Abstract
As demonstrated by means of DNA nanoconstructs[1], as well as DNA functionalization of nanoparticles[2-4] and micrometre-scale colloids[5-8], complex self-assembly processes require components to associate with particular partners in a programmable fashion. In many cases the reversibility of the interactions between complementary DNA sequences is an advantage[9]. However, permanently bonding some or all of the complementary pairs may allow for flexibility in design and construction[10]. Here, we show that the substitution of a pair of complementary bases by a cinnamate group provides an efficient, addressable, UV light-based method to covalently bond complementary DNA. To show the potential of this approach, we wrote micrometre-scale patterns on a surface via UV light and demonstrate the reversible attachment of conjugated DNA and DNA-coated colloids. Our strategy enables both functional DNA photolithography and multi-step, specific binding in self-assembly processes.
The highly specific, thermoreversible base-pair interactions that are formed by complementary DNA strands play an important role in biology and have recently been exploited to selectively bind and create a host of potentially important nanostructures such as branched junction motifs containing crossovers [11]. Using this strategy, functional systems including crystals [4,10,12-16], motors/robots [15,16], computers [17,18], replicators [9,19], and machines [20] have been fabricated. For nanotechnology applications as well as biological assays, however, cross-linking some or all of these bonds permanently can provide stability to higher order constructs, thereby introducing a robust static self-assembly strategy. For instance, this strategy has been demonstrated to be powerful in providing structural information on biomacromolecules when standard techniques were not applicable [21].
Several different cross-linking methods have been investigated. The most common methods employ the cross-linking agent psoralen that forms covalent links to thymines upon exposure to UV light [22]. Unfortunately, the cross-linking efficiency for psoralen in solution with DNA is relatively low (effective cross section σ = 5×10−6 nm2, percent of crosslinking ∼ σΦt where Φ is photon flux in photons/(nm2×s), and t is time in seconds) in comparison to other cross-linking agents (which will be further discussed vide infra), otherwise it requires the attachment of the psoralen group at the 5′-TpA site (T-A sequence) of a DNA ‘sticky end’ [23]. Alternative UV cross-linking agents for DNA that allow for higher efficiency, increased specificity, and flexibility in sequence placement are being pursued. The use of p-carbamoylvinyl phenol nucleoside (p-CVP) [24] and 3-cyanovinylcarbazole nucleoside (CNVK) [25] as UV cross-linking agents have been popularized as a result of higher cross-linking efficiencies than psoralen (∼3 × 10−5 nm2 and 1.3 × 10−4 nm2 for p-CVP and CNVK, respectively). For cross-linking to take place, in these cases, p-CVP requires specific placement next to an adjacent adenine base while CNVK must be adjacent to a pyrimidine.
Here we report a highly selective and efficient interstrand cross-linking methodology using the p-CVP group, herein referred to as cinnamate, where two cinnamate groups are placed directly across from each other on complementary strands. We demonstrate that this strategy increases cross-linking specificity and efficiency and that introducing these cinnamate-modified DNA strands onto surfaces allows for the realization of a photolithography application.
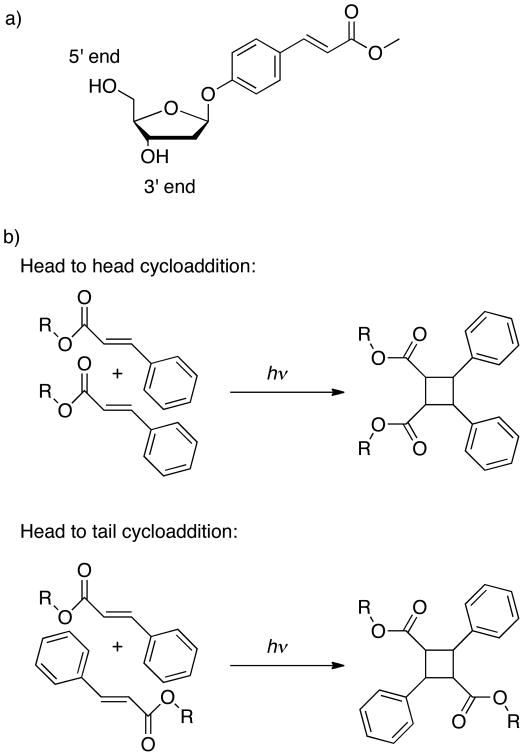
Figure 1 shows the cinnamate-containing nucleoside and the two cycloaddition products that can occur upon exposure of the cinnamate to 360-390 nm UV light. Synthetic procedures are described in the Supplementary Section.S1. The size of the cinnamate-based artificial base is comparable to that of natural purines or pyrimidines, i.e. its structural perturbation is comparable to the effect of one mismatched base pair [see Supplementary Section.S2]. The cinnamate nucleoside was incorporated into DNA strands resulting in the key building blocks for the guided self-assembly and UV cross-linking units of DNA-functionalized colloids.
Figure 1. Schematic representation of cinnamate-containing nucleoside and the cycloaddition products.
a) Cinnamate-containing nucleoside. b)Schematic representation of the cycloaddition between 2 cinnamate groups (only the E isomers are shown) and their potential configurations.
