Abstract
The constant high energy demand of neurons makes them rely heavily on their mitochondria. Dysfunction of mitochondrial energy metabolism leads to reduced ATP production, impaired calcium buffering, and generation of reactive oxygen species. There is strong evidence that mitochondrial dysfunction results in neurodegeneration and may contribute to the pathogenesis of Huntington’s disease (HD). Studies over the past few years have implicated an impaired function of peroxisome proliferator-activated receptor (PPAR)-γ coactivator-1α (PGC-1α), a transcriptional master co-regulator of mitochondrial biogenesis, metabolism and antioxidant defenses, in causing mitochondrial dysfunction in HD. Here we have attempted to discuss in a nutshell, the key findings on the role of PGC-1α in mitochondrial dysfunction in HD and its potential as a therapeutic target to cure HD.
Keywords: PGC-1α, PPARs, SIRT1, SIRT3, mitochondrial dysfunction, Huntington’s disease, oxidative stress, energy metabolism, mitochondrial dynamics, mitophagy, neurodegeneration and therapeutics
1. Introduction
Huntington’s disease (HD) is a dominantly inherited neurodegenerative disorder caused by the expansion of a CAG repeat in the gene encoding the protein huntingtin, leading to expression of mutant huntingtin with expanded polyglutamine repeats [1]. The expansion of polyglutamine repeats results in acquisition of an altered conformation by mutant huntingtin, which in turn causes the protein to aggregate. The function of normal huntingtin protein has not been fully elucidated yet, but it is known to be associated with synaptic vesicles and microtubules, and is an essential scaffold protein regulating axonal transport of vesicles including brain-derived neurotrophic factor (BDNF) [2–7]. The huntingtin protein was recently shown to play a role linking the glycolytic enzyme GAPDH to vesicles, to supply energy from glycolysis for fast axonal transport [8]. Both, a gain-of-function (for mutant huntingtin) and a loss-of-function (for normal huntingtin) hypothesis have been put forward to explain HD pathogenesis. Patients with HD have CAG repeat lengths above 36, with variable penetrance of repeat lengths 36–39 and complete penetrance above 39 repeats; longer repeat lengths (>60) have been associated with juvenile-onset HD [9]. Disease manifestations can begin at any time in life; the most common age range of onset is between 30 and 50 years old, although it occurs in children and the elderly as well. The juvenile variant of HD usually results from paternal transmission and is associated with increased severity as well as with a more rapid progression of the disease.
HD is characterized by progressive motor impairment, personality changes, psychiatric illness and gradual intellectual decline. Pathologically, there is a preferential and progressive loss of the medium spiny neurons (MSNs) in the striatum, as well as cortical atrophy, and degeneration of other brain regions later in the disease. There are no currently available treatments to delay disease onset or retard its progression, and the focus of medical care is limited to symptom management and maximizing function. Transcriptional dysregulation, protein aggregation, mitochondrial dysfunction and enhanced oxidative stress have been implicated in the disease pathogenesis. A key feature of HD patients is pronounced weight loss, despite sustained caloric intake. Deficits in energy expenditure have been linked with mitochondrial dysfunction in HD. The evidence for mitochondrial dysfunction in HD has been reviewed earlier [10–12]; we have summarized a few key findings in the following discussion.
2. Mitochondrial dysfunction in HD
There is extensive evidence for bioenergetic deficits and mitochondrial dysfunction in HD, such as a pronounced weight loss despite sustained caloric intake, nuclear magnetic resonance spectroscopy showing increased lactate in the cerebral cortex and basal ganglia, decreased activities of oxidative phosphorylation (OXPHOS) complexes II and III, and reduced aconitase activity in the basal ganglia, abnormal mitochondrial membrane depolarization in patient lymphoblasts, abnormal ultrastructure of mitochondria in cortical biopsies obtained from patients with both juvenile and adult-onset HD, and pathologic grade dependent reductions in numbers of mitochondria in HD postmortem brain tissue; and in striatal cells from mutant huntingtin-knock-in mice, both mitochondrial respiration and ATP production are significantly impaired (Reviewed in [11]). It was shown that ATP production is decreased as a function of CAG repeat length in human HD lymphoblastoid cell lines [13]. Studies of mitochondria isolated from HD patients and mice indicated that HD mitochondria depolarize at decreased calcium ion levels, and mutant huntingtin protein may directly interact with mitochondria to exert this effect [14–16].
We showed that the phenotypic and neuropathologic features of HD can be modeled in rodents and primates, with the mitochondrial toxin 3-nitropropionic acid (3-NP) [17]. We and others have shown impaired brain creatine kinase activity and significant alterations in levels of high-energy phosphate intermediates in transgenic mouse models of HD [18, 19]. We also found a reduction in numbers and size of mitochondria identified by a reduction in immunohistochemical markers for cytochrome oxidase 2, superoxide dismutase 2 and cytochrome c (Cyt c) in the preferentially vulnerable striatal calbindin-positive neurons in moderate-to-severe grade HD patients, which worsened with increasing disease severity [20]. Using electron microscopy, we showed small degenerating and/or degenerated mitochondria and a reduced mitochondrial density in the striatum of the R6/2 transgenic mouse model of HD [21]. Abnormal mitochondrial morphology was seen in neurons expressing mutant huntingtin exon1-Q46 or Q97, as opposed to normal mitochondrial morphology observed in wild-type huntingtin exon1-Q17 expressing neurons [22]. Abnormal morphology and an increased mitochondrial fragmentation were also observed in fibroblasts from juvenile and adult-onset HD patients [22]. We also showed irregular mitochondrial morphology and distribution in muscles of NLS-N171-82Q and R6/2 transgenic mouse models of HD [21, 23].
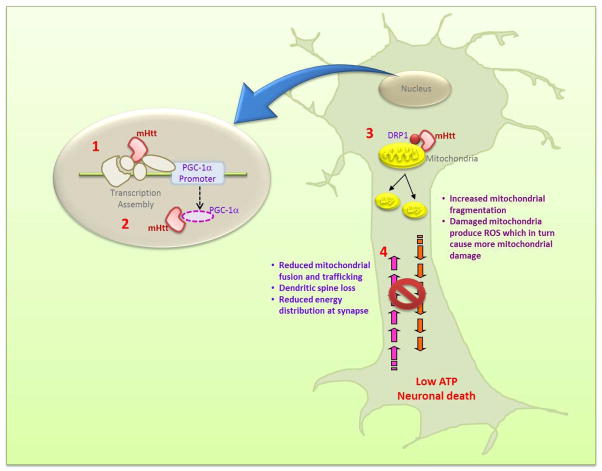
We and others have shown abnormalities in mitochondrial dynamics: Dynamin related protein 1 (Drp1, involved in mitochondrial fission) was found to be significantly increased and mitofusin 1 (Mfn1, involved in mitochondrial fusion) was significantly decreased [20, 22, 24, 25]. A direct interaction of mutant huntingtin with mitochondria or with various protein complexes has also been proposed to play an important role in disease pathogenesis, by regulating mitochondrial fission-fusion events, mitochondrial trafficking along axons and dendrites, and mitochondrial distribution [22, 25–29] (Figure 1). Expression of mutant huntingtin causes abnormal mitochondrial ultrastructure, impaired calcium buffering, bioenergetic defects and mitochondrial DNA (mtDNA) deletions, all of which may be a consequence of a failure to maintain a balance between mitochondrial fission and fusion.
Figure 1.
Mutant huntingtin (mHtt) causes mitochondrial dysfunction in HD by multiple mechanisms. Mutant huntingtin: (1) binds to PGC-1α promoter reducing its expression; (2) binds to PGC-1α directly reducing its activity, which in turn results in reduced expression of PGC-1α target genes such as those involved in mitochondrial biogenesis and antioxidant defense; (3) binds to DRP1 more tightly than the wild-type protein, tripping the balance of mitochondrial fission-fusion processes in favor of fission; and (4) interferes with axonal transport of mitochondria. The net result of these impairments is low ATP at nerve terminals which culminates into neuronal death.
