Abstract
Innate immunity involving antimicrobial peptides represents an integrated and highly effective system of molecular and cellular mechanisms that protects host against infections. One of the most frequent hospital-acquired pathogens, Staphylococcus aureus, capable of producing proteolytic enzymes, which can degrade the host defence agents and tissue components. Numerous antimicrobial peptides derived from chromogranins, are secreted by nervous, endocrine and immune cells during stress conditions. These kill microorganisms by their lytic effect at micromolar range, using a pore-forming mechanism against Gram-positive bacteria, filamentous fungi and yeasts. In this study, we tested antimicrobial activity of chromogranin A-derived peptides (catestatin and cateslytin) against S. aureus and analysed S. aureus-mediated proteolysis of these peptides using HPLC, sequencing and MALDI-TOF mass spectrometry. Interestingly, this study is the first to demonstrate that cateslytin, the active domain of catestatin, is active against S. aureus and is interestingly resistant to degradation by S. aureus proteases.
Introduction
Chromogranins (Cgs) constitute the predominant family of proteins enclosed in secretory vesicles of chromaffin cells. [1] They are naturally processed to produce numerous peptides with various biological activities. [2]–[4] During the past decade, our group characterized several new antimicrobial peptides (AMPs) derived from chromogranin A (CgA) [5]–[8] and chromogranin B (CgB). [3], [9] These peptides are released by stimulated chromaffin cells of adrenal medulla and also by activated polymorphonuclear neutrophils (PMNs). [5], [7] Sequences of these peptides are highly conserved during evolution, suggesting that they are well-integrated in innate immune system. [10] Among these AMPs: chromofungin (CHR, CgA47–66) and catestatin (CAT, CgA344–364), derived from bovine CgA, activate PMNs and induce a calcium influx into immune cells. [11].
Staphylococcus aureus is the most frequently isolated pathogen in Gram-positive sepsis, often involved in blood clotting disorders and destruction of endocardial tissue. [12] S. aureus has developed several mechanisms to avoid immune response including resistance to AMPs, [13] impairment of phagocyte recruitment, [14] escape from neutrophil extracellular traps, [15] interference with complement, [16] neutrophil lysis, resistance to oxidative burst [17] and non-specific binding and degradation of immunoglobulins. [18] The AMPs evasion mechanisms deployed by S. aureus include proteolytic degradation by extracellular proteases of three major catalytic classes, namely metallo-, serine- and papain-like cysteine proteases. [19] The expression of proteolytic enzymes is controlled directly by global regulators of virulence factors such as agr, sar [20], [21] and indirectly by RsbU that controls Sigma(B) activity.[22] Moreover, SarA is also a regulator of methicillin resistance factor (fmtA).[23] It has been previously reported that S. aureus metallo-protease aureolysin can cleave and inactivate human cathelicidin LL-37, thereby contributing to bacterial escape from the innate immune system. [13].
As staphylococci easily colonize skin and epithelia, regardless of the expression of antimicrobial Cgs-derived peptides, [24] we aimed to investigate the antimicrobial effects of CAT and its shorter fragment cateslytin (CTL, CgA344–358) against S. aureus. Taking into account the difference in the activity of these peptides, using HPLC and proteomic analysis (sequencing and MALDI-TOF mass spectrometry), we examined the degradation of these CgA-derived peptides by staphylococcal proteases released into bacterial supernatants. Here, we report the relationship between peptide sequences and their sensitivity to bacterial proteases, as well as the possibility to use them as new antimicrobial agents in combination with antibiotics.
Materials and Methods
Preparation and analysis of synthetic antimicrobial peptides
Synthetic peptides were prepared on an Applied Biosystems 433A peptide synthesizer (Foster City, USA), using the stepwise solid-phase approach with 9-fluorenylmethoxycarbonyl (Fmoc) chemistry. Sequences of the synthetic peptides are as follows: bCAT (bCgA344–364: RSMRLSFRARGYGFRGPGLQL), hCAT (hCgA352–372: SSMKLSFRARGYGFRGPGPQL), bCTL (bCgA344–358: RSMRLSFRARGYGFR) and LL-37 (hCAP134–170 LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES). Then, the synthetic peptides were purified by a Dionex HPLC system (Ultimate 3000; Sunnyvale, CA USA) on a Macherey Nagel Nucleosil RP 300–5C18 column (10×250 mm; particle size 5 µm and pore size 100 nm). Synthetic peptides were analysed by an automated Edman sequencing on an Applied Sequencing System Procise (Applied Biosystems, Foster City, USA). [9] Finally, MALDI-TOF mass measurements were carried out on an Ultraflex™ TOF/TOF (BrukerDaltonics, USA), as previously described. [25].
Isolation and characterization of Staphylococcus aureus strains
Different S. aureus strains were used to demonstrate the peptide antimicrobial activity and subsequently, the peptide degradation: strains ATCC 25923, ATCC49775, S1 and S2, were provided by the Institute of Bacteriology, Strasbourg, France. S1 was isolated from the blood of an 83 y. o. patient and S2 was isolated from the sputum of a 12 days old neonate. After isolation and identification, S. aureus strains were assessed for their susceptibility to various antibiotics, using the agar disc diffusion method. [26] S1 was found Methicillin resistant (MRSA) and is also resistant to Amoxicillin, Oxacillin, Amikacin, Tobramycin, Fluoroquinolones, Erythromycin and Clindamycin. However, it was susceptible to Gentamicin, Synercid, Co-trimoxazole, Rifampicin, Fusidic acid, Vancomycin, Teicoplanin and Linezolid. In contrast, S2 was found to be Methicillin susceptible (MSSA) and is sensitive to all the antibiotics tested.