Initially, we incorporated the cinnamate groups into DNA sequences and investigated whether or not having two cinnamate groups directly across from each other inhibits hybridization of complementary strands. We demonstrate that the hybridization was not effected (vide infra). Next, we determined whether this new strategy allows for the successful cross-linking of complementary strands. Finally, we introduced the functionalized DNA onto a gold surface as well as onto colloids to determine if this strategy allows for the realization of a photolithographic process.
Two of the DNA strands used in this study (strands A and B) consist of two major parts: an inert section and a “sticky end”. The inert section, the longer portion of the strand, was a double strand. A short single strand of DNA, a “sticky end”, was placed at the 3′ end of the inert section. The sticky end was used as the directing and recognition unit to bind to a complementary sticky end of the desired particles [5-8, 26] and surfaces [27] used in our studies. The inert sections of strands A and B (sequences shown below) consist of identical sequences and therefore hybridize with the same complementary strand (CS). The sticky ends of the two strands, however, contain complementary cinnamate-modified sequences. The sequences used are:
(A) 5′-ATC GCT ACC CTT CGC ACA GTC AAT CCA GAG AGC CCT GCC TTT CAT TAC GAC CAA GT-Cross-linker-T ATG A 3′
(B) 5′-ATC GCT ACC CTT CGC ACA GTC AAT CCA GAG AGC CCT GCC TTT CAT TAC GAT CAT A-Cross-linker-AC TTG G 3′
(CS) 5′-TCG TAA TGA AAG GCA GGG CTC TCT GGA TTG ACT GTG CGA AGG GTA GCG AT-3′
We inserted the cinnamate nucleoside at the 7th nucleoside position (5′->3′) within the 11-base sticky end of the A strand, as well as the associated 6th nucleoside position along the B strand. Such 11-base sticky ends were used in earlier studies [5-8,26,27] and have been well characterized. Synthetically the cinnamate group could be placed at any place, however the crosslinking efficiency is increased when the two cinnamate groups are held in close proximity, and placing the cinnamates toward the center of the complementary sticky ends gives a slight advantage. The A and B DNA strands were annealed with an equal number of CS strands to hybridize and form a construct with a 50 nucleotide-pair rigid duplex followed by an 11 nucleotide sticky end.
We first investigated whether the cinnamate-functionalized DNA strand binds selectively with its complementary strand. A denaturing gel (see Methods), Figure 2a, dehybridizes noncovalently-bound DNA strands without disruption of the cross-linked strands. Lanes 1 through 3 were loaded with the DNA strands without UV exposure. Lanes 4 through 6 were loaded with the DNA strands that were exposed to 350 nm UV light for five minutes (refer to Figure 2a caption for complete experimental details). As shown in lanes 1 through 3, after running all of these strand combinations on the denaturing gel, only single stranded DNA was obtained demonstrating that these strands are held together by reversible hydrogen bonds. Upon UV exposure (10 Photons/sec-nm2, see Methods for UV calibration) for 5 minutes, however, we observed substantial cross-linking between the complementary strands A and B (Lane 6) which increased after 15 minutes of UV light exposure (lane 9). As a control, strand A with CS (lane 4) and strand B with CS (lane 5) were also exposed to UV light but did not exhibit any new band corresponding to cross-linked product. Also, although A and B are complementary strands, no cross-linking is observed without UV exposure, as shown in lane 3, since no new band for the hybridized product is visible. These results demonstrate that two cinnamate-modified DNA strands allow for cross-linking only when the two cinnamate nucleosides are held in juxtaposition by flanking complementary sequences in the sticky ends. The efficiency and time dependence of cinnamate cross-linking is shown in the Supplementary Section.S3. The effective linking cross-section was found to be 3×10−5 nm2.
Figure 2. Efficient and specific crosslinking with cinnamate-based DNA strands, and the application on colloids.
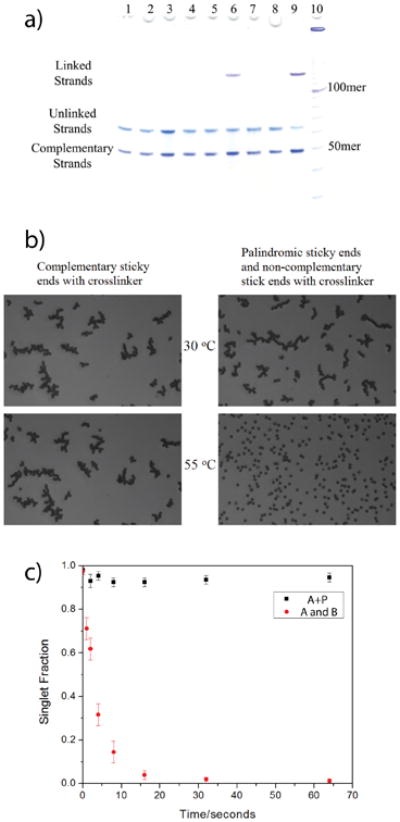
a) A 10% denaturing gel running in 1× TBE buffer at 55 °C. Lane 1 is the strands A and CS without UV exposure. Lane 2 contains the strands B and CS without UV exposure. Lane 3 contains the strands A, B, and CS without UV exposure. Lane 4 contains strands A and CS exposed to 350 nm UV light for 5 minutes. Lane 5 contains strand B and CS exposed to 350 nm UV light for 5 minutes. Lane 6 contains strands A, B, and CS exposed to 350 nm UV light for 5 minutes. Lane 7 contains strands A and CS exposed to 350 nm UV light for 15 minutes. Lane 8 contains strands B and CS exposed to 350 nm UV light for 15 minutes. Lane 9 contains strands A, B, and CS exposed to 350 nm UV light for 15 minutes. Lane 10 contains the 10 bp DNA Ladder.
