Abstract
Glial cell line-derived neurotrophic factor (GDNF) binds a coreceptor GDNF family receptor α1 (GFRα1) and forms a signaling complex with the receptor tyrosine kinase RET. GDNF-GFRα1-RET signaling activates cellular pathways that are required for normal induction of the ureteric bud (UB) from the Wolffian duct (WD). Failure of UB formation results in bilateral renal agenesis and perinatal lethality. Gfrα1 is expressed in both the epithelial and mesenchymal compartments of the developing kidney while Ret expression is specific to the epithelium. The biological importance of Gfrα1's wider tissue expression and its role in later kidney development are unclear. We discovered that conditional loss of Gfrα1 in the WD epithelium prior to UB branching is sufficient to cause renal agenesis. This finding indicates that Gfrα1 expressed in the nonepithelial structures cannot compensate for this loss. To determine Gfrα1's role in branching morphogenesis after UB induction we used an inducible Gfrα1-specific Cre-deletor strain and deleted Gfrα1 from the majority of UB tip cells post UB induction in vivo and in explant kidney cultures. We report that Gfrα1 excision from the epithelia compartment after UB induction caused a modest reduction in branching morphogenesis. The loss of Gfrα1 from UB-tip cells resulted in reduced cell proliferation and decreased activated ERK (pERK). Further, cells without Gfrα1 expression are able to populate the branching UB tips. These findings delineate previously unclear biological roles of Gfrα1 in the urinary tract and demonstrate its cell-type and stage-specific requirements in kidney development.
Keywords: Gfrα1, Ret, kidney development, branching morphogenesis
1. Introduction
The development of the metanephric kidney, the permanent mammalian kidney, requires ureteric budding, reciprocal interactions between the ureteric bud (UB) epithelium and the metanephric mesenchyme (MM), branching morphogenesis and nephrogenesis. Defects in these processes can result in hypoplasia with low nephron endowment which is a risk factor for the development of hypertension and chronic kidney disease in adulthood (Abitbol and Ingelfinger, 2009). Global gene knockout studies have helped identify a number of genes that are important in kidney development (www.gudmap.org)(Costantini and Kopan, 2010; Dressler, 2009; Little et al., 2010). Further investigation of these developmental genes in a cell and time-specific manner are necessary to gain insights into how these genes and downstream pathways can be manipulated to alter the course of kidney diseases resulting from their deficiencies.
Glial cell line-derived neurotrophic factor (GDNF) coreceptor α1 (GFRα1) is a major protein that is essential for early UB induction and kidney development as global Gfrα1-knockout mice die at birth due to renal agenesis (Cacalano et al., 1998; Enomoto et al., 1998). This has hampered further studies of the role of Gfrα1 in later renal development. GFRα1 is the high affinity co-receptor for GDNF. GFRα1 is anchored to the cell surface by glycosylphophatidyl inositol (GPI) linkage. It localizes to lipid rafts within the cell membrane and the binding of GDNF recruits the receptor tyrosine kinase rearranged during transfection (RET). Among the four GFRα coreceptors (GFRα1-4) and four GDNF family ligands (GFLs, GDNF, Neurteurin, Atremin and Persephin), the receptor complex consisting of dimers of GFRα1-GDNF with RET is the physiologically relevant signaling unit during kidney and enteric nervous system development (Saarma, 2000). This three protein complex of GDNF, GFRα1, and RET activates the phosphorylation of specific tyrosine residues in the intracellular domain of RET. This results in activation of downstream signaling pathways such as PLCγ, PI3K, and/or MAPK that have been shown to be necessary for early embryonic kidney development (Jain, 2009; Jain et al., 2006a).
Gfrα1 mRNA is expressed in both the ureteric epithelium and the MM during branching morphogenesis (Enomoto et al., 2004; Golden et al., 1999; Towers et al., 1998). Thus, it is more widely expressed than either GDNF or Ret during kidney development. The biological significance of this wider and persistent expression after UB induction is not clear. A number of studies in vitro suggest that GFRα1/GDNF/RET tyrosine kinase signaling has a role in branching morphogenesis, a process that occurs after UB induction - reviewed in (Costantini and Shakya, 2006). However, evidence in vivo for the role of this pathway in later metanephric development is lacking. Another interesting aspect of Gfrα1 is that it can act in trans in neurological tissues (Ledda et al., 2002; Paratcha et al., 2001). The physiological role of trans-signaling by Gfrα1 in kidney development is not known.
Here we generated unique Gfrα1 reporter and Cre-deletor strains to conditionally examine Gfrα1's role in the epithelia cell in pre and post UB kidney development. We found that Gfrα1 was expressed in the mesonephric mesenchyme and Wolffian duct (WD) before UB induction and in the MM and UB epithelium during branching morphogenesis. Expression of Gfrα1 in the urinary epithelium is critical for UB induction and has a modest role after UB induction during early branching morphogenesis.
2. Results
2.1. Gfrα1 is expressed early in kidney development and throughout branching morphogenesis
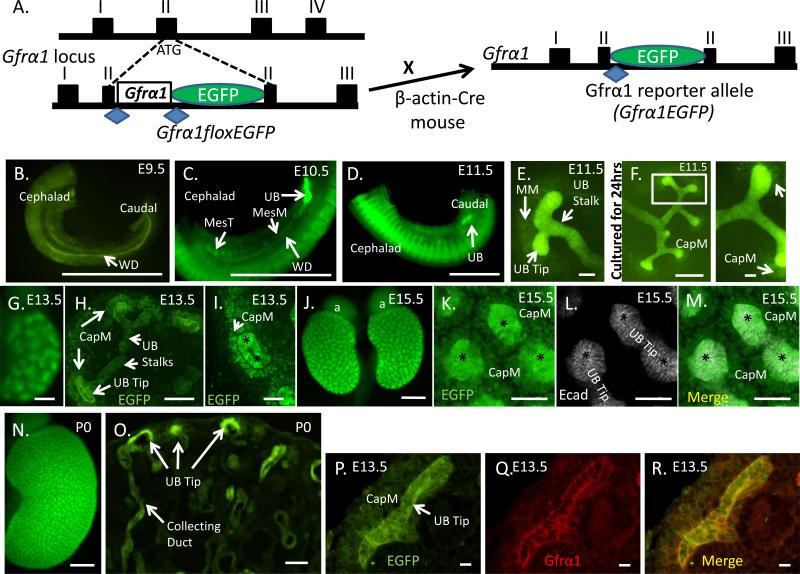
To obtain a clear understanding of Gfrα1 expression in the urinary tract during development we generated a Gfrα1 reporter mouse that expresses the enhanced green fluorescent protein (EGFP) from the Gfrα1 locus (Gfrα1EGFP/+) (Uesaka et al., 2007) (Figure 1A). Robust EGFP signal, indicative of Gfrα1 expression, was identified throughout the developing WD at E9.5 (Figure 1B). At E10.5, Gfrα1 expression was identified in the WD with greater localization to the caudal UB budding domain, as well as in the mesonephric mesenchyme and the cranial mesonephric tubules (future epididymis in males) (Figure 1C). At E11.5, the T-shape stage of branching morphogenesis, Gfrα1-positive cells were present in the UB stalk and in the distal UB tips (Figure 1D and E). There was faint expression of Gfrα1 in the metanephric mesenchyme (MM) at E11.5 (Figure 1E) that becomes more prominent as the MM condenses around the UB tip (Figure 1F). At E13.5, consistent with previously reported mRNA expression of Gfrα1, we detected Gfrα1-positive cells in the “cap” mesenchyme in addition to the ureteric epithelium (Figure 1G-I) (Enomoto et al., 2004; Golden et al., 1999; Towers et al., 1998). At E15.5 strong EGFP expression was seen in the developing kidney by whole mount immunohistochemistry (Figure 1J). High resolution images (Figure 1K-M) confirm both UB tip and cap mesenchyme expression. Thereafter, Gfrα1 continues to be expressed in the developing kidney both in the ureteric stalk (collecting duct), bud tip and the surrounding cap mesenchyme (Figure 1N-O). We performed double label immunohistochemistry with anti-Gfrα1and anti-EGFP antibodies to confirm that EGFP expression recapitulated Gfrα1 expression (Figure 1P-R). These observations support that Gfrα1 has a broader expression in the metanephric kidney than either Ret or Gdnf (Enomoto et al., 2004; Golden et al., 1999; Sainio et al., 1997; Towers et al., 1998) and demonstrate that Gfrα1 is expressed before UB induction in both the Wolffian Duct (WD) and the surrounding mesonephric mesenchyme.
