Abstract
Sterile immunity against live Plasmodium infection can be achieved by immunization with radiation attenuated sporozoites. This protection is known to be mediated in part by antigen-specific memory CD8+ T cells, presumably those residing in the liver. We characterized and compared the transcriptional profile of parasite-specific memory CD8+ T cells residing in the liver and spleen after immunization of mice with irradiated sporozoites. Microarray-based expression analysis of these memory CD8+ T cells indicated that liver resident memory cells display a distinct gene expression profile. We found major differences in the expression of immune function genes as well as genes involved in the cell cycle, cell trafficking, transcription and intracellular signaling. Importantly, the malaria parasite-induced liver resident CD8+ T cells display a transcriptional profile different to that described for CD8+ T cells following other microbial challenges.
Keywords: CD8+ T cells, memory, malaria, liver-stage, sporozoites, Plasmodium
Introduction
CD8+ T cells are crucial components of the protective immune responses against the malaria parasite Plasmodium [1]. Numerous studies have shown that CD8+ T cells specific for the circumsporozoite protein (CS), expressed by Plasmodium sporozoites and the early phase of development within hepatocytes, can efficiently block the ability of the parasite to progress to the next stage of the life cycle [2]. This anti-CS CD8+ T cell response is initiated by dendritic cells in regional lymph nodes draining the skin area where sporozoites are introduced during mosquito blood meal or after needle inoculation [3]. A few days after priming, activated CD8+ T cells egress from the lymph nodes and disseminate to different peripheral organs where they establish residency. Months after immunization with sporozoites, memory CD8+ T cells specific for the CS epitopes can be detected in lymphoid as well as non-lymphoid organs, including the liver and the spleen [4]. During malaria infection, CD8+ T cells present within the liver can rapidly eliminate liver-stage parasites by the recognition of parasite epitopes presented by hepatocytes [5]. Significantly, tissue-resident CD8+ T cells are considered to be a critical component in the protective response to a number of intracellular pathogens [6].
Naïve CD8+ T cells can develop into effectors with a heterogeneous array of functional activities. This is true even if the effector cells develop from a single naïve precursor, suggesting that this diversity may in part result from the influence of tissue-associated microenvironments. In support of this, previous studies have suggested that memory CD8+ T cells residing in the gut [7] and skin [8], are different in surface phenotype and functional properties from those residing in lymphoid organs. Presumably these differences reflect differential gene expression. Gene expression profiling of tissue-derived CD8+ memory T cells may provide important insights into immunity and vaccine development against intracellular pathogens.
In this study, we compared spleen- and liver-resident memory CD8+ T cells specific for the H2-Kd restricted P. yoelii epitope SYVSAEQI. Epitope-specific naïve TCR transgenic CD8+ T cells were adoptively transferred into naive mice which were subsequently immunized with irradiated parasites. This approach allowed us to compare the gene transcription profile of the sporozoite-induced memory CD8+ T cell populations that have identical TCRs but differ solely in their organ of residency. We report a large number of differentially expressed genes, some of which may critically influence tissue trafficking, activation status, effector function and the maintenance of tissue-associated memory.
Results
CS-specific memory CD8+ T cells from spleen and liver display different transcriptional profiles
A low number (5 × 103) of naïve Thy1.1+ CD8+ T cells specific for the P. yoelii H2-Kd restricted epitope SYVSAEQI [9] were transferred to naïve Thy1.2+ recipient mice which were then immunized intradermally with irradiated P. yoelii sporozoites (Fig.1A). Forty-five days after immunization, the expanded antigen-specific memory CD8+ T cells, all of which were CD44hi (Fig. 1B) were purified from the spleen and liver by cell sorting resulting in >95 % purified population that were CD8+ Thy1+ (Fig. 1C). RNA harvested from these cells was then used to perform gene expression analysis using mouse exon 1.0 microarray chips (Affymetrix).
Figure 1. Experimental design and cell isolations.
(A) Naive CD8Py transgenic cells (Thy1.1+) were adoptively transferred into BALB/c mice (Thy1.2+) which were then immunized with irradiated P. yoelli sporozoites. 30-45 days later, Thy1.1+ CD8+ T cells were isolated by FACS. (B) Expression of CD44 on Thy1.1+ CD8+ T cells day 45 after immunization. (C) FACS plots showing the T cell populations prior to and after cell sorting. Number indicates percentage among total lymphocytes. Data are representative of three independent cell isolations.
A total of 588 genes were differentially expressed (FDR q-value = 0.05, absolute fold change of 1.8) between naïve and memory CD8+ T cells isolated from the spleen (spleen-PyCD8). Similarly, when comparing naive cells to liver-derived memory CD8+ T cells (liver-PyCD8) using an identical cutoff, 545 differentially expressed genes were identified (Fig. 2A). Principal Component Analysis (PCA) of the microarrays results showed a distinct segregation between naïve CD8+ T cells, spleen-PyCD8 and liver-PyCD8 (Fig. 2B). These results indicate a divergent gene expression pattern displayed by these two tissue-derived memory CD8+ T cell populations of identical TCR specificity.
