Abstract
Immediate early transcription is an integral part of the neuronal response to environmental stimulation and serves many brain processes including development, learning, triggers of programmed cell death, and reaction to injury and drugs. Following a stimulus, neurons express a select few genes within a short period of time without undergoing de novo protein translation. Referred to as the ‘gateway to genetic response’, these immediate early genes (IEGs) are either expressed within a few minutes of stimulation or later within the hour. In neuronal IEGs that are expressed rapidly, productive elongation in response to neuronal activity is jump-started by constitutive transcription initiation together with RNA polymerase II stalling in the vicinity of the promoter. IEGs expressed later in the hour do not depend on this mechanism. On the basis of this Polymerase II poising, we propose that the immediate early genes can be grouped in two distinct classes: the rapid and the delayed IEGs. The possible biological relevance of these classes in neurons is discussed.
The phrase ‘Immediate early genes’ was initially coined by virologists and was used exclusively to describe viral regulatory factors transcribed de novo by host cells immediately within two minutes after viral integration (Jayaraman, 1972). Any subsequent transcription was, at that time, termed delayed-early transcription and referred to downstream genes on the circular viral plasmid. Decades later, the threshold of two minutes faded as the phrase ‘immediate-early’ was adopted widely by cell biologists (Cochran et al., 1983). This alteration in meaning was based on the recognition that many of the genes induced by viral integration were also induced immediately by extracellular signals such as growth factors (Greenberg and Ziff, 1984, Mulcahy et al., 1985). The mRNAs of most of these genes were detected at their peaks between 30 minutes and an hour (Curran et al., 1985). This observation led to incorporation of the hour into the general definition of IEGs, which are usually defined as genes that can be expressed without de novo protein synthesis. Henceforth, most IEG studies in neurons were largely restricted to the hour long time-frame.
As the molecular mechanisms underlying long-term memory were becoming known to neurobiologists, one important realization was that long lasting synaptic changes such as long-term potentiation (LTP) relied on mechanisms distinct from the shorter lasting forms (Bliss and Collingridge, 1993). Depending on persistence, LTP has been classified as LTP1, LTP2 or LTP3 (Raymond, 2007). The fast decaying LTP1, also known as the early- or E-LTP, is protein and mRNA-synthesis independent. LTP2, the intermediary phase, is protein synthesis-dependent but independent of gene transcription. Presumably, LTP2 could be supported from existing pools of mRNA and protein in the cell, but LTP3, the longest lasting form of LTP, seemed to require de novo mRNA synthesis followed by their translation within a very short ‘critical window’ of time (Montarolo et al., 1986, Nguyen et al., 1994, Frey et al., 1996); if gene transcription was blocked outside of this window, consolidation of L-LTP was unaffected. Thus the immediate induction of genes was proposed to be an integral step in the consolidation of LTP. Identification of these immediate early genes, therefore, was widely considered to be a priority for understanding how synaptic changes could last for extended periods (Curran and Morgan, 1985, Greenberg et al., 1985, Morgan and Curran, 1986, Morgan et al., 1987). Consistent with the idea that immediate transcription can occur in neurons, several IEGs such as cfos were found to be strongly induced within a matter of minutes by such diverse stimuli as seizure activity, electrical stimulation and injury (Dragunow and Robertson, 1987a, b, Morgan et al., 1987, Saffen et al., 1988). Due to their reliable temporal signature, IEG expression is now commonly used to track neuronal activity in specific areas of the brain in response to specific behaviors. For example, IEGs can be induced in the visual cortex by exposing dark-reared animals to light and in hippocampal neurons in response to an animal exploring a novel environment (Worley et al., 1990, Guzowski et al., 1999). Thus, it appeared that IEG expression was a major early step in the neuronal genetic response to many different physiological processes.