Figure 1. Gfrα1 expression during embryonic kidney development.
(A) Schematic illustrating part of the Gfrα1 locus with conditional allele (Gfrα1floxEGFP) knocked-in exon II. Cre-mediated recombination excises Gfrα1 cDNA flanked by loxP sites (blue diamonds) and expresses EGFP to generate the Gfrα1EGFP reporter allele. (B) Direct visualization of EGFP signal which represents Gfrα1 expression at the indicated time points throughout kidney development. At E9.5, Gfrα1 is expressed in the entire Wolffian duct (WD). (C) E10.5, Gfrα1 expression remains in the WD but shows strong expression in the site of the future ureteric bud (UB). It is also present in the mesonephric mesenchyme (MesM) and the mesonephric tubules (MesT). (D, low power; E, high power) At E11.5 strong Gfrα1 expression is seen in the induced UB and in the stalk. Gfrα1 is also expressed, although at much lower levels, in the metanephric mesenchyme (MM). (F) Metanephric kidney harvested at E11.5 and cultured for 24 hours of continues to show expression in the ureteric epithelium with strong expression in the UB tips and weaker in the cap mesenchyme (CapM) condensing around the UB tips. Boxed region is displayed at higher magnification (right). (G-I) Gfrα1 is expressed in E13.5 metanephroi at strong levels in the UB tips, and weakly in the stalk and cap mesenchyme. Wholemount direct immunofluorescence image is shown in G, anti-GFP immunofluorescence on paraffin embedded sections in H (low power), and I (high power of H) show UB tip, stalk and CapM expression. The expression in the mesenchyme (arrow) is directly adjacent to and surrounding the distal UB tip (asterisk). (J-M) Gfrα1 expression in E15.5 whole mount kidneys. J, whole mount, and K-M are high power images of EGFP, anti-Ecadherin (Ecad, white, epithelium) and merged immunostained images of UB tip (asterisks) demonstrating Gfrα1 expression (green) in UB tip and cap mesenchyme (CapM). (N,O) At postnatal day 0 (P0), Gfrα1 continues to be expressed in the kidney with high expression in the UB tips. (P-R) Double EGFP-Gfrα1 immunolabeling on paraffin sections at E13.5 confirm that EGFP reporter recapitulates Gfrα1 expression in the UB tip and CapM. Scale bar: B-D, 500 μm; E, 50 μm; F, 250 μm, high power UB tip at 50 μm; G, 200 μm; H, 100 μm; I, 50 μm; J, 500 μm; K-M, 50 μm; N, 500 μm; O, 50 μm; P-R, 10 μm. Native fluorescence was used to visualize Gfrα1 expression in panels B-G, J, N and O. Anti-GFP immunostaining was used in H, I, K and P to visualize Gfrα1 expression.
2.2. Gfrα1 expression in the collecting system primordia is indispensable for metanephric kidney formation
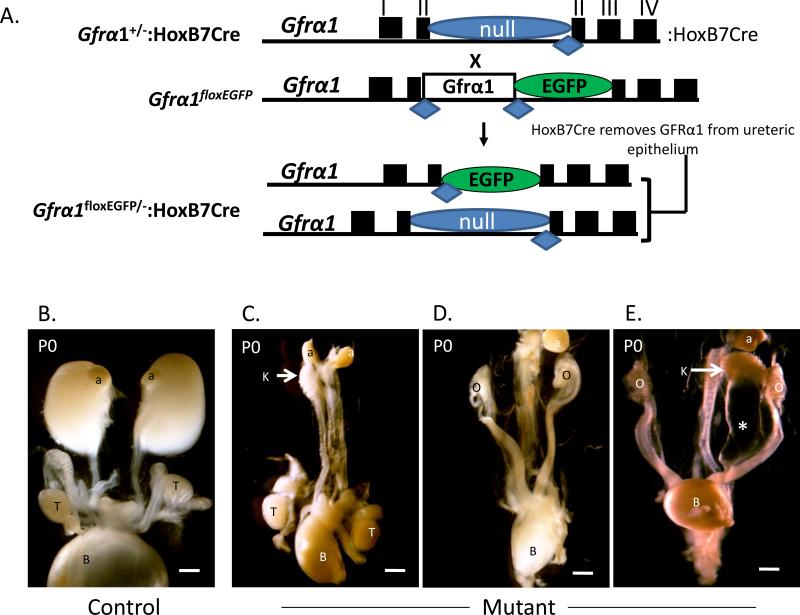
Previous studies have demonstrated that forced expression of Gfrα1 from the Ret locus while eliminating it from non-Ret expressing sites is sufficient for normal kidney development (Enomoto et al., 2004). This suggested that trans-Gfrα1 expression is dispensable if Gfrα1 is expressed in a cell-autonomous manner. However, non-epithelial expression of Gfrα1 in other organ systems has shown that it can act in a paracrine manner, so called “trans-signaling,” to activate Ret signal transduction (Ledda et al., 2002; Paratcha et al., 2001). Therefore, we sought to determine if Gfrα1 from sites other than the urinary epithelium are able to compensate and support normal kidney development. We used the Cre-deletor strain Hoxb7-Cre (Yu et al., 2002) to achieve WD-specific deletion of Gfrα1 in mice harboring a Gfrα1-conditional reporter allele Gfrα1floxEGFP (Uesaka et al., 2007) (Figure 2A). All Gfrα1floxEGFP/-:Hoxb7Cre pups, which have Gfrα1 deleted in the urinary tract epithelium, had kidney defects. The majority had bilateral renal agenesis/aplasia (16/20) and three had unilateral agenesis and contralateral megaureter (Figure 2B and Table 1). These findings reveal unequivocally that Gfrα1 in the WD/urinary epithelium is indispensable and is the mechanism for renal agenesis in global Gfrα1-null mice. Further, trans-Gfrα1 in early stages of kidney development cannot compensate for its loss in the WD.
Figure 2. Gfrα1 expression in the ureteric epithelium is necessary for metanephric kidney development.
(A) Schematic illustrates strategy to specifically delete Gfrα1 in the ureteric epithelium. Exons and loxP sites denoted by rectangles and diamonds, respectively. Mice with only one copy of Gfrα1 (Gfrα1+/-) and the UB epithelium specific Cre (Hoxb7Cre) recombinase were bred with conditional Gfrα1 mice (Gfrα1floxEGFP/floxEGFP). The null allele (“-“) expresses tau-lacZ reporter (see methods). The resulting progeny have either no Gfrα1 in the UB epithelium (Gfrα1floxEGFP/-:Hoxb7Cre, mutant) or retain at least one copy of Gfrα1 (Gfrα1floxEGFP/+:Hoxb7Cre, control). (B) Representative gross images show a littermate control on the left and thereafter a spectrum of renal defects in mutant male and female newborn mice with Gfrα1 deletion in the Hoxb7 domain. In the mutant panel the image on the left shows severe renal hypo/dysplasia and left agenesis in a male. Middle image shows bilateral agenesis in a female. The image on the right shows megaureter (*) and right renal agenesis in a female mouse. Scale bar (1 mm). a- adrenal; b- urinary bladder; k- kidney; o- ovary; t- testis.
Table 1.
Phenotype of Gfrα1floxEGFP/-:Hoxb7Cre versus littermate Controls
| N | Bilateral Agenesis or Severe Hypo/Dysplasia | Bilateral Hypoplasia | Unilateral Agenesis and Contralateral Hydroureter | Normal | |
|---|---|---|---|---|---|
| Gfrα 1floxEGFP/-:Hoxb7Cre | 20 | 16 | 1 | 3 | 0 |
| Gfrα 1floxEGFP/-; Gfrα 1floxEGFP/+:Hoxb7Cre; Gfrα 1floxEGFP/+ | 55 | 0 | 0 | 0 | 55** |
The genotypes and phenotypes of 7 separate litters.
Of the 55 heterozygous one pup had right mild hydronephrosis and the other had right bifurcated ureter.