Antibacterial activity against S. aureus
Different S. aureus strains described above were first pre-cultured aerobically at 37°C for 20 h in a Mueller-Hinton-Broth (MHB) medium, pH 7.3 (Difco Laboratories, Detroit, MI). Bacteria were suspended at absorbance of 0.001 at 620 nm in the MHB medium. Antibacterial activity was tested for 24 h incubation at 37°C with shaking by measuring the inhibition of bacterial growth. Ten µl final volumes (10–200 µg/mL) of synthetic peptides (LL-37, bCAT, hCAT and bCTL) were incubated in microtitration plates with 90 µl of a mid-logarithmic phase culture of bacteria, with a starting absorbance of 0.001 at 620 nm. In the initial inoculi, bacteria were quantified by the agar plate spreading method which was 5×105 colony forming units (CFU)/mL. [27] Tetracycline (10 mg/L) and Cefotaxime (0.1 mg/L) were used as positive controls. Microbial growth was assessed by the increase of absorbance after 24 h incubation at 37°C. [28], [29] The A620 nm value of control cultures growing in the absence of peptide and antibiotics was defined as 100% growth. A620 nm zero with the antibiotics (Tetracycline and Cefotaxime) was taken as 100% inhibition. Absence of bacterial growth was verified by agar plate spreading. Each assay was performed in triplicates.
Killing kinetics
Bacterial strains were first grown in MHB medium as described above. Bacteria killing kinetic activity was measured according to the previously described method, after making few modifications. [30] Initial inoculum was prepared at a concentration of 5×105 CFU/mL with absorbance of 0.001 at 620 nm, which was calculated by agar plate spreading method. [27] Bacteria were incubated with different concentrations of peptides (MIC and 2× MIC), determined by MIC assay via microdilution method. [27] Viable bacterial count was then assessed at different time intervals up to 24 h. Briefly, aliquots were taken at different time intervals and were diluted in phosphate buffer saline (pH 7.4). After appropriate shaking 100 µL of the dilutions were plated on MH agar plates. Plates were incubated at 37°C and colony count was performed to determine CFU/mL after 24 h.
Preparation of S. aureus supernatants
S. aureus strains were precultured and plated on the agar plates and cultivated for 24 h at 37°C. After incubation, one colony per isolate was transferred to 10 mL of the MHB medium and incubated at 37°C for about 30 h to late stationary growth phase. After incubation, cultures were centrifuged at 10,000×g for 15 min and supernatants were filtered using a 0.22 µM Millex®-GV (Millipore, Carrigtwohill, Ireland) to eliminate bacteria. The supernatants were stored at −20°C until further use. In order to check sterility, 1 mL of each supernatant was incubated at 37°C for 48 h. Absence of a colony was interpreted as lack of viable bacteria.
Degradation analysis of the synthetic antimicrobial peptides by S. aureus supernatants
Synthetic peptides catestatin (bovine and human) and cateslytin (bovine) were incubated at 37°C for 24 h, with the different strain supernatants, at a concentration corresponding to the MIC values. Triplicate wells were treated for each concentration of peptide. In parallel, several controls were performed: with and without peptides in MHB medium.
In order to prevent degradation, the above experiment was repeated. For which, protease inhibitor cocktail (complete protease inhibitors except metalloproteases) (Roche Diagnostic GmbH, Mannheim, Germany) supplemented with 100 µM of 1–10 phenanthroline (metalloprotease inhibitor) (Sigma Aldrich GmbH, Steinheim, Germany) was added to bacterial supernatants before the addition of synthetic peptides. The whole suspension was incubated at 37°C for 24 h prior to being analyzed by RP-HPLC.
RP-HPLC purification of CgA-derived peptides after incubation with S. aureus supernatants
The synthetic peptides treated with bacterial supernatants and different controls including culture medium, bacterial supernatants, and synthetic peptides were separated using a Dionex HPLC system (Ultimate 3000; Sunnyvale, CA USA) on a Nucleosil reverse-phase 300–5C18-column (4×250 mm; particle size, 5 µm; porosity, 300 Å) (Macherey Nagel Hoerdt, France). Absorbance was monitored at 214 nm, and the solvent system consisted of 0.1% (v/v) TFA in water (solvent A) and 0.09% (v/v) TFA in 70% (v/v) acetonitrile-water (solvent B). Elution was performed at a flow rate of 700 µL/min, with the gradient as indicated on chromatograms. Fractions were collected with one min time interval. They were subsequently concentrated by evaporation by using a speed-vac, however not allowing them to dry completely.
Automatic Edman sequencing of CgA-derived peptides
The N-terminal sequence of purified peptides was determined by automatic Edman degradation analysis on a Procise microsequencer (Applied Biosystems, Courtaboeuf, France). Samples purified by HPLC were loaded on polybrene-treated glass-fiber filters. Phenylthiohydantoin-amino acids (Pth-Xaa) were identified by chromatography on a C18 column (PTH C-18, 2.1 mm×200 mm). [2].
Proteomic analysis
Mass determination was carried out on a Brucker BIFLEX™ Matrix-Assisted Laser Desorption Ionization – Time-Of-Flight mass spectrometer (MALDI-TOF) (equipped with the high resolution optics (SCOUT™) with X–Y multi-sample probe, a gridless reflector and with the HIMAS™ linear detector). With a maximum accelerating potential of 30 kV, the system can be operated either in linear or the reflective mode. Ionization was carried out with a 337-nm beam from a nitrogen laser with a repetition rate of 3 Hz. The output signal from the detector was digitized at a sampling rate of 250 MHz in linear mode and 500 MHz in reflector mode using a 1 GHz digital oscilloscope (Lecroy model). Brucker supplied software hosted in Sun sparcworkstation was used for data processing and the instrumental control. [31] These studies were realized using the matrix α-cyano-4-hydroxycinnamic acid, obtained from Sigma, and prepared as a saturated solution in acetone. A total of 1–2 µl of the sample matrix aliquot solution was deposited on the probe and dried by ambient air. A thin layer matrix crystal was obtained after rapid spreading and evaporation. [32] A micromolar analyte solution was applied to the matrix and allowed to dry under moderate vacuum. The whole preparation was washed by 1 μl of trifluoroacetic acid (0.5%) aqueous solution. This cleaning procedure helps remove remaining alkaline cations and often leads to an increase in sensitivity.
