Abstract
Background & Aims
Vitamin D protects against colorectal cancer by unclear mechanisms. We investigated the effects of calcitriol (1α,25-dihydroxyvitamin D3, the active form of vitamin D) on levels of different microRNAs (miRs) in colorectal cancer (CRC) cells from humans and xenograft tumors in mice.
Methods
Expression of microRNAs in CRC cell lines was examined using the Ambion mirVana miRNA Bioarray. The effects of calcitriol on expression of miR-627 and cell proliferation were determined by real-time PCR and WST-1 assay, respectively; growth of colorectal xenograft tumors was examined in nude mice. Real-time PCR was used to analyze levels of miR-627 in human colon adenocarcinoma samples and non-tumor colon mucosa tissues (controls).
Results
In HT-29 cells, miR-627 was the only microRNA significantly upregulated by calcitriol. Jumonji domain containing 1A (JMJD1A), which encodes a histone demethylase, was found to be a target of miR-627. By downregulating JMJD1A, miR-627 increased methylation of histone H3K9 and suppressed expression of proliferative factors such as GDF15. Calcitriol induced expression of miR-627, which downregulated JMJD1A and suppressed growth of xenograft tumors from HCT-116 cells in nude mice. Overexpression of miR-627 prevented proliferation of CRC cell lines in culture and growth of xenograft tumors in mice. Conversely, blocking the activity of miR-627 inhibited the tumor suppressive effects of calcitriol in cultured CRC cells and in mice. Levels of miR-627 were decreased in human colon adenocarcinoma samples, compared with controls.
Conclusions
miR-627 mediates tumor-suppressive epigenetic activities of vitamin D on CRC cells and xenograft tumors in mice. The mRNA that encodes the histone demethylase JMJD1A is a direct target of miR-627. Reagents designed to target JMJD1A or its mRNA, or increase the function of miR-627, might have the same antitumor activities of vitamin D without the hypercalcemic side effects.
Keywords: gene regulation, mRNA processing, apoptosis, cancer prevention
Introduction
Low circulating vitamin D levels have been linked to increased risks of cancer by epidemiologic studies, and preclinical research has demonstrated clear antitumor activities of vitamin D against various types of cancers 1. For colon cancer, clinical studies have shown that vitamin D supplementation achieving a level of >82 nM 25(OH)D3 can lower cancer incidence by 50% 2. Further supporting a role of vitamin D in colon cancer, epidemiologic studies have demonstrated close relationships between levels of sunlight exposure 3, 4, serum levels of 25(OH)D3 5–7, vitamin D receptor polymorphisms 8, 9, and colon cancer. Calcitriol (1α,25(OH)2D3), the active form of vitamin D, has been shown to inhibit proliferation, induce differentiation, and activate apoptosis in colon cancer cells 1, 10. However, the mechanism of calcitriol’s antitumor action remains poorly defined and the hypercalcemic side effect of vitamin D limits its use as an anti-cancer agent. Thus, it is critical to identify molecular targets of calcitriol in order to design therapy utilizing the anticancer mechanisms while bypassing its toxic effects.
microRNAs (miRNAs) are small 19–25 nucleotide-long, single-stranded non-coding RNAs that silence their target genes by inhibiting mRNA translation or causing mRNA degradation 11. The miRNAs are transcribed by pol II as long pri-miRNAs and processed to form ~60–70 nucleotide-long pre-miRNAs by Drosha RNase III endonuclease 12. Dicer, another RNase III endonuclease, further processes the pre-miRNAs and the mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) to recognize and bind to target mRNAs. Recent studies have implicated the involvement of miRNAs in many aspects of cancer development, regulating cell cycle 13, differentiation 14, 15, metabolism 16, invasion and metastasis 17, as well as apoptosis 18.
Post-translational modification of the histones (acetylation, methylation, phosphorylation, and ubiquitination) is an important epigenetic mechanism to regulate gene expression. Histone lysine residues (for example, H3K4, H3K9, H3K27, H4K20, etc) can be mono-, di-, or trimethylated. While histone methyltransferases catalyze the methylation of histones, the histone demethylases remove the methylation marks in a site-specific manner 19, 20. In general, H3K9, H3K27, and H4K20 methylation are associated with repression of gene expression, while H3K4, H3K36, and H3K79 methylation are associated with active transcription 21, 22. Two families of histone demethylases have been identified, including the flavin adenine dinucleotide (FAD)-dependent family and the jumonji C (JmjC) domain containing family 23. JMJD1A is a member of the JmjC domain containing family and responsible for demethylation of mono- and dimethylated H3K9 24. Recent studies have shown that JMJD1A enhances colon cancer growth by acting as a mediator of hypoxia-inducible transcription factor (HIF) to induce the expression of hypoxia-inducible genes 25.
To explore the role of microRNAs in the action of calcitriol, we examined the miRNA expression profiles in colon cancer cells treated with calcitriol. We found that calcitriol selectively induces the expression of miR-627. We further demonstrated that miR-627 targets histone demethylase JMJD1A. Our data suggest that miR-627-mediated JMJD1A downregulation is an important intracellular target for calcitriol and mediates epigenetic activities of calcitriol.