Figure 2. Overview of microarray analysis.
(A) Total number of genes that were differentially expressed compared to naive control. (B) Principal component analysis (PCA) to all nine arrays (3 in each group) showing distinct global gene expression in naive, spleen and liver resident memory cells sorted in Figure 1.
A direct comparison of the transcriptional profiles of memory liver-PyCD8 and memory spleen-PyCD8 identified a total of 260 transcripts that were differentially expressed (FDR q-value of 0.1, unadjusted p-value range: 3.9778 × 10−8 to 0.0028 and absolute fold change of 1.8) (Supplemental Table 1). A heatmap generated by hierarchical clustering of the differentially expressed genes shows the unique expression pattern in liver and spleen memory CD8+ T cells (Fig. 3A). The array-based results were validated by RT-PCR of selected genes involved in lymphocyte migration and effector functions (Fig. 3B).
Figure 3. Differential gene expression is observed between liver and spleen resident memory CD8+ T cells.
(A) Differentially expressed genes were selected based on a cutoff of absolute fold change > 1.8 and FDR (q-value) <0.1. Cluster analysis was performed on the differentially expressed genes using hierarchical clustering (B) validation of microarray results assessed by RT-PCR from independently sorted samples.
Gene ontology (GO) analysis
Analysis of gene ontology (GO) suggested that many of the transcripts that were differentially expressed between liver- and spleen-derived CD8+ memory cells are involved in immune responses, as indicated by a high enrichment score (negative log of the p-value by Fisher’s exact test) (Fig. 4A). Amongst genes with known immune function, we observed increased expression of Cd40lg, Fasl, Ifng, Tnfsf10, Cd7 and decreased expression of Ccr2 and Ccr7 in liver, but not in spleen PyCD8 memory cells. In addition, genes involved in other biological processes such as cell cycle, chemokine binding, sugar binding, lysosphingolipid and lysophosphatidic acid receptor activity, were also highly enriched among the differentially expressed gene list.
Figure 4. GO analysis of liver and spleen memory CD8+ T cells.
(A) Overexpression of GO terms. Dotted line represented a enrichment score of 1.3 (p-value of 0.05). Numbers in brackets represented total number of genes in that specific GO term. (B) Fold-changes between liver and spleen memory CD8+ T cells of indicated genes. (C) Expression of Ki67 and (D) IL7R/CD127 between naive, spleen- and liver-memory CD8Py cells one months and five months post immunization. Data are representative of three independent experiments.
Cell cycle genes
Many of the genes encoding cell cycle proteins, such as Ki67, Ccnb2 (Cyclin B2) and Chek1 (Checkpoint kinase 1 homolog), were uniquely up-regulated by the liver-derived CD8+ memory cells (Fig. 4B). The elevated expression of the gene encoding Ki67 by liver-derived memory CD8+ T cells at one month and five months post-immunization, suggested that these cells undergo more vigorous homeostatic proliferation than the cells with identical specificity isolated from the spleen (Fig. 4C). Consistent with this observation, we have observed high levels of homeostatic proliferation by liver-resident memory CD8+ T cells as determined by in vivo BrdU incorporation (data not shown). However, this increased level of homeostatic expansion is independent of changes in the expression levels of homeostatic cytokine receptor genes Il7r or Il15r. While the levels of Il15r expressed in memory liver-PyCD8 and spleen-PyCD8 cells were comparable (liver vs spleen: 1.3 fold), the level of Il7r was found to be lower in liver-CD8Py at both the transcript and protein levels (Fig 4D).
Chemokine and carbohydrate binding
Trafficking to peripheral organs requires the expression of particular combinations of homing and adhesion molecules such as chemokine receptors, integrins and selectins [6]. We observed the down-regulation of the genes encoding classical central memory markers CD62L and CCR7 in liver-CD8Py compared to the spleen-PyCD8. These differences were expected as we were comparing memory T cells harvested from lymphoid and non-lymphoid organs. Notably, we also observed differential expression of genes encoding other chemokine receptors in our dataset. While all of the memory liver-PyCD8 cells expressed CXCR3, there were distinct CXCR3-high and CXCR3-low subpopulations of spleen-PyCD8 memory cells. In addition, liver memory cells appeared to have a slightly increased expression of CXCR6 and a down-regulation of Ccr2, Cxcr4 and Cx3cr1 (Fig. 5A, 5B). CXCR3 has been reported to play a role in the homing of CD8+ T cells to the liver [10], while CXCR6, a receptor for one of the two known transmembrane chemokines CXCL16, appears to be important for liver resident NKT cells homeostasis [11].
