Abstract
Mouse models lacking proteins essential for autophagosome formation have demonstrated that autophagy plays a critical role in T cell development and activation. To better understand the function of autophagy in quiescent and activated lymphocytes, we have generated a mouse deficient in rab7 selectively in T cells and compared the effects of blocking autophagy at an early (atg5−/−) or late (rab7−/−) stage on T cell biology. rab7−/− murine embryonic fibroblasts (MEFs) and T cells generated from these mice exhibit a profound block in autophagosome degradation and are as sensitive as atg5−/− cells to extracellular nutrient limitation. Rab7flox/floxCD4-Cre+ mice lacking the RAB7 protein in both CD4 and CD8 T cells had reduced numbers of peripheral T cells, but this defect was not as severe as in Atg5flox/floxCD4-Cre+ mice despite efficient rab7 deletion and inhibition of autophagic flux. This difference may stem from the reduced ROS generation and enhanced survival of rab7−/− T cells compared with wild-type and atg5−/− T cells in the absence of cytokine stimulation. rab7−/− and atg5−/− T cells exhibited similar defects in proliferation both following antibody-mediated T cell receptor (TCR) cross-linking and using a more physiologic activation protocol, allogeneic stimulation. Interestingly, autophagy was not required to provide building blocks for the upregulation of nutrient transporter proteins immediately following activation. Together, these studies suggest that autophagosome degradation is required for the survival of activated T cells, but that loss of rab7 is better tolerated in naïve T cells than the loss of atg5.
Keywords: RAB7, T cells, autophagy, lysosomal fusion, growth factor withdrawal, ATG5
Introduction
RAB7 is a small GTPase that promotes fusion reactions between late endosomes and lysosomes when in its active, GTP-bound form.1-4 In this role, RAB7 facilitates the lysosomal degradation of plasma membrane proteins and soluble material present in endocytic vesicles. In addition to its function in the endocytic pathway, RAB7 is also essential for autophagosome degradation.5,6 Autophagy is the mechanism by which cells derive nutrients from recycled intracellular material when access to extracellular nutrients is limiting but is also necessary for the turnover of damaged or unnecessary organelles and the clearance of ubiquitinated protein aggregates.7,8 Autophagy begins with the formation of a double-membrane structure that encapsulates the cargo. The membrane of this cup-shaped structure is studded with a cleaved and phosphatidylethanolamine-conjugated form of the MAP1LC3B protein (MAP1LC3B-II) that is often used as a specific marker for autophagosomes. MAP1LC3B-II is present on the inner and outer faces of the autophagosomal membrane and plays a key role in cargo selection.9-12 ATG5 is one of several proteins required for the formation of the autophagosome and for the membrane association of MAP1LC3B-II.13 Once formed, autophagosomes must fuse with lysosomes for their contents to be degraded into components that can be exported to the cytoplasm and recycled. Given its position on the internal membrane of the autophagosome, some MAP1LC3B-II is inaccessible to recycling enzymes and is degraded along with the autophagosomes. As a result, MAP1LC3B-II levels increase when autophagy is induced but then decline as autophagic flux results in autophagosome turnover. Autophagic flux can thus be measured by comparing MAP1LC3B-II levels in the presence or absence of inhibitors of lysosomal proteolysis.14 Although loss of either atg5 or rab7 will block autophagy, cells that lack ATG5 cannot form autophagosomes or produce MAP1LC3B-II,13 while cells deficient in RAB7 function can form but not degrade autophagosomes5,6 leading to the accumulation of MAP1LC3B-II.
Mice lacking essential autophagy proteins have been generated by several groups (reviewed in8). Conventional knockouts of atg3, atg5, atg7, atg9 and atg16l1 appear normal at birth, but die in the neonatal period due to defects in nutritional homeostasis and/or deficiency in the cellular remodeling necessary to adapt to changing developmental and environmental conditions.13,15-18 Deletion of several genes whose products are required for autophagy but also have other functions (becn1, ambra1 and rb1cc1) is embryonic lethal.19-22 Using mouse models, autophagy has been demonstrated to play a key role in several aspects of lymphocyte biology.23,24 Conventional knockouts of atg5 were first used to study the function of autophagy in lymphocytes through the production of fetal liver chimeras.25 More recently, mice deficient in atg5 and atg7 selectively in T cells have been generated eliminating possible effects of the loss of autophagy on engraftment.26,27 atg5 and atg7 T cell knockout (TKO) mice have a very similar phenotype: reduced numbers of peripheral T cells and increased mitochondrial content and ROS production in those that remain. Both prior to and following activation, atg5−/− T cells exhibit a survival defect that leads to decreased cell accumulation. Whether proliferation is also reduced is difficult to tease apart from the survival defect. Current models suggest that atg5−/− T cells die upon activation due to excessive ROS production secondary to the disruption of mitophagy.26,27 Other studies suggest that autophagy is upregulated upon T cell activation and is required to provide energy from internal stores.28 Given that the role of autophagy in T cells is incompletely understood, we generated mice lacking rab7 selectively in T cells and compared the effects of blocking autophagosome formation (atg5−/−) and degradation (rab7−/−) on naïve and activated T cells in vitro and in vivo. To our knowledge, this is the first description of a mouse model with a defect in the late stages of the autophagy pathway.
Results
Construction of a Rab7 conditional allele
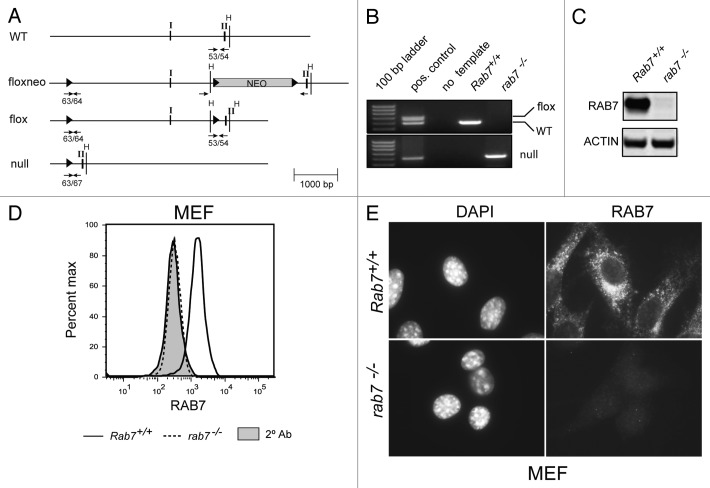
To study the function of RAB7 in T cells, a conditional allele was created. A mouse genomic DNA lambda library was screened and a 10 kb fragment that included the first two exons of Rab7 isolated. LoxP sites were introduced upstream of exon I and at each end of a neomycin cassette used for the selection of an ES cell clone that had undergone homologous recombination to generate a Rab7floxneo allele (Fig. 1A). Mice expressing the Rab7floxneo allele were generated by injecting C57BL/6 blastocysts with this ES cell clone. Rab7+/floxneo mice were crossed with mice transgenic for the Cre recombinase under the control of the protamine promoter that drives Cre expression in spermatids.29 Some offspring of these crosses exhibited incomplete recombination of the three LoxP sites in the Rab7floxneo allele generating the Rab7flox allele that lacks the neomycin resistance cassette (Fig. 1A). The Rab7floxneo allele was hypomorphic, producing less RAB7 protein than the wild-type or Rab7flox allele (data not shown). A null allele was also generated from breedings with Protamine-Cre+ mice. Based on the genotypes of over 75 pups from Rab7+/− crosses, the rab7−/− genotype is embryonic lethal. Breeding cages were inspected daily and dead pups genotyped; no rab7−/− pups were ever recovered. Rab7+/− mice were grossly normal but were observed at less than the expected Mendelian frequency (½ rather than 2/3 of the pups of heterozygous crosses were Rab7+/−). Mice expressing the Rab7flox allele were born at the expected Mendelian frequency and were used in all further studies.