Results
Calcitriol inhibits the proliferation of colon cancer cells
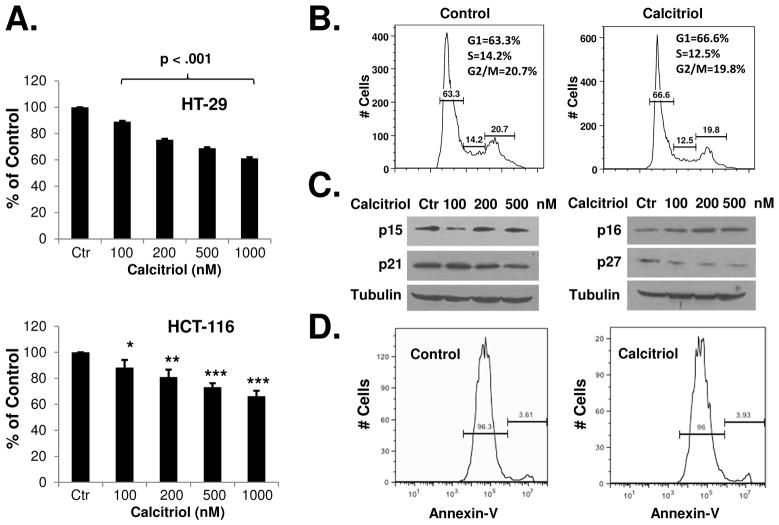
We examined the effects of calcitriol on colon cancer cell growth. As shown in Figure 1A, calcitriol was effective in suppressing the in vitro proliferation of HT-29 and HCT-116 cells. To determine the potential mechanism of calcitriol-induced growth suppression, we analyzed the cell cycle distribution of calcitriol-treated cells. As shown in Figure 1B, calcitriol slightly increased the percentage of cells in the G1 phase of cell cycle (but not statistically significant). To determine if calcitriol caused a general slowdown of the cell cycle, we measured the cell doubling time. After measuring cell numbers at various time points in control and calcitriol-treated cells, we calculated the cell doubling time using an online doubling time calculator (http://www.doubling-time.com/compute.php). As shown in Supplementary Figure 1A, calcitriol increased the cell doubling time by about 3 hours (p < 0.01). We also measured cell cycle progression by a pulse-chase experiment using flow cytometric analysis, which simultaneously detected bromodeoxyuridine (BrdU) labeling and DNA content (propidium iodide labeling). After a brief (30 minutes) pulse labeling with BrdU, the S phase cells (which were labeled with BrdU) were followed as they progressed through the cell cycle (at 4h, 12h, and 16h after labeling), with the cells entering G2 phase and subsequently mitosis. At 16 hours after BrdU labeling, most of the BrdU labeled cells had complete mitosis (G2 and M phases together are usually less than six hours 26) and in the process of a new cycle (anywhere in the G1, S, or G2 phases). As shown in Supplementary Figures 1B and 1C, calcitriol caused a slowdown of the cell cycle: more BrdU-labeled cells were found to lag behind at all three time points among the calcitriol-treated cells than the control cells. Calcitriol may increase the cell doubling time through various mechanisms, for example, by acting on the cell cycle regulators. We examined the effects of calcitriol on the cyclin-dependent kinase inhibitors. As shown in Figure 1C, p15, p21, and p27 were not induced by calcitriol, while p16 was slightly increased (Figure 1C). While it was previously reported that calcitriol induced p21 in HCT-116 cells, a significantly higher dose (1 μM) was used in that study 27. At the lower doses that we used, induction of p16 may contribute to the effects of growth inhibition of calcitriol. Furthermore, using annexin V staining assay, we found that calcitriol induced minimal levels of apoptosis in the cancer cells (Figure 1D).
Figure 1.
Calcitriol inhibits the in vitro growth of colon cancer cells. (A) HT-29 and HCT-116 cells were treated with various doses of calcitriol for 48h and in vitro cell growth was analyzed using WST-1 assay as described in Materials and Methods. (B) HCT-116 cells were treated with 500 nM calcitriol for 48h and cell cycle distribution was analyzed by flow cytometry. The experiment was repeated three times; representative results of three independent experiments were shown. The percentages of cells in each phase of cell cycle (mean ± S.D.) are: Control: G1 (60.6 ± 3.2), S (14.5 ± 0.4), G2 (22.5 ± 1.9); Calcitriol: G1 (65.0 ± 2.7), S (12.8 ± 0.4), G2 (21.2 ± 2.0). (C) HCT-116 cells were treated with various doses of calcitriol for 48h and the expression of cell cycle regulatory proteins was analyzed by western blots. The experiment was repeated three times. Representative images of three independent experiments were shown. (D) HCT-116 cells were treated with 500 nM of calcitriol for 48 hours, apoptosis was determined by annexin V staining and flow cytometry. The experiments have been repeated three times, representative results of three independent experiments were shown. Data shown are mean values + SD (*p<0.05, **p<0.01, ***p<0.001).