2.3. Conditional deletion of Gfrα1 after initial UB budding using a tamoxifen inducible Cre strain
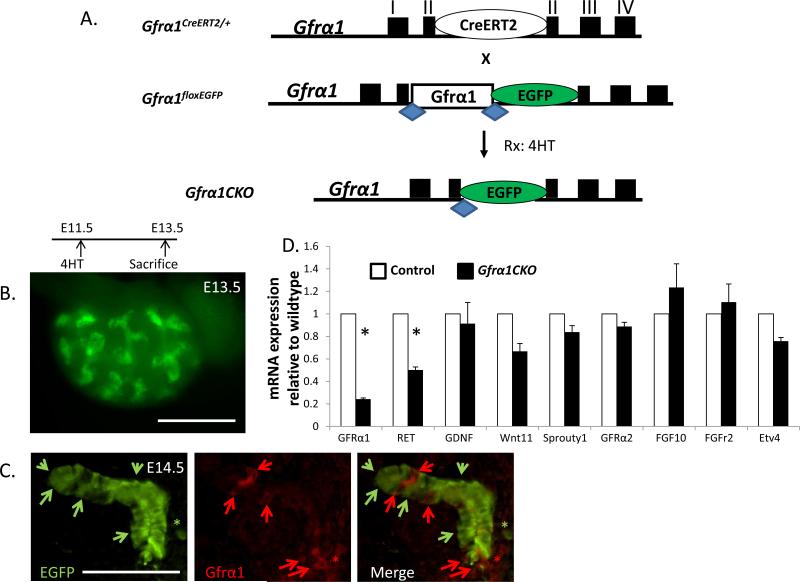
After determining that Gfrα1 is required in the WD during early kidney development and that it is expressed throughout metanephric development we next established an inducible system to delete Gfrα1 after UB induction. To accomplish this, we generated a mouse strain that expresses the tamoxifen inducible Cre recombinase (CreERT2) from the Gfrα1 locus (Gfrα1-CreERT2). The heterozygous Gfrα1CreERT2/+ mice were viable and fertile. The advantage of this mouse line is that a single cross with Gfrα1 conditional mice yields desired progeny expressing only the Gfrα1 conditional allele that can be excised with tamoxifen inducible Cre (Gfrα1floxEGFP/CreERT2) and this will be referred to as Gfrα1CKO (Figure 3A). Furthermore, excision of Gfrα1 is indicated by the EGFP signal which facilitates interpretation of the resulting phenotypes due to Gfrα1 loss (Uesaka et al., 2007). We performed a series of experiments to determine the efficiency of Gfrα1 excision in vivo in the kidney after 4-hydroxytamoxifen (4HT) exposure (see methods). Whole mount live images of Gfrα1CKO embryos 2 days after 4HT injection showed strong EGFP signal indicating successful Gfrα1 excision (Figure 3B). Using combination of E-cadherin (epithelial marker), Gfrα1 and GFP immunostaining on metanephric kidney sections, we were able to determine complete elimination of Gfrα1 in more than 65% of UB tip cells after UB induction (Figure 3C) although the excisional efficiency in the MM was much lower due to weak expression. With a similar level of inducible deletion (70%) in the gut we previously discovered that Gfrα1 is required for survival of enteric neurons as its conditional deletion resulted in complete aganglionosis (Uesaka et al., 2007). We performed quantitative RTPCR analysis and found that total Gfrα1 mRNA was reduced by 76% in E14.5 kidneys after conditional deletion compared to the wild-type kidneys (Figure 3D). The residual Gfrα1 mRNA represents expression of the unexcised Gfrα1 in the UB tip and MM cells.
Figure 3. Generation and characterization of mice with inducible Gfrα1 deletion in vivo after ureteric budding.
(A) Breeding strategy to delete Gfrα1 after ureteric bud (UB) induction using tamoxifen inducible Cre-mediated excision. Mice with the CreERT2 cDNA knocked into the Gfrα1 locus (Gfrα1CreERT2/+) were mated with the Gfrα1 conditional mice (Gfrα1floxEGFP/floxEGFP). Treatment of the embryos with the genotype Gfrα1floxEGFP/CreERT2 (Gfrα1CKO) with 4-hydroxytamoxifen (4HT) excises Gfrα1 resulting in EGFP expression. (B) Gross image shows live EGFP signal in E13.5 kidney dissected 2 days after 4HT treatment illustrating successful Gfrα1 deletion. (C) Double immunofluorescence of E14.5 ureteric buds with anti-GFP (green) and anti-Gfrα1 (red) antibodies illustrate Gfrα1 excision in the majority of UB tip epithelial cells (green arrows). Cells with no excision stain with Gfrα1 (red arrows). Asterisks denote metanephric mesenchyme. (D) mRNA expression of selected genes using quantitative real time PCR on whole kidney lysates of Gfrα1CKO relative to control mice at E14.5. Control n=6 (12 kidneys); Gfrα1CKO n=7 (14 kidneys). Note marked reduction in Gfrα1 expression (mean±s.e.m., *P<0.001, t-test) in Gfrα1CKO kidneys. Scale bar 500 μm in B; 50 μm in C.
2.4. Gfrα1 conditional loss does not alter mRNA levels of several of the genes in the Ret pathway
We next examined if reduced Gfrα1 post UB induction affected expression of positive and negative regulators of Ret signaling and branching morphogenesis: Ret, Gdnf, Wnt11, Sprouty1, Gfrα2, Fgf10, Fgf2r, and Etv4 (Costantini and Kopan, 2010; Majumdar et al., 2003; Pepicelli et al., 1997). Although Ret mRNA expression decreased by about 50%, Gdnf, Wnt11, and Etv4 mRNAs were not significantly affected at E14.5 (Figure 3D). Studies in vitro have shown that Gfrα2 can also bind to Gdnf and activate Ret (Sariola and Saarma, 2003). However, physiological levels of Gfrα2 are not able to compensate for Gfrα1 loss in the urinary tract as Gfrα1 nulls have renal agenesis. We investigated if there were compensatory changes in Gfrα2 expression in the Gfrα1CKO mice that could explain near normal expression of Ret associated downstream genes in Gfrα1CKO mice. There was no difference in Gfrα2 mRNA expression between Gfrα1CKO and wild-type kidneys. Furthermore, there was no significant difference in the expression levels of mRNAs for receptor tyrosine kinase Fgfr2, or its ligand Fgf10 or in the negative tyrosine kinase regulator Sprouty1.
2.5. Gfrα1 loss after UB induction results in a modest decrease in kidney size
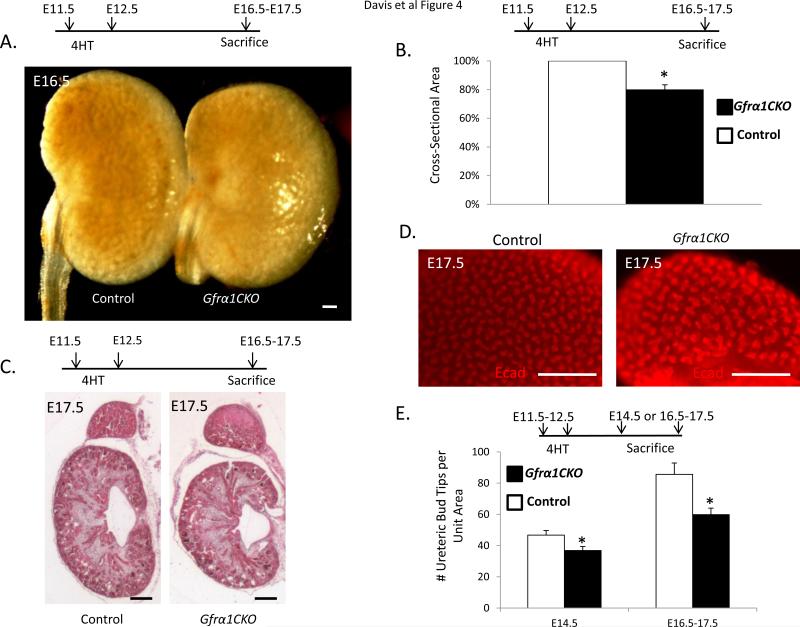
To determine the role of Gfrα1 in the ureteric epithelium after UB induction we treated pregnant mice with 4HT at E11.5-12.5 and analyzed kidneys of the Gfrα1CKO and control littermate embryos at E14.5 or at E16.5-E17.5 (Figure 4A-E). The loss of Gfrα1 resulted in a 20% reduction in kidney size compared to littermate controls at E16.5-17.5 (Figure 4B). Histological assessment of Gfrα1CKO kidneys showed well-defined cortex and medulla (Figure 4C). The nephrogenic zone did not exhibit any overt defects. To further confirm if reduced branching is the cause of hypoplasia we performed whole mount immunohistochemistry using the epithelial marker E-cadherin at different time points (Figure 4D and Supplemental Figure 1). Quantitative analysis of the UB tips at E14.5 and E16.5-17.5 showed a significant, although modest, reduction in the number of UB tips (Figure 4E). Interestingly, a consistent finding in these kidneys were dilated UB ampullae (Figure 4D). This phenotype has also been reported in kidneys that lack Gdnf and Sprouty1 or Ret and Sprouty1 (Michos et al., 2010) suggesting that Gdnf-Gfrα1-Ret signaling after UB induction may be important in maintaining normal ampulla morphology or it may represent a delay in development as the ampulla size decreases with age (Supplemental figure 1).