Figure 1. Generation of a floxed Rab7 allele. (A) Targeting strategy for rab7 deletion in mice. Arabic numbers indicate primer pairs used to specifically amplify the alleles. Roman numerals correspond to exons. WT, wild type; H, HindIII restriction sites; solid triangles, LoxP sites; NEO, neomycin resistance gene. (B) PCR amplification of genomic DNA from MEFs of the indicated genotype. The positive control in the upper panel is tail DNA from a Rab7+/flox mouse and in the lower panel genomic DNA isolated from T cells of a rab7 TKO mouse. (C) Western blot of lysates prepared from Rab7+/+ and rab7−/− MEFs. (D) Rab7+/+ and rab7−/− MEFs were fixed and permeabilized, stained for RAB7, and analyzed by flow cytometry. The secondary antibody alone (2° Ab) reflects nonspecific staining, results from all cells are shown. (E) Rab7+/+ or rab7−/− MEFs were evaluated by immunofluorescence microscopy.
To confirm that rab7 deletion produced the anticipated effect on autophagy, Rab7-/flox MEFs were immortalized with SV40 large T antigen, transduced with retroviruses expressing Cre, cloned, and their genotype analyzed by PCR (Fig. 1B). Rab7+/+ MEFs were prepared in parallel from littermate controls. Loss of the RAB7 protein was also confirmed by western blotting (Fig. 1C). To determine whether rab7 deletion could be confirmed in individual cells, we evaluated Rab7+/+ and rab7−/− MEFs by flow cytometry (Fig. 1D) and immunofluorescence microscopy (Fig. 1E). Using both techniques, RAB7 staining was at background levels (secondary alone) in rab7−/− MEFs. rab7−/− MEFs grew normally under standard culture conditions. In summary, the Rab7flox allele was converted to the rab7null allele in the presence of CRE recombinase leading to loss of the RAB7 protein from cells.
Effects of rab7 deletion in resting T cells
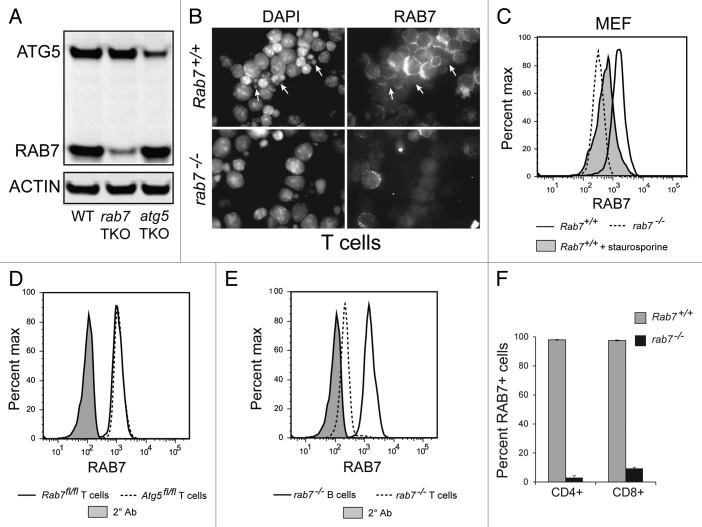
To produce mice lacking rab7 selectively in T cells, Rab7flox/flox mice were crossed to mice expressing the Cre recombinase under the control of the CD4 promoter. CD4 is first expressed in double-positive T cells in the thymus, and thus CD4-Cre leads to deletion of floxed alleles in both CD4- and CD8-positive T cells. The CD4-Cre allele deletes floxed genes later than the Lck-Cre allele used in previous studies,26-28 which we reasoned might result in fewer disruptions in T cell development. Using Lck-Cre mice obtained from Jackson Labs (stock# 003802), rab7 deletion was inefficient and varied greatly between individual animals and thus only results from CD4-Cre animals are reported here. The efficiency of deletion in T cells in Rab7flox/flox-CD4-Cre+ (rab7 TKO) mice was confirmed via western blot. The RAB7 protein level decreased to a similar degree in rab7−/− T cells as the ATG12–ATG5 protein conjugate did in the atg5−/− T cells (Fig. 2A). Loss of RAB7 protein from T cells was also confirmed by immunofluorescence microscopy (Fig. 2B). Interestingly, a population of Rab7+/+ T cells appeared to lack RAB7 in this assay (Fig. 2B, arrows). The nuclei of these apparently rab7-negative cells were condensed and fragmented suggesting that the RAB7 antibody did not recognize RAB7 following the induction of apoptosis and the activation of caspases. To test this idea, MEFs were treated with the classical apoptosis inducer staurosporine for 15 h, incubated with the fixable viability dye eFluor780, and stained for RAB7. As predicted, staurosporine-treated, eFluor780-positive Rab7+/+ MEFs showed reduced RAB7 staining that was intermediate between viable Rab7+/+ and rab7−/− MEFs (Fig. 2C). Similar results were obtained when apoptotic cells were defined as the sub-G1 population following DAPI staining. As a result of these experiments, eFluor780 was included in all cases where intracellular RAB7 levels were evaluated by flow cytometry and the analysis restricted to live cells. Flow cytometry was next used to quantify the deletion of rab7 in T cells isolated from rab7 TKO mice. As expected, Rab7flox/flox T cells lacking Cre and those bearing a wild-type Rab7 allele (Atg5flox/floxRab7+/+) expressed equivalent amounts of RAB7 (Fig. 2D). B cells from rab7 TKO mice also had normal levels of RAB7, while T cells from rab7 TKO mice exhibited a substantial decrease in RAB7 staining (Fig. 2E). rab7 deletion was efficient in both CD4 and CD8 naïve T cells; 97% of CD4 T cells lacked RAB7 while 93% of CD8 lacked RAB7 (Fig. 2F). These studies demonstrate efficient loss of RAB7 protein in T cells in rab7 TKO mice.
Figure 2. Deletion of rab7 in T cells. (A) Freshly isolated purified T cells from WT, rab7 TKO and atg5 TKO mice were analyzed by western blot using the LI-COR Odyssey imager. (B) Rab7+/+ or rab7−/− purified T cells were evaluated for RAB7 expression by immunofluorescence microscopy. Arrows indicate fragmented nuclei. (C) Rab7+/+ and rab7−/− MEFs were treated with 100 nM staurosporine for 15 h, incubated with eFluor780, stained for RAB7, and analyzed by flow cytometry. Live cells are shown except for staurosporine-treated apoptotic cells. (D) Rab7flox/flox and Atg5flox/flox purified T cells were stained for RAB7 and analyzed by flow cytometry. Live cells are shown; the secondary antibody alone (2° Ab) reflects nonspecific staining. (E) Splenocytes from rab7 TKO mice stained for B (B220 positive) and T (CD3 positive) cell markers and RAB7 were analyzed by flow cytometry. Live cells are shown. (F) The deletion of rab7 in CD4- and CD8-positive subsets of purified T cells was evaluated by flow cytometry as in (E). Results from live cells are shown. Using a two-tailed t-test to compare the frequency of RAB7+ cells in Rab7flox/flox and rab7 TKO mice, p < 0.001 for both CD4 and CD8 cells.