Calcitriol selectively induces miR-627 expression and miR-627 mediates the antitumor activity of calcitriol
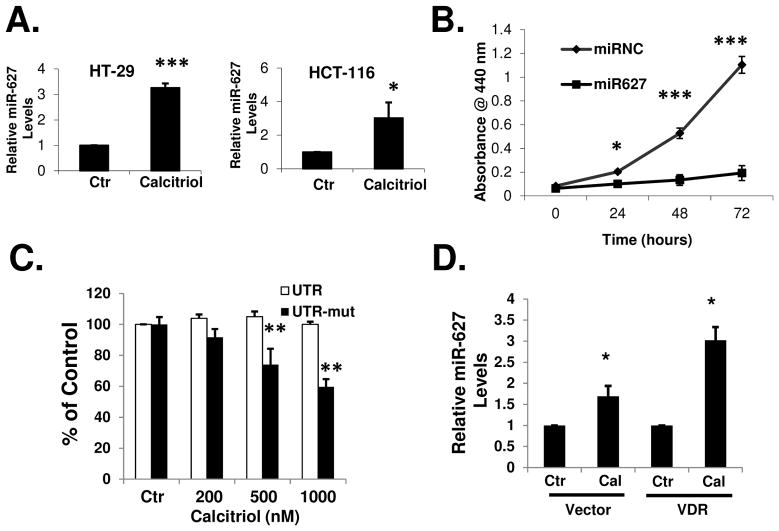
In order to understand the mechanism of how calcitriol inhibits cancer growth, we explored the effects of calcitriol on the expression of the microRNAs in colon cancer cells. We compared microRNA expression profiles in HT-29 cells before and after calcitriol treatment, using the Ambion mirVana miRNA Bioarray. Among the 471 human miRNAs examined, miR-627 is the only microRNA whose expression level was significantly increased by calcitriol (Supplementary Table 1). The upregulation of miR-627 by calcitriol was confirmed by real-time PCR in HT-29 and HCT-116 cells (Figure 2A). We selected miR-627 to further study its role in the antitumor activity of calcitriol. MiR-627 is one of the miRNAs identified in a recent genome-wide miRNA expression study in colon cancer cells 28. To date, there are no studies on the function of miR-627 reported. To determine the effects of miR-627 on cancer proliferation, we transfected synthetic miR-627 mimic into HCT-116 cells. As shown in Figure 2B, miR-627 significantly inhibited cancer cell growth while the negative control miRNA had no effects. Thus, miR-627 may mediate the anti-proliferative activity of calcitriol. To further test this hypothesis, we overexpressed a cDNA fragment containing a miR-627 target sequence (part of the 3′UTR of JMJD1A gene, see text below) in HCT-116 cells. The overexpressed mRNA containing the miR-627 target sequence (driven by a strong CMV promoter) served as a “sponge” to sequester and block the activity of miR-627. We employed this miRNA sponge strategy 29, 30 to create HCT-116 cell lines stably expressing the 3′UTR-derived miR-627 sponge for in vitro and long term in vivo (see text below) studies. When miR-627 was blocked by the sponge, calcitriol failed to suppress the growth of HCT-116 cells (Figure 2C), indicating that induction of miR-627 is critical to the action of calcitriol. In contrast, when the miR-627 binding site was mutated and the sponge did not block miR-627, calcitriol was still able to suppress cancer cell proliferation (Figure 2C). To determine if the vitamin D receptor VDR is required for the activation of miR-627 expression, we examined the effects of calcitriol in SW620 cells, a colon cancer cell line that expresses extremely low level of VDR (undetectable by Northern Blot) 31. As shown in Figure 2D, calcitriol induced miR-627 expression at a level relatively lower than that in HT-29 or HCT-116 cells (comparing to Figure 2A). However, after transfection of SW620 cells with a plasmid to overexpress full length VDR, calcitriol induced miR-627 expression to a level similar to that in HT-29 or HCT-116 cells (Figure 2D).
Figure 2.
Calcitriol induces miR-627 expression in colon cancer cells. (A) HT-29 cells and HCT-116 cells were treated with 100 nM calcitriol for 24h and total RNA was isolated from the cells and real-time PCR analysis was performed as described in Materials and Methods. (B) HCT-116 cells were transfected with 100 nM negative control miRNA or miR-627 mimic. Cell growth was determined at various time points after transfection by WST-1 assay. (C) HCT-116 cells stably expressing the JMJD1A 3′UTR sponge (to block miR-627) or the JMJD1A 3′UTR sponge with miR-627 binding site mutation (ATTC to TGAG) were treated with various doses of calcitriol for 48h and in vitro cell growth was analyzed using WST-1 assay. (D) Untransfected or pCEP4-Flag-VDR transfected SW620 cells were treated with 100 nM calcitriol for 24 hours, total RNA was isolated and real-time PCR analysis was performed. All of the above experiments have been repeated three times, data shown are mean values + S.D. (*p<0.05, **p<0.01, ***p<0.001).
miR-627 Targets JMJD1A
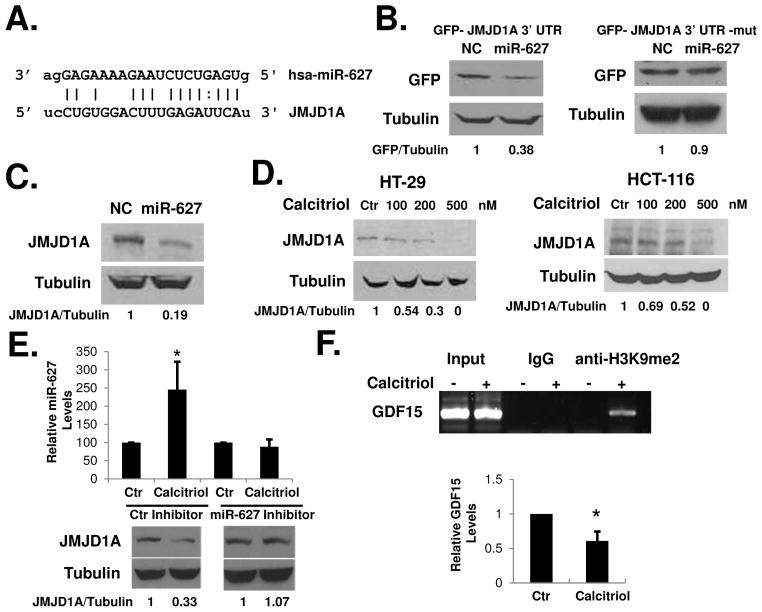
Using the computational methods available at www.microrna.org and miRBase (http://microrna.sanger.ac.uk/), we identified JMJD1A as a potential target for miR-627. The mirSVR score 32 (available at microrna.org) of JMJD1A was relatively high (−1.17). There are about 400 mRNA targets having mirSVR scores similar or higher than that of JMJD1A. We selected JMJD1A for further study because of its known involvement in promoting colon cancer growth (as discussed below). JMJD1A is an iron- and 2-oxoglutarate-dependent dioxygenase which catalyzes the demethylation of mono- and dimethylated H3K9 24. JMJD1A has recently been shown to promote colon cancer growth by mediating the effects of HIF-1α 25. The growth-promoting activity of JMJD1A makes it an interesting target for miR-627. The JMJD1A mRNA contains a 3′UTR sequence that is partially complementary to miR-627 (Figure 3A). When a cDNA fragment containing the 3′UTR sequence of JMJD1A was inserted downstream of the GFP gene in the pEGFP-C1 plasmid and the plasmid was transfected into HCT-116 cells together