–426. [PubMed] [Google Scholar]
- Coalson JJ, Winter VT, Bass LB, Schachter J, Grubbs BG, Williams DM. Chlamydia trachomatis pneumonia in the immune, athymic and normal BALB mouse. Br J Exp Pathol. 1987;68:399–411. [PMC free article] [PubMed] [Google Scholar]
- Dautry-Varsat A, Subtil A, Hackstadt T. Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 2005;7:1714–1722. doi: 10.1111/j.1462-5822.2005.00627.x. [DOI] [PubMed] [Google Scholar]
- Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 2008;4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Nair A, Mekasha S, Alroy J, O’Connell CM, Ingalls RR. Enhanced virulence of Chlamydia muridarum respiratory infections in the absence of TLR2 activation. PLoS One. 2011;6:e20846. doi: 10.1371/journal.pone.0020846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst CR, Bou-Reslan H, Gore BB, Wong K, Grant D, Chalasani S, et al. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS One. 2007;2:e575. doi: 10.1371/journal.pone.0000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MM, Hosono-Fukao T, Tang R, Sugaya N, van Kuppevelt TH, Jenniskens GJ, et al. Direct detection of HSulf-1 and HSulf-2 activities on extracellular heparan sulfate and their inhibition by PI-88. Glycobiology. 2010;20:175–186. doi: 10.1093/glycob/cwp159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Entry mechanisms of Chlamydia trachomatis into non-phagocytic cells. Infect Immun. 2007 doi: 10.1128/IAI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana A, Liu P, Mellone P, Lorenzon L, Vincenzi B, Datta K, et al. HSulf-1 modulates FGF2- and hypoxia-mediated migration and invasion of breast cancer cells. Cancer Res. 2011;71:2152–2161. doi: 10.1158/0008-5472.CAN-10-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Jiang S, Elwell CA, Engel JN. Chlamydia trachomatis co-opts the FGF2 signaling pathway to enhance infection. PLoS Pathog. 2011;7:e1002285. doi: 10.1371/journal.ppat.1002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- Lai JP, Sandhu DS, Shire AM, Roberts LR. The tumor suppressor function of human sulfatase 1 (SULF1) in carcinogenesis. Journal of gastrointestinal cancer. 2008a;39:149–158. doi: 10.1007/s12029-009-9058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, et al. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology. 2008b;47:1211–1222. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna WC, Baldwin RJ, Padva M, Kalus I, Ten Dam G, van Kuppevelt TH, et al. Heparan sulfate 6-O-endosulfatases: discrete in vivo activities and functional co-operativity. Biochem J. 2006;400:63–73. doi: 10.1042/BJ20060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CL, Dierks T. The heparanome--the enigma of encoding and decoding heparan sulfate sulfation. Journal of biotechnology. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Lemjabbar-Alaoui H, van Zante A, Singer MS, Xue Q, Wang YQ, Tsay D, et al. Sulf-2, a heparan sulfate endosulfatase, promotes human lung carcinogenesis. Oncogene. 2010;29:635–646. doi: 10.1038/onc.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kleeff J, Abiatari I, Kayed H, Giese NA, Felix K, et al. Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Molecular cancer. 2005;4:14. doi: 10.1186/1476-4598-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum DH, Tan J, Rosen SD, Werb Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol Cell Biol. 2007;27:678–688. doi: 10.1128/MCB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, Claesson-Welsh L. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem. 2000;275:24653–24660. doi: 10.1074/jbc.M908930199. [DOI] [PubMed] [Google Scholar]
- Mandell GL, Bennett JE, Dolin R. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 7. Vol. 1. Philadelphia, PA: Churchill Livingstone/Elsevier; 2010. online resource (2 v. (cl, 4028, xcvii p.)) [Google Scholar]
- Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy AK, Sharma J, Coalson JJ, Zhong G, Arulanandam BP. Chlamydia trachomatis pulmonary infection induces greater inflammatory pathology in immunoglobulin A deficient mice. Cell Immunol. 2004;230:56–64. doi: 10.1016/j.cellimm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS One. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki S, Hanson SR, Miyaki S, Grogan SP, Kinoshita M, Asahara H, et al. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc Natl Acad Sci U S A. 2010;107:10202–10207. doi: 10.1073/pnas.0913897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JJ, Huillard E, Robinson AE, Ward A, Lum DH, Polley MY, et al. Heparan sulfate sulfatase SULF2 regulates PDGFRalpha signaling and growth in human and mouse malignant glioma. The Journal of clinical investigation. 2012;122:911–922. doi: 10.1172/JCI58215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GA, Rank RG. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect Immun. 2009;77:3284–3293. doi: 10.1128/IAI.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank RG. Chlamydial diseases. In: Fox J, Barthold S, Newcomer C, Smith A, Quimby F, Davisson M, editors. The mouse in biomedical research. 2. San Diego, CA: Academic press; 2006. pp. 325–348. [Google Scholar]
- Rasmussen-Lathrop SJ, Koshiyama K, Phillips N, Stephens RS. Chlamydia-dependent biosynthesis of a heparan sulphate-like compound in eukaryotic cells. Cell Microbiol. 2000;2:137–144. doi: 10.1046/j.1462-5822.2000.00039.x. [DOI] [PubMed] [Google Scholar]
