Abstract
Previous work showed that cell imprinting in a polydimethylsiloxane (PDMS) film produced artificial receptors to cells by template-assisted rearrangement of functional groups on the surface of the polymer thin film which facilitated cell capture in the polymer surface indentations by size, shape, and most importantly chemical recognition. We report here that inactivation of cells by treatment with formaldehyde (4%), or glutaraldehyde (2%), or a combination of the two leads to markedly improved capture selectivity (a factor of 3) when cells to be analyzed are inactivated in the same manner. The enhanced capture efficiency compared to living cells results from two factors: (1) rigidification of the cell surface through crosslinking of amine groups by the aldehyde; and (2) elimination of chemicals excreted from living cells which interfere with the fidelity of the cell imprinting process. Moreover, cell inactivation has the advantage of removing biohazard risks associated with working with virulent bacteria. These results are demonstrated using different strains of mycobacterium tuberculosis.
Keywords: Cell imprinting, cell inactivation, cell sorting, diagnosis, tuberculosis
Cell imprinting is a recently developed technology that captures the structural and chemical information of cells on a polymer surface through template-assisted assembly of functional groups.1-5 A polymer is cured around template cells that are removed subsequently, leaving complementary cavities that not only spatially fit but also chemically recognize the target cells.3 The cell-imprinted materials exhibit specific chemical affinity to the original template cells, thereby functioning as artificial receptors. Such receptors are much less expensive to produce and more durable than natural receptors; consequently, these cell-imprinted polymer films could potentially be broadly used in cell sorting assays. One promising potential application is the detection of pathogens causing infectious diseases, in particular, as a diagnostic tool for tuberculosis.
Mycobacterium tuberculosis is one of the three leading causes of morbidity and mortality in humans.6 Quick and accurate diagnoses of tuberculosis are the key to choosing proper antimicrobial treatment and preventing further spread. However, one major challenge is that the tubercle bacilli in patient sputum samples are at variable concentrations and embedded in a complex mixture of oropharyngeal contaminating microorganisms. To distinguish the suspected pathogen from the other constituents of the sample, microscopic examination combined with differential staining of the acid-fast bacilli on a glass slide is most commonly performed worldwide. However, this strategy suffers from low sensitivity and therefore misses about 50% of the cases. To achieve a conclusive diagnosis, a selective microbiological cell culture is performed, which requires sophisticated biosafety facilities and is limited by the slow growth rate of microorganisms.7
We are working on a potential solution based on cell-imprinted polymer thin films, which may overcome the limitations mentioned above. This strategy selectively captures the target cells in a sample onto a cell-imprinted polymer thin film. There have been attempts to use bioactive molecules like antibodies as a stationary phase for extraction to realize a similar strategy.8-10 However, the bioactive molecules are expensive to prepare, fragile to handle, and not easy to store. Alternatively, based on our recent findings,2 we believe cell imprinting could be a promising strategy to achieve this goal.
When live cells are used as the template for imprinting, several major challenges arise.11 First, live cells are fragile during imprinting, and effective means are needed to ensure the conformational integrity of the template cells. Second, living cells can actively adapt to the environment by secreting chemicals, which thereby could detrimentally affect recognition by the cell-imprinted polymer film, causing the capturing selectivity to be decreased. Last, but not the least, the involvement of pathogens in the production of the imprinted material as well as the cell capturing process brings occupational risk of infection to those working with them.
We describe here a method that can effectively overcome the challenges mentioned above. Recently, we discovered that inactivated bacteria can be selectively captured by a polymer film imprinted with the bacteria after having been treated with the same inactivation process, avoiding the use of live virulent bacteria; moreover, the inactivation strategies, especially those that utilize chemical reagents, resulted in better selectivity of capture than when living cells were used. In this study, we present the results we obtained on the effect of inactivation methods on enriching for identification of M. smegmatis, a surrogate of M. tuberculosis, compared with other bacteria, including Escherichia. coli and Staphylococcus aureus. In addition, we propose two mechanisms that help to explain why cell imprinting with inactivated bacteria can be superior to that of live bacteria. Finally, we demonstrate optimization of the inactivation method based on the mechanisms we have proposed.