with pcDNA6.2-GW-miR-627 (to overexpress miR-627), GFP expression was reduced comparing with cells transfected with pEGFP-JMJD1A-3′UTR and pcDNA6.2-GW-negative-control plasmids (Figure 3B, left). The action of the miR-627 was dependent on the miRNA binding site within the JMJD1A 3′UTR, since GFP expression was reduced to a much lesser degree by the miRNA when the binding site was mutated (Figure 3B, right). To further confirm that miR-627 targets JMJD1A, we overexpressed miR-627 in HCT-116 cells using the pcDNA6.2-GW-miR-627 vector. Overexpression of miR-627 decreased JMJD1A protein in the cells (Figure 3C). If miR-627 targets JMJD1A, then calcitriol should decrease JMJD1A since it induces miR-627. Indeed, treatment with calcitriol decreased JMJD1A protein levels in a dose dependent manner in HT-29 and HCT-116 cells (Figure 3D). Conversely, transfection of HCT-116 cells with an LNA-modified synthetic inhibitor anti-miR-627 effectively blocked calcitriol-induced miR-627 and the decrease of JMJD1A (Figure 3E). Since JMJD1A demethylates both di- or mono-methylated histone H3 lysine 9 (which are associated with repressed gene promoters), by decreasing JMJD1A, calcitriol may increase H3K9 methylation on the promoters of JMJD1A target genes, such as GDF15 25. To determine the effects of calcitriol on histone methylation, we performed chromatin immunoprecipitation (ChIP) assay on the promoter region of the GDF15 gene. Calcitriol induced a significant increase of H3K9me2 on the promoter of GDF15 (Figure 3F). As a result, GDF15 mRNA expression was suppressed by calcitriol (Figure 3F). We also determined the effects of calcitriol on two additional histone methylation markers. As shown in Supplementary Figure 2A, calcitriol increased histone H3K27 methylation and decreased histone H3K4 methylation on the promoter of GDF15. Since H3K27 methylation is associated with repression of gene expression and H3K4 methylation is associated with active transcription 21, 22, the observed changes in these two markers may collectively influence GDF15 expression. It has been shown recently that JMJD1A promotes cell cycle progression through the G1/S transition by activating HOXA1 and CCND1 (encoding cyclin D1) expression 33. Using real-time PCR, we found that these JMJD1A target genes were also downregulated by calcitriol (Supplementary Figure 2B). To further determine the role of miR-627 suppression of JMJD1A in colon cancer growth, we specifically knocked down JMJD1A using siRNA. As shown in Supplementary Figure 3A, siRNA knockdown of JMJD1A significantly inhibited the proliferation of the HCT-116 cells. On the other hand, when we overexpressed JMJD1A in the HCT-116 cells, we found that cell proliferation was stimulated. More importantly, the suppressive effect of miR-627 on cell proliferation was abolished by JMJD1A overexpression (Supplementary Figure 3B).
Figure 3.
miR-627 targets JMJD1A. (A) Predicted duplex formation between human JMJD1A 3′UTR and miR-627. (B) HCT-116 cells were transfected with GFP-JMJD1A 3′UTR (left) or GFP-JMJD1A 3′UTR-mut (right) plasmids together with plasmids expressing miR-627 or a negative control miRNA. Western blotting was performed with anti-GFP and anti-tubulin antibodies. Relative protein levels were quantified and shown under the gel. The experiment was repeated three times. The mean protein levels ± S.D. are: left panel, NC (1.0 ± 0), miR-627 (0.375 ± 0.007); right panel, NC (1.0 ± 0), miR-627 (0.83 ± 0.09). (C) HCT-116 cells were transfected with pcDNA6.2-GW/EmGFP-miR-negative control or pcDNA6.2-GW/EmGFP-miR627 plasmids. 48 hours after transfection, cell lysates were analyzed by western blots with the indicated antibodies. The experiment was repeated three times. The mean protein levels ± S.D. are: NC (1.0 ± 0), miR-627 (0.26 ± 0.13). (D) HT-29 and HCT-116 cells were treated with various doses of calcitriol for 48h, and western blots were done using the indicated antibodies. Relative protein levels were quantified and shown under the gel. The experiment was repeated three times. (E) HCT-116 cells were transfected with 100 nM LNA-modified miRNA inhibitor specific to miR-627 or the negative control inhibitor, and treated with 500 nM calcitriol for 48h. Total RNA and cell lysates were analyzed by real-time PCR for miR-627 and western blots with the indicated antibodies. The experiment was repeated three times. The mean protein levels ± S.D. are: left panel, Ctr (1.0 ± 0), Calcitriol (0.33 ± 0.08); right panel, Ctr (1.0 ± 0), Calcitriol (1.07 ± 0.17). (F) HCT-116 cells were treated with 500 nM calcitriol for 48 hours. ChIP assay was performed as described in Materials and Methods, using primers specific for the GDF15 promoter and the indicated antibodies. Total RNA was isolated and analyzed for GDF15 expression by real-time PCR. The experiment was repeated three times. Representative images of three independent experiments were shown.