b) Micrographs of the aggregation of DNA-coated one micron colloidal particles. Particles are aggregated at 30 °C for one hour, exposed to ∼360 nm UV light for 15 seconds, and then heated to 50 °C for 15 minutes. The left column contains complementary A particles and B particles with cinnamate. The right column contains A-P particles with cinnamate in non-complementary A strands.
c) Fraction of non-aggregated particles as a function of UV exposure time for A particles and B particles (red) and the A-P particles (black) with cross-linkers.
Next, we modified the aforementioned strands to be able to functionalize colloidal particles (see Methods for DNA-coated colloids). We used 1μm streptavidin-coated poly(styrene) particles for these experiments. We added biotin (and tetraethylene glycolas the terminating group at the 5′ end) to sequences A and B in order to attach the DNA to the streptavidin-coated particles. A similar construct was fabricated where the sticky ends were self-complementary palindromic sequences (P: 5′-AATCATGATT-3′) lacking cinnamate. The P DNA sequences were designed so that the particles have a lower melting temperature (∼40 °C [26], ‘normal buffer’ solution, defined in methods section) than the A and B complementary pair (∼45 °C, in normal buffer). We then fabricated five sets of particles functionalized with different combinations of the cinnamate-functionalized DNA strands: A, B, P, A+P, and B+P, where the A+P and B+P functionalized particles have half their surface randomly coated with P strands and the other half with A or B strands [26]. We carried out four sets of experiments using these particles to investigate if it is possible to bind them permanently via a combination of DNA hybridization and UV light-triggered cross-linking.
In the experiments described below, the sample was first annealed for one hour by cooling it slowly from 55 °C to room temperature, followed by exposure to UV light (16 seconds at 60 photons/sec-nm2) at room temperature. The sample was then heated to 55 °C for 10 minutes. The P-coated particles completely dissociated upon heating (Supplementary Figure.S5), indicating that UV exposure did not link these DNA functionalized particles. The mixture of A functionalized particles and B functionalized particles, and the mixture of A+P functionalized particles and B+P functionalized particles showed minimal dissociation (Figure 2b bottom left and Supplementary Figure.S6). UV irradiation successfully cross-linked the cinnamate modified paired strands. The palindrome strands (P) did not interfere with the cinnamate cross-linking. The most revealing experiment had only particles A+P (Figure 2b bottom right) that nearly completely dissociated after heating. Although the cinnamate groups on the non-complementary strands of two particles were held together within a range of ∼20 nm, UV exposure was not effective for cross-linking in this case.
Figure 2c compares the fraction of unbound particles for the equal mixture of complementary cinnamate modified A and B functionalized particles to the A+P functionalized particles. This is a particularly sensitive measure since these particles are typically joined by approximately 150 sticky-end pairs containing cinnamate [26]. If any pair of them were to be cross-linked, the complementary strands would remain stuck together. As demonstrated in Figure 2C, the singlet fraction for the A and B particle mixture is minimal in comparison to the A+P particles. These data suggest that the selectivity of the cinnamate to itself in complementary vs non-complementary strands is ∼300:1.