Figure 5. Fold changes of genes in specific GO terms in liver and spleen memory CD8+ T cells.
Fold-change of genes of (A) chemokine and cytokine receptors and (B) carbohydrate binding. (C) Protein expression of indicated chemokine receptor genes. Data are representative of two independent experiments. (D) Fold-change of genes in transcription and (E) in intracellular signaling. (F) Protein expression of Eomes and T-bet among naive-, spleen- and liver-memory CD8Py cells. Data are representative of three experiments.
Besides CD62L (L-selectin), many carbohydrate-binding molecules such as members of the killer cell lectin-like receptor (KLR) family, were also selectively down-regulated on liver-PyCD8 memory cells when compared to either spleen-PyCD8 memory cells or naive cells: Klrc1(NKG2A), Klrc2 (NKG2C), Klrc3 (NKG2E), Klrd1 (CD94), Klre1, Klrg1(KLRG1), Klrk1 (NKG2D) and Klrg1 (KLRG1) (Fig. 5C). It is noteworthy that expression of the gene encoding KLRG1, a marker of terminal effector T cells as well as a predictor of memory potential [12], was significantly lower in liver-PyCD8 compared to spleen-PyCD8 (liver Vs spleen:−6.5 fold). The differential expression of these surface markers, particularly chemokine receptors, may be important not only for the egress of T cells from lymphoid organs, but also for the trafficking of activated T cells to the liver.
Transcription factors
There were several notable differences between liver-PyCD8 compared to spleen-PyCD8 memory cells in their expression of the genes encoding transcription factors (Fig. 5D). KLF2, which is known to regulate T cell trafficking [13] in part through the regulation of synthesis of sphingosine 1 phosphate (S1P) receptor and CD62L. Consistent with this, we observed a concomitant decrease in the expression of Cd62l and three members of the S1P receptor family (S1pr1, S1pr4 and S1pr5) in the liver. The decreased expression of these molecules may allow the egress of activated T cells from lymph nodes [14]. Another transcription factor that showed differential expression pattern between liver and spleen memory CD8+ T cells is the T-bet/Tbx21 homolog Eomes, known to be involved in the regulation and maturation of effector T cells [15]. While liver-PyCD8 T cells expressed lower levels of Eomes than their splenic counterpart, both spleen- and liver-derived memory cells expressed homogeneously high levels of Tbx21/T-bet (Fig. 5E).
The gene Zeb2 (SIP1) was dramatically up-regulated on spleen-PyCD8 memory cells when compared to liver-PyCD8 or naive cells (liver vs spleen: −9.4 fold). The function of SIP1 in lymphocytes is unknown, but recently it has been described to be important for the expression of molecules that control the signaling in mast cells [16].
Other pathways of interest
Spleen-PyCD8 memory cells preferentially expressed genes associated with G-protein coupled signaling, small GTPase mediated signaling, and genes involved in cellular signaling cascades (Fig. 5F). These genes include Rasgrp2 (encode a RAS releasing protein), Arhgef18 (encode a Rho/Rac GEF protein) and Rap2a (encode a RAS releasing protein). However, the level of Rgs1, which encodes for a regulator of G-protein signaling, was significantly up-regulated in liver-PyCD8 memory cells (4.4 fold liver > spleen). Interestingly, recent studies from our laboratory indicate that G-protein coupled signaling is critically involved in the elimination of parasite-infected hepatocytes by CD8+ T cells (unpublished). The gene Dusp6 (Dual specificity phosphatase 6, also known as MAP kinase phosphatase), a known negative regulator of the Ras/MAPK pathway, was also found to be specifically up-regulated on liver-PyCD8 memory T cells.
Liver-PyCD8 memory T cells represent a distinct memory subpopulation
To determine whether liver resident memory CD8+ T cells share a gene expression profile with memory populations described in previous studies, we performed gene set enrichment analysis (GSEA) which uses enrichment scores to determine the relative enrichment of up or down-regulated gene set in a ranked list of genes[17][18].