miR-627 is Induced by Calcitriol in vivo and Mediates the Antitumor Activity of Calcitriol
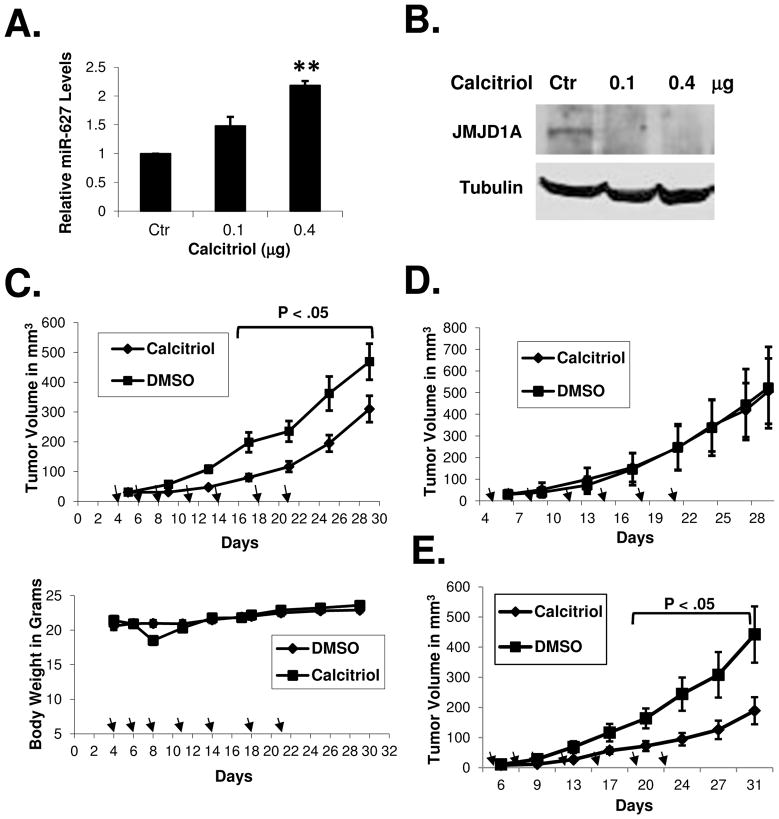
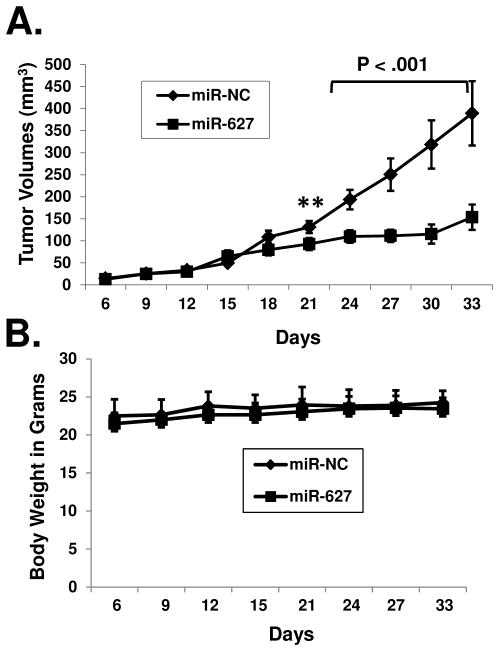
To determine if calcitriol induces miR-627 expression in vivo, we established human colon cancer xenografts in nude mice and treated the mice with calcitriol. As shown in Figures 4A and 4B, calcitriol induced miR-627 expression in vivo in the tumor xenografts while the level of JMJD1A was decreased. At the dose of 0.4 μg (which does not induce hypercalcemia in mice 34), calcitriol suppressed tumor growth in the nude mice, without inducing significant toxicity (measured by the loss of body weight), as shown in Figures 4C. We did not observe any symptoms of toxicity (lethargy, decreased feeding, etc). We blocked the activity of miR-627 by overexpressing the JMJD1A 3′UTR sponge which can sequester miR-627, thus calcitriol was no longer able to decrease JMJD1A (Supplementary Figure 4). As shown in Figure 4D, calcitriol failed to suppress tumor growth in the nude mice when miR-627 was blocked by the sponge. In contrast, when the miR-627 binding site within the 3′UTR sequence was mutated, calcitriol was still able to decrease JMJD1A (Supplementary Figure 4) and suppress tumor growth in nude mice (Figure 4E).
Figure 4.
Calcitriol induces miR-627 expression in tumor xenografts and miR-627 mediates the antitumor activity of calcitriol. (A and B) Nude mice bearing HT-29 xenografts were treated with calcitriol at i.p. doses of 0.1 and 0.4 μg daily for two days. Tumor samples were collected 24h after the second dose and analyzed by real-time PCR (A) and western blotting (B) with the indicated antibodies. The experiments have been repeated three times, data shown are mean values + SD (**p<0.01). (C) Nude mice bearing HT-29 (n=9 per group) xenografts were treated with calcitriol (i.p. doses of 0.4 μg, seven times as indicated by arrows). The same volume of DMSO was administered to the control groups at the same schedule. Tumor volumes and body weights were measured as described in Materials and Methods. (D) Nude mice bearing HCT-116-3′UTR xenografts (transfected with pRNAT-CMV3.2/Puro-3′UTR to stably express the JMJD1A 3′UTR sponge to block miR-627) were treated with calcitriol (i.p. doses of 0.4 μg, seven times as indicated by arrows, the same schedule as that in (C) or DMSO for the control groups, (n=5 per group). Tumor volumes were measured. (E) Nude mice bearing HCT-116-3′UTR-mut xenografts (transfected with pRNAT-CMV3.2/Puro-3′UTR-mut to stably express the JMJD1A 3′UTR sponge with the miR-627 binding site mutated) were treated with calcitriol or DMSO for the control group (n=7 per group) as described in (D). Tumor volumes were measured at indicated time points.
miR-627 Suppresses in vivo Colon Cancer Growth
To determine the effects of miR-627 on in vivo tumor growth, we established HCT-116 cell lines stably expressing negative control miRNA or miR-627. After transplanted into nude mice as xenografts, tumors stably expressing miR-627 exhibited significantly slower growth compared to tumors expressing the negative control miRNA (Figure 5A). There was no significant change in body weight among the two groups of mice (Figure 5B).
Figure 5.