RAB7 is required for autophagic flux
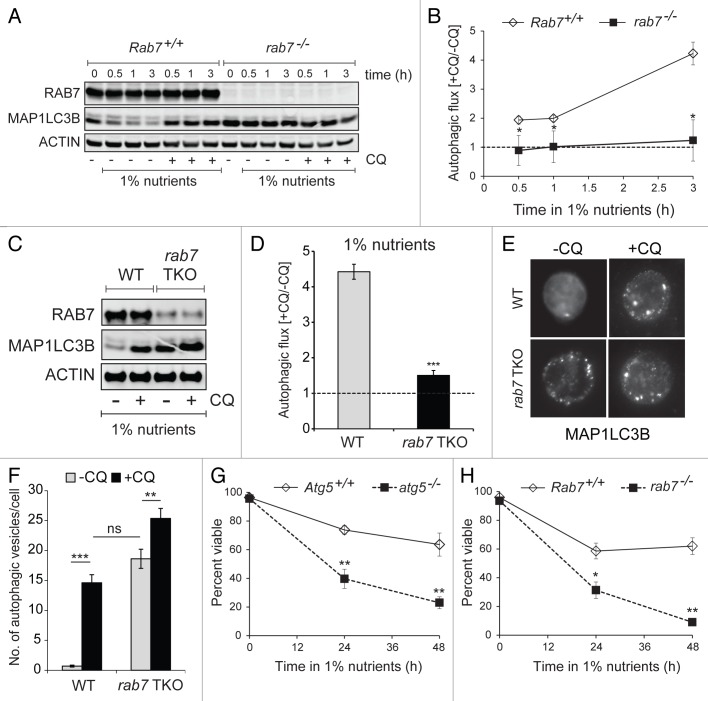
Previous studies using RNAi and dominant-negative mutants have indicated that RAB7 is required for the fusion of autophagosomes with lysosomes.5,6 We confirmed that RAB7 plays an essential role in autophagy by performing autophagic flux assays. By blocking lysosomal acidification, chloroquine stabilizes MAP1LC3B-II levels and allows the estimation of autophagic flux through the comparison of MAP1LC3B-II levels in the presence and absence of the drug.14 If RAB7 is essential for autophagosome degradation, no further increase in MAP1LC3B-II levels would be expected upon the addition of chloroquine. To test this prediction, we measured autophagic flux in MEFs where RAB7 deletion was complete (Fig. 1C). Autophagy was induced by incubation in low nutrient medium and flux assessed by measuring MAP1LC3B-II levels in the presence or absence of chloroquine by western blot. In nutrient-stressed Rab7+/+ MEFs, MAP1LC3B-II accumulated in the presence of chloroquine (Fig. 3A) and autophagic flux increased over time (Fig. 3B). In contrast, MAP1LC3B-II levels in rab7−/− MEFs were initially elevated and not affected by nutrient stress or chloroquine exposure (Fig. 3A); no autophagic flux was observed (Fig. 3B). Thus, elimination of rab7 completely blocked autophagic flux as predicted.
Figure 3. RAB7 is required for autophagic flux. (A) Rab7+/+ or rab7−/− MEFs were cultured in medium containing 1% the normal amount of amino acids and glucose and 10% FCS ± chloroquine (CQ, 50 µM) for the indicated interval and MAP1LC3B-II (lower band) levels evaluated by western blotting using a LI-COR Odyssey imager. The blot shown is representative of 3 independent experiments. (B) Autophagic flux was calculated as the ratio of the amount of MAP1LC3B-II detected with or without chloroquine. Averages from 3 independent experiments ± SEM are shown. (C) Freshly purified splenic T cells were cultured in 1% nutrient medium ± 50 µM chloroquine for 3 h and evaluated as in (A). The blot shown is representative of 3 independent experiments. (D) Autophagic flux was measured in purified T cells as described for (B). Averages from 3 independent experiments ± SEM are shown. (E) Fluorescent microscopy showing MAP1LC3B staining in purified T cells activated for 24 h then placed in 1% nutrient medium for 3 h with or without chloroquine (50 µM). (F) The number of autophagosomes per cell was counted visually in the cells from (E). At least 25 cells from two independent experiments were evaluated for each condition. Means ± SEM (n = 25) are shown. (G) The viability of Atg5+/+ and atg5−/− MEFs in 1% nutrient medium was measured by vital dye (DAPI) exclusion and flow cytometry. (H) As in (G), but using Rab7+/+ and rab7−/− MEFs. Means ± SEM from 3 independent experiments are shown. In (B, D and F–H), t-tests were performed: *p < 0.05; **p < 0.01; ***p < 0.001.
Autophagic flux was next measured in freshly purified naïve T cells under similar conditions. Robust autophagy was observed in nutrient-restricted Rab7+/+ T cells (Fig. 3C and D). As in MEFs, MAP1LC3B-II levels in rab7−/− T cells in the absence of chloroquine were as high as in nutrient-stressed Rab7+/+ T cells treated with chloroquine and autophagic flux in rab7−/− T cells was nearly absent. Autophagosome content was also measured by immunofluorescence. Activated cells were used because naïve cells have very little cytosol. After 3 h of chloroquine treatment in low nutrient medium, the number of autophagosomes in Rab7+/+ T cells increased as expected (Fig. 3E). As many autophagosomes were present in untreated rab7−/− T cells as chloroquine-treated Rab7+/+ cells. The addition of chloroquine caused a small further accumulation in autophagosomes in rab7−/− T cells. This result is likely explained by the fact that purified T cells were approximately 90% pure (10% non-T cells that would not express CD4-Cre) and that 3% to 7% of the T cells failed to delete RAB7 (Fig. 2F). To confirm that the loss of ATG5 or RAB7 blocked autophagy-dependent cell survival to an equivalent extent, the viability of rab7−/− and atg5−/− MEFs under nutrient stress was measured. After 48 h in low nutrient medium, ~60% of the Atg5+/+ and Rab7+/+ MEFs were alive while only 25% and 10% of atg5−/− and rab7−/− MEFs, respectively, were still viable (Fig. 3G and H). Taken together, these results confirm that RAB7 is required for autophagosome degradation and that the loss of rab7 or atg5 disrupts the ability of cells to survive under nutrient stress to a similar extent.
Phenotype of rab7 TKO mice
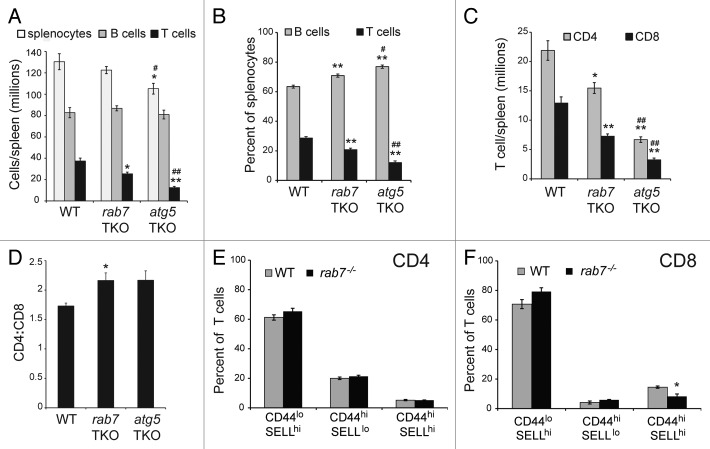
The effect of autophagy disruption on T cells has been analyzed using T cells that are deficient in atg5 or atg7.25-27 To determine whether interrupting autophagy at an early or late stage had differential effects on T cells, we compared our rab7 TKO mice to Atg5flox/flox-CD4-Cre+ (atg5 TKO) animals. Overall, atg5 TKO and rab7 TKO mice had similar phenotypes, but defects were more severe in atg5 TKO than rab7 TKO animals despite similar deletion efficiencies (Fig. 2A). The absolute number of T cells was reduced by 32% in rab7 TKO mice relative to wild-type controls, while it was reduced by 67% in atg5 TKO mice (Fig. 4A). These results are consistent with previous reports of a 58% to 75% reduction in T cell numbers in atg7 TKO mice and a 52% decrease in atg5 TKO mice.26,27 As expected, B cell numbers were not different among the three genotypes, although the percentage of lymphocytes that were B cells increased in rab7 TKO and atg5 TKO animals concomitant with the loss of T cells (Fig. 4A and B). The numbers of both CD4 and CD8 T cells were decreased by the loss of rab7 or atg5 (Fig. 4C). Despite the slightly more efficient deletion of rab7 in CD4 cells (Fig. 2F), the reduction in CD8 T cell numbers in rab7 TKO mice was more pronounced leading to an elevated CD4:CD8 ratio (Fig. 4D). The CD4:CD8 ratio was also increased in atg5 TKO mice as has been previously reported.25,27 Interestingly, while CD44hiSELLlo cell number was elevated in atg5 or atg7 TKO mice in these earlier studies, no change in CD44 or SELL staining was observed in T cells from rab7 TKO mice (Fig. 4E and F). In summary, the negative effects of rab7 deletion on naïve T cells were less severe than the effects of deleting atg5 (Fig. 4A–C).