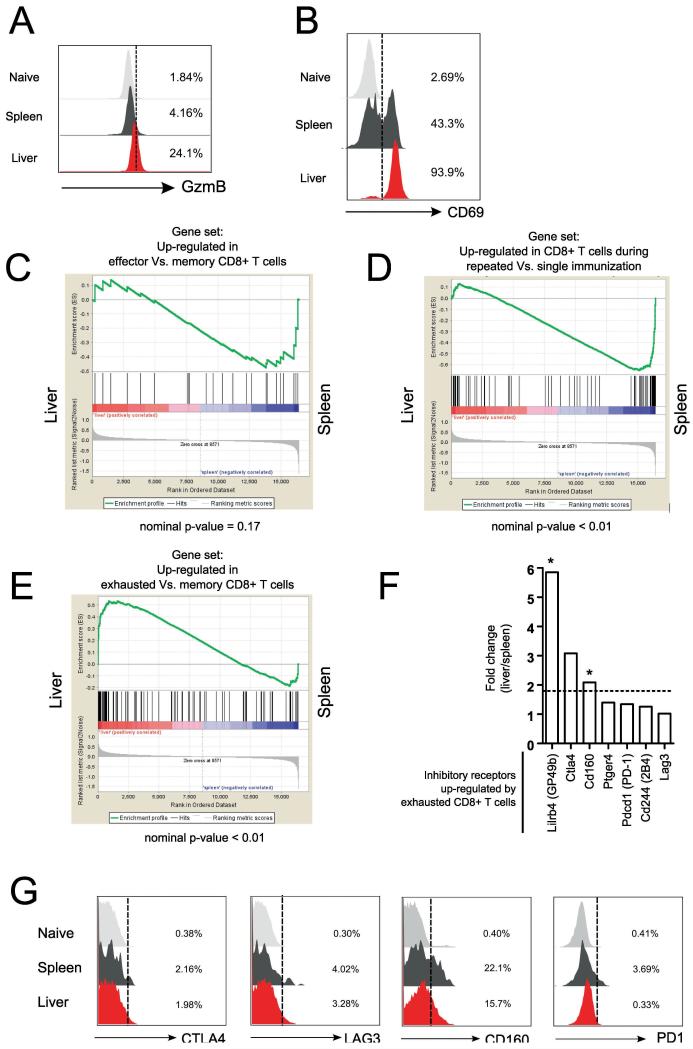
The higher levels of production of the effector protein GZMB (Fig. 6A) and CD69 (Fig. 6B), a marker of recent TCR engagement, suggested that the transcriptional profile of memory resident liver CD8+ T cells may correspond to an ‘effector’ phenotype. To explore this possibility, we compared the gene signature of liver-PyCD8 memory cells to the expression profile previously described for effector CD8+ T cells induced by LCMV infection [19]. GSEA [18] revealed that parasite-induced liver-PyCD8 memory cells did not resemble the profile described for effector cells induced by LCMV as they do not have a significant enrichment of genes that were uniquely up-regulated in the anti-LCMV effector CD8+ T cells (Fig. 6C).
Figure 6. GSEA analysis of liver-resident memory CD8Py T cells.
Protein expression of (A) GZMB and (B) CD69 among spleen and liver memory CD8Py. (C-E) GSEA was performed to determine whether up-regulated effector genes set [19] (C), repeated immunization gene set [21] (D) and exhaustion associated gene set [22] (E) show specific enrichment to liver memory CD8Py induced by irradiated sporozoite immunization. (F) Fold changes (as liver/spleen) of selected exhaustion markers in sorted Thy1.1+CD8+ memory cells from mice immunized with irradiated P. yoelii sporozoites. Dotted lines represent fold-change cutoff = 1.8, * indicates FDR < 0.1 (G) Histograms showing expression of CTLA-4, LAG3, CD160 and PD1. Plots are gated on Thy1.1+ CD8Py. Data are representative of three experiments.
As it is known that CS antigens can persist for nearly two months after immunization with intact, radiation-attenuated parasites [20], we hypothesized that the distinctive phenotype of liver-PyCD8 T cells could be the result of continuous antigen stimulation or T cell exhaustion. Therefore, we compared the transcriptional profile displayed by liver memory CD8+ T cells with the profile of T cells subjected to repeated stimulation with Listeria [21] and those in a state of exhaustion after chronic infection of LCMV [22]. GSEA revealed that malaria sporozoite-induced liver memory CD8+ T cells do not display a gene enrichment profile similar to that displayed by memory cells subjected to repeated antigenic stimulation (Fig. 6D). Moreover, we found that the liver memory population shares some transcriptional signature displayed by exhausted T cells (Fig. 6E). Importantly, we did not observe transcriptional up-regulation or protein expression of the hallmark inhibitory markers such as PD1, CTLA4 and LAG3 (Fig.6F, 6G). On the other hand, the transcript levels of inhibitory receptors CD160 as well as Lilrb4/Gp49b (Fig. 6F), a member of the leukocyte immunoglobulin-like receptors (LILRs/LIRs) which contains an ITIM motif and is known to be involved in the regulation of cytotoxic responses in T cells and NK cells [23], were significantly higher among liver memory CD8+ T cells when compared to spleen memory cells. Taken together, these results suggest that the liver resident CD8+ T cells phenotype induced by immunization with irradiated Plasmodium sporozoites is distinctive from the transcriptional profiles of memory CD8+ T cells described in other microbial challenge systems.