Figure 4. Conditional loss of Gfrα1 during branching morphogenesis causes renal hypoplasia.
Time course of 4HT induction and sacrifice is indicated above each panel. (A) Gross image of Gfrα1CKO at E16.5, 4 days after 4HT treatment of pregnant mothers shows smaller Gfrα1CKO kidneys versus littermate control (Gfrα1floxEGFP/+). (B) Bar graph shows cross-sectional area of the Gfrα1CKO metanephros are less than that of littermate controls (6 separate litters, control n=11, Gfrα1CKO n=11; mean±s.e.m., *P<0.001 by t-test). (C) E17.5 Hematoxylin and Eosin stained sections depict preserved kidney architecture with well-formed cortex and medulla in Gfrα1CKO mice. (D) Whole mount E-cadherin staining highlights the ureteric branching pattern in control and Gfrα1CKO mice. Note decreased branching in Gfrα1CKO and an apparent increase in ampullae dilation. (E) Bar graph illustrates reduced number of UB tips in Gfrα1CKO kidneys compared to controls (mean±s.e.m.; E14.5 Control n=4, Gfrα1CKO n=4; *P<0.03. E16.5-17.5 Control n=11, Gfrα1CKO n=9; *P<0.008). Scale bar 200 μm for all images.
2.6. Conditional Gfrα1 loss post UB-induction affects UB tip proliferation and ERK activity
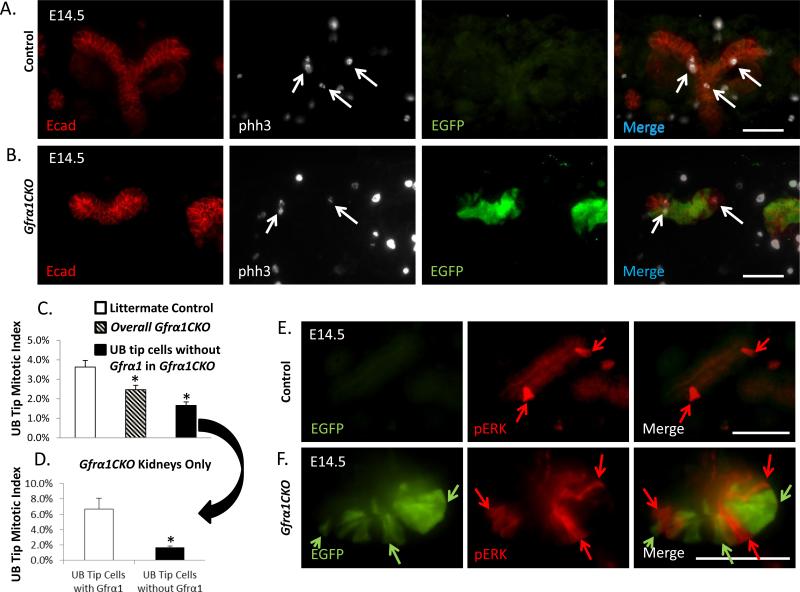
To determine if altered proliferation is the cause of reduced branching and hypoplasia we performed anti-phosphohistone H3 immunohistochemistry on E14.5 kidney sections (Figure 5A-B). The overall UB tip mitotic index was significantly higher (3.6%±0.3%) in the control animals compared with Gfrα1CKO mice (2.5%±0.2%) (Figure 5C). This observation suggests that the reduction in kidney size is due to reduced UB tip proliferation in Gfrα1CKO mice. To directly assess the impact of Gfrα1 loss on cell proliferation we determined the mitotic index in UB tip cells that had no Gfrα1 (1.7% ±0.2%) and cells in which Gfrα1 remained (6.3%±1.4%) in only Gfrα1CKO mice (Figure 5D). This result supports that UB tip proliferation is only partly dependent on Gfrα1 and the cells in which Gfrα1 is not completely excised are able to compensate with increased proliferation. This compensation is consistent with the observation that we find less number of recombinant cells in the nephrogenic zone at E16.5 (Supplemental figure 2). Analysis of cell death by TUNEL staining did not reveal any significant difference between the control and Gfrα1CKO groups (data not shown). We also examined phosphorylated-ERK (pERK) activity in Gfrα1CKO mice because MAPK is one of the key pathways activated by Gdnf-Gfrα1-Ret and it regulates cell proliferation during UB development (Hoshi et al., 2012). In agreement with previous observations, the percentage of pERK-positive cells was low in the branching UB tips at E14.5 in wildtype mice (Hu et al., 2011). This is in contrast to a high percentage of pERK-positive cells in Wolffian duct epithelium at E10.5 (Hoshi et al., 2012) (Supplemental figure 3A-B). We did not find a significant difference in the number of pERK-positive UB tip cells between controls (17.7%±2.7%) and Gfrα1CKO (15.4%±1.9%) mice (Supplemental Figure 3C-E). Direct EGFP visualization in conjunction with pERK immunostaining showed that pERK was absent virtually in all the cells in which Gfrα1 was conditionally deleted compared to controls (Figure 5E, F; Supplemental figure 3D). We quantified the number of pERK-positive cells in Gfrα1CKO UB tips which have lost Gfrα1 (green) (1.5%±0.5%) versus adjacent cells in which Gfrα1 remained (37.1%±3.6%) (Supplemental Figure 3C). These findings suggest that Gfrα1 is a major regulator of ERK activity in UB tip cells.
Figure 5. Loss of Gfrα1 post UB induction results in reduced UB tip proliferation and decreased phospho-ERK activity.
(A-B) Representative immunohistochemistry images of UB tips on paraffin sections of control (Gfrα1floxEGFP/+) and mutant (Gfrα1CKO) mice for E-cadherin (Ecad, epithelial marker), phosphohistone-H3 (phh3, proliferation marker), and green fluorescent protein (EGFP, represents Gfrα1-negative cells). Arrows indicate phh3 staining in epithelial cells. (C) Graph illustrates decreased overall UB tip proliferation in Gfrα1CKO kidneys and in cells with successful Gfrα1 excision (black bar). Proliferation as evaluated by quantification of mitotic index from phh3 immunolabeled UB tips from E14.5 metanephric kidneys (2 days after tamoxifen administration) in littermate control and Gfrα1CKO mice. (D) Cells in the UB tips of Gfrα1CKO mice that have escaped Gfrα1 excision exhibit higher proliferation rate compared to cells that have lost Gfrα1 (Control n=7, Gfrα1CKO n=11; mean±s.e.m.,*P<0.01 by t-test). (E-F) Double-labeled Immunofluorescence images of UB tip cells on frozen section at E14.5 with anti-GFP denoting Gfrα1 deletion (green, arrows) and antiphospho-ERK (red, arrows) in control (E) and 4HT-treated Gfrα1CKO (F) mice. Merged image of mutant (F) on far right, illustrates limited overlap between green and red cells supporting the lack of p-ERK activity in EGFP (Gfrα1 excised) positive cells. Scale bar 50 μm in all images.