Figure 4. Phenotype of rab7 TKO mice. (A and B) Splenocytes were isolated from mice of the indicated genotypes and the absolute numbers (A) and percentages (B) of B (B220 positive) and T (CD3 positive) cells determined. (C) As in (A), but T cells were divided into CD4- or CD8-positive subsets based on surface staining by flow cytometry. (D) The ratio of CD4:CD8 T cells in spleens from the indicated genotypes were calculated. In (A–D), live cells gates were applied. (E and F) CD44 and SELL levels were measured on live-gated CD4+ (E) and CD8+ (F) splenocytes from wild-type or rab7 TKO mice. In (A–F), means from at least 3 independent experiments are shown, error bars are SEM. A t-test was performed to compare either of the knockout T cells to wild-type (denoted by *) or rab7 TKO and atg5 TKO (denoted by #). * or #p < 0.05; ** or ##p < 0.005.
Mitochondrial mass and ROS level increase less in rab7−/− than in atg5−/− T cells
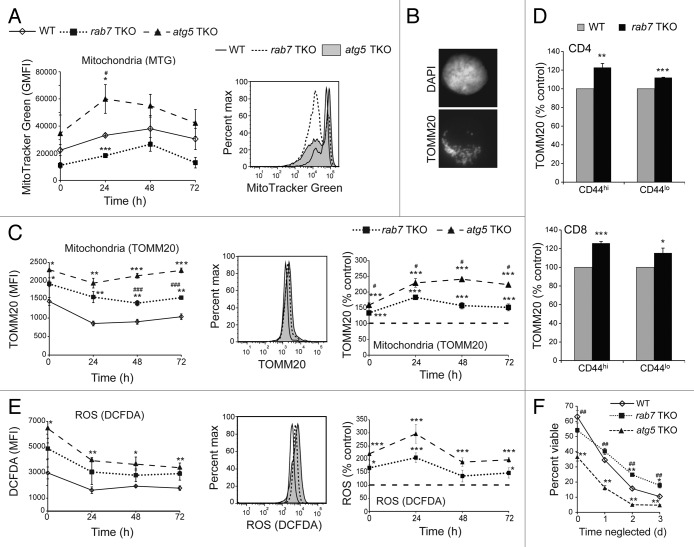
ATG5-deficient T cells have increased mitochondrial mass and ROS levels.25,27 Consistent with these reports, MitoTracker Green (MTG) staining was increased in live T cells isolated from atg5 TKO animals relative to wild-type controls (Fig. 5A). As RAB7-deficient cells should have a similar defect in mitochondrial turnover due to the block in autophagosome degradation (Fig. 3), we expected a similar elevation of MTG staining in rab7−/− T cells. Interestingly, MTG staining in live-gated cells was more markedly biphasic and shifted toward the lower peak in rab7−/− cells such that the MFI was reduced relative to wild type. We reasoned that MTG staining of mitochondria present in autophagosomes might be reduced if the dye did not efficiently cross the autophagosome double membrane. As a second method of quantifying mitochondrial mass, we stained cells with an antibody to the mitochondrial outer membrane protein TOMM20 which produced a classic mitochondrial staining pattern (Fig. 5B). Cells were also stained with DAPI to allow live cell gating (sub-G1 cells were excluded from the analysis). As measured by TOMM20 staining and flow cytometry, mitochondrial mass was increased in rab7−/− T cells compared with wild-type T cells (Fig. 5C). Increased mitochondrial mass was observed in both CD44hi and CD44lo subpopulations of rab7−/− CD4+ and CD8+ T cells when compared with wild-type controls (Fig. 5D). However, rab7−/− T cells exhibited reduced mitochondrial mass relative to atg5−/− T cells (Fig. 5C). If fewer mitochondria accumulate in rab7−/− T cells than in atg5−/− T cells, ROS production may be similarly reduced. Consistent with the TOMM20 staining results, rab7−/− T cells had elevated ROS levels relative to wild-type cells, but reduced ROS levels relative to atg5−/− T cells (Fig. 5E). Together, these results suggest that blocking autophagy at a later stage through RAB7 deletion limited the generation of ROS increasing the viability of rab7−/− T cells relative to atg5−/− cells that do not form autophagosomes.
Figure 5.rab7−/− T cells accumulate fewer mitochondria and produce less ROS than atg5−/− T cells. (A) Purified T cells from WT, rab7 TKO and atg5 TKO mice were cultured in complete T cell medium for the indicated times before staining with MTG. The average geometric mean fluorescence intensity (GMFI) in live cells is graphed; a representative histogram is shown. (B) Wild-type activated purified T cells fixed and immunostained for TOMM20. (C) As in (A), but fixed and permeabilized cells were immunostained for TOMM20. Sub-G1 cells were excluded from the analysis. Data are presented both as MFI and normalized to the wild-type control; a representative histogram is also shown. (D) TOMM20 levels were measured by flow cytometry in CD44hi and CD44lo splenocytes. (E) As in (A), but live cells were stained with CM-H2DCFDA. Data are presented both as MFI and normalized to the wild-type control; a representative histogram is also shown. (F) Purified T cells of the indicated genotypes were left unstimulated in complete media for 3 d at a concentration of 1 × 106 cells/ml. Viability was measured at the indicated time points by vital dye exclusion and flow cytometry. For (A and C–F) means ± SEM are shown from at least 3 independent experiments. T-tests were performed to compare either of the knockout T cells to wild type (denoted by *) or rab7 TKO and atg5 TKO (denoted by #). * or #p < 0.05; ** or ##p < 0.01; *** or ###p < 0.001. In (E), ROS levels in rab7−/− T cells were intermediate between atg5−/− and wild-type T cells in all 4 experiments at all time points. The P values when comparing ROS levels in rab7−/− and atg5−/− T cells as percent control were 0.13, 0.06, 0.10, 0.06 at 0, 24, 48 and 72 h, respectively.
The finding that naïve rab7−/− T cells had lower levels of ROS than atg5−/− T cells suggested that the larger number of peripheral T cells in rab7 than atg5 TKO mice (Fig. 4A) might reflect an increased ability of rab7−/− T cells to withstand cytokine deprivation in vivo. To test this idea, we measured the viability of neglected atg5- and rab7-deficient T cells. Freshly isolated atg5−/− T cells exhibit decreased viability suggesting that they have a survival defect in vivo (Fig. 5F).25,27 rab7−/− T cells, however, were nearly as viable as controls upon isolation. When T cells are cultured without stimulation, they undergo a growth factor withdrawal-like death termed “death by neglect.”30 In the absence of growth factor stimulation, cells rely on autophagy for survival.31 Consistent with this, atg5−/− T cells also exhibited decreased survival during in vitro neglect assays (Fig. 5F). rab7−/− T cells, in contrast, were resistant to cell death in the absence of stimulation. This finding is consistent with our previous work showing that inactivation of RAB7 protects cells from growth factor withdrawal but not extracellular nutrient limitation (Fig. 3H).32 In summary, RAB7-deficient T cells do not accumulate mitochondria and ROS to the same degree as atg5−/− T cells and are more viable upon isolation and under conditions of neglect. These findings may partially explain the increased numbers of splenic T cells in rab7 TKO mice compared with atg5 TKO animals.