Given the likely relevance of IEGs to neuron function, therefore, identification of every neuronal IEG was soon attempted using subtractive hybridization and differential cDNA cloning (Nedivi et al., 1993, Qian et al., 1993, Yamagata et al., 1993). Based on these screens, the number of neuronal activity-induced IEGs was initially estimated to be around 500–1000 (Nedivi et al., 1993). Subsequently however, the number of activity-induced IEGs was found to be closer to 30–40 (Lanahan and Worley, 1998). A substantial number of these IEGs were transcription factors and, as such, are expected to regulate expression of downstream gene products. Additionally though, neurons express a small number of unique IEGs that are not transcription factors. These neuronal IEGs, often referred to as ‘effector IEGs’, perform a range of functions outside the nucleus and many localize to synaptic sites and regulate synaptic function (Leslie and Nedivi, 2011). Because IEGs have such an important role in neuronal physiology, understanding the mechanisms leading to their induction could lead to important insights into neuronal contexts in which different genes are induced.
Many routes to neuronal IEG induction
Under physiological conditions, neurons perceive environmental signals in the form of both electrical activity through synapses and neuromodulatory signals through the release of compounds such as growth factors. In the case of excitatory synapses in the brain, binding of glutamate to ligand-gated ion channels leads to Ca2+ influx through NMDA-type glutamate receptors. In addition, glutamate also binds to and opens AMPA-type glutamate receptors, which generate a post-synaptic potential that depolarizes the membrane, allowing further influx of Ca2+ through voltage-gated ion channels. Calcium, a strong inducer of neuronal and non-neuronal IEGs (Greenberg et al., 1986), is thought to be sensed locally near the membrane and the signal then relayed to the nucleus via signaling cascades that require post-synaptic second messengers and cAMP- and calcium-dependent protein kinases (Greer and Greenberg, 2008, Kandel, 2012). Phosphorylation of constitutively expressed transcription factors could then bind to specific regulatory elements in a gene promoter or enhancer and initiate gene transcription by recruiting RNA Polymerase II (Pol II) (Greer and Greenberg, 2008, West and Greenberg, 2011). This ‘synapse-to-nucleus’ signaling model is built on two lines of evidence. Firstly, activity-induced gene transcription is typically sensitive to antagonists of NMDA receptors, a major source of synaptic Ca2+ influx in neurons. Secondly, and more recently, signaling molecules have now been imaged en route to the nucleus in response to synaptic activity. These molecules include widely studied MAP and CaM kinases and more recently AIDA-1 and CRTC1 (Martin et al., 1997, Deisseroth et al., 1998, Jordan et al., 2007, Ch’ng et al., 2012). Such spatial translocation of signaling molecules is well characterized in non-neuronal cells where the nucleus is not far from the membrane signaling events and thus the mechanism is well suited to mediate immediate cellular response. However, neuronal nuclei are often located several hundred microns away from distal synapses and signaling molecules may not translocate to the nucleus fast enough to explain very fast IEG induction in response to neuronal activity or behavior that begins almost instantaneously: several studies have reported behavioral task-induced IEG products in two minutes (Guzowski et al., 1999, Guzowski et al., 2001).
To explain signal transduction suitable for the fastest IEG expression, we have previously proposed that the signal for LTP-related gene transcription may be relayed to the nucleus via action potentials through calcium (Adams and Dudek, 2005, Saha and Dudek, 2008). Integrated post-synaptic potentials create trains of action potentials that depolarize the membrane to allow Ca2+ influx through NMDA receptors and voltage-gated channels. Because the synaptic depolarization travels along the membrane in a flash, Ca2+ influx can occur throughout the neurons and at somatic membranes in proximity of the nucleus, thus requiring very little time for the signal to travel from distal synapses. At that point, Ca2+ can either act at plasma membrane microdomains associated with L-type Ca2+ channels, or else diffuse in to the somatic cytosol to trigger signaling cascades such as the Ca2+-Calmodulin and MAP kinase pathways (Hardingham et al., 2001, Wheeler et al., 2008). Alternatively, incoming Ca2+ can act directly in the nucleus where it can potentially signal gene transcription (Zhang et al., 2009). Along these lines, several studies have shown that action potentials can trigger important signaling cascades, induce gene transcription and rescue long term potentiation from decay without synaptic involvement (Eshete and Fields, 2001, Dudek and Fields, 2002, Alarcon et al., 2006, Adams et al., 2009, Schoenenberger et al., 2009).