2.7. Explant cultures of metanephric kidneys with near complete Gfrα1 loss show reduction in branching morphogenesis
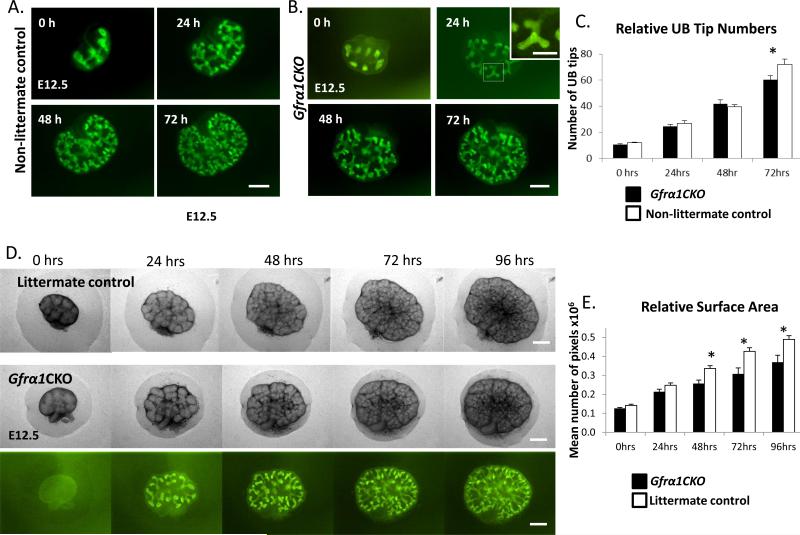
Although the experiments above, with Gfrα1 excision in vivo show a modest role in early branching and proliferation and the potential compensation by the cells with residual Gfrα1, the mild phenotype could be a result of incomplete excision. The excision in vivo was even less efficient (about 50%) at later developmental time points (E13.5-15.5 or E15.5-E17.5) and we did not see any effect on kidney size or number of glomeruli on postnatal day of life 1 (Supplemental Figure 4A-D). More efficient Gfrα1 excision was limited due to embryonic toxicity from doses of 4HT greater than that used above. Therefore, to further analyze the ability of the kidney to undergo branching morphogenesis in the absence of Gfrα1 we established a kidney explant culture system with 4HT in the culture media. This provided excisional efficiency that was greater than 95% in the UB tip cells (Figure 6 and Figure 7B). Additionally, the culture system allowed direct visualization of branching over time. Surprisingly even with near complete excision of Gfrα1 from the ureteric epithelium the branching pattern appeared normal and Gfrα1-null cells were able to colonize the UB tips. To further quantify branching deficits in Gfrα1CKO cultures we compared UB tip numbers of Gfrα1CKO and control reporter mice that express fluorescence reporter (Gfrα1-EGFP or Hoxb7Cre - Rosa-YFP) to facilitate visualization of the branching tips during time-lapse culture (Figure 7A-B). At 72 hours of culture we observed a 16% decrease in the number of UB tips in Gfrα1CKO compared to controls (Figure 6C). To overcome a potential confound of non-littermate reporter controls used above we also compared explant growth between the Gfrα1CKO and control littermates by comparing their surface areas using phase contrast images at various time points during the culture. We used surface area for comparison as the littermates do not have a fluorescent reporter to facilitate quantification of UB tips. We observed a modest reduction (25%) in relative surface area beginning at 48 hours of culture (Figure 7C, D). Therefore, despite potent deletion of Gfrα1 in the UB epithelium there was only a modest effect on branching morphogenesis.
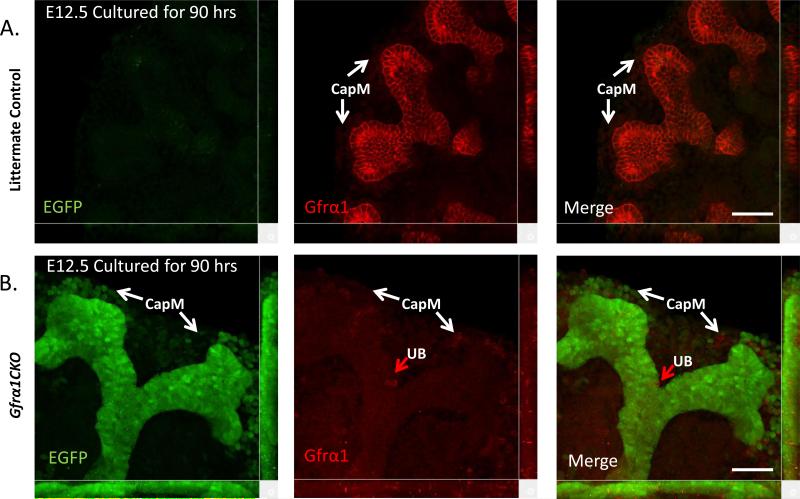
Figure 6. Robust excision of Gfrα1 in UB epithelium metanepheroi cultured ex-vivo.
Confocal microscopy images obtained from EGFP (green) and Gfrα1 (red) double immunofluorescence labeling of control (A) or Gfrα1CKO (B) kidneys cultured for 90 hours in presence of 4HT. Note robust green signal and only a few Gfrα1-labeled (red arrow) cells in Gfrα1CKO panels indicate highly efficient Gfrα1 excision in the UB tip. There is only partial excision of Gfrα1 in the cap mesenchyme (CapM). Scale bar 50 μm.
Figure 7. Metanephric kidney culture experiments show a modest decrease in branching in Gfrα1CKO mice.
(A-B) Fluorescent images of metanephroi from (A) control (Gfrα1EGFP/+) and (B) Gfrα1CKO embryos dissected at E12.5 and cultured in presence of 4HT at the indicated time points in hours (h). To ensure controls and Gfrα1CKO mice had similar number of UB tips at the beginning of the culture pregnant mothers were treated with 4HT the night before sacrifice to induce EGFP expression. Inset in B at 24h is higher magnification of the area depicted by white square and it shows presence of EGFP positive cells in almost the entire branching UB tip indicating near complete excision. Note Gfrα1CKO kidneys continue to branch and green cells colonize the tips. (C) Bar graph shows comparison between the number of UB tips in cultured metanephroi of Gfrα1CKO mice and controls at indicated time points. A mild reduction in branching is seen at 72 hours. Results are represented as mean±s.e.m (Control n=11, Gfrα1CKO n=9. *P<0.05 by t-test). (D) Phase images of littermate control (Gfrα1floxEGFP/+) metanephroi compared to phase and fluorescent images of Gfrα1CKO dissected at E12.5, cultured in the presence of 4HT. (E) Bar graph shows comparison between surface areas (mean number of pixels per metanephros ±s.e.m; *P<0.01 by t-test) of Gfrα1CKO (n= 7) and littermate controls (n=12) at indicated time points. Scale bar: 250 μm (inset scale bar 125 μm).
3. Discussion
Gfrα1 is an important protein for early development of many organ systems including initiation of the UB for normal metanephric kidney development in the mouse. Our studies address two critical aspects of Gfrα1 in the urinary system by utilizing novel and innovative Gfrα1 reporter, conditional, and tamoxifen inducible Cre mouse strains. We provide conclusive evidence that prior to UB budding Gfrα1 in non-ureteric epithelial structures such as the mesonephric mesenchyme cannot signal in trans and rescue UB budding. Therefore, Gfrα1 activity in the WD is critical for UB induction. Second, by eliminating Gfrα1 from the ureteric epithelium post UB induction we demonstrate that Gfrα1 loss results in mild metanephric hypoplasia. This suggests branching is only partially dependent on Gfrα1 signals. Our studies also provide evidence that the mechanism of reduced branching in Gfrα1CKO mice is due to reduction in UB proliferation. Finally we note that pERK activity in UB tip cells is dependent on Gfrα1 as cells without Gfrα1 had almost no pERK signal.
Among the Gfrα coreceptors, Gfrα1 is the most relevant for kidney development because its germline deletion causes renal agenesis and no kidney defects are reported in other Gfrα-null mice (Airaksinen and Saarma, 2002; Airaksinen et al., 1999; Nishino et al., 1999; Rossi et al., 1999). Based on this and studies in vitro showing severe effects on branching in kidneys cultured in the presence of Gdnf antibodies we expected that loss of Gfrα1 would cause drastic defects in branching (Towers et al., 1998; Vega et al., 1996). Therefore it was surprising that its loss in the majority of Gfrα1CKO UB tips in vivo and near complete loss in the UB tip in explants cultures showed only moderate hypoplasia. This is in contrast to a critical role of Gfrα1 in UB induction and suggests that branching morphogenesis may rely on factors other than Gfrα1. One possibility is continued activation of Ret by other Gfrαs such as Gfrα2 that is also reportedly expressed in the kidney (Golden et al., 1999). There is no evidence in vivo that supports compensation of Gfrα1 loss by Gfrα2 in the kidney or enteric nervous system. We did not see any overt changes in Gfrα2 expression in the Gfrα1CKO mice that would suggest Gfrα2 is compensating for Gfrα1 loss post UB induction. Definitive proof to rule out Gfrα2's compensatory role would require double Gfrα1CKO/Gfrα2-null mice. Another possibility is Ret independent roles of GFLs as GFLs can activate NCAM in the absence of RET in the nervous system (Paratcha et al., 2003). This may be unlikely as no other GFL or Gfrα coreceptor or NCAM knockout have a renal phenotype (Airaksinen and Saarma, 2002; Paratcha et al., 2003) and multiple double homozygous on Gfrα1CKO background are required for definitive confirmation.