Inhibition of autophagosome production and degradation produces similar effects in activated T cells
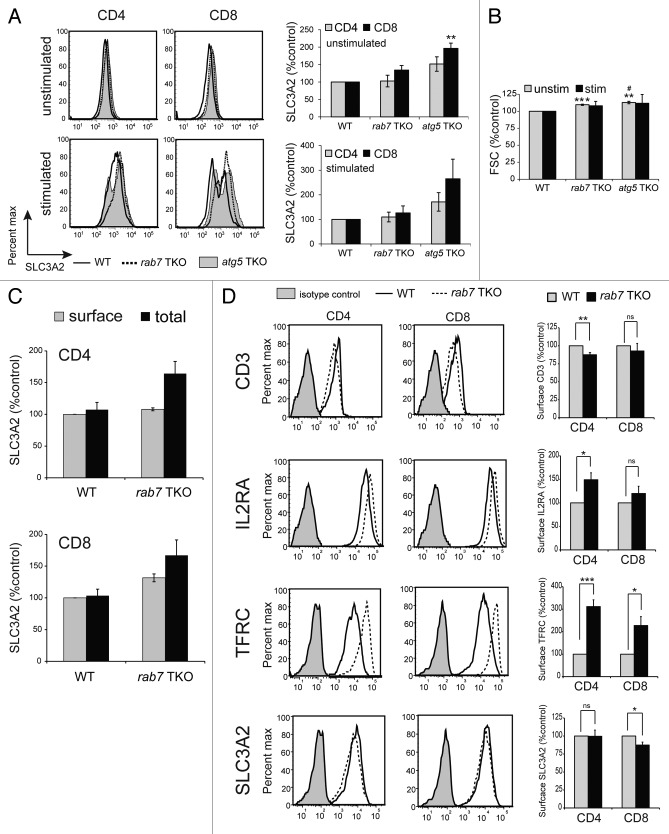
The functional role of the catabolic process of autophagy in blasting T cells is incompletely understood. As naïve T cells express very low levels of nutrient transporters,33-35 their access to nutrients immediately following activation is likely to be quite limited. We therefore hypothesized that autophagy might be required immediately following TCR ligation to recycle nutrients from intracellular sources to support the dramatic upregulation of early activation markers which include nutrient transporters that provide access to extracellular nutrients. To test this idea, T cells from all three genotypes were either stimulated with plate-bound anti-CD3 and soluble anti-CD28 or left untreated for 24 h. SLC3A2 (also known as 4F2 heavy chain or CD98), an early activation marker and amino acid transporter associated protein, was upregulated as well or even more efficiently in both autophagy-deficient genotypes compared with controls (Fig. 6A). atg5−/− T cells expressed higher levels of SLC3A2 on the cell surface than either rab7−/− or wild-type controls. This unexpected result might occur if atg5−/− T cells were larger due to the accumulation of material that would have been degraded if the autophagy pathway were intact. Using forward scatter to estimate cell size, atg5−/− T cells were about 10% larger than controls both before and after stimulation (Fig. 6B). As rab7−/− T cells showed the same increase in forward scatter, the increased SLC3A2 in atg5−/− T cells cannot be attributed solely to changes in cell size. Although upregulation of SLC3A2 after activation is a delayed response, it remained possible that its upregulation was due in part to trafficking of preformed intracellular SLC3A2 to the cell surface. To test this idea, we compared surface and total SLC3A2 levels in naïve peripheral T cells. All SLC3A2 was on the surface of both CD4+ and CD8+ wild-type T cells (Fig. 6C). In rab7−/− T cells, intracellular SLC3A2 was detected, consistent with the degradation defects expected in the absence of RAB7. This internal SLC3A2 may be trafficked to the cell surface upon activation but is not sufficient to account for the 10-fold increase in surface SLC3A2 levels that occurs upon activation. In summary, as autophagy-deficient T cells upregulated SLC3A2 normally, autophagy was not required to generate the amino acids and energy necessary for the synthesis of early activation markers, at least under standard in vitro activation conditions.
Figure 6. Effect of rab7 deletion on surface markers. (A) Surface SLC3A2 levels were measured by flow cytometry in live, purified T cells of the indicated genotypes following 24 h of incubation in complete medium (unstimulated) or following activation with CD3/CD28 antibodies (stimulated). A representative histogram and a graph of the mean ± SEM at least 3 independent experiments is shown. (B) Mean FSC ± SEM from the experiments in (A) is graphed. (C) Surface and total SLC3A2 expression was measured in splenic T cells from wild-type or rab7 TKO mice. Analysis was restricted to live (surface SLC3A2) or nonapoptotic (total SLC3A2) cells. Means ± SEM from 3 independent experiments are shown. Differences between surface and total SLC3A2 levels within a genotype did not reach statistical significance at p < 0.05. (D) Purified T cells from WT or rab7 TKO mice were activated with CD3 or CD28 antibodies for 65 h before staining with the indicated surface proteins and CD4/CD8; the analysis was restricted to live cells. The mean ± SEM from four independent experiments is graphed, a representative histogram is shown. T-tests were performed to compare either of the knockout T cells to wild-type (denoted by *) or rab7 TKO and atg5 TKO (denoted by #). * or #p < 0.05; **p < 0.01; ***p < 0.001.
When RAB7 is deleted, endocytic cargo destined for the lysosome is instead recycled.32 The effect of RAB7 deletion on surface levels of IL2RA/CD25, the T cell receptor (CD3), and the nutrient transporters TFRC/CD71 (the transferrin receptor) and SLC3A2 was measured in activated cells at 65 h. If endocytic degradation is disrupted in rab7−/− T cells, surface expression of these molecules might be higher than in control cells. In fact, minor decreases in SLC3A2 and CD3 surface levels were observed that reached statistical significance in CD8 and CD4 rab7−/− T cells, respectively (Fig. 6D). As SLC3A2 levels were slightly elevated on rab7−/− cells at 24 h after activation (Fig. 6A), this decrease might be a consequence of decreased cellular health at later time points when cell death increased. A mild upregulation of IL2RA was seen in CD4 rab7−/− T cells and there was a small but not statistically significant upregulation in CD8 T cells. IL2 supports the survival and growth of activated T cells, and increased IL2RA expression may provide an advantage to activated rab7−/− T cells. In contrast to the relatively minor effects on SLC3A2, CD3 and IL2RA, TRFC surface expression was strikingly increased in activated rab7−/− T cells. Iron is required for cell growth but also catalyzes the production of ROS.36 Thus, cellular iron uptake is tightly regulated, and the impact of elevated TRFC levels on activated cells is likely complex and context dependent but could also counter-balance the negative effects of the loss of autophagy on the growth and survival of rab7−/− T cells. TRFC levels were below the detection limit in both wild-type and rab7−/− naïve T cells, and any differences that might exist could not be quantified (data not shown). Together, these results suggest that deletion of rab7 had relatively minor effects on the endocytic trafficking of surface molecules in activated T cells with the exception of a surface protein with an unusually rapid rate of recycling (TRFC).