Results and Discussion
The bacterial model system used in these experiments was chosen to mimic the identification of M. tuberculosis bacilli (MTB), an extremely slow-growing bacterium(one division every 20 hours) that causes millions of infections per year worldwide.12 A major challenge for TB control programs is the lack of a rapid and low-cost diagnostic strategy. TB infections are most commonly diagnosed by acid-fast staining smears of sputum samples.13 The work described here is a preliminary exploration of a potential strategy that could selectively capture MTB cells from a patient sample to a small region of a test slide, thereby increasing the sensitivity of a smear-based TB diagnosis.
In our preliminary experiment, PDMS films imprinted with living M. smegmatis showed preferential binding of living M. smegmatis as compared to E. coli and S. epidermidis. However, the use of virulent bacteria during the production of the cell-imprinted polymer thin films and the cell-capture process may bring risk of infection, which could be a major hurdle for implementation of this method. The biohazard risk could be fully removed if the imprinting-capture process is based on inactivated bacteria, which have lost their biological activity but still retains their shape and surface characteristics.
We screened both physical (UV) and chemical inactivation methods to determine the most appropriate method for our cell-imprinting strategy and compared these procedures to imprinting with live bacteria (Figure 1A). In our experiments, we carefully chose chemicals that are already confirmed by literature to be effective in killing MTB. The chemicals we have tested include 75%v/v ethanol, 5%wt bleach, 1%wt hydrogen peroxide, and 4%wt formaldehyde; the concentrations were chosen according to literature reports on sufficient dosages to kill MTB.14 Also, as UV has been widely used for sterilization 15 including killing Mycobacteria in milk,16 UV was also tested to inactivate the bacteria in our experiments. Every reagent effectively inactivated M. smegmatis, E.coli and S. aureus at the tested concentration.
Figure 1.
Bright-field images of M.smegmatis before and after inactivation. (A) untreated M.smegmatis and inactivated M.smegmatis using (B) 1% hydrogen peroxide for 10 min, (C) 4% formaldehyde for 10 min and (D) 254 nm UV from a 6 W mercury lamp for 5 min. The cells were stained using Kinyoun’s acid-fast stain.17
After inactivation, the morphology of bacteria was inspected under the microscope. It was found that ethanol (not shown), bleach (not shown) and hydrogen peroxide (Figure 1B) dissolved or thinned the cell wall of M. smegmatis, while formaldehyde kept the cell morphology largely intact (Figure 1C). Also, we found UV light with a 254 nm peak emission to be effective in inactivating M. smegmatis and led to no detectable change to the cell morphology (Figure 1D).
Figure 1 shows that the morphology of the cells is maintain upon exposure to UV or formaldehyde but not hydrogen peroxide. Using inactivated cells, the capturing performance of the cell-imprinted PDMS films were tested. Fluorescently tagged bacteria in PBS were flowed over the imprinted region with the assistance of an array of microfluidic channels3, and the captured bacteria were inspected under a fluorescence microscope. Figure 2 presents the number of counted cells in each inspected area for each different procedure. It was found that the imprinted area captured significantly more bacteria than the non-imprinted area.
Figure 2.

Comparison of cell capturing performance on imprinted PDMS and on native PDMS imprinted with M. smegmatis. Living bacteria and bacteria inactivated by UV or 4% formaldehyde were used for the tests; for each test, the template and the sample were prepared with the same method. The concentration of M. smegmatis, S.aureus, and E.coli in the sample was prepared at a ratio of 1: 3: 3 to facilitate comparison of capture rate.
Furthermore, we found that the selectivity of capturing, defined as the ratio of captured target bacteria to other bacteria, was higher when inactivated bacteria were used in the process, compared to living bacteria In addition, formaldehyde treated bacteria led to better selectivity than when UV treated bacteria were used. Table 1 summarizes the ratios of selectivity in capturing, calculated from the data shown in Fig. 2.