Statistical analysis
The MIC values (µg/mL) are reported as means of three independent experiments. To determine significance, between different peptides (rows) and bacterial strains (columns), one way ANOVA was used. Overall significance and correlation was evaluated by 4×4 factorial ANOVA for independent samples. Significance was accepted at p≤0.05.
Results
Antibacterial activity of CgA-derived peptides against S. aureus
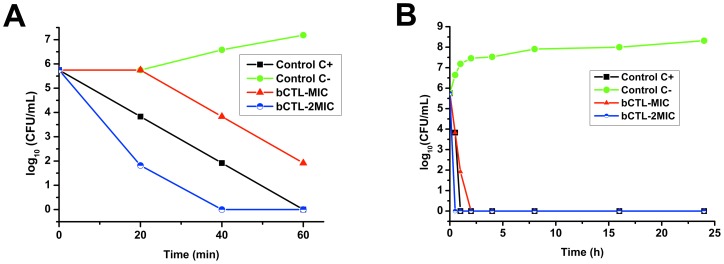
The aim of present study was to examine antibacterial activity of bovine and human CAT (bCgA344–364 and hCgA352–372) and bovine CTL (bCgA344–358), against S. aureus. In comparison with the other peptides tested, CTL was found to be most active against all S. aureus strains tested (ATCC49775, ATCC25923, S1, and S2). Activity of these CgA derived peptides was compared to the well-known C-terminal peptide (LL-37) of hCAP-18 (Table 1). Activity of bCTL was comparable to that of LL-37 (p>0.05) against S1 and S2, but significantly different (p<0.05) for ATCC49775 and ATCC25923. However, MIC (presenting 100% growth inhibition) values for bCAT and hCAT were highly significantly different (p<0.01) that were about 2–4 fold higher than short fragment CTL (Table 1). MIC value for CTL varies between different MRSA and MSSA strains (p<0.05) and can effectively kill bacteria at 45 µg/mL. While both human and bovine CAT have more than 100 µg/mL MIC values. In order to detail CTL antimicrobial activity, we further determined the killing rate of CTL. Time-killing kinetic assays were performed against S. aureus (ATCC 25923). As shown in Figure 1, CTL acts very rapidly against S. aureus. AT concentration 2×MIC CTL present 4 logs bacterial killing at within 20 min of treatment and 100% within 40 min (Figure 1A). Moreover, at MIC, CTL reaches 100% bacterial killing within 60 min of treatment. This killing assay was continued till 24 h (Figure 1B) and bacteria were unable to grow further during 24 h. As CTL is the most active, we hypothesized this fragment is more resistant to proteases produced by S. aureus. In order to demonstrate the stability of CTL compared to human and bovine CAT, we incubated the three peptides with the supernatants of four different S. aureus strains.
Table 1. Antibacterial assays of catestatin (bovine, bCAT and human, hCAT) and cateslytin (bovine, bCTL) against different S. aureus strains, compared to the cathelicidin antimicrobial peptide-18 (LL-37).
| Antimicrobial peptides | MIC (µg/mL) | |||
| S. aureus ATCC49775 | S. aureus ATCC25923 | S. aureus S1 (MRSA) | S. aureus S2 (MSSA) | |
| bCAT | 100 aA | 100 aA | 100 aA | 115 bA |
| hCAT | 125 aB | 135 aB | 130 aB | 150 bB |
| bCTL | 37 aC | 40 aC | 37 aC | 45 bC |
| LL-37 | 30 aD | 30 aD | 30 aC | 35 bC |
Results are presented as MIC (µg/mL) of each peptide against four S. aureus strains. Values represent the means of the triplicate (n = 3) wells. Means with same letters are not significantly different (p<0.05). Small letters (a, b, c) represents significance between different S. aureus strains, while capital letters (A, B, C) represent significantly difference between peptides.
Figure 1. The bacterial killing kinetics of the bCTL against the S. aureus ATCC 25923.
Different concentrations (MIC 40 µg/mL and 2× MIC 80 µg/mL) of bCTL were used. C+, represents antibiotic control (Cefotaxime 0.1 µg/mL + Tetracycline 10 µg/mL) and C-, represents phosphate buffer saline control. (A): S. aureus killing kinetics at time zero to 60 min. (B): S. aureus killing kinetics over 24 h time period.