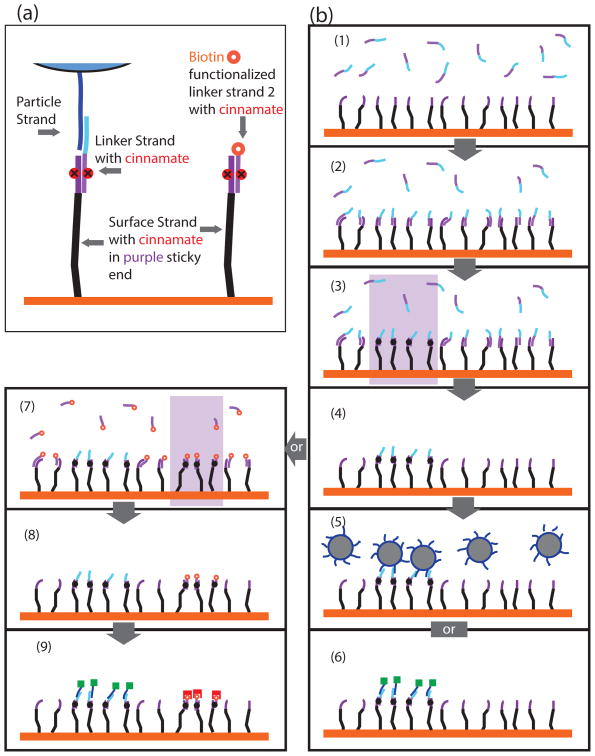
After we demonstrated the efficiency and specificity of the cinnamate-modified DNA strands on particles, we targeted photolithography as an application for these cross-linkable particles, so as to demonstrate the utility of our methodology. The crosslinking can be selectively activated by UV exposure on a substrate, allowing access to multi-functionalized DNA surfaces that are highly desirable for bio-medical applications [28]. We modified our design strategy for the DNA sequences to include a three-component structure: a “surface strand” (SS), a “particle strand” (PS), and a “linker strand” (LS) (sequences included in the caption of Figure 3). The SS was end-functionalized with a disulfide group to allow for attachment to a gold surface [27]. The SS consists of a 50 base-pair DNA backbone (Figure 3a black) and a 22 base-pair sticky-end (Figure 3a purple) containing the cinnamate nucleoside (red). The linker strand has two parts: a) a cinnamate-containing sequence complementary to SS (Figure 3a purple) and b)a functional DNA sequence that is designed to bind specifically to PS (Figure 3a blue). Finally the target material, in our case fluorescently labeled PS or colloidal particles contain PS strands which are complementary to LS (Figure 3a blue). The LS-SS DNA was designed to have a melting temperature without UV radiation of ∼45 °C (0.1μM strand concentration, in normal buffer, see Supplementary Section.S2), which ensures efficient cross-linking at room temperature.
Figure 3. Schematic protocol for DNA Photo-Lithography with cinnamate-based DNA strands.
a) Schematic representations of the DNA constructs and the position of cinnamate cross-linkers. The sequences are: Surface Strand: 5′-RSS-50basesBackbone-TTGAGAAATGC-cinnamate-CGTAAAGAGTT-3′; Linker Strand: 5′-CATCTTCATCCAACTCTTTACG-cinnamate-GCATTTCTCAA-3′; Particle Strand: 5′-GGATGAAGATG-50basesBackbone-BiotinTEG-3′; Linker Strand 2: 5′-BiotinTEG- AACTCTTTACG-cinnamate-GCATTTCTCAA-3′.
b) Procedures to fabricate the multi-functionalized DNA surface. Steps 1 and 2: hybridization of linker-strands (LS) to surface strands (SS) (purple section of DNA strand) by cooling them from 55 °C to 25 °C over a period of 30 minutes; Step 3: permanently UV cross-link SS and LS in selected area (purple section with solid black dot); Step 4: heating the sample to 55°C to de-hybridize unlinked strands and washing of the sample. The functionalized DNA surface is then ready for use, e.g. by reversibly binding colloids (step 5) to the patterned regions, or by use of fluorescently labeled complementary DNA strand (step 6). To add another function to the surface, linker strand 2 (LS2) needs to be hybridized to the SS and then UV cross-linked in the desired region (step 7). The multi-functionalized surface is the ready for use (after being heated to 55°C and washed (step 8)). We can visualize the patterns by red streptavidin binding to biotin in the LS2 patterned region, and green fluorescently labeled PS hybridizing to LS patterned region (step 9).
Figure 3b shows schematically the procedures used for the preparation of the samples for photolithography. First, a gilded surface was coated with SS (see Methods for DNA coated gold surface). Next, LS was hybridized to SS by annealing from 55°C to 25°C over a 30 minute period (steps 1 and 2). Then, the samples were exposed to UV light to permanently cross-link LS to SS in the exposed regions (step 3). Finally, the sample was heated to 55 °C and washed three times with normal buffer to de-hybridize any unlinked strands (step 4). The functionalized DNA surface was then ready to use for the reversible binding of colloids or fluorescently labeled DNA to the patterned regions (steps 5 and 6).
Figure 4a shows images of the surfaces decorated by 1 micron poly(styrene) particles (left) and fluorescently labeled DNA (middle), both within the region exposed with UV light through a “Y-shaped” photomask (right). Similar experiments were carried out using an “NYU” photo mask of different feature sizes (Figure 4b right). Both fluorescent and colloid images show a minimum feature size of ∼2 μm (Figures 4b). (see Methods for light microscope and confocal microscope)
Figure 4. DNA photo-lithographic pattern on surface.
(a) and (b): right: exposed patterns with different feature sizes (∼14μm in (a) and 4μm and 1.5μm in (b)); left: colloid patterns (inverted so unstuck particles have escaped); middle: Green fluorescently labeled conjugate images.
(c) Multi-functionalized patterns of ‘nature materials’ (abbreviated as ‘n m’) in New Times Roman font. Left: Green fluorescently labeled DNA hybridizes to LS patterned in the region ‘materials,’ and red streptavidin binds to LS2 patterned biotin in the region ‘nature’. Right: similar patterning of larger ‘n’ and ‘m’ letters. Separate confocal filtering channels are shown as smaller figures.