Mitochondrial turnover is dependent on autophagy, which declines with age and is frequently dysfunctional in neurodegenerative diseases [30]. Mitochondrial reactive oxygen species (ROS) production and oxidation of mitochondrial lipids have been shown to play a role in autophagy. Mitophagy denotes the degradation of mitochondria through macroautophagy. Through mitophagy, cells regulate mitochondrial number in response to their metabolic state and also implement a quality control system for selective elimination of damaged/defective mitochondria. Changes in mitochondrial dynamics that redistribute defective mitochondrial components, and/or increase fission or decrease fusion, facilitate isolation of damaged mitochondria and their subsequent elimination by mitophagy. Defective mitochondrial fission can yield uneven products, with one depolarized daughter mitochondrion and one hyperpolarized mitochondrion [31]. Such depolarized mitochondria are much less likely to fuse, have reduced levels of OPA1 protein, and are eventually autophagocytosed. Excessive fusion, on the other hand, prevents autophagic mitochondrial degradation and protects mitochondria from massive degradation by starvation-induced autophagy [32, 33]. When mitophagy is compromised, oxidized proteins accumulate, and cellular respiration decreases. As mitochondrial energy production and metabolic pathways supply energy for ion exchange pumps whose function is to maintain an electrochemical gradient across the mitochondrial membrane, defective energy metabolism could translate into an enhanced susceptibility of HD mitochondria to undergo depolarization and eventually mitophagy. A number of studies have evaluated this, and have found that mitochondria from HD patients are exquisitely sensitive to depolarizing stresses. In one study, treatment of HD lymphoblasts with complex IV inhibitors resulted in mitochondrial depolarization and apoptotic cell death involving caspase activation [34]. In another study, electrical measurements of HD lymphoblast mitochondria yielded lower than normal membrane potentials and depolarization in response to modest Ca2+ loads [15]. More recently, Martinez-Vicente et al. [35] have shown that HD pathology is associated with autophagic cargo recognition defects that lead to slower turnover, functional decay and accumulation of damaged mitochondria in the cytoplasm.
3. Oxidative damage in HD
Mitochondria are both targets and important sources of ROS. Increased levels of ROS including superoxide (O2•−), hydrogen peroxide (H2O2), hydroxyl radical (•OH), and reactive nitrogen species such as peroxynitrite (ONOO−) impair cellular function by degrading proteins, lipids, and nucleic acids. It has been shown that oxidative stress stimulates mitochondrial fission; the addition of hydrogen peroxide to cultured cerebellar granule neurons induced mitochondrial fragmentation within one hour of treatment [36]. It was also shown that nitric oxide causes increased mitochondrial fission in neurons, prior to the onset of neuronal loss in a mouse model of stroke [37]. On the other hand, expression of Mfn or a dominant negative Drp1 in cultured neurons, was protective against oxidative insults [36, 37]. The generation of ROS appears to be increased in damaged mitochondria, and in cells with compromised mitochondrial function. Acute exposure to relatively high levels of ROS, especially in the presence of calcium, can induce the mitochondrial permeability transition, uncouple oxidative phosphorylation with catastrophic effects on mitochondrial energetics, and contribute to cytotoxicity via necrosis and/or apoptosis. Oxidative stress within mitochondria can lead to a vicious cycle in which ROS production progressively increases, leading in turn to progressive augmentation of damage. Mitochondrial fission and mitophagy play a critical role in removing damaged mitochondria which otherwise generate further ROS. Another recently recognized source of ROS in HD is nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) [38]. Valencia et al. [38] found higher than normal levels of NOX activity in the postmortem brain of human HD patients compared with controls, in brain synaptosomes of a knock-in mouse model of HD (HD140Q/140Q) and in primary cortical and striatal neurons of HD140Q/140Q mice. High NOX activity in primary cortical and striatal neurons of HD140Q/140Q mice correlated with increased ROS production and neurite swellings. They also found that mutant huntingtin preferentially colocalized at plasma membrane lipid rafts with gp91-phox, a catalytic subunit for the NOX2 isoform; and Rac1, which is required for assembly of NOX2 components at lipid rafts, was also elevated in HD140Q/140Q neurons [38].
The extent and severity of oxidative damage in HD are features well recognized but perhaps underappreciated. Oxidative damage occurs to lipids, proteins and DNA, and it has been suggested that the latter may contribute to CAG repeat expansion during DNA repair [39]. Studies in both HD patients and experimental models of HD support a role for oxidative stress and ensuing mitochondrial dysfunction in mediating the neuronal degeneration observed in HD. Oxidative damage is well documented in plasma of HD patients, HD postmortem brain tissue, lymphoblasts and cerebrospinal fluid [40–45]. Markers of oxidative damage, including heme oxygenase (an inducible isoform that occurs in response to oxidative stress), 3-nitrotyrosine (a marker for peroxynitrite-mediated protein nitration), and malondialdehyde (MDA), are elevated in human HD striatum and cortex as compared with age-matched control brain specimens [41]. Consistent with the immunohistochemical data, biochemical assays in HD patients show significant increases in MDA and 4-hydroxynonenal (HNE) brain levels, which are almost 8-fold greater than in control subjects [46]. Accumulation of lipofuscin (a product of unsaturated fatty acid peroxidation), has been reported in HD brains, and is most prominent in vulnerable striatal neurons [41]. There are marked increases in 8-hydroxy-2′-deoxyguanosine (8-OHdG), a marker of oxidative damage to DNA, in both plasma and urine of HD patients [47, 48].
Chen et al. [47] detected a correlation between lipid peroxidation products in plasma and degree of clinical severity in patients with HD, and proposed using them as a potential biomarker for evaluating treatment efficacy. Increased global oxidative stress (GOS), a reduction in antioxidant systems and carbonylated proteins have all been reported to correlate with disease stage in patients with HD [49]. There is increased DNA fragmentation in HD patients which correlated with CAG repeat length [41, 50–53]. Aldolase C, glial fibrillary acidic protein, tubulin, γ-enolase, and creatine kinase B were found to be the targets of oxidative modification in both striatum and cortex from HD patients [44]. More recently, it was shown that the oxidation of mitochondrial enzymes resulted in decreased catalytic activity in the striatum of HD patients, providing a link to the bioenergetic deficits observed in HD [54]. These authors also provided evidence that pyridoxal kinase and antiquitin 1 oxidation may result in decreased pyridoxal 5-phosphate availability, which in turn is necessary as a cofactor in transaminations, synthesis of glutathione, and synthesis of GABA and dopamine, two neurotransmitters that play a key role in HD pathology. These findings suggest that oxidative stress plays an important role in the pathogenesis of HD.
Additional indirect evidence for oxidative damage in HD comes from the studies of Lim et al. [55] and Tabrizi et al. [56]. Lim et al. [55] investigated dysfunction of Ca2+ homeostasis in mitochondria from striatal neurons of postmortem brains of HD patients. They found that mitochondria in mutant striatal neurons are unable to handle large Ca2+ loads, possibly due to the increased sensitivity to Ca2+ induced permeability transition pore opening, which dissipates the membrane potential, prompting the release of accumulated Ca2+. Harmful ROS, produced by defective mitochondria, increase in mutant cells, particularly if the damage to mitochondria is artificially exacerbated with, for example, complex II inhibitors. Mitochondria in mutant cells are thus particularly vulnerable to stresses induced by Ca2+ and ROS. The observed decrease of cell Ca2+ could be a compensatory attempt to prevent Ca2+ stress that would irreversibly damage mitochondria, and eventually lead to cell death. Furthermore, the severe impairment in the mitochondrial tricarboxylic acid (TCA)-cycle enzyme aconitase in HD brain, has been attributed to Fe-S clusters within the protein, which make it a particularly vulnerable target for free radical–mediated oxidative damage [56]. Aconitase inactivation directly correlates with the generation of superoxide produced by excitotoxicity [57]. There is evidence for a direct link between glutamate/NMDA excitotoxicity and aconitase inhibition via nitric oxide (NO•) and mitochondrial free radical generation.