MiR-627 suppresses in vivo tumor growth. HCT-116 cell lines stably expressing miR-627 or negative control miRNA were established and miR-627 expression was confirmed by real-time PCR. Negative control miRNA (provided by Invitrogen, does not target any known vertebrate gene) or miR-627 expressing cells were transplanted into nude mice to establish tumor xenografts (five mice per group). Tumor volumes and body weights were measured, (**p<0.01).
miR-627 Expression is Decreased in Human Colon Cancer
To determine the expression levels of miR-627 in human colon cancers, we analyzed RNA samples isolated from human colon adenocarcinoma specimens as well as non-tumor colon mucosa tissues (Supplementary Table 2). As shown in Figure 6, miR-627 levels in the tumor specimens were significantly lower than those in the normal tissues (Student’s t-test). Using Pearson’s correlation coefficient test, we found no association between miR-627 expression levels and tumor stages (r= −0.277, p=0.133) or nodal metastases (r=0.181, p=0.236). There was also no correlation between miR-627 expression and tumor grade of differentiation or tumor sites (left or right colon). It is possible that miR-627 expression is decreased during the early stage of carcinogenesis in the colon.
Figure 6.
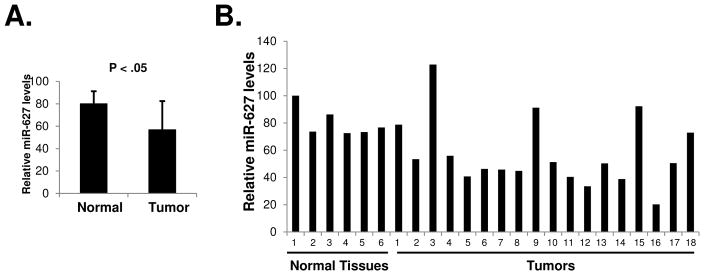
Expression of miR-627 in human colon cancer specimens. (A) Total RNA samples isolated from 18 human colon adenocarcinomas and 6 non-tumor colon mucosa were analyzed for miR-627 expression by real-time PCR. Statistical analysis was done with unpaired two-tailed Student’s t-test. (B) The relative miR-627 levels in individual samples were shown compared to one of the normal colon mucosa tissue.
Discussion
MicroRNAs have been indicated to play important roles in the regulation of cancer cell functions including differentiation, proliferation, apoptosis, and metastasis 35, 36. In this study, we have demonstrated that miR-627 contributes to the anticancer activities of calcitriol in colon cancer by targeting histone demethylase JMJD1A. Through miR-627, calcitriol induces histone methylation and suppresses the expression of growth-promoting genes such as GDF15. Other JMJD1A target genes (such as HOXA1, ADM, and EDN125) may also be downregulated and together contribute to tumor suppression by calcitriol. In the meantime, 3 miRNAs are increased by about 30% by calcitriol and 28 miRNAs are down-regulated by calcitriol by 17% to 63% (Supplementary Table 1). These miRNAs may also contribute to the antitumor activities of calcitriol by either down-regulating oncogenes or up-regulating tumor suppressors. Thus, our data have established a novel epigenetic mechanism of action of vitamin D in colon cancer (as summarized in a model shown in Supplementary Figure 5). This study has also identified miR-627 and JMJD1A as novel targets for the effective treatment of colon cancer.
A recent study involving 2690 participants has demonstrated that the circulating vitamin D concentration is inversely associated with the risk of colon and rectal cancer 37. Our data showed that miR-627 expression is lower in colon cancer specimens than that in the normal colon tissues. Thus, lower blood levels of vitamin D may cause the decrease of miR-627 expression, which may serve as an important mechanism to promote colon cancer growth. At present, we do not know how vitamin D up-regulates miR-627. The miR-627 gene is located on chromosome 15. Vitamin D could induce miR-627 expression directly through the VDR-RXR trans-activating effect on a cis-acting element in the miR-627 promoter (to be identified). Alternatively, vitamin D could act indirectly on some other up-stream regulator(s) of miR-627 transcription, such as the transcription factors that control miR-627 expression. Vitamin D could also affect the processing steps in miR-627 biogenesis (Drosha and Dicer enzymes, etc.). Finally, vitamin D could raise miR-627 levels by increasing its stability.
It has been shown that JMJD1A expression is induced by HIF-1α and JMJD1A enhances hypoxic gene expression to promote tumor growth 25. By targeting JMJD1A, miR-627 may mediate the activity of vitamin D to work against HIF-1α, which is a key factor in promoting tumor growth and survival 38. In the in vivo growth experiments (Figure 5), we observed that the tumors expressing miR-627 grew initially but the growth slowed down significantly after reaching certain sizes. Some tumors even regressed during later days of the experiments. As the tumor growes larger, the center core is subjected to hypoxia and HIF-1α expression is increased to promote angiogenesis, growth, survival, metastasis, and to regulate anaerobic glucose metabolism 39. Several hundred genes are induced in hypoxia and the massive transcriptional reorganization may be the result of actions of the histone modifying enzymes such as JMJD1A. By inducing miR-627 to decrease JMJD1A, vitamin D might counteract HIF-1α and cause the collapse of tumor growth in hypoxia.
Our data indicate that induction of miR-627 expression may serve as an important mechanism of vitamin D in suppression of colon cancer growth. Strategies to deliver miR-627 or directly inhibit JMJD1A may prove to be effective against colon cancer without eliciting hypercalcemia.
Materials and Methods
Cells and Transfection
The cell lines HT-29, HCT-116, and SW620 were purchased from American Type Culture Collection. HCT-116 and SW620 cells were cultured in RPMI1640 media containing 10% fetal bovine serum (FBS). HT-29 cells were cultured in Dulbecco’s Modified Eagle Medium containing 10% FBS. For transient transfection, plasmids were transfected into cells using LipofectamineTMPlus Reagent (Invitrogen) following the manufacturer’s protocol. miRNA mimics and inhibitors were transfected into cells using X-treme GENE siRNA transfection reagent (Roche) following the manufacturer’s protocol.