Nuclear molecular mechanisms: poised for rapid transcription
Certain viruses, like the mouse mammary tumor viruses (MMTV), can incorporate their DNAs into host chromatin and can then undergo rapid transcription bursts in response to glucocorticoids (Ringold et al., 1977, Archer et al., 1994). Likely this rapid response is due to permissive chromatin conformation and accessibility of DNA because the extent of transcription of MMTV proviruses is regulated by the site(s) of host DNA where the viral DNA integrates (Ringold et al., 1979). In physiological circumstances, however, the means to serve very fast gene transcription within minutes, as is seen in neurons after a behavioral task, remained unknown. Until recently, focus in neuronal gene transcription has been on transcription factors as key regulators of transcription leading to synaptic plasticity and long-term memory formation. For example, strong evidence exists favoring a role for cAMP response element-binding protein (CREB), CCAAT enhancer binding protein δ (C/EBPδ) and nuclear factor kappaB (NF-κB) in the regulation of LTP and memory (Dash et al., 1990, Martin and Kandel, 1996, Meffert et al., 2003, Pintchovski et al., 2009, Arguello et al., 2013). Although transcription factors are undoubtedly important for neuronal gene transcription, a focus on them as triggering element assumes that transcription proceeds according to a classical model. The classical model of transcription begins when transcription factors bind to specific sequences in the promoter or enhancer elements of the gene and co-ordinate with remodeling complexes to modify chromatin and assemble the preinitiation complex. Although exact the temporal requirements of chromatin remodeling and preinitiation complex assembly in cells is unknown, an estimate may be made from reconstituted transcription studies in cell-free systems. In these studies, preinitiation complex assembly from purified initiation factors on nucleosome free DNA template required about ten minutes (Serizawa et al., 1997). An additional critical rate-limiting step of the classical model is also the subsequent recruitment and phosphorylation of Pol II to initiate gene transcription. However, a detailed consideration of the kinetics of the very fastest of IEG induction in the context of neuronal activity has led us to suggest inadequacies in this canonical model.
As mentioned already, many neuronal IEG transcripts, like arc, can be detected within two minutes of a stimulating event (Guzowski et al., 1999). Accordingly all of the following presumably must take place prior to those two minutes: 1) signal transduction to the nucleus, 2) post-translational modifications, 3) DNA-binding of transcription factors like CREB, 4) activation of enzymes to remodel chromatin, 5) assembly of preinitiation complexes, 6) recruitment and phosphorylation of Pol II and finally, 7) complete productive elongation to produce a detectable RNA. Furthermore, transcription initiation is often abortive in the first few rounds unless Pol II clears the promoter (Marshall and Price, 1992). Taken together, we proposed that the regulation of rapidly induced immediate genes, and in particular arc, may take place at a level other than that of transcription initiation. We recently tested this hypothesis and learned that immediate gene induction can be achieved either by regulating transcription initiation or by controlling transcription at the level of elongation (Saha et al., 2011).
Some clues into the very fast transcription of IEGs were found in recent work showing that expression of many genes can be regulated after polymerase II recruitment and transcription initiation. RNA polymerase II initiates transcription in these genes, but pauses after transcribing a short piece of mRNA near the promoter (Lis, 1998). Referred to as ‘promoter proximal Polymerase stalling’, this mechanism allows transcription to be regulated after initiation by modulating efficient release of the stalled polymerase into productive elongation. As seen convincingly in the prototypical heat shock protein (hsp) genes in Drosophila, the paused polymerase is released infrequently and inefficiently in the absence of stimuli and the mRNA fails to be productively elongated. However, upon heat shock stimulation, the paused polymerase is released almost immediately to complete successful transcription of the gene (Lis, 1998). This phenomenon of polymerase ‘stalling’ is widespread in the genomes of yeast, Drosophila, Caenorhabditis elegans, and mammals (Radonjic et al., 2005, Guenther et al., 2007, Muse et al., 2007, Baugh et al., 2009). In higher eukaryotes, a major mediator of stalling is the Negative elongation factor (NELF), a four subunit complex composed of NELF-A, B, E and either NELF-C or NELF-D (Narita et al., 2003). Although the exact mechanism of NELF-dependent inhibition remains unknown, RNAi-mediated depletion of Drosophila NELF reduced Polymerase promoter occupancy in most genes (Muse et al., 2007). This observation suggests that NELF is required for promoter-proximal stalling and knocking it down is a suitable tool to study the biological roles of polymerase stalling.