Several of the aforementioned oxidative alterations observed in human HD are recapitulated in transgenic and neurotoxin mouse models of HD, such as 1) increased concentrations of 8-OHdG in the urine, plasma, striatal microdialysates, and isolated brain nuclear DNA, 2) increased immunostaining for 8-OHdG and lipid peroxidation markers (MDA, HNE and 8-iso-prostaglandin) in the striatum, 3) a progressive increase in the level of mtDNA damage in the striatum and cerebral cortex, 4) significantly increased oxidation of a number of key cellular proteins, including the metabolic enzymes creatine kinase, aconitase, neuron-specific enolase, heat shock protein 90, and the voltage-dependent anion channel 1 in the R6/2 transgenic mice; in another N-truncated mutant huntingtin transgenic model of HD, the N171-82Q mice, we found evidence of increased oxidative and nitrosative stress; in full-length knock-in mice, CAG140, 8-OHdG levels are elevated in brain and urine of the animals; and in the 3-NP and malonate toxin and excitotoxin models of HD, increased oxidative damage has been consistently observed [40, 42, 58–61].
mtDNA damage was shown to be associated with oxidative stress and the development of HD pathogenesis in the 3-NP induced and R6/2 transgenic mice models of HD [59]. Recently, Siddiqui et al. [62] showed that the striatal cells expressing mutant huntingtin (the STHdhQ111 striatal cells) show higher basal levels of mitochondrial-generated ROS and mtDNA lesions and a lower spare respiratory capacity than the cells expressing normal huntingtin. Silencing of APE1, the major mammalian apurinic/apyrimidinic (AP) endonuclease that participates in the base excision repair (BER) pathway, caused further reductions of spare respiratory capacity in the mutant huntingtin-expressing cells. They also showed that human postmortem striata exhibit significant increases in the levels of mtDNA and nuclear DNA lesion frequencies and significant mtDNA depletion; and a primary culture of HD diploid skin fibroblast from an HD patient shows significant reductions in spare respiratory capacity compared to control skin fibroblasts [62]. More recently, Wang et al. [63] showed that the YAC128 mouse embryonic fibroblasts exhibit a strikingly higher level of mitochondrial matrix Ca2+ loading and elevated superoxide generation compared with wild-type cells. Blocking mitochondrial Ca2+ uptake abolished elevated superoxide generation demonstrating that mitochondrial oxidative stress seen in these cells is critically dependent on mitochondrial Ca2+ loading in HD cells. Similar results were obtained using neurons from HD model mice and fibroblast cells from HD patients. Mitochondrial Ca2+ loading in HD cells caused a 2-fold higher level of mtDNA damage due to the excessive oxidant generation [63].
4. PGC-1α: The master co-activator
Peroxisome proliferator-activated receptor (PPAR)-γ coactivator-1 (PGC-1) family of coactivators is an extensively regulated group of proteins that are highly responsive to a variety of environmental cues, from temperature to nutritional status, to physical activity. This family of coactivators plays a crucial role in integrating signaling pathways, tailoring them to best suit the changing cellular and systemic milieu. The first and perhaps the best studied member of the PGC-1 family of coactivators is PGC-1α, which was initially identified as a PPARγ-interacting protein from brown fat [64]. PGC-1β is the closest homolog of PGC-1α and shares extensive sequence identity, whereas PGC-related coactivator (PRC) has more limited homology. PGC-1α is a highly versatile transcriptional co-activator that interacts with a broad range of transcription factors involved in a wide variety of biological processes and/or responses including mitochondrial biogenesis, OXPHOS, antioxidant defense, adaptive thermogenesis, glucose/fatty acid metabolism, fiber type switching in skeletal muscle, and heart development [65–67]. PGC-1α forms heteromeric complexes with transcription factors, including nuclear respiratory factors, NRF-1 and NRF-2, and the nuclear receptors, PPARα, PPARδ, PPARγ, estrogen related receptor α (ERRα) [68]. These transcription factors, in turn, regulate the expression of many nuclear-encoded mitochondrial genes, such as Cyt c, complexes I–V and the mitochondrial transcription factor A (Tfam) [69, 70]. Activation of the mitochondrial genes leads to increased enzymatic capacity for fatty-acid β-oxidation, Krebs cycle, and OXPHOS. Importantly, PGC-1α also induces the expression of genes involved in heme biosynthesis, ion transport, mitochondrial protein translation, and protein import and stimulates respiratory function [71, 72]. In light of the above, PGC-1α is aptly termed a master co-regulator of mitochondrial function.
5. Functional consequences of impaired PGC-1α activity in HD
In recent years, impaired PGC-1α expression and/or function has emerged as a common underlying cause of mitochondrial dysfunction in HD. There is substantial evidence for impairment of PGC-1α levels and activity in HD [23, 73–76]. Involvement of PGC-1α in HD was first suggested by the findings that PGC-1α knockout mice exhibit mitochondrial dysfunction, defective bioenergetics, a hyperkinetic movement disorder and striatal degeneration, which are features also observed in HD [77, 78]. Impaired PGC-1α function and levels occur in striatal cell lines, transgenic mouse models of HD and in postmortem brain tissue from HD patients, and interference of mutant huntingtin with the CREB/TAF4 complex was shown to be instrumental in this impairment [73, 74] (Figure 1). We found that mutant huntingtin causes a reduction in TORC1, the most potent transcriptional activator of PGC-1α [79]. Mutant huntingtin also increases transglutaminase (Tgase) activity, which impairs transcription of PGC-1α, whereas Tgase inhibitors reverse this impairment both in vitro as well as in Drosophila [80]. Mice that had PGC-1α knocked-out and mutant huntingtin knockin showed worsened motor performance and earlier degeneration of striatum than mutant huntingtin knockin mice, as well as an increased susceptibility to the mitochondrial toxin 3-NP in HD transgenic mice [73]. Conversely, administration of a lentiviral vector expressing PGC-1α into the striatum of R6/2 mice was neuroprotective in that it increased the mean neuronal volume of medium spiny neurons [73]. Microarray expression data from the caudate nucleus of HD patient postmortem brain tissue showed that there was reduced expression of 24 out of 26 PGC-1α target genes [74]. We showed that the ability to upregulate PGC-1α in response to an energetic stress, produced by administration of a creatine analogue, guanidinopropionic acid was markedly impaired in HD transgenic mice [23, 75]. We also showed that the pathologic grade-dependent significant reduction in numbers of mitochondria in striatal spiny neurons correlated with reductions in PGC-1α and Tfam [20]. Sequence variation in the PGC-1α gene modifies the age of onset of HD [81, 82].
The functional importance of PGC-1α in glial cells in HD was highlighted by the studies of Xiang et al. [83]. They showed that expression of mutant huntingtin in primary oligodendrocytes results in decreased expression of PGC-1α, and decreased expression of myelin basic protein (MBP) and deficient myelination were found in the R6/2 transgenic mouse model of HD [83]. A decrease in MBP and deficient postnatal myelination also occurs in the striatum of PGC-1α knockout mice [83]. A role of PGC-1α in myelination was earlier suggested by studies in PGC-1α knock-out mice showing white matter abnormalities in the striatum as well as significantly reduced expression of myelin-associated oligodendrocyte basic protein (MOBP) [77]. Abnormalities in oligodendrocytes and white matter have been detected by structural MRI or pathological examinations in HD brains [84–90], even in subjects farthest from the predicted disease onset [84, 88, 91], suggesting that disruptions in white matter integrity may represent an early event in HD pathogenesis. Taken together, these results demonstrate another mechanism by which defective PGC-1α function may contribute to the pathogenesis of HD.