Analysis of the proteolytic cleavage of catestatin (h/bCAT) and cateslytin (CTL) in the presence of S. aureus strain supernatants
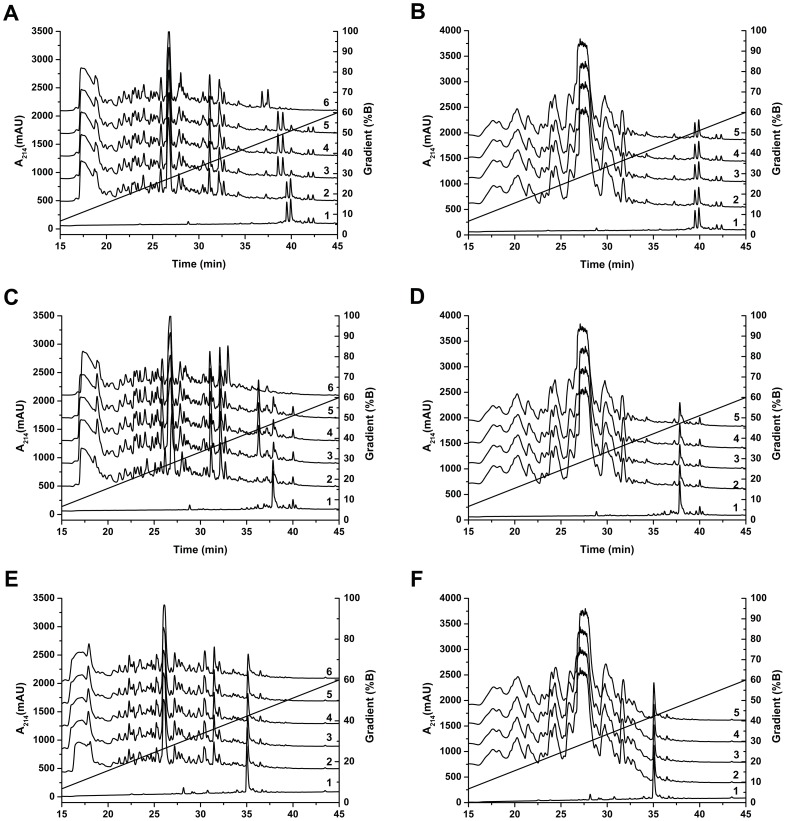
S. aureus agr and sar regulate expression of proteases, which often are modulated by SarA regulator during biofilm formation. [33] These proteases are mostly expressed in the late growth phase and secreted in the extracellular environment to facilitate the bacterial spread. [34] As described above, S. aureus supernatants were prepared by the extended incubation period and are rich in proteases. Four S. aureus strains were tested for these experiments: two of them are referenced strains (ATCC 49775 and ATCC 25923), while two strains (S1 and S2) were isolated from the patients of Strasbourg hospital. S1 is a MRSA (methicillin resistant S. aureus) and S2 is a MSSA (methicillin sensitive S. aureus). The synthetic peptides were incubated with S. aureus supernatants and were separated by RP-HPLC (Figure 2 A, C, E). The HPLC profiles of CAT and CTL are compared with HPLC profiles of MHB medium and S. aureus supernatants. Peaks resulting from the bacterial degradation were analyzed by automatic Edman sequencing and MALDI-TOF. In contrast to bovine and human CAT (Figure 2A, 2C), CTL is not degraded (Figure 2E).
Figure 2. HPLC chromatograms of bCAT, hCAT and CTL alone or with different bacterial strain supernatants, with or without protease inhibitors.
(A): Alignment of the HPLC chromatograms corresponding to: (1) bCAT, (2) bCAT+MHB, (3) bCAT+S49775, (4) bCAT+S25923 (5) bCAT+S1 (6) bCAT+S2. (B): Alignment of the HPLC chromatograms corresponding to: (1) bCAT, (2) bCAT+Pi+S49775, (3) bCAT+Pi+S25923 (4) bCAT+Pi+S1 (5) bCAT+Pi+S2. (C): Alignment of the HPLC chromatograms corresponding to: (1) hCAT, (2) hCAT+MHB, (3) hCAT+S49775, (4) hCAT+S25923 (5) hCAT+S1 (6) hCAT+S2. (D): Alignment of the HPLC chromatograms corresponding to: (1) hCAT, (2) hCAT+Pi+S49775, (3) hCAT+Pi+S25923 (4) hCAT+Pi+S1 (5) hCAT+Pi+S2. (E): Alignment of the HPLC chromatograms corresponding to: (1) bCTL, (2) bCTL+MHB, (3) bCTL+S49775, (4) bCTL+S25923 (5) bCTL+S1 (6) bCTL+S2. (F): Alignment of the HPLC chromatograms corresponding to: (1) bCTL, (2) bCTL+Pi+S49775, (3) bCTL+Pi+S25923 (4) bCTL+Pi+S1 (5) bCTL+Pi+S2.
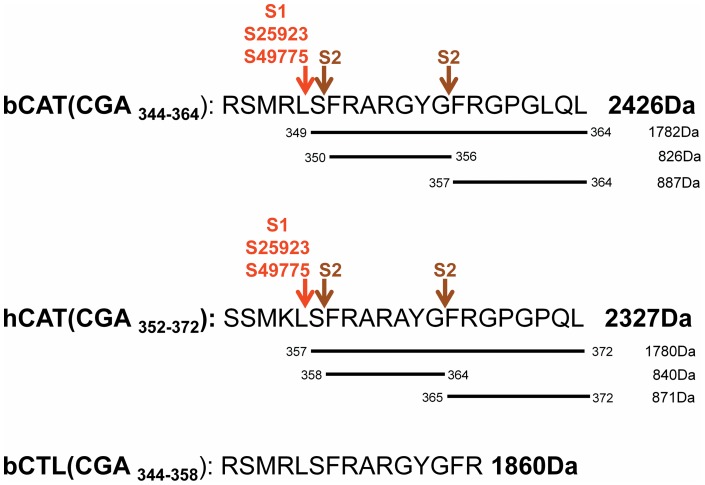
HPLC of bCAT (Figure 2A) indicates 2 major peaks, eluted at 38.7 and 40 min, corresponding to full-length peptide according to sequencing and MALDI-TOF mass spectrometry (2426 Da) (Figure 3). Presence of these 2 isoforms may be related to beta-turn conformation induced by proline residue in position 360 and its isomeric state (cis/trans). [35] In MHB medium, bCAT was not processed. Whereas, in presence of culture supernatants (S49775, S25923 and S1), bCAT was processed to generate fragments eluted at 38.3 and 38.6 min (Figure 2A). Sequencing and MALDI-TOF analysis indicate that these 2 fragments correspond to the isoforms of bCgA349–364 (1782 Da) (Figure 3). In presence of culture supernatant of S2, bCAT was largely cleaved to generate fragments eluted at 28.0, 36.6 and 37.5 min (Figure 2A). Sequencing and MALDI-TOF analysis indicate that these fragments correspond to bCgA350–356 (826 Da) and the 2 isoforms of bCgA357–364 (887 Da) (Figure 3).
Figure 3. The proteolytic cleavage sites induced after treatment of bCAT, hCAT and CTL with different S. aureus strains (ATCC25923, ATCC49775, S1 and S2).