Currently, polymerase stalling is envisioned as a modulator of transcription dynamics and coordinator of gene expression where effective regulation of the stalling duration can bring about rapid and robust gene transcription (Nechaev and Adelman, 2011). However, the correlation between Pol II pausing and rapid gene induction was not tested directly by assessing the induction rate of a gene in the absence of Pol II stalling (Li and Gilmour, 2011). Because polymerase stalling uncouples transcription from the time-consuming initial steps, Saha et al. hypothesized that this mechanism was ideal to serve rapid expression of neuronal IEGs (Saha et al., 2011). Furthermore, because stalled polymerase helps to retain permissive epigenetic signatures on promoters, prompt successive rounds of polymerase recruitment to the accessible chromatin would lead to a robust transcription output. In a direct test of the hypothesis that polymerase stalling plays a critical role in neuronal activity-induced IEG transcription, Saha et al. (Saha et al., 2011) observed a strong relationship between rapid transcription and poised polymerase II. Also note that, upon transcription initiation, transient polymerase stalling occurs in all genes, including delayed IEGs. However, the action of NELF in the case of rapid IEGs prolongs this natural pause and utilizes it to respond rapidly to neuronal activity.
Using the neuronal ‘effector’ IEG, arc, as a model gene in untreated dissociated rat neurons, Saha et al. found polymerase II significantly enriched near the arc transcription start site (TSS) (Saha et al., 2011). This enrichment had hallmarks of transcription that has already been initiated but is stalled: 1) Rpb1, the largest polymerase II subunit, was phosphorylated on serine 5 residues of the C-terminal domain (CTD) heptad repeats (Boehm et al., 2003), 2) The arc promoter region was enriched with histones in a generally permissive state with high levels of trimethylated lysine 4 residues of histone 3. Additionally, 3) NELF-A and NELF-E enrichment aligned with the polymerase peak in chromatin-immunoprecipitation studies. Knock-down of NELF-A or NELF-E abolished promoter proximal polymerase II enrichment. In the absence of poised polymerase in neurons from the knock down of NELFs, rapid induction of arc was severely attenuated at five minutes after an activity trigger. This impairment of rapid IEG transcription in the absence of stalling suggests that the two processes are coupled. Interestingly, the lack of stalling did not reliably affect arc induction 45 minutes after the trigger, suggesting that only the very earliest transcriptional response relies on NELF and the polymerase stalling mechanism (Saha et al., 2011). Because RNA polymerase II is stalled on numerous genes, including non-IEGs, NELF subunit knockdown may have widespread effects (Gilchrist et al., 2008). Saha et. al., therefore verified that neuronal excitability in their NELF-depleted neurons was normal. Thus even with the potential widespread effects of NELF knockdown, no differences in synaptic function or excitability were found.