PGC-1α plays a critical role in mitochondrial biogenesis in muscle, and in influencing whether muscle contains slow-twitch fatigue resistant oxidative fibers (type I and IIA fibers, rich in mitochondria, use oxidative phosphorylation to generate ATP) or fast-twitch fatiguable glycolytic fibers (type IIX and IIB fibers, containing fewer mitochondria, use glycolysis to generate ATP) [92]. PGC-1α levels are normally high in muscle enriched with type I fibers, such as the soleus muscle, and very low in type II fiber rich muscles such as the extensor digitorum longus and the gastrocnemius [92]. In PGC-1α knockout mice, there is a shift from type I and type IIA towards type IIX and IIB muscle fibers which is accompanied by myopathy and exercise intolerance [93]. Using myosin heavy chain immunostaining and succinate dehydrogenase histochemistry, we showed a significant increase of type IIB fibers and a significant reduction of type I fibers in soleus muscle of two transgenic mouse models of HD, consistent with reduced PGC-1α levels and activity [21, 23]. Impaired generation of ATP in muscle and a myopathy occurs in gene-positive individuals at risk for HD, HD patients and HD transgenic mice [94–96]. We observed reduced PGC-1α activity in muscle of transgenic mouse models of HD, and in myoblasts and muscle biopsies from HD patients [21, 23].
PGC-1α also plays a role in the suppression of oxidative stress, and it induces mitochondrial uncoupling proteins and antioxidant enzymes, including copper/zinc superoxide dismutase (SOD1), manganese SOD (SOD2), and glutathione peroxidase (Gpx-1) [67]. Oxidative damage is a well characterized feature which is documented in plasma of HD patients, HD postmortem brain tissue, and in HD transgenic mice [40, 42, 97] (evidence discussed in detail under section 3 earlier in this review). Spongiform degeneration of striatum is a well-known feature of HD and similar lesions occur in SOD2 null mice [67, 98]. In concert with the reduced PGC-1α expression in the brains of R6/2 transgenic mice, we showed that the oxidative stress response genes such as hemeoxygenase-1 (HO-1), Nrf-2 and Gpx1 are also reduced, resulting in increased oxidative stress as shown by an increase in oxidative damage markers such as MDA and 8-OHdG [21].
Brown adipose tissue (BAT) is a highly specialized tissue consisting of lipid droplets surrounded by numerous mitochondria. PGC-1α plays an important role in induction of uncoupling protein 1 (UCP1) in BAT. UCP1 is an intramembrane protein that helps BAT cells generate heat, by a futile cycle in which protons re-enter the mitochondrial matrix, instead of driving the ATP synthase. This process is important in maintaining core body temperature in homeothermic mammals during exposure to cold, and is known as adaptive thermogenesis. In 2006, Weydt et al. [74] reported for the first time that the N171-82Q HD transgenic mice developed progressive hypothermia, beginning at 15–17 weeks of age and significant reductions in body temperature during cold challenge [74]. Although PGC-1α induction in N171-82Q HD mice upon cold exposure was intact, they found that UCP-1 mRNA up-regulation was severely blunted, suggesting that PGC-1α transcriptional activity is impaired in BAT from N171-82Q HD mice. These findings were recapitulated in studies from our laboratory in another transgenic mouse model of HD, the NLS-N171-82Q mice, which were also shown to have temperature dysregulation. The BAT from these mice and the R6/2 transgenic mice showed abnormal lipid-containing vacuoles [21, 75].
6. PGC-1α as a therapeutic target in HD
PGC-1α and PPARs
PGC-1α is now increasingly being recognized as an important therapeutic target for HD. As discussed above, there is plethora of evidence for impaired PGC-1α expression and/or function in HD, therefore pharmacologic/transcriptional activation of PGC-1α pathway is expected to have neuroprotective effects. Indeed, overexpression of PGC-1α was shown to enhance the mitochondrial membrane potential and to reduce mitochondrial toxicity in in vitro models of HD [74]. Lentiviral delivery of PGC-1α to the striatum of R6/2 HD mice completely prevented striatal atrophy at the site of PGC-1αα injection [73]. Another potential approach to activate the PGC-1α pathway, and thereby improve mitochondrial function, is via activation of peroxisome proliferator-activated receptors (PPARs). The PPARs are a subfamily of nuclear receptors, which are ligand-modulated transcription factors that regulate gene-expression programs of metabolic pathways. PPAR agonists increase oxidative phosphorylation capacity in mouse and human cells, and enhance mitochondrial biogenesis. Administration of a PPARγ agonist, thiazolidinedione, was shown to produce beneficial effects on weight loss, mutant huntingtin aggregates and global ubiquitination profiles in R6/2 mice [99]. Earlier, it was shown in STHdhQ111 cells, that PPARγ activation by rosiglitazone prevents the mitochondrial dysfunction and oxidative stress that occurs when mutant striatal cells are challenged with pathological increases in calcium [100]. We recently showed that bezafibrate, which is a pan-PPAR agonist, improved expression of PGC-1α and downstream target genes, improved behavioral deficits, survival, and striatal atrophy and reduced oxidative damage, in the R6/2 transgenic mouse model of HD [21]. There have been some concerns regarding the use of PPAR ligands as therapeutic agents [101, 102]. PPARα mediates the hepatocarcinogenic effect of prolonged use of certain peroxisome proliferators in rodents. However, no carcinogenic effect of peroxisome proliferators has been found in humans, possibly because expression of PPARα is much lower in human liver than in rodent liver or due to other species-specific differences. PPARγ has an anti-proliferative effect in pre-adipocytes and is protective in several malignant cell types. PPARγ ligands can induce terminal differentiation of human liposarcoma cells in vitro and in patients suffering from advanced liposarcoma [103]. PPARγ ligands also promote terminal differentiation of malignant breast epithelial cells in vitro, induce apoptosis and fibrosis of injected breast tumor cells (MCF-7) in mice, and reduce tumor incidence in rats treated with nitrosomethylurea and in mice injected with prostate tumor cells (PC-3) [104]. However, PPARγ ligands have been reported both to promote and to protect against colon cancer in mice and PPARβ has been linked to colon cancer [105].
PGC-1α, SIRT1 and AMPK
Sirtuins (silent information regulators) are members of the NAD+-dependent histone deacetylase family of proteins in yeast, and its homologs in mice and humans participate in a variety of cellular processes, including mitochondrial functions, cellular metabolism, energy metabolism, gluconeogenesis, cell survival, and aging. Although the role of the sirtuins in extending life-span has been contested and it remains unclear whether sirtuins play a direct role in anti-aging effects of caloric restriction, however, the important role of sirtuins as major regulators of metabolism cannot be ignored. The SIRT1 activator, resveratrol, increases the activity of PGC-1α and improves mitochondrial activity as a consequence of its deacetylation of PGC-1α, which increases its effects on liver, fat and muscle metabolism. The metabolic effects of resveratrol were shown to result from competitive inhibition of cAMP-degrading phosphodiesterases, leading to increased cAMP levels, which in turn leads to an increase in intracellular Ca+2 levels and activation of CamKKβ-AMPK pathway. This cascade culminates into an increase in NAD+ and activation of Sirt1 [106]. Resveratrol protects against 3-NP induced motor and behavioral deficits [107, 108]. We showed that resveratrol treatment of the N171-82Q HD transgenic mice, produced increased PGC-1α and reduced the apparent vacuolization in brown adipose tissue and reduced glucose levels, but there were no beneficial effects in the striatum, probably due to poor brain penetration [109]. There has been controversy surrounding the ability of this compound to extend life span in lower organisms, but when tested in regular C57Bl/6 mice, resveratrol though it produced improved health, similar to that produced by dietary restriction and delayed aging related deterioration, however it did not increase life span [110]. Overexpression of SIRT1 improves motor function, reduces brain atrophy and attenuates mutant huntingtin mediated metabolic abnormalities in three different transgenic mouse models of HD [111, 112]. Another sirtuin, which is of particular interest as a target for therapeutic intervention, is SIRT3. It is one of the three sirtuins that are located in mitochondria, where it interacts with Complex I of the respiratory chain and deacetylates several proteins in Complex I. It also increases fatty acid oxidation, SOD2 activity, levels of glutathione, and inhibits activation of the mitochondrial permeability transitions. SIRT3 was shown to be reduced in a cellular model of HD, and induction of SIRT3 with viniferin reversed the depletion and produced neuroprotective effects [113].