Table 1.
selectivity of capture, calculated by dividing the number of captured M. smegmatis by the number of captured E. coli, on the imprints of M. smegmatis.
| Inactivation method | Without inactivation | 4% Formaldehyde | UV Exposure |
|---|---|---|---|
| Selectivity ratio | 7.4 | 13.8 | 9.1 |
Additional experiments were conducted to understand better the effect of inactivation on imprinting and capture performance. A common function of inactivation methods is to terminate the biological activity of cells. We observed that this function effectively eliminated the formation of a thin film on the template cells as shown in Fig. 3. The template for imprinting was prepared by sedimentation of cells to form a single layer on a substrate. When living bacteria were used in the template preparation, formation of a thin film was observed (Figure 3A, C). These thin films, which are thought to be formed by extracellular matrix material secreted by cells in response to their environment, may reduce the recognition effect by blocking the surface of the bacteria. When inactivated cells were used, no thin film was observed on the template (Figure 3B, D). The absence of this thin film ensures that the surfaces of templated bacteria are exposed to the imprinting polymer during the imprinting step. This was confirmed using atomic force microscopy (AFM), which showed that the imprinted surface obtained from inactivated bacteria had more distinguishable features than those from living bacteria (Figure 4). Quantitative measurement showed that the average depth of the cavities from inactivated M. smegmatis was 285±6 nm, while that from living M. smegmatis was 261±7 nm.
Figure 3.
(A, B) SEM images and (C, D) fluorescence images of cell-imprinting templates made with (A, C) living and (B, D) 4% formaldehyde-inactivated M. smegmatis. The cell suspension in PBS was used for seeding the cells to the substrate. After sedimentation of the cells at 4 °C overnight the buffer was removed by centrifugation. The arrow in (A) points to an edge of a thin film peeled from the substrate. The samples in (C, D) were stained with auramine-rhodamine dye, which binds to mycolic acids, a characteristic component in the cell wall and the extracellular matrix of mycobacterium.
Figure 4.
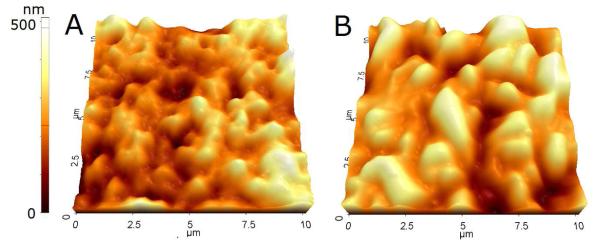
AFM images of PDMS surface imprinted with (A) living and (B) 4% formaldehyde-inactivated M. smegmatis.
However, the absence of a thin film is not the only reason for the observed improvement in selectivity when using inactivated cells. We observed that better selectivity was demonstrated by formaldehyde-inactivated cells as compared to UV-inactivated cells. To interpret the underlying mechanism, we looked into the mechanisms of their inactivation effects. Surface-maintaining inactivation methods can be divided into two categories based on their effects on the cell wall. Physical inactivation methods, such as UV and heating treatment, terminate the biological activity by damaging the DNA molecules and/or denaturing the proteins while generally leaving the surface morphology of the cells unchanged.18 Chemical inactivation methods, particularly, those based on chemical fixatives, involve chemical crosslinking of functional groups on cell surface, primarily amine groups when aldehydes are used. The effect of crosslinking makes the morphology and the surface of cells more rigid. Therefore, chemically fixed cells provide better capture performance than UV-inactivated cells in the cell-imprinting process, because the surface structure of the cells are better preserved and more rigidified. As cell imprinting is based on the recognition of the cell surface, we believe that the chemical fixatives provide superior performance than physical inactivation methods when used for imprinting, as they help to better preserve the surface of the cells.
We carried out experiments with different fixatives to prove this deduction and to find the most effective fixatives for our inactivated cell-imprinting strategy. The fixatives used in these experiments were chosen based on literature reports19 that demonstrated by microscopic inspection that cell morphology was maintained after treatment. Formaldehyde and glutaraldehyde are commonly recommended as the best chemicals to preserve the cell’s surface structures.20, 21 In our experiments, these two aldehydes were used alone or in combination at concentrations below that where significant cell shrinkage is known to occur.22 Figure 5 shows that a combination of 4% formaldehyde and 2% glutaraldehyde was the most effective inactivation solution for cell imprinting. The selectivity ratio of capture under this condition was approximately 20 while with glutaraldehyde or formaldehyde alone was slightly below 15. Without fixative, the selectivity ratio was approximately 7.6.
Figure 5.