Discussion
We report studies describing the transcriptional profiles of memory CD8+ T cells induced by immunization with radiation attenuated Plasmodium sporozoites. We found that antigen-specific memory cells homing to the liver display a profile that differs significantly from that observed in memory cells residing in the spleen. This is a most intriguing finding considering that these two memory populations originate from identical naïve T cell precursors, sharing an identical TCR that was activated simultaneously after a single immunization.
The liver-PyCD8 memory T cells share characteristics with effector memory cells (Tem) described in other systems, such as a lower expression of CD62L and CCR7, markers used to differentiate between central memory (Tcm) and effector memory (Tem) subsets [24]. This finding is also consistent with our previous study which indicated that liver memory cells express lower levels of CD62L and CCR7 [4]. However, it is now widely recognized that substantial subset heterogeneity exists within the memory CD8+ T cell population [25]. Indeed, distinct memory CD8+ T cells residing in different peripheral organs can be further differentiated from one another and from those residing in lymphoid tissues by the selective expression of tissue-specific homing molecules. For example, β7 integrin and CCR9 are preferentially up-regulated on gut-homing T cells [26]. On the other hand, T cells homing to the skin require expression of CLA and CCR10 [27]. More recently it has been shown that brain-, skin- and mucosal-resident T cells selectively up-regulate CD103[28][29][30]. The expression profiling presented here provides clues in identifying genes that may be important in CD8+ T cell homing and development in the liver such as CXCR3 and CXCR6. It is possible that the liver-associated expression of these molecules is influenced by tissue-derived factors and they may play a critical role in homing as well as the survival and functionality of activated T cells, as described in other systems [31]. In addition, CD8+ T cell subsets with different effector function potentials have also been described [32]. Given the differential expression of genes associated with effector function between spleen-PyCD8 and liver-PyCD8 memory cells such as Gzmb and Fasl, it remains an area of future investigation to determine whether these two populations may also show differences regarding their functional properties.
Our study also revealed that several transcription factors are differentially expressed between liver and spleen memory cells. One of them is KLF2, which has been previously described to regulate the expression of CD62L and S1P, both implicated in T cell homing in lymphoid organs [33]. We also found that EOMES, a master regulator of the development of effector CD8+ T cells [15], and Zeb2 (SIP1), a transcription factor that regulates mast cell cytokine secretion [16], are expressed at lower levels in liver resident memory CD8+ T cells. How the expression of these transcription factors impact the differentiation, as well as functional properties of tissue-associated memory T cell development have not been fully investigated.
Importantly, the liver-derived memory T cells express high levels of cell cycle genes such as Ki67 and Chek1. It is possible that a continuous low-level proliferation is partially driven by the extended presence of Plasmodium liver-stage antigens, which are known to persist for at least two months after immunization with radiation-attenuated, non-replicating malaria sporozoites [20]. Continuous Plasmodium antigen presentation may induce some level of persistent T cell activation, as evidenced by the CD69 expression and up-regulation of certain effector molecules transcripts in memory liver-PyCD8+ T cells. While some chronic viral infections, such as LCMV, can lead to T cell exhaustion, we did not observe significant up-regulation of the hallmark exhaustion markers. Clearly, even if the liver resident memory CD8+ T cells experience prolonged presentation of Plasmodium antigen, the resulting transcriptional profile of Plasmodium sporozoite-induced CD8+ T cells did not resemble the gene signature of T cell exposed to repeated exposure to bacterial antigens [21] or CD8+ T cells undergoing exhaustion as a result of chronic viral infections [22]. The lack of a demonstrable inflammatory reaction to the persistent Plasmodium antigen could explain the absence of hallmark exhaustion markers on the antigen-specific liver-PyCD8 T cells.
As noted previously, the liver microenvironment is likely to influence the transcriptional profile of these cells. Tissue residency is known to influence expression of certain markers and it has been proposed that the liver may serve as a ‘graveyard’ for lymphocytes, as apoptotic lymphocytes have been observed to accumulate in the liver [34]. In fact, it is believed that in the liver there is a predominance of immunosuppressive signals [35] and, therefore, we speculate that the different cell types found in this organ, such as Kupffer cells, liver sinusoidal endothelial cells (LSECs) and hepatic stellate cells, could impact the gene expression profile and influence the differentiation of memory CD8+ T cells that reside in the liver microenvironment.