Plasmids Construction, siRNA, miRNA Mimics and Inhibitors
The 3′UTR cDNA fragment and the full-length cDNA of JMJD1A gene and full-length VDR gene were obtained by PCR using EST clones as templates. The PCR primers for the JMJD1A 3′UTR amplified a 120 bp sequence containing the miR-627 target site (Forward: TCTCCCTGCACATTGGAAAT; Reverse: CTCACAAGTTGGGTACAGTT). The 3′UTR cDNA was inserted into pEGFP-C1 plasmid at the 3′ end of the EGFP cDNA (Xho I and EcoR I). The pEGFP-C1-3′UTR plasmid containing a mutation within the miR-627 recognition site (ATTC to TGAG mutation within JMJD1A 3′UTR) was created by PCR using the QuickChange II site-directed mutagenesis kit (Stratagene), following the supplied protocol. For the miR-627 sponge experiments, the 3′UTR cDNA was also inserted into pRNAT-CMV3.2/Puro plasmid (BamH I and Xho I) to create pRNAT-CMV3.2/Puro-3′UTR for stable transfection into HCT-116 cells. The pRNAT-CMV3.2/Puro-3′UTR-mut containing mutation within the miR-627 recognition site (ATTC to TGAG) in the 3′UTR was created by PCR using the QuickChange II site-directed mutagenesis kit (Stratagene). The VDR cDNA was cloned into the pCEP4-Flag vector to express VDR as a Flag-tagged protein. To construct miRNA expression vectors, double strand oligoes containing pre-miR-627 were cloned into pcDNA6.2-GW/EmGFP-miR plasmid (Invitrogen) by directional cloning. For co-transfection of pEGFP-C1-3′UTR and miRNA expression vectors, EmGFP was removed using Dra I digestion to create pcDNA6.2-GW-miR-627. Negative control and JMJD1A siRNAs were purchased from Ambion (# s31588). miRCURY LNA anti-miR-627 inhibitor and negative control inhibitor were purchased from Exiqon. Synthetic miRNA mimics (Pre-miR™ miRNA Precursors) for miRNA-627 (# AM17100) and negative control miR-NC (# AM17110) were purchased from Ambion.
Western Blot Analysis
Cells were lysed in RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS in PBS). Complete protease inhibitor cocktail (Roche) was added to lysis buffer before use. Protein concentration was determined by Bio-Rad DC protein assay (Bio-Rad). Protein samples were subjected to SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked in 5% non-fat milk in PBS overnight and incubated with primary antibody and subsequently with appropriate horse radish peroxidase-conjugated secondary antibody. Signals were developed with ECL reagents (Pierce) and exposure to X-ray films. Anti-JMJD1A polyclonal antibody was kindly provided by Dr. Doug Demetrick at University of Calgary. Anti-β-tubulin anti-GFP antibodies were purchased from Santa Cruz Biotechnology. Anti-p15, p16, p21, and p-27 antibodies were purchased from Cell Signaling Technology. Anti-Flag antibody was purchased from Sigma. Image digitization and quantification were done with UN-SCAN-IT software from Silk Scientific.
Real-time PCR
The miRNA expression was measure by real-time PCR using TaqMan® MicroRNA assay (Cat # RT1560 for miR-627) from Applied Biosystems (Foster city, CA). Total RNA was isolated from cells using mirVana™ miRNA Isolation Kit (Ambion). 5 μg of total RNA was used in reverse transcription reaction. The cDNAs were used as templates to perform PCR on an Applied Biosystems 7500 Real-time PCR System following the manufacturer’s protocol. Relative miRNA expression levels were calculated using 18S RNA as reference. GDF15, CCND1, and HOXA1 mRNA expressions were measured by real-time PCR using the TaqMan® Gene Expression assay (Cat # Hs00171132_m1, Hs0076555_m1, Hs00939046_m1, respectively).
miRNA Expression Profiling
Total RNAs were isolated from HT-29 cells using the mirVana™ miRNA Isolation Kit (Ambion). miRNAs were further concentrated with the flashPAGE™ Fractionator System (Ambion). The miRNAs from control or calcitriol-treated HT-29 cells were labeled with the mirVana miRNA Labeling Kit (Ambion) with Cy3 or Cy5 Post Labeling Reactive Dye (Amersham Biosciences), respectively. The probes were then hybridized to the mirVana miRNA Bioarrays (Ambion) overnight. The array slides were scanned with an Aron 4000B Genepix Microarray Scanner and analyzed with the Genepix software.
Chromatin Immunoprecipitation Assay
HCT-116 cells were treated with 500 nM calcitriol for 48 hours. ChIP assay was performed using the ChIP assay kit from Millipore (Billerica, MA), following the supplied protocol. Immunoprecipitations were performed using anti-H3K9me2, anti-H3K4me3, anti-H3K27me2/3 (Cell Signaling Technology, Danvers, MA) or control IgG antibodies. PCR was performed with the primers designed from the sequences of the GDF15 promoter (5′-CTGTCTCTGGCCGAGGCG AG-3′ and 5′-CACTTACCTTCTGGCGTGAGTATCCGG-3′), covering a ~200 bp fragment that is approximately 100 bp downstream of the transcription start sites of GDF15 gene.
Tumor Xenografts in Nude Mice
Six to eight weeks old female nude mice (Nu/Nu) were purchased from Charles River (Wilmington, MA). The mice were maintained in sterile conditions using the Innovive IVC System from Innovive (San Diego, CA), following the protocol approved by the Institutional Animal Care and Use Committee of North Dakota State University. Tumor xenografts were established by subcutaneous injection of 2×106 cancer cells in the flank area of the mice. Calcitriol and DMSO (as control) were administered by i.p. injection. Two axes of the tumor (L, longest axis; W, shortest axis) were measured with a caliper. Tumor volume was calculated as: V = L × W2/2.
Human Colon Cancer Specimens
Total RNAs isolated from human colon adenocarcinomas and non-tumor colon mucosa tissues were obtained from Department of Pathology, Roswell Park Cancer Institute, and approved by the Institutional Review Board. All RNA samples were isolated using Trizol and checked for RIN on Bioanalyzer.