Selectivity of cell capturing on cell imprints obtained with cells treated by different inactivation methods. The selectivity is calculated by dividing the number of template cells (M.smegmatis) with the number of other cells (E.coli), captured on the imprints of templated cell on PDMS. For each test the template cells and the sample were prepared with the same method.
The optimized cell inactivation formula, a combination of 4% formaldehyde and 2% glutaraldehyde, was further evaluated with various MTB strains (MTB H37Ra, MTB H37 Rv and MTB ΔlprG) to verify its general applicability to virulent cells. Cell culture experiments confirmed effective inactivation of all the strains tested. Table 2 presents the selectivity of cell capture, calculated by dividing the number of template cells with the number of other cells captured on the imprints of templated cells on PDMS. The results confirmed that by inactivation with aldehydes that removes biohazard risks associated with the procedure, the virulent strains of MTB cells, from a mixture containing other cells, could be selectively captured a cell-imprinted polymer thin film.
Table 2.
Selectivity of cell capture, calculated by dividing the number of template cells with the number of other cells (E.coli) captured on the imprints of templated cell on PDMS.
| Templated cell strain | MTB H37Ra | MTB H37Rv | MTB ΔlprG |
|---|---|---|---|
| Selectivity ratio | 15.8 | 16.2 | 12.6 |
Conclusions
We found that if the cells used in creating the template for cell imprinting and the cells present in the sample are inactivated in the same way, inactivated cells could be used with advantage in the imprinting-capturing strategy we proposed previously with living cells. When inactivated bacteria are employed in the entire process, the concern about the occupational risk of using virulent bacteria in preparation and utilization of the imprinted polymer is eliminated. In our previous work, we have found that the mechanism of cell capture with cell-imprinted polymer involves both shape and chemical recognition.3 The inactivation process employed in our current strategy may have played two roles: (1) to terminate the biological activity of the cells and therefore eliminating secretion of extracellular matrix, which helps expose the surface of the inactivated cells to the imprinting polymer; (2) to fix the morphology and surface structure of the inactivated cells, which helps to preserve the structural and chemical information on the cell surface. In general, physical inactivation methods only realize the first, whereas chemical fixatives can achieve both these functions. This work suggests that when cells, especially, pathogenic cells, are used for imprinting, the inactivated form may be a better choice not only for better selectivity but also for increased biosafety.
Experimental
Materials
M. smegmatis mc2 155, MTB H37Ra, MTB H37 Rv, MTB ΔlprG, E. coli (ATCC25922) and S. aureus (ATCC 29213) were obtained from the Stanford University medical center clinical microbiology laboratory strain collection. Poly(dimethyl-siloxane) (PDMS) was obtained from GE Silicone. CellTracker Orange, Auramine-Rhodamine stain and Kinyoun’s acid-fast stain kit were purchased from Invitrogen and Sigma Aldrich, respectively, and used following the protocols provided by the vendors. All other chemicals were purchased in analytical or higher grade from Sigma Aldrich or VWR.
Cell Handling. Each bacterial strain was cultured on an LB agar plate at 37 °C. Before the experiment, fresh culture was harvested and rinsed using PBS 1x (pH 7.4) by centrifuging for 10 min at 1200 G and 4 °C. The rinsed cells were subjected to a mild centrifugation for 3 min at 200 G to remove clumps. OD600 was used to measure the cell density in a suspension. Unless specified, the cell suspension of each strain was diluted to be 109 cell/mL for making templates and 107 cell/mL for preparing the sample. For visualization, we stained the sample with Cell-Tracker Orange (excitation wavelength 541 nm; fluorescence emission wavelength 565 nm) following the protocol suggested by the vendor.
Inactivation of the cells. Cells in suspension were inactivated with physical (UV) or chemical means (75%v/v ethanol, 5%wt bleach, 1%wt hydrogen peroxide, 4%wt formaldehyde and 2%wt glutaraldehyde in PBS). Inactivation with UV was carried out in microcentrifuge tubes under 6W 254 nm UV (UVG11, UVP) for 1 min. Treatment with reagents was approached by suspending the cells in a buffer solution with the reagents and kept at room temperature for 10 min, followed by resuspension in PBS. Inactivation was confirmed by testing for colony formation.