The experimental molecular masses obtained after MALDI-TOF analysis, are indicated.
Moreover, the HPLC profile of hCAT (Figure 2C) showed one major peak at 38 min that corresponds to the full-length peptide according to sequencing and MALDI-TOF mass spectrometry (2327 Da) (Figure 3). In presence of MHB medium, hCAT was not cleaved. However, after incubation with S49775, S25923 and S1, hCAT was partially processed to generate a fragment eluted at 36 min. Sequencing and MALDI-TOF analysis indicated that this fragment corresponds to hCgA357–372 (1780 Da) (Figure 3). In presence of supernatant S2, hCAT was completely cleaved to generate fragments eluted at 28.5 and 33 min (Figure 2C). Sequencing and MALDI-TOF analysis indicated that these fragments correspond to hCgA358–364 (840 Da) and hCgA365–372 (871 Da) (Figure 3).
Finally, the shortest peptide bCTL (bCTL, CgA344–358), is eluted as a single peak (Figure 2E) and is not degraded by any of the staphylococcal strains tested.
Interestingly, incubation of synthetic peptides with bacterial supernatants supplemented with protease inhibitors, prevents the peptide degradation (Figure 2B, D, F). HPLC profiles of bCAT (Figure 2B: chromatogram 2–5) indicate that it is not cleaved by any of the four strain supernatants tested in presence of protease inhibitors. Similar results were obtained for hCAT (Figure 2D: chromatogram 2–5). In addition, CTL remains unaffected by S. aureus supernatants with or without protease inhibitors (Figure 2E, F).
The proteolytic cleavage sites of bCAT and hCAT obtained after incubation with S49775, S25923 and S1 were identical (Figure 3). They corresponded to the peptide bonds L348−S349 and L356–S357 for bCAT and hCAT, respectively. With S2, they correspond to the peptide bonds S349−F350+G356−F357 and S357−F358+G364−F365 for bCAT and hCAT, respectively. It is important to point out that the short peptide bCTL (bCgA344–358) derived from CAT was not cleaved by S. aureus proteases (Figure 2E, F) demonstrating that CTL resists to the proteolytic cocktail produced by S. aureus.
Discussion
Due to continuous resistance development by S. aureus, the interest for natural AMPs is increasing because some of them display potent antibacterial activity. However, secondary reactions such as hemolytic activity, [36] limit their use as systemic anti-infective drugs. Until recently, no such reaction was reported for the antimicrobial Cgs-derived peptides in the literature. Several CgA-derived peptides were previously characterized to display antimicrobial properties, [9] however, data concerning killing of S. aureus is not reported. CAT was previously reported, to be active against different bacterial strains at micromolar range. [24] CTL is well characterized for antimicrobial activity against Micrococcus luteus, few yeasts and fungal strains. Moreover, it is not hemolytic for human erythrocytes. [5]. In the present study, for the first time, we brought to light the antibacterial activity of CTL against different strains of S. aureus with a MIC value of 37–45 µg/mL (Table 1).
In vivo, a cross talk is established via dynamics of the interactions between host and bacteria. Many bacterial strains express a variety of proteases, ranging from non-specific and powerful enzymes that degrade many proteins involved in innate immunity, to proteases that are extremely specific in their mode of action. [37] Here, we questioned whether S. aureus proteases can cleave the three CgA-derived peptides studied. This proteolytic degradation could explain the 3–4 fold difference observed in antibacterial activity. To address this issue, we hypothesized that the human and bovine CAT peptides are degraded by the staphylococcal proteases and that CTL resists to the degradation and maintains its activity. Therefore, we tested the effect of four staphylococcal supernatants and demonstrated that bovine and human CAT are similarly degraded to generate inactive fragments, while CTL is not degraded. For bovine and human CAT, the cleavage sites induced by the supernatants are different. For S2 these correspond to the cleavage of S-F/G-F. Whereas, for ATCC49775, ATCC25923, and S1, it corresponds to cleavage of L-S (Figure 3). Upon cleavage by S. aureus proteases, the CAT peptides are no longer active against S. aureus. Previous data showed that bovine CgA344–351 and CgA348–358 display an antimicrobial activity against Micrococcus luteus at 20 µM, suggesting that L348/356 is important for its antibacterial activity. [5] Furthermore, in the protease inhibition assay, we demonstrated that degradation of human and bovine CAT can be prevented by the use of protease inhibitors. CTL maintains its position either with or without protease inhibitors indicating that proteases have no effect on the CTL.
The biosynthesis of CTL results by the action of prohormone convertases PC1/2 present in the granular matrix of chromaffin cells. [2], [38] The prohormone thiol protease (PTP) was also reported to be essential for CAT synthesis (bCgA344–364) by cleaving D-R and L-R. [38] In addition, in chromaffin secretory vesicles, the cysteine protease cathepsin L (CTSL) [39] generates CTL by the additional cleavage R-G of CAT. CAT fragment is known to activate the neutrophils by inducing extra-cellular calcium influx in human neutrophils via calmodulin-regulated calcium independent phospholipase A2. [11] Here, we demonstrate that CTL is resistant to the degradation by staphylococcal proteases, which strengthens the involvement of this CgA-derived domain in innate immunity. [10], [40].