A revised model: rapid and delayed IEGs
In order to determine the role of Pol II stalling in the transcription of other neuronal IEGs, Saha et al. identified by microarray all IEGs that responded to a neuronal activity-inducing protocol in rat cortical neuron cultures. Subsequently, next-gen sequencing of ChIP material (ChIP-seq) was used to study the level of stalling in the identified IEGs. Similar to what was found with arc, the complement of IEGs that are expressed with very short latencies was characterized as having polymerase enriched in proximity to their promoters. Moreover, their early expression was sensitive to NELF knockdown but their later expression remained unaffected. The group of IEGs having a longer latency for detection was found to be largely devoid of such polymerase enrichment near their promoter (Saha et al., 2011). Based on these data, Saha et al. proposed that IEGs can be divided into two subtypes: rapid IEGs, defined as IEGs with products detected within minutes after stimulation, and delayed IEGs, the products of which are not detected until later in the hour. Not surprisingly, all of these rapid IEGs had polymerase bound to the promoter regions and the majority of delayed genes did not. A promoter was considered to be bound to Pol II if it was represented in the ChIP-seq data significantly more than in quiescent intergenic regions of the genome. Although promoters of some delayed IEGs were considered bound by the polymerase, the cases were few, and did not differ from the percentage of genes in the genome as a whole. To quantitatively assess stalling from ChIP-seq data, a stalling index was calculated by normalizing the number of polymerase ‘hits’ in the promoter region by the number of ‘hits’ within the gene body (Muse et al., 2007). For example, a low calculated stalling index can be a reflection of active transcription with a relative abundance of polymerase in the gene body, or it may reflect polymerase paucity near the promoter. Because basal expression of IEGs is typically very low, small stalling indices in the very few polymerase-bound delayed IEGs is reflective of a lack of stalling at their promoters (Saha et al., 2011). In contrast, the rapid IEGs were found to have high stalling indices. The average stalling index of all rapid IEGs was significantly more than the average stalling index of all genes in the rat genome. This suggested a molecular hallmark of those genes predisposed for immediate response. Thus the classification of rapid- and delayed-IEGs coincides with an abundance of RNA polymerase stalled near gene promoters, indicative of an actual biological underpinning to these IEG subcategories (Saha et al., 2011).
IEGs are often key regulators of the neuronal genome that allow signaling pathways to access specific sets of downstream genes (Dragunow, 1996). In addition, several effector IEGs are now known to participate directly in several aspects of neuronal functions. Despite such a diverse array of functions, the IEGs can now be grouped in sub-categories based on the latency of their expression. This raises the question as to whether there exists a functional role for of such a gene group that responds so rapidly to neuronal activity.
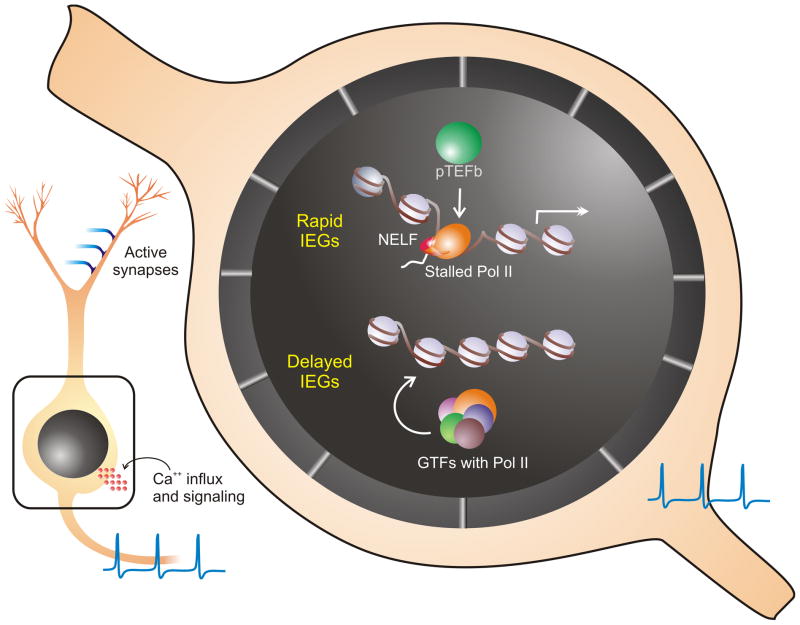
From the neuron’s perspective, a parsimonious strategy for synchronous gene induction in response to neuronal activity or other such fast signal would be to have a common mechanism used by all genes that respond first and fast. In the case of transcription and translation, factors could be required immediately to build upon and consolidate brief synaptic events, such as LTP. These ‘urgent’ genes would be loaded with stalled polymerase to deliver almost instantaneous bursts of gene transcription (Figure 1). The trigger for these rapid IEGs could be served by action potential driven signaling pathways. In contrast, with their products apparently not urgently required at the synapse or to rapidly initiate transcription cascades, the delayed IEGs likely undergo a more classical mode of transcription that includes the initiation process (Figure 1). Signals for expression of these genes could be conveyed by relatively slower mechanisms such as synapse-to-nucleus signaling pathways.