Template preparation. A10-μL cell suspension (approximately 109 cells/mL) was spread on the surfaces of pre-cleaned microscope slides and kept in a covered Petri dish at 4 °C for 8 h. After the excess water was removed by centrifugation at 1500 G for 1 min, the substrate was dried at 60 °C for 2h and rinsed with water before use as the template.
Stamp Fabrication. Optimization of the imprinting protocol was conducted and discussed in our previous work2. Briefly, we diluted a PDMS curing mixture (monomer/cross-linker = 10:1) using cyclohexane to a volume ratio of 2:1 and spin-coated this solution onto a microscope slide (30 s at 1500 rpm). After precuring the PDMS at 80 °C for 4 min, we pressed the template stamp into the prepolymer and kept it at 37 °C for 8 h. Then we peeled off the template slide and cleaned the imprinted polymer film by immersing it in a Petri dish filled with distilled water and sonicating it for 5 min. The substrates were then inspected with a scanning probe microscope (XE-70, Park Systems) under noncontact AFM (tapping) mode, using etched silicon cantilevers (resonance frequency ~300 kHz, tip radius <10 nm) with medium-low tip oscillation damping ~15%) and a scan rate of 0.2 Hz.
Cell capture. After cleaning with water, the imprinted surface is used to selectively capture the template cells in each of the cell suspensions, which have been treated with the same method for making the inactivated cells used in the template. A microfluidic channel was employed to assist passing the cell suspension over the imprinted surface. PDMS chips containing an array of microchannels were fabricated via standard soft lithography. Each channel was 30-μm high and 100-μm wide. The total volume of the channels was about 1 μL. The chip was reversibly bonded to the imprinted substrate by the adhesion between PDMS surfaces without heat or plasma treatment. A pipet tip was inserted into the inlet of the channel as a reservoir and was filled with cell suspension. A syringe was connected to the outlet of the channel to draw the cell suspension through the channel via negative pressure. For each test, a 50-μL cell suspension (OD600 = 0.2) was infused at a flow velocity of 0.2 mm/s. Then 50-μL of PBS was used to rinse the channel at the same flow rate. The imprinted area of the chip was inspected under a confocal microscope (TCS SP2, Leica).
ACKNOWLEDGMENTS
This work was supported by the Stanford Consortium for Innovation, Design, Evaluation and Action (C-IDEA) [NIH RC4TW008781].
References
- 1.Aherne A, Alexander C, Payne MJ, Perez N, Vulfson EN. Bacteria-Mediated Lithography of Polymer Surfaces. J. Am. Chem. Soc. 1996;118:8771–8772. [Google Scholar]
- 2.Schirhagl R, Hall EW, Fuereder I, Zare RN. Separation of Bacteria with Imprinted Polymeric Films. Analyst. 2012;137:1495–1499. doi: 10.1039/c2an15927a. [DOI] [PubMed] [Google Scholar]
- 3.Ren K, Zare RN. Chemical Recognition in Cell-Imprinted Polymers. ACS Nano. 2012;6:4314–4318. doi: 10.1021/nn300901z. [DOI] [PubMed] [Google Scholar]
- 4.Schirhagl R, Ren KN, Zare RN. Surface-Imprinted Polymers in Microfluidic Devices. Sci. China Chem. 2012;55:1–15. [Google Scholar]
- 5.Schirhagl R, Qian J, Dickert FL. Immunosensing with Artificial Antibodies in Organic Solvents or Complex Matrices. Sensors and Actuators B: Chemical. 2012;173:585–590. [Google Scholar]
- 6.The Top 10 Causes of Death. World Health Organization. 2011. Fact sheet N°310.