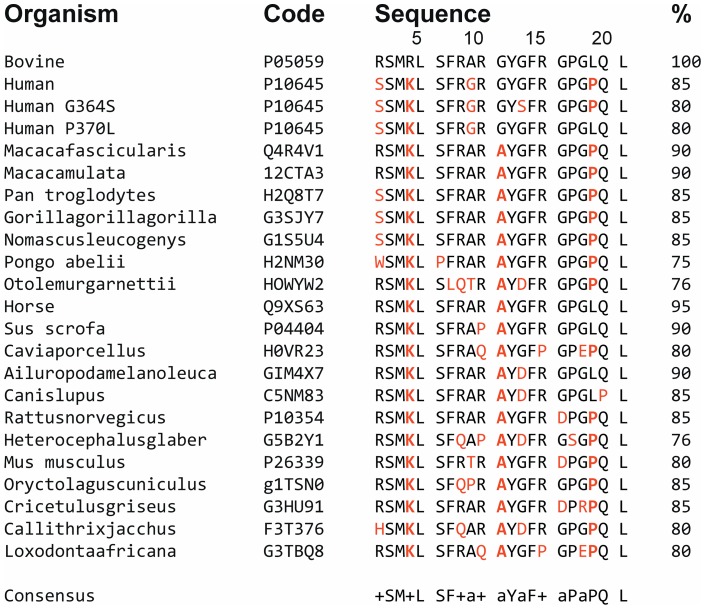
The primary structures of CAT and CTL are highly conserved during evolution (Figure 4). Arginine ratio of these peptide sequences is important, since it modulates the interaction with negatively charges of the microorganism's membranes. In addition, it was previously shown that arginine residues have a high tendency to interact with the lipids as suggested for other peptides, such as HIV-1 transcriptional activator Tat protein. [41], [42] For hCAT, bCAT, and bCTL the arginine ratios are 15%, 23%, and 33% respectively. High arginine ratio of bCTL supports a strong interaction with the negatively charged lipid bilayer as compared to the both CAT. Structure activity relationship of CTL with bacterial membrane is also demonstrated by recent experiments concerning the CTL derived peptide with a hydrophobic N-terminal end FLE-CTL (FLE-RSMRLSFRARGYGFR) (5R). With the help of HPLC and antimicrobial assays, we have shown that this synthetic peptide is inactive against S. aureus at 400 µg/mL and interacts strongly with bacterial membrane. In contrast, the shorter peptide that is lacking the C-terminal end (YGFR) of CTL: FLE-RSMRLSFRARG, displays antibacterial activities at 200 µg/mL (data not shown). Thus, CTL sequence possesses the cationic amphipathic features to exhibit potent antimicrobial activities.
Figure 4. Sequence alignment of bovine catestatin (CgA344–364) with corresponding fragments from several species.
For each position predominant identical residues are indicated in bold letters. Homology sequence is indicated (%).The data base used is UniProtKB. (+, basic residue; a, for A/G/T/P).
Concerning its secondary structure, an aggregated antiparallel beta sheet structure was previously reported for CTL [43] by CD (Circular Dichroism) and ATR (Attenuated total reflectance) experiments, whereas hCAT and bCAT form short helical structures (residue 7 to residue 11) in the presence of high concentrations of DPC, as confirmed by CD and NMR (Nuclear magnetic resonance) experiments. [44] CTL adopts a major β-sheet character only on negatively charged membranes, whereas it is essentially unstructured in water. These β-sheet formations give a much more stability to peptide than the helical formation reported for CAT. [44] In addition, the (HR-MAS) 1H NMR analysis of CTL indicates that the arginine and hydrophobic residues are in close proximity, thus creating a deep penetration of charged residues into the membrane. [42] Therefore, the electrostatic interaction between positively charged arginine residues and negatively charged lipids appears to be responsible for binding of CTL to the lipid bilayer and the aromatic residues stabilize the lipid-peptide interaction.
In our group, we recently evaluated the synergistic effect of three CgA-derived peptides (CAT, CTL and amidated CTL) with antibiotics and demonstrated that this co-treatment induce a reduction of the antibiotics concentration used and can potentiate their activities. Antimicrobial assays were carried out by combining the AMPs at concentration below the MIC. The comparison was made with antibiotic or peptide separately at the same doses. For all these experiments, we evaluated the Fractional Inhibitory Concentration (FIC) of CgA-derived peptides combined with Minocyclin or Voriconazole. FIC corresponds to a synergistic effect in the range of ≤0.5, an additive effect when it is between >0.5 and <2, and an antagonistic effect when it is >2. [45] For the combination of amidated CTL and Minocyclin, we obtained a FIC of 0.37 against S. aureus, and for CTL and Voriconazole, we obtained a FIC of 0.25, and 0.5 against Candida albicans and Candida tropicalis respectively. [10] Regarding these data, one could imagine a mechanism in which, the peptides could favor destabilization of bacterial membrane, thus allowing the antibiotics to rapidly penetrate inside bacterial cells to reaching their site of action. These in vitro results are likely to occur in vivo, especially during systemic inflammation. Indeed, sepsis triggers numerous changes in proteases and inhibitors activity. [46] These regulations have been strongly related with sepsis severity. [47] Under such influences, CAT and CTL could even be difficult to release, owing to structural modifications of CgA, [48] due to sepsis-related oxidative stress. These modifications include oxidation of methionine, aromatic residues, glycanes, phosphorylation and also aggregation of the complete protein that might prevent the processing of CgA to produce active antimicrobial peptides. The structure-function relationship of AMPs is also very important to unravel the regular pattern of antibacterial activity. It has been previously demonstrated, that selective end tagging or short hydrophobic stretches can be, added to increase AMPs activity. [49], [50] Recently, we studied the properties of a modified CTL by inserting cysteine at C-terminal. This modified CTL was used to develop antimicrobial polymer coatings via alternative deposition of polyelectrolytes conjugated to CTL-C. [51] Activity of CTL is not altered, even with this modification or when inserted into PEM (Polyelectrolyte multilayer) coatings. Moreover, it is non-toxic for human gingival fibroblast. [51].
To conclude, for the first time we present the involvement of staphylococcal proteases on the cleavage of antimicrobial CgA-derived peptides. As the CTL domain is resistant to staphylococcal proteases, it constitutes a promising natural antibacterial peptide for further studies.
Acknowledgments
We are grateful for the generous gift of bacterial strains from the bacteriology Institute of Civil Hospital of Strasbourg. Authors thank Cosette Betcha and Ouria Tahar for their excellent technical assistance. We also acknowledge Azhar Ayyaz Pirzado and Adnan Khan Niazi for their help in writing.