Another reason for modest effect is that the cells where Gfrα1 is not completely eliminated are able to partially compensate as seen by their increased proliferation and pERK activity. This suggests extraordinary capacity of unaffected UB tip cells to compensate. This is unlike the enteric nervous system where Gfrα1 loss in 70% of neurons led to complete aganglionosis of the colon (Uesaka et al., 2007). To minimize this confound, the explant culture system was used and near complete excision of Gfrα1 in the UB epithelium was achieved and yet only a modest role of Gfrα1 was observed indicating that the kidney relies on multiple signaling mechanisms for branching morphogenesis. The reduction in kidney size between Gfrα1CKO and littermate control was 25% in explant culture versus 20% in vivo despite more potent deletion in the culture system. This may be due to a threshold of Gfrα1 levels required in the UB tip beyond which additional loss does not significantly impact kidney growth during branching.
We cannot exclude the possibility that Gfrα1 from the metanephric mesenchyme (MM) (trans-signaling) could compensate for Gfrα1 loss in the UB tips during branching morphogenesis as the excision efficiency in the MM was low due to weak expression in the MM compared to that in the UB tip. Although, we believe trans-signaling is unlikely because: 1) during pre UB induction Gfrα1 in mesonephric mesenchyme does not compensate for its loss; and 2) elimination of all trans Gfrα1 expression did not affect kidney development (Enomoto et al., 2004). To completely rule out a role of trans-Gfrα1 signaling in branching morphogenesis experiments with inducible Cre strains with potent activity in both the MM and UB are needed.
Numerous receptor tyrosine kinases (RTK) are expressed in the metanephric kidney that may have a role in later kidney development including receptors for FGF ligands and the RTK Met (Bates, 2011; Song et al., 2011). Therefore, a more likely scenario for the mild hypoplasia is compensation by other RTKs. Recent studies have identified non-Ret pathways involved in kidney development and branching morphogenesis. One study has shown that FGF10/FGFR2 tyrosine kinases signaling can compensate for branching morphogenesis in the absence of Ret or GDNF and Spry1 (Michos et al., 2010). The β-catenin pathway was also suggested as an alternative mechanism for Ret-independent branching (Kiefer et al., 2010). In support of redundant pathway involvement we noted that expression of several downstream mRNA targets of Ret was not changed in Gfrα1CKO kidneys. Although a positive feedback mechanism among Ret signaling components has been reported (Barak et al., 2011) (Pepicelli et al., 1997) the mechanism of reduced Ret mRNA transcripts observed in the Gfrα1CKO mice needs further investigation.
Our results show that Gfrα1's importance in ureteric epithelium during kidney development decreases with time. It is critical in UB induction where its loss causes agenesis and mildly important during early branching where its loss results in modest hypoplasia. These different roles of Gfrα1 during preUB and postUB induction are novel observations to our knowledge. Studies investigating genes known to be important in kidney ontogeny at different developmental stages are limited. Among these WT1 has differential functions at different developmental stages (Hu et al., 2011). Additionally, Gfrα1's importance at different stages during development depends on the organ system. In the mouse gut, at E11.5-E13.5 Gfrα1 is critical for enteric neuron proliferation, after E15.5 it has an essential role for neuron survival only in the colon, and after postnatal day 5 it is not essential (Uesaka et al., 2007). Therefore, in addition to a time-specific requirement, organs have differential susceptibility to Gfrα1 loss.
We previously reported that Ret signaling is a major regulator of pERK in the Wolffian duct during UB bud formation (Hoshi et al., 2012) . Even post-UB induction Gfrα1 is a major regulator of ERK1/2 activity as there was near complete loss of pERK in cells that had no Gfrα1. An important observation from our time-lapse kidney culture experiments is that EGFP-positive cells were maintained throughout the branching distal UB tips. Previous studies using a chimeric system demonstrated that cells lacking Ret are less competent to colonize the UB tips compared to wild-type cells (Chi et al., 2009). The presence of Gfrα1-null cells in the branching UB tips suggests that Gfrα1 is not necessary for tip colonization.
In conclusion, our studies demonstrate that Gfrα1, a critical component of the Ret signaling complex, is required in the WD epithelium during early UB induction and cannot be compensated by trans-activity from nonepithelial tissues during this developmental stage. It has a modest role in branching that is associated with decreased cell proliferation and ERK activity. Our results provide further support for redundant pathways other than Gdnf-Gfrα1-Ret signaling to ensure on-going branching morphogenesis. Ultimately further studies with Gdnf and Ret conditional models are needed to establish the role of the entire Gdnf-Gfrα1-Ret signaling complex after budding in vivo.
4. Experimental Procedures
4.1. Animals
Institution approved protocols were followed for all animal studies and were compliant with National Institutes of Health guidelines. For timed pregnancies, the day of the copulatory plug was defined as embryonic day 0.5 (E0.5). To use EGFP as a surrogate for Gfrα1 expression (Gfrα1 reporter) we bred Gfrα1floxEGFP/+ mice (Uesaka et al., 2007) with β-actinCre mice (Meyers et al., 1998). This deleted the floxed Gfrα1 allele and expressed the EGFP reporter allele from cells that express β-actinCre (Gfrα1EGFP/+). Genotyping of the reporter Gfrα1EGFP/+ was performed with forward primer P1 (Uesaka et al., 2007) and reverse primer P8428 (Jain et al., 2006b).
For experiments to determine the cell autonomous roles of Gfrα1 in the epithelial lineage we bred Gfrα1 heterozygous mice (Gfrα1+/-) in which one of the alleles has been knocked out and instead tau-lacZ (TLZ) is knocked-in and expressed from the Gfrα1 locus (Enomoto et al., 2004) with mice that express the ureteric epithelium specific Cre under the control of the Hoxb7 promoter- Hoxb7Cre [tg(Hoxb7-cre)13Amc; MGI:2675121] (Yu et al., 2002). The Gfrα1+/-:Hoxb7Cre mice were bred with the Gfrα1 conditional mice (Gfrα1floxEGFP/floxEGFP) creating Gfrα1floxEGFP/-:Hoxb7Cre mice with no Gfrα1 expression in Hoxb7 lineage. For some organ culture experiments Hoxb7Cre mice were bred with Rosa-YFP [Gt(ROSA)26Sortm1(EYFP)Cos; MGI ID: 2449038] mice to facilitate visualization of the UB tips in the embryos (Soriano, 1999; Yu et al., 2002).
To express the tamoxifen-inducible Cre allele from Gfrα1 locus (Gfrα1-CreERT2) we knocked-in CreERT2 cDNA into the Gfrα1 locus using the same strategy as was previously used to target this locus in generating Gfrα1 null and conditional mice. Briefly, the CreERT2 gene cassette was inserted into the second exon of the previously described Gfrα1 targeting vector (Enomoto et al., 2004; Uesaka et al., 2007),the Gfrα1-CreERT2 construct was injected into embryonic stem cells, cells that underwent homologous recombination were selected, injected into blastocysts and high percentage chimeras were bred to obtain Gfrα1-CreERT2 heterozygous mice. Heterozygous Gfrα1-CreERT2 were bred and Homozygous Gfrα1-CreERT2 mice were born with renal agenesis similar to the expected phenotype in Gfrα1 nulls further confirming successful integration into the Gfrα1 locus (Cacalano et al., 1998; Enomoto et al., 1998). Detailed characterization of this Gfrα1-CreERT2 mouse strain will be provided elsewhere. Genotyping was performed using the common forward primer P1, reverse wild-type primer P2 (Uesaka et al., 2007) and reverse CreERT2 primer SJ33 (5’ACTCGTTGCATCGACCGGTAATGCAGGC3’). For inducible deletion of Gfrα1 post UB induction the Gfrα1CreERT2/+ mice were bred with the Gfrα1floxEGFP/floxEGFP mice. To induce Cre recombinase mediated deletion pregnant mice were injected (intraperitoneal) with 4-hydroxytamoxifen (4HT) (Sigma) (a cumulative dose of 3mg/30g) dissolved in autoclaved corn oil. A single dose of 4HT at E11.5 or E12.5 or divided over two injections at E11.5 or E12.5 did not affect the final phenotype therefore these data were combined. Doses greater than 3mg/30g resulted in toxicity. We observed Gfrα1 excision 8-10 hours after 4HT injection as judged by EGFP immunofluorescence. For late excision experiments pregnant mice were injected with a cumulative dose of 3mg/30g at or between E13.5-15.5 or E15.5-17.5. Gfrα1EGFP/+, Gfrα1CreERT2/+, and Gfrα1floxEGFP/floxEGFP mice were maintained on a pure C57BL/6 background.