- 7.Gilbert GL. Molecular Diagnostics in Infectious Diseases and Public Health Microbiology: Cottage Industry to Postgenomics. Trends Mol. Med. 2002;8:280–287. doi: 10.1016/s1471-4914(02)02349-3. [DOI] [PubMed] [Google Scholar]
- 8.Lahann J, Balcells M, Lu H, Rodon T, Jensen KF, Langer R. Reactive Polymer Coatings: A First Step toward Surface Engineering of Microfluidic Devices. Anal. Chem. 2003;75:2117–2122. doi: 10.1021/ac020557s. [DOI] [PubMed] [Google Scholar]
- 9.Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH. Functionalized Conducting Polymer Nanodots for Enhanced Cell Capturing: The Synergistic Effect of Capture Agents and Nanostructures. Adv. Mater. 2011;23:4788–4792. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 10.Kretzer JW, Lehmann R, Schmelcher M, Banz M, Kim KP, Korn C, Loessner MJ. Use of High-Affinity Cell Wall-Binding Domains of Bacteriophage Endolysins for Immobilization and Separation of Bacterial Cells. Appl. Environ. Microbiol. 2007;73:1992–2000. doi: 10.1128/AEM.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenik M, Seifner A, Krassnig S, Seidler K, Lieberzeit PA, Dickert FL, Jungbauer C. Sensors for Bioanalytes by Imprinting-Polymers Mimicking Both Biological Receptors and the Corresponding Bioparticles. Biosens. Bioelectron. 2009;25:9–14. doi: 10.1016/j.bios.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Tuberculosis. World Health Organization. 2012. Fact sheet N°104.
- 13.Chan ED, Heifets L, Iseman MD. Immunologic Diagnosis of Tuberculosis: A Review. Tuber. Lung Dis. 2000;80:131–140. doi: 10.1054/tuld.2000.0243. [DOI] [PubMed] [Google Scholar]
- 14.Best M, Sattar SA, Springthorpe VS, Kennedy ME. Efficacies of Selected Disinfectants against Mycobacterium-Tuberculosis. J. Clin. Microbiol. 1990;28:2234–2239. doi: 10.1128/jcm.28.10.2234-2239.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JCH, Ossoff SF, Lobe DC, Dorfman MH, Dumais CM, Qualls RG, Johnson JD. Uv Inactivation of Pathogenic and Indicator Microorganisms. Appl. Environ. Microbiol. 1985;49:1361–1365. doi: 10.1128/aem.49.6.1361-1365.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaghy J, Keyser M, Johnston J, Cilliers FP, Gouws PA, Rowe MT. Inactivation of Mycobacterium Avium Ssp. Paratuberculosis in Milk by Uv Treatment. Lett. Appl. Microbiol. 2009;49:217–221. doi: 10.1111/j.1472-765X.2009.02644.x. [DOI] [PubMed] [Google Scholar]
- 17.Fusillo MH, Burns HD. Simultaneous Auramine and Kinyoun Stain for Screening Smears for Acid-Fast Bacilli. Am. J. Clin. Pathol. 1968;49:753–754. [PubMed] [Google Scholar]
- 18.Bintsis T, Litopoulou-Tzanetaki E, Robinson RK. Existing and Potential Applications of Ultraviolet Light in the Food Industry - a Critical Review. J. Sci. Food Agric. 2000;80:637–645. doi: 10.1002/(SICI)1097-0010(20000501)80:6<637::AID-JSFA603>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 19.Maunsbac Ab. Influence of Different Fixatives and Fixation Methods on Ultrastructure of Rat Kidney Proximal Tubule Cells. I. Comparison of Different Perfusion Fixation Methods and of Glutaraldehyde Formaldehyde and Osmium Tetroxide Fixatives. J. Ultrastruct. Res. 1966;15:242–282. doi: 10.1016/s0022-5320(66)80109-0. [DOI] [PubMed] [Google Scholar]
- 20.Doughty MJ, Bergmanson JPG, Blocker Y. Impact of Glutaraldehyde Versus Glutaraldehyde Formaldehyde Fixative on Cell Organization in Fish Corneal Epithelium. Tissue Cell. 1995;27:701–712. doi: 10.1016/s0040-8166(05)80025-4. [DOI] [PubMed] [Google Scholar]
- 21.Chao Y, Zhang T. Optimization of Fixation Methods for Observation of Bacterial Cell Morphology and Surface Ultrastructures by Atomic Force Microscopy. Appl. Microbiol. Biotechnol. 2011;92:381–392. doi: 10.1007/s00253-011-3551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doughty MJ, Bergmanson JPG, Blocker Y. Shrinkage and Distortion of the Rabbit Corneal Endothelial Cell Mosaic Caused by a High Osmolality Glutaraldehyde-Formaldehyde Fixative Compared to Glutaraldehyde. Tissue Cell. 1997;29:533–547. doi: 10.1016/s0040-8166(97)80054-7. [DOI] [PubMed] [Google Scholar]