Funding Statement
The authors thank the University of Strasbourg, the Inserm for financial support and the Faculty of Odontology and Higher Education Commission (HEC) of Pakistan for the PhD grant of RA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Helle KB (2010) Regulatory peptides from chromogranin A and secretogranin II: putative modulators of cells and tissues involved in inflammatory conditions. Regul Pept 165: 45–51. [DOI] [PubMed] [Google Scholar]
- 2. Metz-Boutigue MH, Garcia-Sablone P, Hogue-Angeletti R, Aunis D (1993) Intracellular and extracellular processing of chromogranin A. Determination of cleavage sites. Eur J Biochem 217: 247–257. [DOI] [PubMed] [Google Scholar]
- 3. Strub JM, Garcia-Sablone P, Lonning K, Taupenot L, Hubert P, et al. (1995) Processing of chromogranin B in bovine adrenal medulla. Identification of secretolytin, the endogenous C-terminal fragment of residues 614-626 with antibacterial activity. Eur J Biochem 229: 356–368. [DOI] [PubMed] [Google Scholar]
- 4. Montero-Hadjadje M, Vaingankar S, Elias S, Tostivint H, Mahata SK, et al. (2008) Chromogranins A and B and secretogranin II: evolutionary and functional aspects. Acta Physiol (Oxf) 192: 309–324. [DOI] [PubMed] [Google Scholar]
- 5. Briolat J, Wu SD, Mahata SK, Gonthier B, Bagnard D, et al. (2005) New antimicrobial activity for the catecholamine release-inhibitory peptide from chromogranin A. Cell Mol Life Sci. 62: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lugardon K, Chasserot-Golaz S, Kieffer AE, Maget-Dana R, Nullans G, et al. (2001) Structural and biological characterization of chromofungin, the antifungal chromogranin A-(47–66)-derived peptide. J Biol Chem 276: 35875–35882. [DOI] [PubMed] [Google Scholar]
- 7. Lugardon K, Raffner R, Goumon Y, Corti A, Delmas A, et al. (2000) Antibacterial and antifungal activities of vasostatin-1, the N-terminal fragment of chromogranin A. J Biol Chem. 275: 10745–10753. [DOI] [PubMed] [Google Scholar]
- 8. Strub JM, Goumon Y, Lugardon K, Capon C, Lopez M, et al. (1996) Antibacterial activity of glycosylated and phosphorylated chromogranin A-derived peptide 173–194 from bovine adrenal medullary chromaffin granules. J Biol Chem 271: 28533–28540. [DOI] [PubMed] [Google Scholar]
- 9. Metz-Boutigue MH, Goumon Y, Lugardon K, Strub JM, Aunis D (1998) Antibacterial peptides are present in chromaffin cell secretory granules. Cell Mol Neurobiol 18: 249–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aslam R, Atindehou M, Lavaux T, Haikel Y, Schneider F, et al. (2012) Chromogranin A-derived peptides are involved in innate immunity. Curr Med Chem 19: 4115–4123. [DOI] [PubMed] [Google Scholar]
- 11. Zhang D, Shooshtarizadeh P, Laventie BJ, Colin DA, Chich JF, et al. (2009) Two chromogranin a-derived peptides induce calcium entry in human neutrophils by calmodulin-regulated calcium independent phospholipase A2. PLoS One 4: e4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohbayashi T, Irie A, Murakami Y, Nowak M, Potempa J, et al. (2011) Degradation of fibrinogen and collagen by staphopains, cysteine proteases released from Staphylococcus aureus. Microbiology 157: 786–792. [DOI] [PubMed] [Google Scholar]
- 13. Sieprawska-Lupa M, Mydel P, Krawczyk K, Wojcik K, Puklo M, et al. (2004) Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother 48: 4673–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Postma B, Poppelier MJ, van Galen JC, Prossnitz ER, van Strijp JA, et al. (2004) Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J Immunol 172: 6994–7001. [DOI] [PubMed] [Google Scholar]
- 15. Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, et al. (2005) Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci U S A 102: 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rooijakkers SH, Ruyken M, Roos A, Daha MR, Presanis JS, et al. (2005) Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol 6: 920–927. [DOI] [PubMed] [Google Scholar]
- 17. Karavolos MH, Horsburgh MJ, Ingham E, Foster SJ (2003) Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology 149: 2749–2758. [DOI] [PubMed] [Google Scholar]
- 18. Prokesova L, Potuznikova B, Potempa J, Zikan J, Radl J, et al. (1995) Cleavage of human immunoglobulins by proteinase from Staphylococcus aureus. Adv Exp Med Biol 371A: 613–616. [DOI] [PubMed] [Google Scholar]
- 19. Dubin G (2002) Extracellular proteases of Staphylococcus spp. Biol Chem 383: 1075–1086. [DOI] [PubMed] [Google Scholar]
- 20. Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A (1993) The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun 61: 3879–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chien Y, Manna AC, Projan SJ, Cheung AL (1999) SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem 274: 37169–37176. [DOI] [PubMed] [Google Scholar]
- 22. Palma M, Cheung AL (2001) sigma(B) activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect Immun 69: 7858–7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao Y, Verma V, Belcheva A, Singh A, Fridman M, et al. (2012) Staphylococcus aureus methicillin-resistance factor fmtA is regulated by the global regulator SarA. PLoS One 7: e43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Radek KA, Lopez-Garcia B, Hupe M, Niesman IR, Elias PM, et al. (2008) The neuroendocrine peptide catestatin is a cutaneous antimicrobial and induced in the skin after injury. J Invest Dermatol 128: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sizova D, Charbaut E, Delalande F, Poirier F, High AA, et al. (2007) Proteomic analysis of brain tissue from an Alzheimer's disease mouse model by two-dimensional difference gel electrophoresis. Neurobiol Aging 28: 357–370. [DOI] [PubMed] [Google Scholar]
- 26. Davis MF, Peterson AE, Julian KG, Greene WH, Price LB, et al. (2013) Household Risk Factors for Colonization with Multidrug-Resistant Staphylococcus aureus Isolates. PLoS One 8: e54733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards (2003) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically–Eighth Edition: Approved Standard M07-A8.: NCCLS, Wayne, PA, USA.