Figure 1. The revised model.
Synaptic activity at distal synapses can induce action potentials, which leads to Ca2+ influx in the soma to engage signaling cascades to the nucleus. Within the nucleus, genes respond to these incoming signals in two ways. The delayed IEGs undergo transcription initiation, which includes recruitment of general transcription factors (GTFs), Pol II and acquisition of histone modifications. Thus, they manifest relatively slower expression kinetics. The rapid IEGs are poised for rapid transcription within 2–5 minutes of activity due to the NELF-mediated Pol II stalling near their promoters. The presence of positive transcription elongation factor b (P-TEFb) promotes the entry of stalled Pol II into the productive elongation phase. Moreover, the stalling of the pioneer Pol II facilitates the recruitment of additional Pol II to these promoters pre-loaded with GTFs and active histone marks. Newly recruited Pol II can then re-initiate successive rounds of transcription resulting in a quick burst of the gene product. The exact signaling pathway leading up to Pol II productive elongation remains unknown. Also note that, upon transcription initiation, transient polymerase stalling can occur in all genes, including delayed IEGs. However, rapid IEGs are ‘armed’ to prolong this natural pause and utilize it to respond rapidly.
Another correlative speculation can be derived from the concept of the ‘critical window’ of transcription necessary for consolidation of synaptic plasticity (Montarolo et al., 1986, Nguyen et al., 1994, Frey et al., 1996). These studies have shown that transcription inhibitors are effective in blocking long term synaptic changes only if administered before or during stimulation but importantly, they are ineffective if applied even shortly after the trigger. Presumably then, transcriptional events in that brief window of time, which are now known to depend on a poised polymerase mechanism, could be necessary for such long-term alterations in synaptic strength.
Conclusion
The reliable and rapid timing of IEG expression is utilized by several contemporary techniques. Imaging techniques such as ‘cellular compartment analysis of temporal activity by fluorescent in situ hybridization’ (catFISH) uses probes for IEG pre-mRNA for tracking neuronal activity in time (Guzowski and Worley, 2001). Recently though, IEG transcription has also been used as the basis of a technology for reactivating memory traces across specific neuronal circuits. By knocking in cfos promoter-driven artificial stimulation systems, such as channelrhodopsin or Designer Receptors Exclusively Activated by Designer Drugs (DREADDS), ensembles of neurons that had been activated naturally by an animal’s behavior can be made to express these genetically encoded receptors, which are then used to reactivate the same neuronal ensemble artificially (Garner et al., 2012, Liu et al., 2012). Thus, the distinct behavioral roles of circuits activated in ways that induce rapid and delayed IEGs may be probed in vivo by designing similar studies comparing artificial stimulation systems driven by the cfos promoter (a rapid IEG) with those driven by a delayed IEG (cox2, for example).
The multiple mechanisms of IEG transcription described in this review present us with a new opportunity to dissect the functional roles of the different classes of IEGs. With now an appreciation for the very earliest of the dynamics, we can expect to gain a better understanding of the parameters necessary for any relationship between IEG expression and their physiological tasks on hand. For example, regionally selective reduction of rapid IEG expression by local NELF knock out could reveal the role of these IEGs in a particular brain region. The actual function of the rapid response in the wider context of cognition, therefore, will require further study.
Highlights.
Immediate Early Genes (IEGs) can be induced with either rapid or delayed responses.
RNA Pol II is poised in proximity of the promoters of arc and other rapid IEGs.
Rapid induction of arc and other rapid IEGs is dependent on poised Pol II.