Recently Wakim et al. [36] reported a comparative analysis of the phenotypic and transcriptional profiles between antigen-specific brain-resident CD8+ memory cells and memory cells from the spleen. There are notable similarities between liver- and brain-resident CD8+ memory cells including the down-regulation of the expression of the key regulators of T cell differentiation, Eomes and Tcf-1, and the up-regulation of the genes encoding the inhibitory receptors CTLA4 and PD1 as well as the gene encoding granzyme B (Gzmb). An area that appears to distinguish tissue-resident memory CD8+ cells from memory cells in the spleen is the apparent need for alternate survival factors for long-term maintenance. Liver- and brain-resident memory cells have reduced expression of the IL-7 receptor and, in the case of brain-resident memory cells, are unresponsive to IL-7 or IL-15 stimulation. Also of note is that neither of these tissue-resident memory cell population demonstrates an expression profile to suggest that they have developed to a state of exhaustion. While there are a number of similarities, important differences in gene expression are also noted. In liver-PyCD8+ T cells the genes encoding CXCR3 and CXCR6 were selectively up-regulated and it is likely these differences reflect tissue-specific demands for trafficking. Another area where liver-resident and brain-resident CD8+ memory T cells appear to differ is in their potential to proliferate. In contrast to the liver-PyCD8+ T cells reported here that have gene expression profiles indicating high levels of homeostatic proliferation, the brain-resident CD8+ memory cells had a significantly reduced capacity to proliferate, even after antigen challenge. Taken as a whole, the results from Wakim et al. and our study reinforce the notion that tissue-resident memory cells are a distinct differentiation fate from the central memory and the circulating memory pool. In addition, a comparison of the transcriptional and phenotypic profiles of the liver- and brain-resident cells predicts that each tissue will harbor a memory population with a distinct expression and functional profile.
Our study is the first to describe the transcriptional profile of antigen-specific memory CD8+ T cells against pre-erythroytic stage of Plasmodium parasites. We find major transcriptional differences between spleen and liver memory cells, which indicate that tissue residency plays an important part in memory differentiation and development of T cells induced by irradiated malaria sporozoites. This observation has implications for vaccine development as many studies use peripheral blood mononuclear cells (PBMCs) or splenocytes to evaluate vaccine-induced responses. It is possible that the evaluation of T cell responses obtained from splenocytes or PBMCs may not entirely reflect the functional and transcriptional properties of liver-resident effector memory cells. The distinctive gene signature of the liver-resident memory CD8+ T cell population may provide important insights into immunity and vaccine development against liver-stage Plasmodium parasites.
Materials and Methods
Animals
Five- to eight-week old female BALB/c were purchased from Taconic Farms (Hudson, NY). Generation of the T cell receptor (TCR) transgenic mice specific for H2-Kd SYVPSAEQI epitope of Plasmodium yoelii was previously described [9]. TCR transgenic mice were bred and maintained in the Thy1.1+ background at the Johns Hopkins animal facility. Experiments involving mice were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University.
Adoptive transfer and immunization
One day before immunization, 5000 naïve TCR transgenic Thy1.1+ CD8+ T cells were adoptively transferred to Thy1.2+ recipient mice. P. yoelii 17XNL sporozoites were obtained from salivary glands of female Anopheles stephensi and irradiated at 40 000 rad as described [9]. For immunization, 5 × 104 irradiated sporozoites were injected intradermally at the base tail.
Antigen-specific CD8+ T cell isolation
Tissues were harvested from mice 30 – 45 days after immunization. Single-cell suspensions of lymphocytes from spleens were obtained by grinding the tissues between the ground ends of two microscope slides and then filtering them through 100 μm pore size nylon mesh. To isolate intrahepatic lymphocytes, livers were perfused with 10 ml ice-cold HBSS and the liver pellet was suspended in 35% percoll followed by centrifugation at 500 G for 20 minutes at room temperature. To improve sorting efficiency, antigen-specific Thy1.1+ CD8+ T cells were first enriched using PE-anti-Thy1.1 (Clone OX-7) antibodies (BD Bioscience, San Diego, CA), MACS anti-PE-microbeads and MACS LS separation columns (Miltenyi Biotech, Auburn, CA) at 4° C. The enriched cell fraction was then sorted using a FACsAria (BD Biosciences) and the viability of the sorted cells was verified by flow cytometry.
RNA isolation and microarray
RNA was extracted from purified memory Thy1.1+CD8+ T cells immediately after cell sorting using the RNAqueous Micro kit according to the manufacturer’s protocol (Ambion, Austin, TX). cDNA was synthesized using WT-Ovation Exon Module version 1 and the FL-Ovation cDNA Biotin Module version 2 (NuGEN Technologies, San Carlos, CA). The cDNA probe was then hybridized onto Mouse Exon 1.0 Affymetrix chips at the JHSPH Genomic Analysis and Sequencing Core.