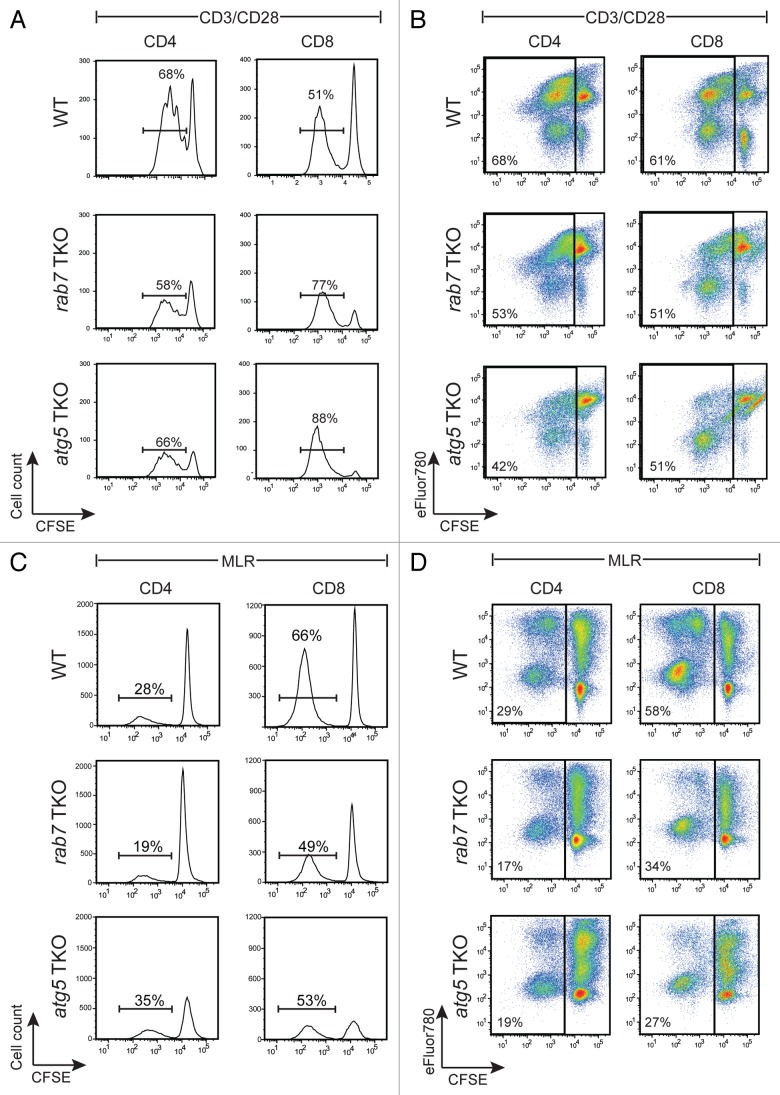
The ability of rab7−/− and atg5−/− T cells to proliferate in response to ligation of CD3 and CD28 was next evaluated. T cells were isolated from 8- to 12-week-old age- and sex-matched mice of each genotype, labeled with the cytosolic dye CFSE, and added to plates coated with anti-CD3 antibodies in the presence of soluble anti-CD28. At 65 h post-stimulation, cells were stained for CD4 and CD8 and proliferation measured by CFSE dilution. Both rab7−/− and atg5−/− T cells exhibited a similar defect in proliferation in this assay (Fig. 7A and B). About 53% of rab7−/− and 42% of atg5−/− CD4 T cells divided under these experimental conditions as opposed to 68% of wild-type CD4 T cells. Similar results were obtained in CD8 T cells, where 51% of rab7−/− and atg5−/− T cells divided as opposed to 61% of the wild-type CD8 T cells (Fig. 7B). Intracellular staining confirmed that ≥ 70% of the T cells from rab7 TKO mice that had diluted CFSE were in fact rab7−/− (not shown). Autophagy-deficient T cells that failed to divide were more prone to die than nondividing wild-type cells (Fig. 7A and B). TCR cross-linking provides a very strong activation stimulus that may create an artificially high dependence on autophagy by generating increased cellular stress. To examine the proliferation of autophagy-deficient T cells in a more physiologic setting, a mixed lymphocyte reaction (MLR) was performed. In an MLR, the 1 to 10% of T cells that recognize nonself MHC will be activated. T cells were isolated from wild-type, rab7 TKO, and atg5 TKO C57BL/6 mice and mixed with mitomycin C treated splenocytes from Balb/c mice. rab7−/− T cells again had a proliferation defect of similar magnitude to that seen in atg5−/− T cells (Fig. 7C and D). As expected in this assay, CD8 T cells proliferated more than CD4 cells for all three genotypes. Although cell accumulation was reduced in autophagy-deficient T cells, significant dilution of CFSE was observed demonstrating that these cells are capable of multiple rounds of cell division. As seen with anti-CD3/CD28 antibodies, nondividing atg5−/− T cells were minimally viable; the survival defect in non-dividing rab7−/− T cells in the MLR was, however, was less severe. In summary, activated rab7−/− T cells display similar survival defects to atg5−/− T cells suggesting that autophagic flux and not simply autophagosome formation is required for normal T cell accumulation following activation.
Figure 7. Deletion of rab7 or atg5 produced similar defects in activated T cells. (A) Freshly purified, CFSE-labeled splenic T cells were activated with plate-bound anti-CD3 and soluble anti-CD28 antibodies. At 65 h, cells were surface-stained for CD4 and CD8 and then incubated with eFluor780 prior to analysis by flow cytometry. CFSE levels in live-gated CD4+ or CD8+ populations are shown and the percent of live cells that diluted CFSE is indicated. (B) As in (A), but without live gating; percentage of all cells that proliferated (diluted CFSE) is shown. (C and D) CFSE-labeled purified T cells isolated from age- and sex-matched wild-type (WT), rab7 TKO, and atg5 TKO mice were mixed with mitomycin C-treated splenocytes from a Balb/c mouse. After 84 h of incubation, cells were surface-stained for CD4 and CD8, labeled with eFluor780, and CFSE dilution evaluated in the H-2Dd-negative (C57BL/6) population. Live (C) or total (D) cells are shown; percentages indicate the fraction of cells that diluted CFSE. In (A–D), a representative experiment is shown; these experiments were repeated at least 3 times with similar results.
Discussion
In mice, disrupting autophagosome formation reduces the number of peripheral T cells and leads to defects in naïve and activated T cell survival.25-27 Here, we found that T cells that could form but not degrade autophagosomes due to the loss of RAB7 were also present at reduced numbers in vivo and failed to accumulate normally upon activation in vitro (Figs. 4 and 7). While deletion of either atg5 or rab7 was similarly efficient (Fig. 2A) and led to a near complete block in autophagic flux (Fig. 3),37 differences in the phenotypes of atg5 and rab7 TKO mice were noted. The reduction in the number of peripheral T cells in rab7 TKO animals was less severe than observed in atg5 or atg7 TKO mice (Fig. 4A and C),26,27 and the viability of naïve rab7−/− T cells was near normal upon isolation and elevated after neglect (Fig. 5F). Together, these differences may have created less of a stimulus for homeostatic proliferation (Fig. 4E and F) than seen in other autophagy-deficient mice where CD44hiSELLlo cells increase.