Delayed IEGs lack poised Pol II in most cases and are NELF knockdown insensitive.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JP, Dudek SM. Late-phase long-term potentiation: getting to the nucleus. Nat Rev Neurosci. 2005;6:737–743. doi: 10.1038/nrn1749. [DOI] [PubMed] [Google Scholar]
- Adams JP, Robinson RA, Hudgins ED, Wissink EM, Dudek SM. NMDA receptor-independent control of transcription factors and gene expression. Neuroreport. 2009;20:1429–1433. doi: 10.1097/WNR.0b013e3283311db6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon JM, Barco A, Kandel ER. Capture of the late phase of long-term potentiation within and across the apical and basilar dendritic compartments of CA1 pyramidal neurons: synaptic tagging is compartment restricted. J Neurosci. 2006;26:256–264. doi: 10.1523/JNEUROSCI.3196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer TK, Lee HL, Cordingley MG, Mymryk JS, Fragoso G, Berard DS, Hager GL. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol Endocrinol. 1994;8:568–576. doi: 10.1210/mend.8.5.8058066. [DOI] [PubMed] [Google Scholar]
- Arguello AA, Ye X, Bozdagi O, Pollonini G, Tronel S, Bambah-Mukku D, Huntley GW, Platano D, Alberini CM. CCAAT Enhancer Binding Protein delta Plays an Essential Role in Memory Consolidation and Reconsolidation. J Neurosci. 2013;33:3646–3658. doi: 10.1523/JNEUROSCI.1635-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Uzgil B, Lin P, Avliyakulov NK, O’Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran BH, Reffel AC, Stiles CD. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983;33:939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- Curran T, Bravo R, Muller R. Transient induction of c-fos and c-myc in an immediate consequence of growth factor stimulation. Cancer Surv. 1985;4:655–681. [PubMed] [Google Scholar]
- Curran T, Morgan JI. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229:1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- Dash PK, Hochner B, Kandel ER. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- Dragunow M. A role for immediate-early transcription factors in learning and memory. Behav Genet. 1996;26:293–299. doi: 10.1007/BF02359385. [DOI] [PubMed] [Google Scholar]
- Dragunow M, Robertson HA. Generalized seizures induce c-fos protein(s) in mammalian neurons. Neurosci Lett. 1987a;82:157–161. doi: 10.1016/0304-3940(87)90121-2. [DOI] [PubMed] [Google Scholar]
- Dragunow M, Robertson HA. Kindling stimulation induces c-fos protein(s) in granule cells of the rat dentate gyrus. Nature. 1987b;329:441–442. doi: 10.1038/329441a0. [DOI] [PubMed] [Google Scholar]
- Dudek SM, Fields RD. Somatic action potentials are sufficient for late-phase LTP-related cell signaling. Proc Natl Acad Sci U S A. 2002;99:3962–3967. doi: 10.1073/pnas.062510599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshete F, Fields RD. Spike frequency decoding and autonomous activation of Ca2+-calmodulin-dependent protein kinase II in dorsal root ganglion neurons. J Neurosci. 2001;21:6694–6705. doi: 10.1523/JNEUROSCI.21-17-06694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490 ( Pt 3):703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, Mayford M. Generation of a synthetic memory trace. Science. 2012;335:1513–1516. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Greene LA, Ziff EB. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J Biol Chem. 1985;260:14101–14110. [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Worley PF. Cellular compartment analysis of temporal activity by fluorescence in situ hybridization (catFISH) Curr Protoc Neurosci. 2001;Chapter 1(Unit 1):8. doi: 10.1002/0471142301.ns0108s15. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Arnold FJ, Bading H. A calcium microdomain near NMDA receptors: on switch for ERK-dependent synapse-to-nucleus communication. Nat Neurosci. 2001;4:565–566. doi: 10.1038/88380. [DOI] [PubMed] [Google Scholar]