Our particles have a gravitational height, kBT/mg, of approx. 1 micron and hence sediment toward the bottom. In order to determine if the particles attach to the surface by DNA hybridization, after allowing the particles to hybridize with the complementary LS (Supplementary Movie.S1), we inverted the sample. The particles remain on the surface even after inversion of the sample verifying that the particles were attached to the surface by DNA hybridization between PS and LS (Figure 4a & b left). Since the binding between LS and the colloids is thermoreversible, the colloids diffuse away from the patterned regions of the surface upon heating to 55 °C (See Methods for temperature controlling stage and Supplementary Movie.S2). Additionally, cooling the sample leads to re-association(Supplementary Movie.S3). We confirmed the effective cross-section, 3×10−5 nm2, of the cinnamate by colloid-surface melting temperatures (Supplementary Section.S4).
To make a spatially dependent multi-functionalized surface and demonstrate the versatility of our method, we first carried out the photolithography as described above (steps 1 through 6), then repeated steps 3 and 4 on a different region of the gold surface using modified linker strands (steps 7 and 8). The new linker strand 2 (LS2) contains a functional molecule (biotin in this case) at its 5′ end unlike the single stranded sticky-end of LS. The newly functionalized region of the gold surface with LS2 strands was visualized by the conjugation of red fluorescent streptavidin (see Methods Section for fluorescent imaging).The sticky LS in the first exposure region remained functional as tested by the addition of fluorescently labeled PS strands (step 9).
Figure 4c shows the result of our multi-functionalization experiment, after step 9 of Figure 3b. Green fluorescently labeled PS strand sticks to the LS patterned region (letters of ‘materials’ in Figure 4c left) by complementary DNA hybridization. Red streptavidin binds to LS2 patterned biotin region (letters of ‘nature’ in Figure 4c left). The distinct colors in separate filtering channels confirmed the successful patterning of two different functions without interference (See Methods Section for confocal imaging).
Finally, we investigated the pattern resolution (Figure 5a). We exposed surfaces that were decorated with fluorescently labeled DNA and colloids to UV irradiation for different durations of time through an inverted “U” photomask. The light intensity and particle concentration were analyzed through a cut across the two straight legs of the inverted “U”. Looking at the intensity profile (bottom row), the black curves represent the relative particle concentration, the green curves represent the intensity of green fluorescent light from the confocal images, and the red curves are a linear rescaling of the exposed peaks based on estimated coverage (from left to right: 8%, 15%, 28%, and 48%, see Supplementary Section.S4) (The last panel is the exposure profile (bottom row)). The images and curves show that shorter exposure time (5 seconds) led to weak fluorescent light and non-uniformity in colloidal patterns (Figure 5a (left)) due to inhomogeneous cross-linking. Increasing exposure time yielded a stronger fluorescent signal and complete colloidal patterning, but it also lowered the pattern resolution by producing undesired cross-linking in the peripheral regions (Figure 5a right). An exposure time of ten seconds gave a good balance between uniform functionalization and high resolution.
Figure 5. Resolution of DNA photo-lithography.
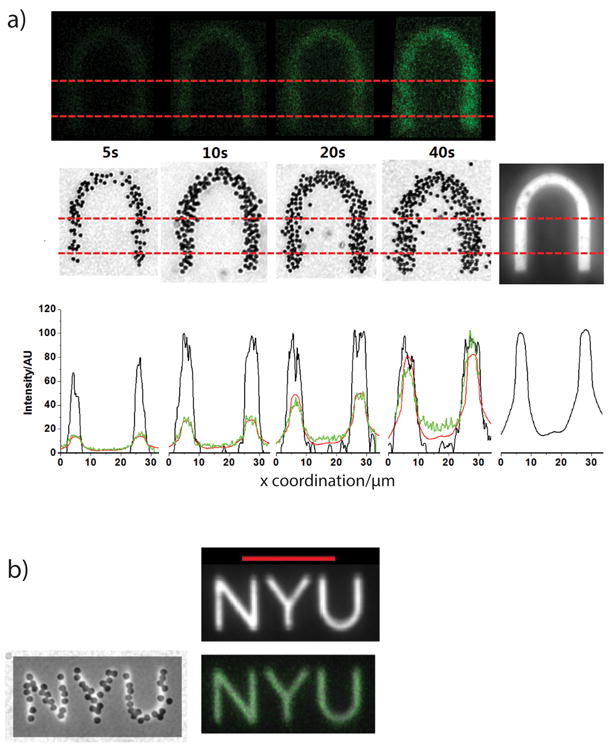
a) Surface coated with cinnamate-modified DNA strands followed by exposure to UV light through an inverted “U” shaped photomask for different durations of time. (First row) fluorescent images, (second row) colloids' images, (last image in second row) exposed pattern, (bottom row) intensity profile (green curve for first row, black curve for second row, red curve are rescaling of the exposed pattern) between dashed red lines along the x axis.
b) Superimposed colloidal images and fluorescent images with exposed pattern respectively with a resolution of ∼2μm for the smallest feature and ∼1μm for pattern edges.