4.2. Immunological Analysis
Paraformaldehyde fixed (PFA) whole GU tract or metanephric kidneys were dissected, blocked (PBS containing 0.2% non-fat dry milk, 1% BSA, 0.3%Triton X-100) overnight at 4 °C followed by primary antibody incubation for 2 days at 4 °C, washed (PBS with 0.3% Triton X-100) and then incubated with secondary antibody overnight at 4 °C. Specimens were visualized with Eclipse 80i (Nikon) compound upright microscope or SMZ1500 dissecting microscope (Nikon). Images were captured using CoolSnapES camera (Roper Scientific), processed, and analyzed with NIS-Elements (Nikon) and Adobe Photoshop CS3 software. Confocal microscopy images were obtained on the Eclipse 80i (Nikon) equipped with D-EclipseC1 confocal system (Nikon). For immunohistochemistry on paraffin-embedded tissue sections antigen retrieval was used by boiling the slides immersed in EDTA (1mM) or citrate buffer (pH 6.0) for 15 minutes. In some cases of Gfrα1 immunohistochemistry on paraffin sections, antigen unmasking was done overnight by immersion in 20μM Tris-Chloride (pH 9.0) at 70 °C. Blocking, antibody staining, image capture and analysis were performed as described above. Frozen sections were analyzed in a similar fashion. For cryoprotection, previously fixed tissues were immersed in 30% sucrose for at least 24 hours. Specimens were embedded in Optimal Cutting Temperature (OCT) compound (Suskura Finetek) and stored at -80 °C Cryosections (10 microns) were cut in series on a cryostat (Leica CM 1850). Slides were fixed in fresh 4% PFA for 6-10 minutes at room temperature before starting the immunostaining procedure described above.
Primary antibodies used were: GFP (chicken, 1:500 dilution, Aves); Gfrα1 (goat, 1:50, Neuromics); E-cadherin (goat, 1:100, R&D); phospho-histone H3 (mouse, 1:100, Cell Signaling); phospho-Erk 1/2 (rabbit, 1:100, Cell Signaling); Pax-2 (rabbit, 1:100, Convance); WT1 (mouse, 1:100, Dako); Six2 (rabbit, 1:100, Proteintech). Secondary antibodies (Jackson Immuno Research) used were: DyeLight-488-conjugated chicken (1:1000); Cy3 conjugated IgG (goat or rabbit or mouse, 1:100); Cy5 conjugated IgG (goat or rabbit or mouse 1:100).
4.3. Quantitative real-time PCR analysis
E14.5 whole kidneys were dissected in sterile PBS, immediately placed in 1ml of Trizol (Invitrogen), and homogenized (PowerGen 125) for RNA extraction according to manufacturer's instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed in triplicate with Applied Biosystems 7900 HT Sequence Detection System using Fast SYBR Green technology. All primer sets were designed using Primer Express 3.0 software (Applied Biosystems) and purchased from Integrated DNA Technologies (IDT®) (sequences listed in supplemental table 1). Glyceraldehydes-3-phosphate dehydrogenase (GAPDH) was used for normalization. PCR parameters were: initial degeneration at 95°C for 5 minutes followed by 40 cycles of denaturation at 95°C for 10 seconds, annealing/extension at 60 °C for 30 seconds. Primer validation was done using dissociation curve analysis to exclude primer dimer formation prior to and after all the experiments. Analysis was performed by absolute quantitification method and levels were expressed after normalization with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression (QTmeansample/ QTmean GAPDH). Statistical significance was determined using the Student t-test.
4.4. Histomorphic Analysis
Mouse GU tracts were dissected at indicated ages, fixed in fresh 4% PFA. Serial histological analysis with hematoxylin and eosin (H&E) staining was performed as previously described (Jain et al., 2004). Glomerular counts were performed on paraffin sections as previously described (Jain et al., 2006a). Briefly, kidneys were sectioned in their entirety; all glomeruli were counted from H&E sections at 120 micron intervals. Cross-sectional area was calculated on whole metanephric kidneys from mutant and littermate controls with NIS-Elements BR 3.2 (Nikon) software (Michos et al., 2010). Whole mount ureteric tip counts were performed on E-cadherin (IHC) stained kidneys. Using a SMZ1500 dissecting microscope, images of mutant (Gfrα1CKO) and littermate controls were captured under the same magnification using CoolSnapES camera (Roper Scientific). Ureteric bud tips appearing within a single unit area of a grid placed over the center of the kidney were counted in a blinded fashion by two individuals (the results were averaged) using the NIS-Elements software (Nikon).
4.5. Proliferation and activated-Erk quantification and apoptosis assay
Proliferation analysis was performed by measuring the mitotic index of ureteric tip epithelial cells at E14.5 on paraformaldehyde fixed paraffin or OCT embedded sections. Briefly, sections were double or triple immune-stained for phospho-histone H3 (phh3), E-cadherin, and EGFP. Phospho-histone H3 cells double stained with E-cadherin and/or EGFP were counted on Gfrα1CKO and controls. For overall UB tip proliferation (mitotic index), pHH3 cells within a discrete UB tip were counted against the total number of epithelial cells per tip (pHH3 and E-cadherin+/E-cadherin+). To quantify proliferation of Gfrα1 negative cells (express EGFP reporter), pHH3 and EGFP double positive cells were counted and compared against the total number of EGFP positive cells per UB tip (pHH3+EGFP+/EGFP+). Sections from 7 control and 11 mutant kidneys were analyzed. Total counts were >2500 UB tip cells in the control and >4000 UB tip cells in the mutant. Quantification of ERK activity was obtained at E14.5 on paraformaldehyde fixed paraffin or OCT embedded sections in the same manner as above. Sections were double (pERK and EGFP) or triple immune-stained (pERK, E-cadherin, and EGFP or pERK, Gfrα1, and EGFP). Sections from 4 control and 5 mutant kidneys were analyzed. Total counts were >1400 UB tip cells in the controls and >2000 in the mutants. Apoptosis analysis was performed using TUNEL staining on paraffin sections as previously described (Jain et al., 2006a).
4.6. Metanephric organ cultures
Metanephric kidneys were dissected at the indicated time points and cultured. Culture was performed on BD Falcon 0.4μm pore polyethylene terephthalate (PET) tack-etched membrane inserted into 6 well culture plate as previously described and 4HT (final concentration 1μM) was added to the medium in inducible deletion experiments (Rogers et al., 1991). Time-lapse microscopy was performed in a specialized culture chamber with 5% CO2 air mixture. FITC and phase images were captured every 30 minutes (HQ2, Photometrics) with respective filters. Images were processed using Metamorph software. For quantification of ureteric bud tips we cultured E12.5 metanephric kidneys for 72 hr and all tips were counted in a blinded fashion from images at the indicated times during culture. Kidneys with 9-13 UB tips at the beginning of culture from each genotype were used for this analysis. To facilitate visualization of the UB tips at the start of culture pregnant mothers were injected with 4HT (intraperitoneal) at E11.5. There was no appreciable leakiness of Cre without 4HT as seen by no EGFP signal when 4HT injection in vivo was omitted (Figure 7D). Culture media was changed every 48 hours. Surface area was determined by number of pixels present in manual traces of the outline of the phase contrast image using Metamorph software at the time points indicated. Each kidney at each time point was traced three times and the average was used.
5. Statistical analysis
Sample size was three or more for each genotype at each age reported. Statistical significance for all studies was determined by the Students t-test.
Supplementary Material
Highlights.
Gfrα1 expressed in the ureteric epithelium is required for ureteric bud induction.
Conditional loss of Gfrα1 after ureteric bud induction results in renal hypoplasia.
Gfrα1 regulates epithelial cell proliferation and phosphorylated-ERK.
Acknowledgments
We thank Angela Lluka and Amanda Knoten for excellent technical assistance. We are grateful to Dr. Jeff Milbrandt for resources to generate the Gfrα1CreERT2 mice. We thank Drs. Feng Chen, Keith Hruska, Paul Austin and Paul Hmiel for valuable scientific discussions. Part of this research was presented by T.K. Davis during a fellowship platform presentation at the annual American Pediatric Society of Nephrology in 2010.
Sources of Support: This work was supported by core facilities provided by the National Institutes of Health (NIH) George M. O'Brien Center for Kidney Disease Research (P30-DK079333) to Washington University, and NIH grants DK081644 and DK082531 (S. J.). T.K. Davis was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award T32-HD043010 from the National Institute of Child Health and Human Development (NICHD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Statement of Disclosure.
We disclose that there are no conflicts of financial interest.