The antioxidant action of PGC-1α
Another potential mechanism by which PGC-1α confers neuroprotection is by its antioxidant activity. It is well known that PGC-1α plays an important role in the suppression of oxidative stress, and it induces mitochondrial uncoupling proteins and antioxidant enzymes, including SOD1, SOD2 (MnSOD), and Gpx-1 [67]. The mitochondrial antioxidant enzymes form the first line of defense against mitochondrial ROS, including SOD2, the enzyme that scavenges superoxide anion to produce H2O2, and peroxiredoxin III (Prx3), V (Prx5), mitochondrial thioredoxin (Trx2), and mitochondrial thioredoxin reductase (TrxR2). PGC-1α was found to be directly associated with the regulatory promoter sequences of SOD2, UCP-2 and Prx5. Importantly, UCP-2 has already been proposed to be a direct target of PGC-1α transcriptional regulation [72]. PGC-1α controls expression of SIRT3 in mitochondria, which in turn activates SOD2 by deacetylating it and reduces ROS [114–117]. The ability of SIRT3 activation to enhance fatty acid oxidation may also be beneficial [118]. Therefore, therapeutic approaches targeting PGC-1α may be beneficial both in improving mitochondrial function and biogenesis as well as in restoring the expression of antioxidant enzymes and ameliorating oxidative damage in HD. Agents which can transcriptionally activate the Nrf-2/ARE pathway, leading to increased expression of antioxidant enzymes, and chaperone proteins, as well as reduce the production of oxidants by iNOS and cyclooxygenase-2, are also promising agents for neuroprotection in HD [60]. An advantage of these approaches is that they modulate endogenous neuroprotective pathways, and that they involve catalytic processes which are not consumed but are regenerative, allowing ongoing antioxidant effects. Mitochondrial medicine and transcriptional modulation of antioxidant pathways therefore hold great promise for the development of neuroprotective therapies to ameliorate or halt the progression of HD.
Recently Tsunemi et al. [119] showed that increased PGC-1α function could ameliorate neuronal loss and some of the neurological symptoms of HD; by crossing mice inducibly overexpressing PGC-1α with a transgenic mouse model of HD. The authors found that PGC-1α overexpression virtually eradicates aggregates of mutant huntingtin protein in the brains of the HD mice. They further showed that PGC-1α’s ability to induce clearance of mutant huntingtin protein aggregates stems from its capacity to switch on the expression of TFEB, a master regulatory transcription factor that activates genes in the autophagy-lysosome pathway of protein turnover [119, 120]. These findings highlight the important role of PGC-1α in maintaining mitochondrial quality control, in accelerating mitochondrial biogenesis and increasing ATP generation.
7. Conclusions and Therapeutic Implications
There is now incontrovertible evidence that a deficiency of PGC-1α plays a role in the pathogenesis of HD. There are reduced levels of PGC-1α mRNA in striated cell lines with mutant huntingtin, transgenic mouse models of HD and in HD postmortem brain tissue. Furthermore one study showed that 24/26 PGC-1α co-activated genes were reduced in expression in HD postmortem brain tissue. The impairment of PGC-1α expression appears to be due to mutant huntingtin mediated interference with CREB/TAF4 mediated activation of the PGC-1α promoter, and we showed that there was also reduced TORC1, the most potent transcriptional activator of PGC-1α [79]. Increased transglutaminase activity occurs in HD, and it is a transcriptional modifier that reduces PGC-1α expression [80].
We showed that energy depletion produced by administration of guanidinopropionic acid leads to activation of AMPK, and up regulation of PGC-1α in both brain and muscle of normal mice, but the activation of PGC-1α and its downstream genes was completely blocked in a transgenic mouse model of HD [23, 75]. Similarly the ability of energy depletion to activate PGC-1α in myoblasts from HD patients was impaired. Transgenic mouse models of HD show features which are seen in PGC-1α deficient mice, including impaired temperature regulation, and lipid vacuolation of brown adipose tissue, reduced type I oxidative muscle fibers, and striatal degeneration. Furthermore, we showed that there were reduced numbers of mitochondria in striatal medium sized spiny neurons in HD brains, and that these reductions correlated with reduced levels of PGC-1α mRNA and protein. We showed for the first time, that an alternatively spliced short isoform of PGC-1α (38 kDa) is preferentially depleted in HD postmortem brain tissue and HD transgenic mice at early stages of the disease [76]. Increased expression of PGC-1α reduces atrophy of striatal neurons in transgenic mouse models of HD, reduces loss of striatal neurons and markedly reduces HD aggregates by increasing TFEB mediated autophagy.
There are a number of pharmacologic approaches to increasing PGC-1α including activation of PPAR nuclear receptors. Thiazolidinediones including pioglitazone activate PPARs, and increase PGC-1α, which exerts neuroprotective effects in both cellular and transgenic mouse models of HD [121, 122]. We showed that administration of the pan-PPAR agonist bezafibrate was efficacious in improving rotarod performance and survival, as well as striatal atrophy and atrophy of striatal medium spiny neurons [21]. It also ameliorated the depletion of type I muscle fibers and lipid vacuolation of brown adipose tissue. The numbers of mitochondria were depleted in both muscle fibers and striatal neurons of the R6/2 HD transgenic mice. Bezafibrate treatment produced a significant increase in numbers of mitochondria in both muscle fibers and in striatal neurons in R6/2 HD transgenic mice as determined using electron microscopy. There was also a reduction in immunostaining for MDA, and MDA levels by HPLC were found to be reduced in the striatum of R6/2 mice after bezafibrate treatment [21].
A number of other approaches to modulating PGC-1α and ameliorating mitochondrial dysfunction also have great promise. Activation of SIRT1 results in deacetylation of PGC-1α which increases its activity. Increased SIRT1 is neuroprotective in transgenic mouse models of HD, while a deficiency exacerbates the phenotype and reduces survival of HD transgenic mice [111, 112]. Another approach to activating sirtuins is to increase NAD+ levels which can be achieved with administration of nicotinamide precursors, such as nicotinamide riboside [123]. This has the advantage of activating both SIRT1 and SIRT3, leading to increased PGC-1α, as well as induction of antioxidant enzymes, and SIRT3 mediated increases in SOD2 and mitochondrial reduced glutathione.
Another consequence of PGC-1α deficiency is reduced expression of antioxidant enzymes and increased oxidative damage, which occurs in both HD transgenic mice and in postmortem brain tissue and body fluids of HD patients. A number of mitochondria targeted antioxidants are being developed. Administration of both coenzyme Q10 and creatine exert antioxidant and neuroprotective effects in transgenic mouse models of HD, and they are now in phase III clinical trials in HD. A particularly interesting approach is using the antioxidant TEMPOL linked to gramicidin (XJB-5-131), which localizes it to the inner mitochondrial membrane [124]. Administration of XJB-5-131 to a transgenic mouse model of HD resulted in improvement in mitochondrial function, reduced generation of ROS, reduced loss of striatal neurons, and amelioration of behavioral deficits [125].
Another approach to ameliorating oxidative damage is to activate the Nrf2/ARE transcriptional pathway, which leads to increased expression of hemeoxygenase 1, NADPH-oxidoreductase, antioxidant enzymes, heat shock proteins, and enzymes which synthesize glutathione. We showed that administration of the triterpenoids CDDO-ethylamide and CDDO-trifluoroethylamide reduced oxidative stress, improved motor impairment, reduced striatal atrophy and increased survival in a transgenic mouse model [60]. Another activator of the Nrf2/ARE pathway is dimethyl fumarate which showed efficacy in phase III clinical trials in multiple sclerosis. It produced neuroprotective effects in two different transgenic mouse models of HD [126].