- Jayaraman R. Transcription of bacteriophage T4 DNA by Escherichia coli RNA polymerase in vitro: identification of some immediate-early and delayed-early genes. J Mol Biol. 1972;70:253–263. doi: 10.1016/0022-2836(72)90537-2. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- Leslie JH, Nedivi E. Activity-regulated genes as mediators of neural circuit plasticity. Prog Neurobiol. 2011;94:223–237. doi: 10.1016/j.pneurobio.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gilmour DS. Promoter proximal pausing and the control of gene expression. Curr Opin Genet Dev. 2011;21:231–235. doi: 10.1016/j.gde.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Kandel ER. Cell adhesion molecules, CREB, and the formation of new synaptic connections. Neuron. 1996;17:567–570. doi: 10.1016/s0896-6273(00)80188-9. [DOI] [PubMed] [Google Scholar]
- Martin KC, Michael D, Rose JC, Barad M, Casadio A, Zhu H, Kandel ER. MAP kinase translocates into the nucleus of the presynaptic cell and is required for long-term facilitation in Aplysia. Neuron. 1997;18:899–912. doi: 10.1016/s0896-6273(00)80330-x. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Montarolo PG, Goelet P, Castellucci VF, Morgan J, Kandel ER, Schacher S. A critical period for macromolecular synthesis in long-term heterosynaptic facilitation in Aplysia. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Role of ion flux in the control of c-fos expression. Nature. 1986;322:552–555. doi: 10.1038/322552a0. [DOI] [PubMed] [Google Scholar]
- Mulcahy LS, Smith MR, Stacey DW. Requirement for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells. Nature. 1985;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, Endoh M, Yamada T, Handa H. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- Pintchovski SA, Peebles CL, Kim HJ, Verdin E, Finkbeiner S. The serum response factor and a putative novel transcription factor regulate expression of the immediate-early gene Arc/Arg3.1 in neurons. J Neurosci. 2009;29:1525–1537. doi: 10.1523/JNEUROSCI.5575-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361:453–457. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Raymond CR. LTP forms 1, 2 and 3: different mechanisms for the “long” in long-term potentiation. Trends Neurosci. 2007;30:167–175. doi: 10.1016/j.tins.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Ringold GM, Shank PR, Varmus HE, Ring J, Yamamoto KR. Integration and transcription of mouse mammary tumor virus DNA in rat hepatoma cells. Proc Natl Acad Sci U S A. 1979;76:665–669. doi: 10.1073/pnas.76.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringold GM, Yamamoto KR, Bishop JM, Varmus HE. Glucocorticoid-stimulated accumulation of mouse mammary tumor virus RNA: increased rate of synthesis of viral RNA. Proc Natl Acad Sci U S A. 1977;74:2879–2883. doi: 10.1073/pnas.74.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffen DW, Cole AJ, Worley PF, Christy BA, Ryder K, Baraban JM. Convulsant-induced increase in transcription factor messenger RNAs in rat brain. Proc Natl Acad Sci U S A. 1988;85:7795–7799. doi: 10.1073/pnas.85.20.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Dudek SM. Action potentials: to the nucleus and beyond. Exp Biol Med (Maywood) 2008;233:385–393. doi: 10.3181/0709-MR-241. [DOI] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger P, Gerosa D, Oertner TG. Temporal control of immediate early gene induction by light. PLoS One. 2009;4:e8185. doi: 10.1371/journal.pone.0008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H, Tsuchihashi Z, Mizumoto K. The RNA polymerase II preinitiation complex formed in the presence of ATP. Nucleic Acids Res. 1997;25:4079–4084. doi: 10.1093/nar/25.20.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DG, Barrett CF, Groth RD, Safa P, Tsien RW. CaMKII locally encodes L-type channel activity to signal to nuclear CREB in excitation-transcription coupling. J Cell Biol. 2008;183:849–863. doi: 10.1083/jcb.200805048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley PF, Cole AJ, Murphy TH, Christy BA, Nakabeppu Y, Baraban JM. Synaptic regulation of immediate-early genes in brain. Cold Spring Harb Symp Quant Biol. 1990;55:213–223. doi: 10.1101/sqb.1990.055.01.023. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. Expression of a mitogen-inducible cyclooxygenase in brain neurons: regulation by synaptic activity and glucocorticoids. Neuron. 1993;11:371–386. doi: 10.1016/0896-6273(93)90192-t. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5:e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]