References
- Abitbol CL, Ingelfinger JR. Nephron mass and cardiovascular and renal disease risks. Semin Nephrol. 2009;29:445–54. doi: 10.1016/j.semnephrol.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–94. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- Airaksinen MS, Titievsky A, Saarma M. GDNF family neurotrophic factor signaling: four masters, one servant? Mol Cell Neurosci. 1999;13:313–25. doi: 10.1006/mcne.1999.0754. [DOI] [PubMed] [Google Scholar]
- Barak S, Ahmadiantehrani S, Kharazia V, Ron D. Positive autoregulation of GDNF levels in the ventral tegmental area mediates long-lasting inhibition of excessive alcohol consumption. Transl Psychiatry. 2011;1:e60. doi: 10.1038/tp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates CM. Role of fibroblast growth factor receptor signaling in kidney development. Am J Physiol Renal Physiol. 2011;301:F245–51. doi: 10.1152/ajprenal.00186.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacalano G, Farinas I, Wang LC, Hagler K, Forgie A, Moore M, Armanini M, Phillips H, Ryan AM, Reichardt LF, Hynes M, Davies A, Rosenthal A. GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron. 1998;21:53–62. doi: 10.1016/s0896-6273(00)80514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, Mendelsohn C, Costantini F. Ret-Dependent Cell Rearrangements in the Wolffian Duct Epithelium Initiate Ureteric Bud Morphogenesis. Developmental Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Shakya R. GDNF/Ret signaling and the development of the kidney. Bioessays. 2006;28:117–27. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H, Araki T, Jackman A, Heuckeroth RO, Snider WD, Johnson EM, Jr., Milbrandt J. GFR alpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron. 1998;21:317–24. doi: 10.1016/s0896-6273(00)80541-3. [DOI] [PubMed] [Google Scholar]
- Enomoto H, Hughes I, Golden J, Baloh RH, Yonemura S, Heuckeroth RO, Johnson EM, Jr., Milbrandt J. GFRalpha1 expression in cells lacking RET is dispensable for organogenesis and nerve regeneration. Neuron. 2004;44:623–36. doi: 10.1016/j.neuron.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Golden JP, DeMaro JA, Osborne PA, Milbrandt J, Johnson EM. Expression of Neurturin, GDNF, and GDNF Family-Receptor mRNA in the Developing and Mature Mouse. Experimental Neurology. 1999;158:504–528. doi: 10.1006/exnr.1999.7127. [DOI] [PubMed] [Google Scholar]
- Hoshi M, Batourina E, Mendelsohn C, Jain S. Novel mechanisms of early upper and lower urinary tract patterning regulated by RetY1015 docking tyrosine in mice. Development. 2012;139:2405–15. doi: 10.1242/dev.078667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Gao F, Tian W, Ruteshouser EC, Wang Y, Lazar A, Stewart J, Strong LC, Behringer RR, Huff V. Wt1 ablation and Igf2 upregulation in mice result in Wilms tumors with elevated ERK1/2 phosphorylation. J Clin Invest. 2011;121:174–83. doi: 10.1172/JCI43772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S. The many faces of RET dysfunction in kidney. Organogenesis. 2009;5:1–14. doi: 10.4161/org.5.4.10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Encinas M, Johnson EM, Jr., Milbrandt J. Critical and distinct roles for key RET tyrosine docking sites in renal development. Genes Dev. 2006a;20:321–33. doi: 10.1101/gad.1387206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Golden JP, Wozniak D, Pehek E, Johnson EM, Jr., Milbrandt J. RET Is Dispensable for Maintenance of Midbrain Dopaminergic Neurons in Adult Mice. J. Neurosci. 2006b;26:11230–11238. doi: 10.1523/JNEUROSCI.1876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Naughton CK, Yang M, Strickland A, Vij K, Encinas M, Golden J, Gupta A, Heuckeroth R, Johnson EM, Jr., Milbrandt J. Mice expressing a dominant-negative Ret mutation phenocopy human Hirschsprung disease and delineate a direct role of Ret in spermatogenesis. Development. 2004;131:5503–13. doi: 10.1242/dev.01421. [DOI] [PubMed] [Google Scholar]
- Kiefer SM, Robbins L, Stumpff KM, Lin C, Ma L, Rauchman M. Sall1-dependent signals affect Wnt signaling and ureter tip fate to initiate kidney development. Development. 2010;137:3099–3106. doi: 10.1242/dev.037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda F, Paratcha G, Ibanez CF. Target-derived GFRalpha1 as an attractive guidance signal for developing sensory and sympathetic axons via activation of Cdk5. Neuron. 2002;36:387–401. doi: 10.1016/s0896-6273(02)01002-4. [DOI] [PubMed] [Google Scholar]
- Little M, Georgas K, Pennisi D, Wilkinson L, Peter K. Chapter Five - Kidney Development: Two Tales of Tubulogenesis, Current Topics in Developmental Biology. Academic Press; 2010. pp. 193–229. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–3185. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–41. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, Licht JD, Martin GR, Costantini F. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J, Mochida K, Ohfuji Y, Shimazaki T, Meno C, Ohishi S, Matsuda Y, Fujii H, Saijoh Y, Hamada H. GFR alpha3, a component of the artemin receptor, is required for migration and survival of the superior cervical ganglion [In Process Citation]. Neuron. 1999;23:725–36. doi: 10.1016/s0896-6273(01)80031-3. [DOI] [PubMed] [Google Scholar]
- Paratcha G, Ledda F, Baars L, Coulpier M, Besset V, Anders J, Scott R, Ibanez CF. Released GFRalpha1 potentiates downstream signaling, neuronal survival, and differentiation via a novel mechanism of recruitment of c-Ret to lipid rafts. Neuron. 2001;29:171–84. doi: 10.1016/s0896-6273(01)00188-x. [DOI] [PubMed] [Google Scholar]
- Paratcha G, Ledda F, Ibanez CF. The neural cell adhesion molecule NCAM is an alternative signaling receptor for GDNF family ligands. Cell. 2003;113:867–79. doi: 10.1016/s0092-8674(03)00435-5. [DOI] [PubMed] [Google Scholar]
- Pepicelli CV, Kispert A, Rowitch DH, McMahon AP. GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev Biol. 1997;192:193–8. doi: 10.1006/dbio.1997.8745. [DOI] [PubMed] [Google Scholar]
- Rogers SA, Ryan G, Hammerman MR. Insulin-like growth factors I and II are produced in the metanephros and are required for growth and development in vitro. J Cell Biol. 1991;113:1447–53. doi: 10.1083/jcb.113.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J, Luukko K, Poteryaev D, Laurikainen A, Sun YF, Laakso T, Eerikainen S, Tuominen R, Lakso M, Rauvala H, Arumae U, Pasternack M, Saarma M, Airaksinen MS. Retarded growth and deficits in the enteric and parasympathetic nervous system in mice lacking GFR alpha2, a functional neurturin receptor. Neuron. 1999;22:243–52. doi: 10.1016/s0896-6273(00)81086-7. [DOI] [PubMed] [Google Scholar]
- Saarma M. GDNF - a stranger in the TGF-beta superfamily? Eur J Biochem. 2000;267:6968–71. doi: 10.1046/j.1432-1327.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Sainio K, Suvanto P, Saarma M, Arumae U, Lindahl M, Davies JA, Sariola H. Glial Cell-line derived neurotrophic factor is a morphogen for the ureteric bud epithelium. Development. 1997;20:4077–4087. doi: 10.1242/dev.124.20.4077. [DOI] [PubMed] [Google Scholar]
- Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci. 2003;116:3855–62. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- Song R, El-Dahr SS, Yosypiv IV. Receptor tyrosine kinases in kidney development. J Signal Transduct. 2011;2011:869281. doi: 10.1155/2011/869281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain [letter] [In Process Citation]. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Towers PR, Woolf AS, Hardman P. Glial cell line-derived neurotrophic factor stimulates ureteric bud outgrowth and enhances survival of ureteric bud cells in vitro. Exp Nephrol. 1998;6:337–51. doi: 10.1159/000020541. [DOI] [PubMed] [Google Scholar]
- Uesaka T, Jain S, Yonemura S, Uchiyama Y, Milbrandt J, Enomoto H. Conditional ablation of GFRalpha1 in postmigratory enteric neurons triggers unconventional neuronal death in the colon and causes a Hirschsprung's disease phenotype. Development. 2007;134:2171–81. doi: 10.1242/dev.001388. [DOI] [PubMed] [Google Scholar]
- Vega Q, Worby C, Lechner M, Dixon J, Dressler G. Glial cell line-derived neurotrophic factor activates the receptor tyrosine kinase RET and promotes kidney morphogenesis. Proc Natl Acad Sci USA. 1996;93:10657–10661. doi: 10.1073/pnas.93.20.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129:5301–12. doi: 10.1242/dev.129.22.5301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.