In summary, there is substantial evidence that a deficiency of PGC-1α directly contributes to the mitochondrial dysfunction and oxidative damage which play an important role in HD pathogenesis. A number of new approaches are available which may address these deficits, and lead to effective neuroprotective agents to slow or halt the progression of HD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Group, H.s.D.C.R. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 3.Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N, DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- 4.Caviston JP, Holzbaur EL. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godin JD, Colombo K, Molina-Calavita M, Keryer G, Zala D, Charrin BC, Dietrich P, Volvert ML, Guillemot F, Dragatsis I, Bellaiche Y, Saudou F, Nguyen L, Humbert S. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Colin E, Zala D, Liot G, Rangone H, Borrell-Pages M, Li XJ, Saudou F, Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelieres FP, Marco S, Saudou F. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 2013;152:479–491. doi: 10.1016/j.cell.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Johri A, Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Ther. 2012;342:619–630. doi: 10.1124/jpet.112.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 12.Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011;121:493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seong IS, Ivanova E, Lee JM, Choo YS, Fossale E, Anderson M, Gusella JF, Laramie JM, Myers RH, Lesort M, MacDonald ME. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 14.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 15.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 16.Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beal MF, Brouillet E, Jenkins BG, Ferrante RJ, Kowall NW, Miller JM, Storey E, Srivastava R, Rosen BR, Hyman BT. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang SF, Hennessey T, Yang L, Starkova NN, Beal MF, Starkov AA. Impaired brain creatine kinase activity in Huntington’s disease. Neurodegener Dis. 2011;8:194–201. doi: 10.1159/000321681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mochel F, Durant B, Meng X, O’Callaghan J, Yu H, Brouillet E, Wheeler VC, Humbert S, Schiffmann R, Durr A. Early alterations of brain cellular energy homeostasis in huntington disease models. J Biol Chem. 2012;287:1361–1370. doi: 10.1074/jbc.M111.309849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Moody JP, Edgerly CK, Bordiuk OL, Cormier K, Smith K, Beal MF, Ferrante RJ. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johri A, Calingasan NY, Hennessey TM, Sharma A, Yang L, Wille E, Chandra A, Beal MF. Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2012a;21:1124–1137. doi: 10.1093/hmg/ddr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, Ciammola A, Squitieri F, Beal MF. Impaired PGC-1alpha function in muscle in Huntington’s disease. Hum Mol Genet. 2009;18:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirendeb U, Reddy AP, Manczak M, Calkins MJ, Mao P, Tagle DA, Reddy PH. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum Mol Genet. 2011;20:1438–1455. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa V, Giacomello M, Hudec R, Lopreiato R, Ermak G, Lim D, Malorni W, Davies KJ, Carafoli E, Scorrano L. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol Med. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bossy-Wetzel E, Petrilli A, Knott AB. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy PH, Mao P, Manczak M. Mitochondrial structural and functional dynamics in Huntington’s disease. Brain Res Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johri A, Chaturvedi RK, Beal MF. Hugging tight in Huntington’s. Nat Med. 2011a;17:245–246. doi: 10.1038/nm0311-245. [DOI] [PubMed] [Google Scholar]
- 29.Shirendeb UP, Calkins MJ, Manczak M, Anekonda V, Dufour B, McBride JL, Mao P, Reddy PH. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shacka JJ, Roth KA, Zhang J. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front Biosci. 2008;13:718–736. doi: 10.2741/2714. [DOI] [PubMed] [Google Scholar]
- 31.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, Lawler JF, Jr, Greenamyre JT, Snyder SH, Ross CA. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med. 1999;5:1194–1198. doi: 10.1038/13518. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jahani-Asl A, Cheung EC, Neuspiel M, MacLaurin JG, Fortin A, Park DS, McBride HM, Slack RS. Mitofusin 2 protects cerebellar granule neurons against injury-induced cell death. J Biol Chem. 2007;282:23788–23798. doi: 10.1074/jbc.M703812200. [DOI] [PubMed] [Google Scholar]
- 37.Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valencia A, Sapp E, Kimm JS, McClory H, Reeves PB, Alexander J, Ansong KA, Masso N, Frosch MP, Kegel KB, Li X, DiFiglia M. Elevated NADPH oxidase activity contributes to oxidative stress and cell death in Huntington’s disease. Hum Mol Genet. 2013;22:1112–1131. doi: 10.1093/hmg/dds516. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 41.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington’s disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stack EC, Matson WR, Ferrante RJ. Evidence of oxidant damage in Huntington’s disease: translational strategies using antioxidants. Ann N Y Acad Sci. 2008;1147:79–92. doi: 10.1196/annals.1427.008. [DOI] [PubMed] [Google Scholar]
- 43.Klepac N, Relja M, Klepac R, Hecimovic S, Babic T, Trkulja V. Oxidative stress parameters in plasma of Huntington’s disease patients, asymptomatic Huntington’s disease gene carriers and healthy subjects: a cross-sectional study. J Neurol. 2007;254:1676–1683. doi: 10.1007/s00415-007-0611-y. [DOI] [PubMed] [Google Scholar]
- 44.Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 45.Tasset I, Sanchez F, Tunez I. The molecular bases of Huntington’s disease: the role played by oxidative stress. Rev Neurol. 2009;49:424–429. [PubMed] [Google Scholar]
- 46.Stoy N, Mackay GM, Forrest CM, Christofides J, Egerton M, Stone TW, Darlington LG. Tryptophan metabolism and oxidative stress in patients with Huntington’s disease. J Neurochem. 2005;93:611–623. doi: 10.1111/j.1471-4159.2005.03070.x. [DOI] [PubMed] [Google Scholar]
- 47.Chen CM, Wu YR, Cheng ML, Liu JL, Lee YM, Lee PW, Soong BW, Chiu DT. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington’s disease patients. Biochem Biophys Res Commun. 2007;359:335–340. doi: 10.1016/j.bbrc.2007.05.093. [DOI] [PubMed] [Google Scholar]
- 48.Hersch SM, Gevorkian S, Marder K, Moskowitz C, Feigin A, Cox M, Como P, Zimmerman C, Lin M, Zhang L, Ulug AM, Beal MF, Matson W, Bogdanov M, Ebbel E, Zaleta A, Kaneko Y, Jenkins B, Hevelone N, Zhang H, Yu H, Schoenfeld D, Ferrante R, Rosas HD. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2’dG. Neurology. 2006;66:250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- 49.Tunez I, Sanchez-Lopez F, Aguera E, Fernandez-Bolanos R, Sanchez FM, Tasset-Cuevas I. Important Role of Oxidative Stress Biomarkers in Huntington’s Disease. J Med Chem. 2011;54:5602–5606. doi: 10.1021/jm200605a. [DOI] [PubMed] [Google Scholar]
- 50.Braak H, Braak E. Allocortical involvement in Huntington’s disease. Neuropathol Appl Neurobiol. 1992;18:539–547. doi: 10.1111/j.1365-2990.1992.tb00824.x. [DOI] [PubMed] [Google Scholar]
- 51.Tellez-Nagel I, Johnson AB, Terry RD. Studies on brain biopsies of patients with Huntington’s chorea. J Neuropathol Exp Neurol. 1974;33:308–332. doi: 10.1097/00005072-197404000-00008. [DOI] [PubMed] [Google Scholar]