The negative effects of blocking autophagy are often attributed to increased ROS production from mitochondria that fail to be cleared by mitophagy.38 In the absence of RAB7, mitochondria that fail to be turned over should accumulate primarily inside autophagosomes rather than in the cytosol. The double autophagosomal membrane might separate these engulfed mitochondria from their substrates and thereby decrease ROS production and detrimental effects. While neglected rab7−/− T cells did display elevated ROS levels relative to wild-type controls, rab7−/− T cells produced lower levels of ROS than atg5−/− T cells (Fig. 5E). ROS elevation may still occur in rab7−/− T cells because autophagosome accumulation eventually inhibits autophagosome formation. Alternatively, fatty acids might still be accessible to engulfed mitochondria, or enhanced iron access in rab7−/− cells might increase ROS production. Interestingly, ROS production was well correlated with mitochondrial mass (Fig. 5C and E). It was unexpected that rab7−/− T cells accumulated fewer mitochondria than atg5−/− T cells. Experiments using the live cell mitochondrial dye MTG suggested that there was a dramatic reduction in mitochondrial mass in rab7−/− T cells to below wild-type levels (Fig. 5A). However, MTG may not effectively penetrate the autophagic vesicles in the rab7-deleted T cells. TOMM20 staining in fixed and permeabilized cells should accurately reflect mitochondrial mass even if mitochondria have been engulfed by autophagosomes. TOMM20 quantification indicated that mitochondrial mass in rab7−/− T cells was increased over wild-type T cells as expected but reduced relative to atg5−/− T cells (Fig. 5C). It is possible that increased access to iron due to increased surface TRFC levels in rab7−/− T cells initiates a feedback loop that limits mitochondrial biogenesis. Although the TRFC signal was at background levels in naïve wild-type or rab7−/− T cells (data not shown), it may be that TRFC levels are elevated but below the limit of detection. Regardless of the mechanism behind the reduced mitochondrial mass, reduced ROS generation may contribute to the increased survival of neglected naïve rab7−/− T cells (Fig. 5F) and the increased numbers of peripheral T cells in rab7 TKO mice compared with atg5 TKO animals (Fig. 4).26,27
While the loss of autophagy reduces T cell fitness by altering organelle homeostasis, a role for autophagy in supplying nutrients to activated T cells has also been suggested.28 Our initial hypothesis was that autophagy was necessary immediately after activation to supply building blocks and energy for the dramatic and immediate upregulation of nutrient transporter expression in quiescent cells that have limited access to extracellular nutrients. However, SLC3A2 surface levels increased normally in autophagy-deficient T cells after activation (Fig. 6A) arguing against this model. Recent reports demonstrate that the proteasome is a key source of amino acids for protein synthesis and thus proteasomal degradation may supply the substrates for the synthesis of SLC3A2 and other early activation markers in the presence or absence of an intact autophagy pathway.39
One plausible explanation for the increased number of T cells in the spleens of rab7 TKO mice relative to atg5 TKO mice is that, in addition to its role in organelle turnover (Fig. 5),26,27 autophagy is required to maintain nutrient homeostasis in naïve T cells. The availability of growth and survival signals defines the number of peripheral T cells present in vivo;40 cytokines also regulate nutrient access by preventing the lysosomal degradation of nutrient transporter proteins.32,41 Although transporter levels were at the limit of detection in naïve cells and therefore difficult to compare following neglect, TRFC levels were significantly increased in activated rab7−/− T cells relative to controls (Fig. 6D). TRFC is distinct from the other surface molecules evaluated in that it undergoes repetitive cycles of endocytosis and recycling.36 Thus, TRFC repeatedly passes the checkpoint that determines whether an internalized molecule is recycled or degraded in the lysosome through a RAB7-dependent step. Other surface proteins, including SLC3A2, which promotes amino acid transport across the plasma membrane, were much less affected by rab7 deletion under our experimental conditions. Increased TRFC expression could lead to increased access to iron which might facilitate mitochondrial metabolism and growth and, along with the milder upregulation of IL2RA, contribute to the slightly increased survival of activated rab7−/− T cells relative to atg5−/− T cells (Fig. 7B). While the effects of RAB7 on endocytic trafficking to the lysosome somewhat complicates the comparison of the effects of a late and early block in autophagy, this caveat is present in all experiments performed with inhibitors of lysosomal degradation; chloroquine in particular has broad effects on cells.42-45 Most genes also have pleiotropic effects; Atg5, for example, has additional roles beyond autophagosome formation.46 The simplest interpretation of the finding that activated rab7−/− and atg5−/− T cells have similar defects is that blocking autophagy at an early or late stage has rather similar effects on cells. In summary, our observations suggest that blocking autophagy in T cells at an early or late stage is similarly detrimental in activated cells, but blocking autophagosome degradation by eliminating RAB7 has fewer negative effects in naïve T cells. In conditions where it is beneficial therapeutically to block autophagy, for example in cancer therapy, it may therefore be preferable to inhibit autophagosome degradation rather than formation to minimize ROS generation and toxicity in normal, quiescent cells.
The rab7 conditional knockout described here is the first mouse model with a genetic defect that allows autophagosome formation but not degradation. Rab7flox/flox animals should be of great utility in future studies of vesicular trafficking and autophagy. For example, it would be interesting to determine whether the loss of rab7 interferes with adipogenesis similar to what has been seen following atg5 or atg7 deletion.47,48 The floxed Rab7 model system might also be used to address whether the loss of autophagosome degradation is as toxic to neurons as is the loss of autophagosome formation given that protein aggregates should also be sequestered in autophagosomes in rab7- but not atg5 and atg7-deficient cells.49,50 rab7 deletion in at least two cell types, fibroblasts and T cells, was well tolerated. Thus, rab7−/− cell lines generated from these animals should also be useful for future studies.
Materials and Methods
Antibodies and reagents
T-cell purification was performed using a negative selection kit (Stem Cell Technologies, 19751). Antibodies were obtained from the following sources: RAB7 (9367), MAP1LC3B (4108, stronger reactivity with MAP1LC3B-II than MAP1LC3B-I) and ATG5 (Cell Signaling Technologies, 8540S); TOMM20 (Sigma-Aldrich, WH0009804M1-100UG); ACTIN (Santa Cruz Biotechnology, sc-1615), APC-CD4 (100411), PerCP/Cy5.5-CD8a (100733), PE-SLC3A2 (128207), Biotin-anti-mouse H-2Dd (110603), Purified CD3 (100314), FITC-SELL (104405), PE-Cy7-CD44 (103029), Fc block (101319), BV421-Donkey anti-rabbit (406410), Streptavidin-PE/Cy7 (all from Biolegend, 405206); CD28 (16028185), PE/Cy7-B220 (25045282), FITC-CD3e (eBioscience, 11003382), Alexa488-IL2RA (eBioscience, 53-0251-82); PE-TRFC (eBioscience, 12-0711-82); PE-Rat IgG1 isotype control (BD PharMingen, 551979); FITC-mouse IgG1 isotype control (Biolegend, 400110); anti-hamster IgG (H+L) (Vector Laboratories, Inc., AI-9100), Alexa488-goat anti-rabbit (Life Technologies, A-11034). eFluor 780 (65086514) was from eBioscience, CFSE (34554), CM-H2DCFDA (C6827) and Mitotracker Green (M-7514) were purchased from Life Technologies, staurosporine was from Enzo (ALX-350360G001) and PE-Donkey anti-mouse IgG (H+L) was from Jackson Immuno Research Laboratories, Inc. (715-116-151). Secondary reagents (goat anti-rabbit and donkey anti-goat, 92632211 and 92632214, respectively) were purchased from LI-COR. All other chemicals were obtained from Sigma-Aldrich or EMD Biosciences.
Generation of knockout mice
A 10 kb fragment that included the first two exons of Rab7 was isolated from a mouse genomic DNA lambda library and used to produce a targeting vector that deletes exons I and II of Rab7. Constructs were confirmed by sequencing at each cloning step. 129/Sv x 129/Sv-CP ES cells were electroporated with the targeting construct and stable integrants selected with neomycin. A Rab7+/floxneo ES cell clone (J11) was identified by Southern blotting. The functionality of the LoxP sites was confirmed by introducing the Cre recombinase and screening clones by Southern blotting. The J11 clone was injected into C57BL/6 blastocysts in the Transgenic Mouse Facility at UC Irvine. Of the 3 chimeric male mice produced, one transmitted the floxed allele to approximately half his progeny. The neomycin resistance cassette used for selection was removed by crossing Rab7+/floxneo mice to a Protamine-Cre line that expresses CRE in spermatids (generously provided by Grant MacGregor).29 The rab7null allele was also produced by breeding onto the Protamine-Cre line. The Rab7flox mice used in these studies were backcrossed with C57BL/6J mice for > 10 generations. The animals will be available from Jackson Labs (stock #0021589, B6.Cg(129)-Rab7tm1.1Ale/J). Atg5+/flox mice on a C57BL/6 background were kindly supplied by Noboru Mizushima (Department of Physiology and Cell Biology, Tokyo Medical and Dental University, Tokyo, Japan) via Craig Walsh (University of California, Irvine). All animal protocols were approved by the Institutional Animal Care and Use Committee of University of California, Irvine.