Figure 5b shows the pattern image superimposed on the colloid and fluorescent images. The line width is ∼1.5 μm in the exposure pattern and ∼2 μm in the fluorescent image, which suggests at least a 1μm resolution on the edges. Though this resolution is slightly lower than standard UV photolithography in vacuum (resolution of ∼0.8 μm) [29, 30] or direct laser writing [31], the resolution obtained using this technique is competitive to other solution-based methods using DNA for photolithography [32]. This multi-functionalized DNA technique with micron-scale resolution might prove useful in DNA chips for gene identification and for directing colloidal self-assembly.
In summary, we have introduced a simple, highly selective, and efficient method to covalently bind complementary DNA strands in solution and on surfaces using UV irradiation triggered cross-linking as an optional separate step. Since DNA is a functional material, these techniques can also be used to bind reversibly and/or permanently, via DNA linkers, an assortment of molecules, proteins, and nanostructures. Potential applications range from the simple permanent joining of particles, to advanced self-assembly protocols that assemble support scaffolds, and also a multi-dimensional system in which a support scaffold is first assembled, a structure formed and crosslinked on the scaffold followed by reversible disassembly of the scaffold. Using this new methodology, we have developed a new photolithography method capable of functionally and chemically patterning surfaces down to a 1μm resolution. Our photolithographic technique opens the door for the possibility to write chemical and physical patterns, or to make high-resolution multi-functionalized DNA surfaces for genetic detection [33] or DNA computing [34]. Future studies using total internal reflection microscopy may allow for the exposure of only a few hundred nanometers of a particle to UV light. This may enable chemically different patches to be printed on micron-sized colloidal particles for medical and soft matter research uses in a more controlled manner [35,36].
Methods
Synthesis of p-carbamoylvinyl phenol nucleoside
The cinnamate-functionalized phosphoramidite was obtained in four high yielding steps (Supplementary Section.S1). Briefly, the glycosylation of the cinnamate derivative with Hoffer's α-chloro saccharide was achieved using cesium carbonate. Sodium methoxide was employed in order to remove the p-toluyl protecting groups, which afforded free hydroxyl groups at the 3′ and 5′ positions of the saccharide. Finally, the 5′-OH was protected with dimethoxytritylchloride followed by the conversion of the 3′-OH to a phosphoramidite. The cinnamate-containing phosphoramidite was incorporated into a DNA oligonucleotide using published procedures [37].
UV calibration
UV power output was measured with a SiC photodiode with a peak sensitivity at 340 nm. The power output one cm from the UV lamp was ∼0.6 mW/cm2 or ∼10 photons/sec-nm2. For Hg lamp (Leica 106z lamp housing with 50W mercury burner) with ×100 Leica air objective on a Leica DMRXA microscope with a Leica Type A filter cube, power output is ∼60 photons/sec-nm2 as in the cross-linking experiment on the particles. For the external Hg lamp that was used for the photo-lithography experiments (Leica EL6000) with a ×63 Leica air objective, the power output is ∼30 mW/cm2 or ∼500 photons/sec-nm2.
Denaturing Gels
The gels contained 8.3 M urea and were run at 55 °C. The running buffer consisted of 89 mM Tris, HCl, pH 8.0, 89 mM boric acid, and 2 mM EDTA (TBE). The sample buffer consisted of 10 mM NaOH, 1 mM EDTA, a trace amount of Xylene Cyanol FF, and bromophenol blue tracking dye. Gels were run on a Hoefer SE 600 electrophoresis unit at 55 °C (31 V/cm, constant voltage). When further denaturation was required, a 6% acrylamide gel solution containing 7.0 M urea and 41 % formamide, or a 4% acrylamide gel solution containing 6.8 M urea and 47 % formamide were substituted for a regular denaturing gel.
DNA coated gold surface
A cover slip (2.5 cm × 2.5 cm) was cleaned with acetone, plasma etched using a SPI supplies Plasma Prep II, and coated with 5 nm of chromium and 40 nm of gold (SIGMA-ALDRICH 99.999%) using a BAL-TEC MCS 010 Multi Control System evaporator. An end-functionalized thiol surface-strand (72 base pairs) was annealed to its complementary strand (49 base pair) by heating it to 95 °C and then cooling it to 25 °C resulting in the formation of a rigid double stranded backbone. The sample was then incubated on the gold surface for 12 hours under the following conditions: 25 °C normal buffer (10mM PB , 50mM NaCl, 1% w/w F127, pH-7.4) with a thiol DNA concentration of 40 μM. The small incubation chamber was sealed with vaccuum grease to avoid evaporation. After incubation, excess strands were washed away using normal buffer at 55 °C. This procedure was repeated three times. The resulting surface had a DNA density of ∼3000 μm−2 as determined by radioactive measurement [27].