- 28. Bulet P, Dimarcq JL, Hetru C, Lagueux M, Charlet M, et al. (1993) A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J Biol Chem 268: 14893–14897. [PubMed] [Google Scholar]
- 29. Wu M, Hancock RE (1999) Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J Biol Chem 274: 29–35. [DOI] [PubMed] [Google Scholar]
- 30. Pag U, Oedenkoven M, Papo N, Oren Z, Shai Y, et al. (2004) In vitro activity and mode of action of diastereomeric antimicrobial peptides against bacterial clinical isolates. J Antimicrob Chemother 53: 230–239. [DOI] [PubMed] [Google Scholar]
- 31. Pons AM, Zorn N, Vignon D, Delalande F, Van Dorsselaer A, et al. (2002) Microcin E492 is an unmodified peptide related in structure to colicin V. Antimicrob Agents Chemother. 46: 229–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kussmann M, Lassing U, Sturmer CA, Przybylski M, Roepstorff P (1997) Matrix-assisted laser desorption/ionization mass spectrometric peptide mapping of the neural cell adhesion protein neurolin purified by sodium dodecyl sulfate polyacrylamide gel electrophoresis or acidic precipitation. J Mass Spectrom 32: 483–493. [DOI] [PubMed] [Google Scholar]
- 33. Mrak LN, Zielinska AK, Beenken KE, Mrak IN, Atwood DN, et al. (2012) saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS One 7: e38453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, et al. (2001) Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183: 7341–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpe S (1995) Proline motifs in peptides and their biological processing. Faseb J 9: 736–744. [PubMed] [Google Scholar]
- 36. Conlon JM, Mechkarska M, Prajeep M, Sonnevend A, Coquet L, et al. (2012) Host-defense peptides in skin secretions of the tetraploid frog Silurana epitropicalis with potent activity against methicillin-resistant Staphylococcus aureus (MRSA). Peptides 37: 113–119. [DOI] [PubMed] [Google Scholar]
- 37. Potempa J, Pike RN (2009) Corruption of innate immunity by bacterial proteases. J Innate Immun 1: 70–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee JC, Taylor CV, Gaucher SP, Toneff T, Taupenot L, et al. (2003) Primary sequence characterization of catestatin intermediates and peptides defines proteolytic cleavage sites utilized for converting chromogranin a into active catestatin secreted from neuroendocrine chromaffin cells. Biochemistry 42: 6938–6946. [DOI] [PubMed] [Google Scholar]
- 39. Biswas N, Rodriguez-Flores JL, Courel M, Gayen JR, Vaingankar SM, et al. (2009) Cathepsin L colocalizes with chromogranin a in chromaffin vesicles to generate active peptides. Endocrinology 150: 3547–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sternberg EM (2006) Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nat Rev Immunol 6: 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Henriques ST, Melo MN, Castanho MA (2006) Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem J 399: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ziegler A (2008) Thermodynamic studies and binding mechanisms of cell-penetrating peptides with lipids and glycosaminoglycans. Adv Drug Deliv Rev 60: 580–597. [DOI] [PubMed] [Google Scholar]
- 43. Jean-Francois F, Castano S, Desbat B, Odaert B, Roux M, et al. (2008) Aggregation of cateslytin beta-sheets on negatively charged lipids promotes rigid membrane domains. A new mode of action for antimicrobial peptides? Biochemistry 47: 6394–6402. [DOI] [PubMed] [Google Scholar]
- 44. Sugawara M, Resende JM, Moraes CM, Marquette A, Chich JF, et al. (2010) Membrane structure and interactions of human catestatin by multidimensional solution and solid-state NMR spectroscopy. Faseb J 24: 1737–1746. [DOI] [PubMed] [Google Scholar]
- 45. Serra P, Brandimarte C, Martino P, Carlone S, Giunchi G (1977) Synergistic treatment of enterococcal endocarditis: in vitro and in vivo studies. Arch Intern Med 137: 1562–1567. [PubMed] [Google Scholar]
- 46. Kwiecinski J, Josefsson E, Mitchell J, Higgins J, Magnusson M, et al. (2010) Activation of plasminogen by staphylokinase reduces the severity of Staphylococcus aureus systemic infection. J Infect Dis 202: 1041–1049. [DOI] [PubMed] [Google Scholar]
- 47. Michel A, Agerer F, Hauck CR, Herrmann M, Ullrich J, et al. (2006) Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J Bacteriol 188: 5783–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Strub JM, Sorokine O, Van Dorsselaer A, Aunis D, Metz-Boutigue MH (1997) Phosphorylation and O-glycosylation sites of bovine chromogranin A from adrenal medullary chromaffin granules and their relationship with biological activities. J Biol Chem 272: 11928–11936. [DOI] [PubMed] [Google Scholar]
- 49. Pasupuleti M, Schmidtchen A, Chalupka A, Ringstad L, Malmsten M (2009) End-tagging of ultra-short antimicrobial peptides by W/F stretches to facilitate bacterial killing. PLoS One 4: e5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Malmsten M, Kasetty G, Pasupuleti M, Alenfall J, Schmidtchen A (2011) Highly selective end-tagged antimicrobial peptides derived from PRELP. PLoS One 6: e16400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cado G, Aslam R, Séon L, Garnier T, Fabre R, et al.. (2013) Self-defensive biomaterial coating against bacteria and yeasts: polysaccharide multilayer film with embedded antimicrobial peptide. Adv functional materials [DOI: 10.1002/adfm.201300416].