Microarray analysis
Hybridization was done for three biological replicates composed of cells isolated from pools of 10-12 immunized mice for each replicate. Microarray data analysis was performed using Partek Genomic Suite software (St. Louis, MO) version 6.6. Core probeset was used for probeset filtering. Probeset-level expression was normalized using the robust multi-array (RMA) method, signal was log2 transformed and probeset summarization was done by median polish. For gene expression analysis, exon expression was first summarized to gene level using mean. A false discovery rate (q-value) of 0.05 (unadjusted p-value range: 3.9778 × 10-8 to 0.0028) was chosen and genes with an absolute fold-change of 1.8 were selected for gene ontology (GO) analysis. GO enrichment was performed using Fisher’s exact test. Terms containing fewer than 10 genes or greater than 500 genes were not included in the analysis. GO terms with an enrichment score >1.3 (which corresponded to negative log of the p-value 0.05) were considered significant. Gene ontology (GO) ANOVA was also performed using Partek software and those with FDR of q-value < 0.05 were included in the analysis. GSEA V.2 algorithm/platform was used for gene set enrichment analysis (GSEA) (Broad Institute) and was performed as described [18] after converting probe set identifiers to gene symbols. Genes were ranked according to t-tests and 1000 permutations on the gene set were performed. Enrichment was considered significant if FDR was less than 0.05.
Quantitative PCR
cDNA was synthesized from RNA obtained from independently sorted samples. RNA was reverse transcribed into cDNA using the RT2 First Strand kits (SAbioscience) following the manufacturer’s protocol. Quantitative real-time PCR was performed using SYBR Green PCR Master Mix (SAbioscience) on an ABI 7300 real time PCR instrument. Fold-change was normalized to naive control samples using the following housekeeper genes: Gusb, Hprt, Hsp90ab1, Gapdh and Actb.
Antibodies and Flow Cytometry
Antibodies were purchased from eBioscience unless otherwise noted. The following fluorochrome-conjugated antibodies were used: APC-conjugated CD8 (clone 53–6.7), KLRG1 (clone 2F1); FITC-conjugated Ki67 (BD), PD-1 (clone J43), 2B4 (clone eBio244F4) and Granzyme B (clone 16G6); PE-conjugated Thy1.1 (clone His51), CTLA-4(clone UC10-4B9), CD160 (clone eBioCNX46-3), NKG2D (clone A10), Eomes (clone Dan11mag) and T-bet (clone eBio4B10). Intracellular staining with anti-Eomes and anti-T-bet antibodies were performed using Foxp3 staining buffer set (eBioscience) following the manufacturer’s protocol. All results were collected with CellQuest software on a FACSCalibur (Becton Dickinson).
Supplementary Material
Acknowledgements
We thank Alan Aderem, Stephen Ramsey and Michael Overstreet for insightful discussions and Anne Jedlicka and the JHSPH Genomic Analysis and Sequencing Core for technical assistance. This work is supported by NIH grant number AI44375 (FZ). The authors thank the Bloomberg Family Foundation for continued support. S. Tse was supported by a pre-doctoral fellowship from the Johns Hopkins Malaria Research Institute.
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
Reference
- 1.Overstreet MG, Cockburn IA, Chen Y, Zavala F. Protective CD8+ T cells against Plasmodium liver stages: immunobiolog of an “unnatural” immune response. Immunol Rev. 2008;225:272–282. doi: 10.1111/j.1600-065X.2008.00671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero P, Maryanski JL, Corradin G, Nussenzweig RS, Nussenzweig V, Zavala F. Cloned cytotoxic T cells recognize an epitope in the circumsporozoite protein and protect against malaria. Nature. 1989;341:323–326. doi: 10.1038/341323a0. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarty S, Cockburn IA, Kuk S, Overstreet MG, Sacci JB, Zavala F. CD8+ T lymphocytes protective against malaria liver stages are primed in skin-draining lymph nodes. Nat Med. 2007;13:1035–1041. doi: 10.1038/nm1628. [DOI] [PubMed] [Google Scholar]
- 4.Morrot A, Hafalla JCR, Cockburn IA, Carvalho LH, Zavala F. IL-4 receptor expression on CD8+ T cells is required for the development of protective memory responses against liver stages of malaria parasites. J Exp Med. 2005;202:551–560. doi: 10.1084/jem.20042463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cockburn IA, Tse S-W, Radtke AJ, Srinivasan P, Chen Y-C, Sinnis P, et al. Dendritic Cells and Hepatocytes Use Distinct Pathways to Process Protective Antigen from Plasmodium in vivo. PLoS Pathog. 2011;7:e1001318. doi: 10.1371/journal.ppat.1001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol. 2009;9:153–161. doi: 10.1038/nri2496. [DOI] [PubMed] [Google Scholar]
- 7.Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting Edge: Gut Microenvironment Promotes Differentiation of a Unique Memory CD8 T Cell Population. J Immunol. 2006;176:2079–2083. doi: 10.4049/jimmunol.176.4.2079. [DOI] [PubMed] [Google Scholar]