Preparation of DNA-coated colloids
1μm Streptavidin-covered poly(styrene) particles were purchased from Invitrogen (Dynabeads MyOne Streptavidin C1). Biotinylated particle strands with a double stranded backbone were incubated with these particles for one hour under the following conditions: 25 °C, light shaking, normal buffer, particle volume fraction 0.05 %, and a biotinylated strand concentration of 0.5 μM. Particles were washed by centrifugation and re-suspensed in normal buffer. This process was repeated three times in order to wash away excess strands. The resulting surface had a DNA density of ∼6400 μm−2 as determined by radioactive measurement [26].
Light microscope
A Leica DMRXA microscope with a Qimaging Retiga 1300 camera and a Leica external Hg lamp was used to observe colloidal assembly and to perform the photolithography experiments. A Leica Type A filter cube was used to provide 365 nm wavelength UV light for the cinnamate cross-linking. Black field masks were purchased from FineLine imaging and placed at the aperture conjugated to sample image.
Temperature controlling stage
A temperature stage was built on a light microscope to provide fast in-situ temperature control. Briefly, 1000 Ω ITO glass was placed on a 3mm thick copper plate, two ends of which were connected to peltiers (2.5 cm by 2.5 cm) and then to a thermal sink with constant temperature. We were able to control and detect the temperature with <0.5 °C relative error using a LakeShore DRC 93C Temperature Controller and LakeShore PT-111 temperature sensor.
Confocal microscope and fluorescent imaging
A Leica DM6000 CS confocal microscope was used to take all fluorescent images. Excitations with 488 nm and 631 nm were used. Streptavidin with Alexa Fluror 488® (Invitrogen) was used to dye the biotinylated complementary DNA (‘particle strand’) that was conjugated to the DNA sticky end in the LS pattern. Streptavidin with Alexa Fluror 633® (Invitrogen) was used to directly conjugate and visualizebiotin-patterned region. A high salt concentration buffer (500 mM NaCl) was used to stabilize the DNA interaction.
Supplementary Material
Acknowledgments
This research has been partially supported by the MRSEC Program of the National Science Foundation under Award Number DMR-0820341 for the cinnamate-functionalized phosphoramidite, NASA NNX08AK04G for microscopy, and DOE-BES-DE-SC0007991 to PMC for data acquisition and analysis, as well as by the following grants to NCS for DNA synthesis and characterization: GM-29554 from the National Institute of General Medical Sciences, CTS-0608889 and CCF-0726378 from the National Science Foundation, 48681-EL and W911NF-07-1-0439 from the Army Research Office, and N000140910181 and N000140911118 from the Office of Naval Research. J.Romulus acknowledges support through the Margaret Strauss Kramer Graduate Student Fellowship in Chemistry.
Footnotes
Author Contributions: L.F. designed and performed experiments, analyzed data and wrote the paper; J. Romulus and M.L. synthesized the cinnamate-containing phosphoramidite and wrote the paper; R.S. incorporated the cinnamate in the DNA strands, performed gel experiments, analyzed data, and wrote the paper; J.Royer performed experiments, analyzed data, and wrote the paper; K.W. performed experiments, analyzed data, and wrote the paper; Q.X. performed experiments and analyzed data; N.C.S. initiated and directed the project, designed experiments, analyzed data, and wrote the paper; M.W. initiated and directed the project, designed experiments, and wrote the paper; P.M.C. initiated and directed the project, designed experiments, analyzed data, and wrote the paper.
Contributor Information
Lang Feng, Email: lang.feng@nyu.edu.
Nadrian C. Seeman, Email: ned.seeman@nyu.edu.
Marcus Weck, Email: marcus.weck@nyu.edu.
Paul Chaikin, Email: chaikin@nyu.edu.
References
- 1.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 3.Alivisatos AP, Johnsson KP, Peng K, Wilson TE, Loweth CJ, Bruchez MP, Jr, Schultz PG. Organization of ‘nanocrystal molecules’ using DNA. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 4.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 5.Biancaniello PL, Kim AJ, Crocker JC. Colloidal interactions and self-assembly using DNA hybridization. Phys Rev Lett. 2005;94:058302. doi: 10.1103/PhysRevLett.94.058302. [DOI] [PubMed] [Google Scholar]
- 6.Valignat MP, Theodoly O, Crocker JC, Russel WB, Chaikin PM. Reversible self-assembly and directed assembly of DNA-linked micrometer-sized colloids. Proc Nat Acad Sci USA. 2005;102:4225–4229. doi: 10.1073/pnas.0500507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers PH, Michel E, Bauer CA, Vanderet S, Hansen D, Roberts BK, Calvez A, Crews JB, Lau KO, Wood A, Pine DJ, Schwartz PV. Selective, Controllable, and Reversible Aggregation of Polystyrene Latex Microspheres via DNA Hybridization'. Langmuir. 2005;21:5562–5569. doi: 10.1021/la046790y. [DOI] [PubMed] [Google Scholar]