- 52.Butterworth NJ, Williams L, Bullock JY, Love DR, Faull RL, Dragunow M. Trinucleotide (CAG) repeat length is positively correlated with the degree of DNA fragmentation in Huntington’s disease striatum. Neuroscience. 1998;87:49–53. doi: 10.1016/s0306-4522(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 53.Portera-Cailliau C, Hedreen JC, Price DL, Koliatsos VE. Evidence for apoptotic cell death in Huntington disease and excitotoxic animal models. J Neurosci. 1995;15:3775–3787. doi: 10.1523/JNEUROSCI.15-05-03775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorolla MA, Rodriguez-Colman MJ, Tamarit J, Ortega Z, Lucas JJ, Ferrer I, Ros J, Cabiscol E. Protein oxidation in Huntington disease affects energy production and vitamin B6 metabolism. Free Radic Biol Med. 2010;49:612–621. doi: 10.1016/j.freeradbiomed.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 55.Lim D, Fedrizzi L, Tartari M, Zuccato C, Cattaneo E, Brini M, Carafoli E. Calcium homeostasis and mitochondrial dysfunction in striatal neurons of Huntington disease. J Biol Chem. 2008;283:5780–5789. doi: 10.1074/jbc.M704704200. [DOI] [PubMed] [Google Scholar]
- 56.Tabrizi SJ, Cleeter MW, Xuereb J, Taanman JW, Cooper JM, Schapira AH. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann Neurol. 1999;45:25–32. doi: 10.1002/1531-8249(199901)45:1<25::aid-art6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 57.Patel M, Day BJ, Crapo JD, Fridovich I, McNamara JO. Requirement for superoxide in excitotoxic cell death. Neuron. 1996;16:345–355. doi: 10.1016/s0896-6273(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 58.Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, Calabrese V, Cini C, De Marco C, Butterfield DA. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol Cell Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Acevedo-Torres K, Berrios L, Rosario N, Dufault V, Skatchkov S, Eaton MJ, Torres-Ramos CA, Ayala-Torres S. Mitochondrial DNA damage is a hallmark of chemically induced and the R6/2 transgenic model of Huntington’s disease. DNA Repair (Amst) 2009;8:126–136. doi: 10.1016/j.dnarep.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stack C, Ho D, Wille E, Calingasan NY, Williams C, Liby K, Sporn M, Dumont M, Beal MF. Triterpenoids CDDO-ethyl amide and CDDO-trifluoroethyl amide improve the behavioral phenotype and brain pathology in a transgenic mouse model of Huntington’s disease. Free Radic Biol Med. 2010;49:147–158. doi: 10.1016/j.freeradbiomed.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bogdanov MB, Andreassen OA, Dedeoglu A, Ferrante RJ, Beal MF. Increased oxidative damage to DNA in a transgenic mouse model of Huntington’s disease. J Neurochem. 2001;79:1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- 62.Siddiqui A, Rivera-Sanchez S, del Castro MR, Acevedo-Torres K, Rane A, Torres-Ramos CA, Nicholls DG, Andersen JK, Ayala-Torres S. Mitochondrial DNA damage is associated with reduced mitochondrial bioenergetics in Huntington’s disease. Free Radic Biol Med. 2012;53:1478–1488. doi: 10.1016/j.freeradbiomed.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang JQ, Chen Q, Wang X, Wang QC, Wang Y, Cheng HP, Guo C, Sun Q, Tang TS. Dysregulation of mitochondrial calcium signaling and superoxide flashes cause mitochondrial genomic DNA damage in Huntington disease. J Biol Chem. 2013;288:3070–3084. doi: 10.1074/jbc.M112.407726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 65.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 66.Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 67.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 68.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 70.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 71.St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 72.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 73.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 74.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 75.Chaturvedi RK, Calingasan NY, Yang L, Hennessey T, Johri A, Beal MF. Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington’s disease following chronic energy deprivation. Hum Mol Genet. 2010;19:3190–3205. doi: 10.1093/hmg/ddq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johri A, Starkov AA, Chandra A, Hennessey T, Sharma A, Orobello S, Squitieri F, Yang L, Beal MF. Truncated peroxisome proliferator-activated receptor-gamma coactivator 1alpha splice variant is severely altered in Huntington’s disease. Neurodegener Dis. 2011b;8:496–503. doi: 10.1159/000327910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 78.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chaturvedi RK, Hennessey T, Johri A, Tiwari SK, Mishra D, Agarwal S, Kim YS, Beal MF. Transducer of regulated CREB-binding proteins (TORCs) transcription and function is impaired in Huntington’s disease. Hum Mol Genet. 2012;21:3474–3488. doi: 10.1093/hmg/dds178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McConoughey SJ, Basso M, Niatsetskaya ZV, Sleiman SF, Smirnova NA, Langley BC, Mahishi L, Cooper AJ, Antonyak MA, Cerione RA, Li B, Starkov A, Chaturvedi RK, Beal MF, Coppola G, Geschwind DH, Ryu H, Xia L, Iismaa SE, Pallos J, Pasternack R, Hils M, Fan J, Raymond LA, Marsh JL, Thompson LM, Ratan RR. Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol Med. 2010;2:349–370. doi: 10.1002/emmm.201000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weydt P, Soyal SM, Gellera C, Didonato S, Weidinger C, Oberkofler H, Landwehrmeyer GB, Patsch W. The gene coding for PGC-1alpha modifies age at onset in Huntington’s Disease. Mol Neurodegener. 2009;4:3. doi: 10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taherzadeh-Fard E, Saft C, Andrich J, Wieczorek S, Arning L. PGC-1alpha as modifier of onset age in Huntington disease. Mol Neurodegener. 2009;4:10. doi: 10.1186/1750-1326-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiang Z, Valenza M, Cui L, Leoni V, Jeong HK, Brilli E, Zhang J, Peng Q, Duan W, Reeves SA, Cattaneo E, Krainc D. Peroxisome-Proliferator-Activated Receptor Gamma Coactivator 1 {alpha} Contributes to Dysmyelination in Experimental Models of Huntington’s Disease. J Neurosci. 2011;31:9544–9553. doi: 10.1523/JNEUROSCI.1291-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gomez-Tortosa E, MacDonald ME, Friend JC, Taylor SA, Weiler LJ, Cupples LA, Srinidhi J, Gusella JF, Bird ED, Vonsattel JP, Myers RH. Quantitative neuropathological changes in presymptomatic Huntington’s disease. Ann Neurol. 2001;49:29–34. [PubMed] [Google Scholar]
- 85.Fennema-Notestine C, Archibald SL, Jacobson MW, Corey-Bloom J, Paulsen JS, Peavy GM, Gamst AC, Hamilton JM, Salmon DP, Jernigan TL. In vivo evidence of cerebellar atrophy and cerebral white matter loss in Huntington disease. Neurology. 2004;63:989–995. doi: 10.1212/01.wnl.0000138434.68093.67. [DOI] [PubMed] [Google Scholar]
- 86.Reading SA, Yassa MA, Bakker A, Dziorny AC, Gourley LM, Yallapragada V, Rosenblatt A, Margolis RL, Aylward EH, Brandt J, Mori S, van Zijl P, Bassett SS, Ross CA. Regional white matter change in pre-symptomatic Huntington’s disease: a diffusion tensor imaging study. Psychiatry Res. 2005;140:55–62. doi: 10.1016/j.pscychresns.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 87.Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, Guttman M, Johnson S, MacDonald M, Beglinger LJ, Duff K, Kayson E, Biglan K, Shoulson I, Oakes D, Hayden M. Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. J Neurol Neurosurg Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Durr A, Craufurd D, Kennard C, Hicks SL, Fox NC, Scahill RI, Borowsky B, Tobin AJ, Rosas HD, Johnson H, Reilmann R, Landwehrmeyer B, Stout JC. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8:791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rosas HD, Lee SY, Bender AC, Zaleta AK, Vangel M, Yu P, Fischl B, Pappu V, Onorato C, Cha JH, Salat DH, Hersch SM. Altered white matter microstructure in the corpus callosum in Huntington’s disease: implications for cortical “disconnection”. Neuroimage. 2010;49:2995–3004. doi: 10.1016/j.neuroimage.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nopoulos PC, Aylward EH, Ross CA, Mills JA, Langbehn DR, Johnson HJ, Magnotta VA, Pierson RK, Beglinger LJ, Nance MA, Barker RA, Paulsen JS. Smaller intracranial volume in prodromal Huntington’s disease: evidence for abnormal neurodevelopment. Brain. 2011;134:137–142. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bartzokis G, Lu PH, Tishler TA, Fong SM, Oluwadara B, Finn JP, Huang D, Bordelon Y, Mintz J, Perlman S. Myelin breakdown and iron changes in Huntington’s disease: pathogenesis and treatment implications. Neurochem Res. 2007;32:1655–1664. doi: 10.1007/s11064-007-9352-7. [DOI] [PubMed] [Google Scholar]