- 8.Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, et al. Long-Lived Epithelial Immunity by Tissue-Resident Memory T (TRM) Cells in the Absence of Persisting Local Antigen Presentation. Proc Natl Acad Sci U S A. 2012;109:7037–7042. doi: 10.1073/pnas.1202288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sano G, Hafalla JCR, Morrot A, Abe R, Lafaille JJ, Zavala F. Swift Development of Protective Effector Functions in Naive Cd8+ T Cells against Malaria Liver Stages. J Exp Med. 2001;194:173–180. doi: 10.1084/jem.194.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hokeness KL, Deweerd ES, Munks MW, Lewis CA, Gladue RP, Salazar-Mather TP. CXCR3-Dependent Recruitment of Antigen-Specific T Lymphocytes to the Liver during Murine Cytomegalovirus Infection. J Virol. 2007;81:1241–1250. doi: 10.1128/JVI.01937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Thorlacius H, Johnston B, Staton TL, Xiang W, Littman DR, et al. Role for CXCR6 in Recruitment of Activated CD8+ Lymphocytes to Inflamed Liver. J Immunol. 2005;174:277–283. doi: 10.4049/jimmunol.174.1.277. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Krüppel-like factors in leukocyte development, function, and disease. Blood. 2010;116:4404–4414. doi: 10.1182/blood-2010-05-285353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pappu R, Schwab SR, Cornelissen I, Pereira JP, Regard JB, Xu Y, et al. Promotion of Lymphocyte Egress into Blood and Lymph by Distinct Sources of Sphingosine-1-Phosphate. Science. 2007;316:295–298. doi: 10.1126/science.1139221. [DOI] [PubMed] [Google Scholar]
- 15.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, et al. Control of Effector CD8+ T Cell Function by the Transcription Factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 16.Barbu EA, Zhang J, Berenstein EH, Groves JR, Parks LM, Siraganian RP. The Transcription Factor Zeb2 Regulates Signaling in Mast Cells. J Immunol. 2012;188:6278–6286. doi: 10.4049/jimmunol.1102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haining WN, Wherry EJ. Integrating Genomic Signatures for Immunologic Discovery. Immunity. 2010;32:152–161. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and Functional Profiling of Memory CD8 T Cell Differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 20.Cockburn IA, Chen Y-C, Overstreet MG, Lees JR, Van Rooijen N, Farber DL, et al. Prolonged antigen presentation is required for optimal CD8+ T cell responses against malaria liver stage parasites. PLoS Pathog. 2010;6:e1000877. doi: 10.1371/journal.ppat.1000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wirth TC, Xue H-H, Rai D, Sabel JT, Bair T, Harty JT, et al. Repetitive Antigen Stimulation Induces Stepwise Transcriptome Diversification but Preserves a Core Signature of Memory CD8+ T Cell Differentiation. Immunity. 2010;33:128–140. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wherry EJ, Ha S-J, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Gu X, Laouar A, Wan J, Daheshia M, Lieberman J, Yokoyama WM, et al. The gp49B1 Inhibitory Receptor Regulates the IFN-γ Responses of T Cells and NK Cells. J Immunol. 2003;170:4095–4101. doi: 10.4049/jimmunol.170.8.4095. [DOI] [PubMed] [Google Scholar]
- 24.Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 25.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation Directs Memory Precursor and Short-Lived Effector CD8(+) T Cell Fates via the Graded Expression of T-bet Transcription Factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berlin C, Berg EL, Briskin MJ, Andrew DP, Kilshaw PJ, Holzmann B, et al. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74:185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 27.Reiss Y, Proudfoot AE, Power CA, Campbell JJ, Butcher EC. CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. J Exp Med. 2001;194:1541–1547. doi: 10.1084/jem.194.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T Cells Persisting Within the Brain After Local Infection Show Functional Adaptations to Their Tissue of Residence. Proc Natl Acad Sci U S A. 2010;107:17872–17879. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 30.Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 Transcription-Factor Deficiency in T Cells Results in Unrestrained Cytokine Production and Upregulation of Bystander Chemokine Receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parish IA, Kaech SM. Diversity in CD8(+) T cell differentiation. Curr Opin Immunol. 2009;21:291–297. doi: 10.1016/j.coi.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 34.Crispe IN, Dao T, Klugewitz K, Mehal WZ, Metz DP. The liver as a site of T cell apoptosis: graveyard, or killing field? Immunol Rev. 2000;174:47–62. doi: 10.1034/j.1600-0528.2002.017412.x. [DOI] [PubMed] [Google Scholar]
- 35.Crispe IN. The Liver as a Lymphoid Organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 36.Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, et al. The Molecular Signature of Tissue Resident Memory CD8 T Cells Isolated from the Brain. J Immunol. 2012;189:3462–3471. doi: 10.4049/jimmunol.1201305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.