Statistical Analysis
Differences between the mean values were analyzed for significance using the unpaired two-tailed Student’s t-test for independent samples. Correlation significance was assessed using Pearson’s correlation coefficient test. P 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by the National Institutes of Health (CA130062 and RR015566 to B.G.), and North Dakota EPSCoR (EPS-0814442 to B.G.).
Abbreviations
- miRNA
microRNA
- 3′UTR
3′ untranslated region
- Calcitriol
1α,25-dihydroxyvitamin D3
- JMJD1A
jumonji domain containing 1A
- GDF15
growth and differentiation factor 15
- H3K9me2
dimethylated lysine 9 of histone H3
- GFP
green fluorescent protein
Footnotes
Disclosures: The authors have no potential conflicts to disclose.
Author Contributions: S.K.R.P. conducted experiments, Q.Z. conducted experiments, B.G. designed the experiments, conducted experiments, and wrote the manuscript, Y.M.R. and C.M. provided RNA samples from human colon cancer and normal tissues.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 2.Gorham ED, Garland CF, Garland FC, et al. Vitamin D and prevention of colorectal cancer. J Steroid Biochem Mol Biol. 2005;97:179–94. doi: 10.1016/j.jsbmb.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 3.Garland CF, Garland FC. Do sunlight and vitamin D reduce the likelihood of colon cancer? Int J Epidemiol. 1980;9:227–31. doi: 10.1093/ije/9.3.227. [DOI] [PubMed] [Google Scholar]
- 4.Robsahm TE, Tretli S, Dahlback A, et al. Vitamin D3 from sunlight may improve the prognosis of breast-, colon- and prostate cancer (Norway) Cancer Causes Control. 2004;15:149–58. doi: 10.1023/B:CACO.0000019494.34403.09. [DOI] [PubMed] [Google Scholar]
- 5.Feskanich D, Ma J, Fuchs CS, et al. Plasma vitamin D metabolites and risk of colorectal cancer in women. Cancer Epidemiol Biomarkers Prev. 2004;13:1502–8. [PubMed] [Google Scholar]
- 6.Garland CF, Comstock GW, Garland FC, et al. Serum 25-hydroxyvitamin D and colon cancer: eight-year prospective study. Lancet. 1989;2:1176–8. doi: 10.1016/s0140-6736(89)91789-3. [DOI] [PubMed] [Google Scholar]
- 7.Wactawski-Wende J, Kotchen JM, Anderson GL, et al. Calcium plus vitamin D supplementation and the risk of colorectal cancer. N Engl J Med. 2006;354:684–96. doi: 10.1056/NEJMoa055222. [DOI] [PubMed] [Google Scholar]
- 8.Egan JB, Thompson PA, Ashbeck EL, et al. Genetic polymorphisms in vitamin D receptor VDR/RXRA influence the likelihood of colon adenoma recurrence. Cancer Res. 2010;70:1496–504. doi: 10.1158/0008-5472.CAN-09-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenab M, McKay J, Bueno-de-Mesquita HB, et al. Vitamin D receptor and calcium sensing receptor polymorphisms and the risk of colorectal cancer in European populations. Cancer Epidemiol Biomarkers Prev. 2009;18:2485–91. doi: 10.1158/1055-9965.EPI-09-0319. [DOI] [PubMed] [Google Scholar]
- 10.Evans SR, Schwartz AM, Shchepotin EI, et al. Growth inhibitory effects of 1,25-dihydroxyvitamin D3 and its synthetic analogue, 1alpha,25-dihydroxy-16-ene-23yne-26,27-hexafluoro-19-nor-cholecalcifero l (Ro 25–6760), on a human colon cancer xenograft. Clin Cancer Res. 1998;4:2869–76. [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Jima DD, Jacobs C, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–94. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 17.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 18.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 19.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 20.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–51. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–79. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 23.Lim S, Metzger E, Schule R, et al. Epigenetic regulation of cancer growth by histone demethylases. Int J Cancer. 2010 doi: 10.1002/ijc.25538. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 24.Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–95. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Krieg AJ, Rankin EB, Chan D, et al. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–53. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper GM. The Cell: A Molecular Approach. 2 2000. The Eukaryotic Cell Cycle. [Google Scholar]
- 27.Liu G, Hu X, Chakrabarty S. Vitamin D mediates its action in human colon carcinoma cells in a calcium-sensing receptor-dependent manner: downregulates malignant cell behavior and the expression of thymidylate synthase and survivin and promotes cellular sensitivity to 5-FU. Int J Cancer. 2010;126:631–9. doi: 10.1002/ijc.24762. [DOI] [PubMed] [Google Scholar]
- 28.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–50. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, et al. Generation of miRNA sponge constructs. Methods. 2012;58:113–7. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Palmer HG, Gonzalez-Sancho JM, Espada J, et al. Vitamin D(3) promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of beta-catenin signaling. J Cell Biol. 2001;154:369–87. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho HS, Toyokawa G, Daigo Y, et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G(1) /S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer. 2011 doi: 10.1002/ijc.26501. [DOI] [PubMed] [Google Scholar]
- 34.Muindi JR, Modzelewski RA, Peng Y, et al. Pharmacokinetics of 1alpha,25-dihydroxyvitamin D3 in normal mice after systemic exposure to effective and safe antitumor doses. Oncology. 2004;66:62–6. doi: 10.1159/000076336. [DOI] [PubMed] [Google Scholar]
- 35.Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–82. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- 36.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–92. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 37.Lee JE, Li H, Chan AT, et al. Circulating levels of vitamin D and colon and rectal cancer: the Physicians’ Health Study and a meta-analysis of prospective studies. Cancer Prev Res (Phila) 2011;4:735–43. doi: 10.1158/1940-6207.CAPR-10-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maynard MA, Ohh M. The role of hypoxia-inducible factors in cancer. Cell Mol Life Sci. 2007;64:2170–80. doi: 10.1007/s00018-007-7082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–85. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.