- 8.Leunissen ME, Dreyfus R, Cheong FC, Grier DG, Sha R, Seeman NC, Chaikin PM. Switchable self-protected attractions in DNA-functionalized colloids. Nature Mater. 2009;8:590–595. doi: 10.1038/nmat2471. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Sha R, Dreyfus R, Leunissen ME, Maass C, Pine DJ, Chaikin PM, Seeman NC. Self-replication of information-bearing nanoscale patterns. Nature. 2011;478:225–228. doi: 10.1038/nature10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leunissen ME, Dreyfus R, Sha RJ, Wang T, Seeman NC, Pine DJ, Chaikin PM. Towards self-replicating materials of DNA-functionalized colloids. Soft Matter. 2009;5:2422–2430. [Google Scholar]
- 11.Kallenbach NR, Ma RI, Seeman NC. An immobile nucleic acid junction constructed from oligonucleotides. Nature. 1983;305:829–831. [Google Scholar]
- 12.Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou Pe, Ginell Sl, Mao C, Seeman Nc. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macfarlane RJ, Lee BY, Jones MR, Harris N, Schatz CG, Mirkin CA. Nanoparticle Superlattice Engineering with DNA. Science. 2011;334:204–208. doi: 10.1126/science.1210493. [DOI] [PubMed] [Google Scholar]
- 14.Winfree E, Liu FR, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 15.Sherman WB, Seeman NC. DNA Walking Biped. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- 16.Yurke B, Turberfield AJ, Mills APJ, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 17.Lewin DI. DNA computing. Comput Sci Eng. 2002;4:5–8. [Google Scholar]
- 18.Qian L, Winfree E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 19.Schulman R, Winfree E. Synthesis of crystals with a programmable kinetic barrier to nucleation. Proc Nat Acad Sci USA. 2007;104:15236–15241. doi: 10.1073/pnas.0701467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu H, Chao J, Xiao SJ, Seeman NC. A Proximity-Based Programmable DNA Nanoscale Assembly Line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wassarman D, Steitz J. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 22.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Hélène C. Sequence-specific photo-induced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Proc Nat Acad Sci USA. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand cross-links in human cells. EMBO Reports. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura Y, Itoa Y, Fujimotoa K. Interstrand photocross-linking of DNA via p-carbamoylvinyl phenol nucleoside. Bioorg Med Chem Lett. 2005;15:1299–1301. doi: 10.1016/j.bmcl.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 25.Yoshimura Y, Fujimoto K. Ultrafast reversible photo-cross-linking reaction: Toward in situ DNA manipulation. Org Lett. 2008;10:3227–3230. doi: 10.1021/ol801112j. [DOI] [PubMed] [Google Scholar]
- 26.Dreyfus R, Leunissen ME, Sha R, Tkachenko AV, Seeman NC, Pine DJ, Chaikin PM. Simple Quantitative Model for the Reversible Association of DNA Coated Colloids. Phys Rev Lett. 2009;102:048301. doi: 10.1103/PhysRevLett.102.048301. [DOI] [PubMed] [Google Scholar]
- 27.Xu Q, Feng L, Sha R, Seeman NC, Chaikin PM. Subdiffusion of a Sticky Particle on a Surface. Phys Rev Lett. 2011;106:228102. doi: 10.1103/PhysRevLett.106.228102. [DOI] [PubMed] [Google Scholar]
- 28.Xia D, Yan J, Hou S. Fabrication of Nanofluidic Biochips with Nanochannels for Applications in DNA Analysis. Small. 2012;8:2787–2801. doi: 10.1002/smll.201200240. [DOI] [PubMed] [Google Scholar]
- 29.Maalouf A, Gadonna M, Bosc D. An improvement in standard photolithography resolution based on Kirchhoff diffraction studies. J Phys D Appl Phys. 2009;42:015106. [Google Scholar]
- 30.Gorzolnik B, Mela P, Möller M. Nano-structured micropatterns by combination of block copolymer self-assembly and UV photolithography. Nanotechnology. 2006;17:5027–5032. [Google Scholar]
- 31.Varghese B, Cheong FC, Sindhu S, Yu T, Lim CT, Valiyaveettil S, Sow CH. Size selective assembly of colloidal particles on a template by directed self-assembly technique. Langmuir. 2006;22:8248–8252. doi: 10.1021/la060732q. [DOI] [PubMed] [Google Scholar]
- 32.Naiser T, Mai T, Michel W, Ott A. Versatile maskless microscope projection photolithography system and its application in light-directed fabrication of DNA microarrays. Rev Sci Instrum. 2006;77:063711. [Google Scholar]
- 33.Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SPA. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 34.Liu QH, Wang LM, Frutos AG, Condon AE, Corn RM, Smith LM. DNA computing on surfaces. Nature. 2000;403:175–179. doi: 10.1038/35003155. [DOI] [PubMed] [Google Scholar]
- 35.Pregibon DC, Toner M, Doyle PS. Multifunctional encoded particles for high-throughput biomolecule analysis. Science. 2007;315:1393–1396. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- 36.Jiang S, Chen Q, Tripathy M, Luijten E, Schweizer KS, Granick S. Janus particle synthesis and assembly. Adv Mater. 2010;22:1060–1071. doi: 10.1002/adma.200904094. [DOI] [PubMed] [Google Scholar]
- 37.Caruthers MH. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230:281–85. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.