- Ratzka A, Kalus I, Moser M, Dierks T, Mundlos S, Vortkamp A. Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Developmental dynamics: an official publication of the American Association of Anatomists. 2008;237:339–353. doi: 10.1002/dvdy.21423. [DOI] [PubMed] [Google Scholar]
- Rosmarin DM, Carette JE, Olive AJ, Starnbach MN, Brummelkamp TR, Ploegh HL. Attachment of Chlamydia trachomatis L2 to host cells requires sulfation. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1120244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnati M, Coltrini D, Caccia P, Dell’Era P, Zoppetti G, Oreste P, et al. Distinct role of 2-O-, N-, and 6-O-sulfate groups of heparin in the formation of the ternary complex with basic fibroblast growth factor and soluble FGF receptor-1. Biochem Biophys Res Commun. 1994;203:450–458. doi: 10.1006/bbrc.1994.2203. [DOI] [PubMed] [Google Scholar]
- Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- Stephens RS, Poteralski JM, Olinger L. Interaction of Chlamydia trachomatis with mammalian cells is independent of host cell surface heparan sulfate glycosaminoglycans. Infect Immun. 2006;74:1795–1799. doi: 10.1128/IAI.74.3.1795-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Raymond L, Rockey DD, Fischer E, Hackstadt T, Caldwell HD. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc Natl Acad Sci U S A. 1996;93:11143–11148. doi: 10.1073/pnas.93.20.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Rosen SD. Functional consequences of the subdomain organization of the sulfs. J Biol Chem. 2009;284:21505–21514. doi: 10.1074/jbc.M109.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V, Maus E, Sigar IM, Ramsey KH, Shukla D. Role of heparan sulfate in sexually transmitted infections. Glycobiology. 2012 doi: 10.1093/glycob/cws106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C, Apodaca G, Engel J. Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect Immun. 1997;65:758–766. doi: 10.1128/iai.65.2.758-766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Wuppermann FN, Hegemann JH, Jantos CA. Heparan sulfate-like glycosaminoglycan is a cellular receptor for Chlamydia pneumoniae. J Infect Dis. 2001;184:181–187. doi: 10.1086/322009. [DOI] [PubMed] [Google Scholar]
- Yabushita H, Noguchi Y, Habuchi H, Ashikari S, Nakabe K, Fujita M, et al. Effects of chemically modified heparin on Chlamydia trachomatis serovar L2 infection of eukaryotic cells in culture. Glycobiology. 2002;12:345–351. doi: 10.1093/glycob/12.5.345. [DOI] [PubMed] [Google Scholar]
- Yue X, Li X, Nguyen HT, Chin DR, Sullivan DE, Lasky JA. Transforming growth factor-beta1 induces heparan sulfate 6-O-endosulfatase 1 expression in vitro and in vivo. J Biol Chem. 2008;283:20397–20407. doi: 10.1074/jbc.M802850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zautner AE, Jahn B, Hammerschmidt E, Wutzler P, Schmidtke M. N-and 6-O-sulfated heparan sulfates mediate internalization of coxsackievirus B3 variant PD into CHO-K1 cells. J Virol. 2006;80:6629–6636. doi: 10.1128/JVI.01988-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Stephens RS. Mechanism of C. trachomatis attachment to eukaryotic host cells. Cell. 1992;69:861–869. doi: 10.1016/0092-8674(92)90296-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total RNA was isolated from HeLa cells. SULF1 and SULF2 mRNA levels were quantified by qRT-PCR analysis and normalized to GAPDH mRNA.
HeLa cells were transiently transfected with the vector expressing Myc-tag alone (Vector) or Myc-tagged SULF1 (HS1), Myc-tagged SULF2 (HS2), or a catalytically inactive version of Myc-tagged SULF1 (IHS1) and then infected with C. muridarum. At 24 hpi, cells were fixed and stained with antibodies to Myc (red), to visualize intracellular and extracellular SULFs, and to the Chlamydial Major Outer Membrane Protein (MOMP; Green) to visualize vacuoles. Bacterial and host DNA were detected by DAPI staining (blue). White arrows point to cells expressing Myc-tagged HS1, HS2, or IHS1. Note the presence of multiple vacuoles in the vector or IHS1-transfected cells (yellow arrows) and the decreased number of vacuoles per cell in SULF1- and SULF2-transfected cells compared to vector-transfected and IHS-transfected cells (MOMP staining; also see Fig 1C for quantitation). Scale bar = 20 microns
(A) Total RNA was isolated from HaCaT cells. SULF1 and SULF2 mRNA levels were quantified by qRT-PCR and normalized to GAPDH mRNA levels. (B) HaCaT cells were transduced with lentivirus encoding scrambled shRNA or two different shRNAs targeting SULF2 (SULF2A and SULF2B) and selected for puromycin resistance. Total RNA was isolated from shRNA-transduced cells and hsulf1 mRNA level was quantified by qRT-PCR. mRNA expression was normalized to GAPDH and then expressed as a fold change compared to scrambled shRNA-transduced cells.
Wild type mice were intranasally infected with 5×106 IFU of C. muridarum. (A) Infectious progeny were recovered from whole lungs at 5 and 48 hpi, stored frozen, and titered by quantitation of vacuole formation upon secondary infection of HeLa cells for 24 hrs. Whereas very few infectious progeny were recovered at 5 hpi, abundant viable progeny were detectable at 48 hpi. (B) H&E staining of representative lung section reveals abundant inflammatory infiltrates in Chlamydia-infected lungs at 48 hpi.