- 92.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 93.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 94.Gizatullina ZZ, Lindenberg KS, Harjes P, Chen Y, Kosinski CM, Landwehrmeyer BG, Ludolph AC, Striggow F, Zierz S, Gellerich FN. Low stability of Huntington muscle mitochondria against Ca2+ in R6/2 mice. Ann Neurol. 2006;59:407–411. doi: 10.1002/ana.20754. [DOI] [PubMed] [Google Scholar]
- 95.Kosinski CM, Schlangen C, Gellerich FN, Gizatullina Z, Deschauer M, Schiefer J, Young AB, Landwehrmeyer GB, Toyka KV, Sellhaus B, Lindenberg KS. Myopathy as a first symptom of Huntington’s disease in a Marathon runner. Mov Disord. 2007;22:1637–1640. doi: 10.1002/mds.21550. [DOI] [PubMed] [Google Scholar]
- 96.Turner C, Cooper JM, Schapira AH. Clinical correlates of mitochondrial function in Huntington’s disease muscle. Mov Disord. 2007;22:1715–1721. doi: 10.1002/mds.21540. [DOI] [PubMed] [Google Scholar]
- 97.Johri A, Beal MF. Antioxidants in Huntington’s disease. Biochim Biophys Acta. 2012b;1822:664–674. doi: 10.1016/j.bbadis.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hinerfeld D, Traini MD, Weinberger RP, Cochran B, Doctrow SR, Harry J, Melov S. Endogenous mitochondrial oxidative stress: neurodegeneration, proteomic analysis, specific respiratory chain defects, and efficacious antioxidant therapy in superoxide dismutase 2 null mice. J Neurochem. 2004;88:657–667. doi: 10.1046/j.1471-4159.2003.02195.x. [DOI] [PubMed] [Google Scholar]
- 99.Chiang MC, Chen CM, Lee MR, Chen HW, Chen HM, Wu YS, Hung CH, Kang JJ, Chang CP, Chang C, Wu YR, Tsai YS, Chern Y. Modulation of energy deficiency in Huntington’s disease via activation of the peroxisome proliferator-activated receptor gamma. Hum Mol Genet. 2010;19:4043–4058. doi: 10.1093/hmg/ddq322. [DOI] [PubMed] [Google Scholar]
- 100.Quintanilla RA, Jin YN, Fuenzalida K, Bronfman M, Johnson GV. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J Biol Chem. 2008;283:25628–25637. doi: 10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Viscomi C, Bottani E, Civiletto G, Cerutti R, Moggio M, Fagiolari G, Schon EA, Lamperti C, Zeviani M. In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metab. 2011;14:80–90. doi: 10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yatsuga S, Suomalainen A. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum Mol Genet. 2012;21:526–535. doi: 10.1093/hmg/ddr482. [DOI] [PubMed] [Google Scholar]
- 103.Demetri GD, Fletcher CD, Mueller E, Sarraf P, Naujoks R, Campbell N, Spiegelman BM, Singer S. Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-gamma ligand troglitazone in patients with liposarcoma. Proc Natl Acad Sci U S A. 1999;96:3951–3956. doi: 10.1073/pnas.96.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suh N, Wang Y, Williams CR, Risingsong R, Gilmer T, Willson TM, Sporn MB. A new ligand for the peroxisome proliferator-activated receptor-gamma (PPAR-gamma), GW7845, inhibits rat mammary carcinogenesis. Cancer Res. 1999;59:5671–5673. [PubMed] [Google Scholar]
- 105.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 106.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, Chung JH. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kumar P, Padi SS, Naidu PS, Kumar A. Effect of resveratrol on 3-nitropropionic acid-induced biochemical and behavioural changes: possible neuroprotective mechanisms. Behav Pharmacol. 2006;17:485–492. doi: 10.1097/00008877-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 108.Binienda ZK, Beaudoin MA, Gough B, Ali SF, Virmani A. Assessment of 3-nitropropionic acid-evoked peripheral neuropathy in rats: neuroprotective effects of acetyl-l-carnitine and resveratrol. Neurosci Lett. 2010;480:117–121. doi: 10.1016/j.neulet.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 109.Ho DJ, Calingasan NY, Wille E, Dumont M, Beal MF. Resveratrol protects against peripheral deficits in a mouse model of Huntington’s disease. Exp Neurol. 2010;225:74–84. doi: 10.1016/j.expneurol.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 110.Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang M, Wang J, Fu J, Du L, Jeong H, West T, Xiang L, Peng Q, Hou Z, Cai H, Seredenina T, Arbez N, Zhu S, Sommers K, Qian J, Zhang J, Mori S, Yang XW, Tamashiro KL, Aja S, Moran TH, Luthi-Carter R, Martin B, Maudsley S, Mattson MP, Cichewicz RH, Ross CA, Holtzman DM, Krainc D, Duan W. Neuroprotective role of Sirt1 in mammalian models of Huntington’s disease through activation of multiple Sirt1 targets. Nat Med. 2011;18:153–158. doi: 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jeong H, Cohen DE, Cui L, Supinski A, Savas JN, Mazzulli JR, Yates JR, 3rd, Bordone L, Guarente L, Krainc D. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2011;18:159–165. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fu J, Jin J, Cichewicz RH, Hageman SA, Ellis TK, Xiang L, Peng Q, Jiang M, Arbez N, Hotaling K, Ross CA, Duan W. trans-(−)-epsilon-Viniferin increases mitochondrial sirtuin 3 (SIRT3), activates AMP-activated protein kinase (AMPK), and protects cells in models of Huntington Disease. J Biol Chem. 2012;287:24460–24472. doi: 10.1074/jbc.M112.382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, Gius D. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany NY) 2011;3:102–107. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 116.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E, La Spada AR. PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chiang MC, Chern Y, Huang RN. PPARgamma rescue of the mitochondrial dysfunction in Huntington’s disease. Neurobiol Dis. 2012;45:322–328. doi: 10.1016/j.nbd.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 122.Jin YN, Hwang WY, Jo C, Johnson GV. Metabolic state determines sensitivity to cellular stress in Huntington disease: normalization by activation of PPARgamma. PLoS One. 2012;7:e30406. doi: 10.1371/journal.pone.0030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang T, Chan NY, Sauve AA. Syntheses of nicotinamide riboside and derivatives: effective agents for increasing nicotinamide adenine dinucleotide concentrations in mammalian cells. J Med Chem. 2007;50:6458–6461. doi: 10.1021/jm701001c. [DOI] [PubMed] [Google Scholar]
- 124.Jiang J, Kurnikov I, Belikova NA, Xiao J, Zhao Q, Amoscato AA, Braslau R, Studer A, Fink MP, Greenberger JS, Wipf P, Kagan VE. Structural requirements for optimized delivery, inhibition of oxidative stress, and antiapoptotic activity of targeted nitroxides. J Pharmacol Exp Ther. 2007;320:1050–1060. doi: 10.1124/jpet.106.114769. [DOI] [PubMed] [Google Scholar]
- 125.Xun Z, Rivera-Sanchez S, Ayala-Pena S, Lim J, Budworth H, Skoda EM, Robbins PD, Niedernhofer LJ, Wipf P, McMurray CT. Targeting of XJB-5-131 to Mitochondria Suppresses Oxidative DNA Damage and Motor Decline in a Mouse Model of Huntington’s Disease. Cell Rep. 2012;2:1137–1142. doi: 10.1016/j.celrep.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellrichmann G, Petrasch-Parwez E, Lee DH, Reick C, Arning L, Saft C, Gold R, Linker RA. Efficacy of fumaric acid esters in the R6/2 and YAC128 models of Huntington’s disease. PLoS One. 2011;6:e16172. doi: 10.1371/journal.pone.0016172. [DOI] [PMC free article] [PubMed] [Google Scholar]