Primers and PCR
Primer sequences used are given below (locations shown in Fig. 1A):
Primer 53: 5′-CTCACTCACTCCTAAATGG-3′; Primer 54: 5′-TTAGGCTGTATGTATGTGC-3′; Primer 63: 5′-GGGCTGCAGGAATTCGGATAAC-3′; Primer 64: 5′-CCACATTCAAAGACTGTCTCCC-3′; Primer 67: 5′-CATGGTAACAAGTCTGTCGTCC-3′. PCR cycling parameters used: primers 53/54 (wild-type allele produces a 550 bp band, flox allele produces a 580 bp band): 94°C for 5 min, (94°C for 1 min, 55°C for 1 min, 72°C for 2 min) for 35 cycles and 72°C for 10 min. Primers 63/64 (flox or floxneo produce a 500 bp band): 94°C for 5 min, (94°C for 1 min, 61°C for 1 min, 72°C for 2 min) for 35 cycles and 72°C for 10 min. Primers 63/67 (only the null allele produces a 500 bp band): 94°C for 5 min, (94°C for 1 min, 61°C for 1 min, 72°C for 2 min) for 35 cycles and 72°C for 10 min.
Cell culture
atg5−/− MEFs were generously provided by Noboru Mizushima. rab7−/− MEFs were generated from Rab7-/flox embryos and immortalized with SV40 Large T antigen introduced by retroviral transduction. MEFs were maintained in DMEM (Mediatech) supplemented with 10% fetal calf serum (FCS) and antibiotics. For nutrient deprivation experiments, DMEM lacking glucose and amino acids was made from chemical components and supplemented with 10% FCS. This medium was mixed with standard DMEM containing 10% FCS to generate medium containing 1% the normal amount of amino acids and glucose. T cells were isolated from total splenocytes by negative selection with magnetic beads (EasySepTM mouse T Cell Enrichment Kit: Stem Cell Technologies, 19751). Freshly isolated splenocytes/purified T cells were cultured in RPMI 1640 (Mediatech) supplemented with 10% FCS (Sigma-Aldrich), 10 mM HEPES (Mediatech), 55 μM β-mercaptoethanol (Sigma-Aldrich), antibiotics, 1 mM Na-Pyruvate (Mediatech), 1× Non-Essential Amino acids (Lonza) and 1 mM L-glutamine (Mediatech).
Western blotting
Cells were washed twice with cold PBS and lysed in the plate on ice with lysis buffer (2% Triton X-100, 80 mM HEPES, 0.4 mM EDTA, pH 6.6) containing complete protease inhibitor cocktail (Roche), 50 mM sodium fluoride, 100 nM sodium orthovandate, and 50 mM β-glycerophosphate. Protein concentration was quantified using a BCA Assay (Pierce) and an equal amount of protein loaded in each lane of 4–12% NuPAGE gradient gels (Life Technologies). Proteins were transferred to nitrocellulose membranes in NuPAGE transfer buffer containing 20% methanol and exposed to primary antibodies diluted in the blocking buffer recommended by the manufacturer. Following incubation with secondary, blots were scanned with an Odyssey imager (LI-COR Biosciences) and quantified.
Plate-bound activation assay and MLR
Anti-hamster IgG (Vector Laboratories, AI-9100) was coupled to 24 well plates by incubation in 0.1 M carbonate buffer (0.84 g NaHCO3, 0.356 g Na2CO3, in 100 ml of dH2O; pH 9.5) either overnight at 4°C or 1 h at room temperature (RT). Plates were washed with 1× warm HBSS and 1000 ng/ml anti-CD3 (Biolegend, 100314) added prior to a 1 h incubation at 37°C. Plates were washed with 1× HBSS and purified T cells labeled with 4 µM CFSE were added at a concentration of 1 × 106 cells/ml in T cell medium containing 200 ng/ml of soluble anti-CD28. Cells were analyzed after 65 h of incubation in a humidified incubator at 37°C and 5% CO2. For the MLR, T cells were isolated from C57BL/6 mice, labeled with CFSE (5 µM), and co-cultured with fresh Balb/c splenocytes that had been treated with 50 µg/ml mitomycin C for 30 min at 37°C. Cells were analyzed after 84 h of incubation in a humidified incubator at 37°C and 5% CO2.
Flow cytometry
Surface staining was conducted on ice in PBS containing 10% FBS and 0.05% NaN3. For intracellular staining (RAB7), cells were stained for surface antigens, incubated with the vital dye eFluor780 in the absence of NaN3 and serum, fixed in 1% paraformaldehyde (PFA) for 10 min at RT, and permeabilized with IF blocking buffer (PBS containing 0.3% saponin, 10% FBS and 0.05% NaN3) and analyzed on an LSR II flow cytometer (BD Biosciences) using FlowJo software (Treestar). To measure ROS production, 1x106 naïve T cells were washed and re-suspended in 1 ml HBSS. Cells were incubated at 37°C for 30 min then stained with 1 µM CM-H2DCFDA for 8 to10 min at 37°C. Cells were washed with HBSS and analyzed by flow cytometry. To measure mitochondrial mass, 1 × 106 naïve T cells were washed and re-suspended in 1 ml T cell medium lacking FBS. After 30 min at 37°C cells were stained with 100 nM Mitotracker green for 30 min at 37°C. Cells were washed with RPMI and analyzed by flow cytometry. Anti-TOMM20 (1:100) was added to cells fixed in 1% paraformaldehyde for 10 min at RT and permeabilized with IF blocking buffer and incubated overnight at 4°C followed by PE-Donkey anti-mouse IgG secondary (1:100) in IF blocking buffer for 2 h at RT. Analysis was restricted to live cells by DAPI staining. To measure total cellular SLC3A2, cells were fixed in 1% paraformaldehyde for 10 min at RT, Fc receptors blocked for 10 min at RT, and then cells were stained for CD4/8 for 20 min at RT followed by PE-SLC3A2 for 1 h at RT. DAPI was used to exclude apoptotic cells (sub-G1) from the analysis.
Immunofluorescence microscopy
20,000 MEFs were plated in 24 wells on poly-(l-lysine)-coated coverslips, fixed in 1% PFA for 10 min at RT, washed three times with PBS, and incubated for 1 h at RT in IF blocking buffer for permeabilization. Cells were washed with IF wash then incubated with anti-RAB7 or anti-MAP1LC3B (1:100) in IF blocking buffer overnight at 4°C followed by anti-rabbit Alexa488 secondary (1:100) in IF blocking buffer for 2 h at RT. Samples prepared for MAP1LC3B immunostaining were fixed and permeabilized by 100% cold methanol at −20°C for 10 min. Cells were washed with IF wash buffer (PBS with 2% FCS, 0.03% saponin, and 0.05% NaN3). Coverslips were mounted using VectaShield mounting medium with DAPI (Vector Labs) and images collected using a Nikon Eclipse TE2000 fluorescence microscope equipped with a CoolSNAP CCD camera (Photometrics). T cells activated with anti-CD3/anti-CD28 were spun onto glass slides (1000 rpm for 3 min; Shandon Cytospin3) prior to fixation and staining.
Acknowledgments
This work was supported in part by grants to ALE from the NIH (K08 CA100526 and R01 GM089919), the American Cancer Society (RGS 120976-RSG-11-111-01-CDD) and the Gabrielle’s Angel Foundation. Special thanks to Tullia Lindsten for technical expertise and guidance during the generation of the Rab7+/floxneo ES cells and to Tom Fielder for injection of blastocysts and the initial maintenance and breeding of chimeric males. We thank Noboru Mizushima for providing atg5-deficient cell lines and floxed mice, Craig Walsh for the CD4-Cre transgenic mice, Eigen Peralta for preliminary studies with Rab7flox/floxLck-Cre mice, Brian Weist and Bryan Bell for advice and assistance with T cell activation assays, Grant MacGregor for use of the Cytospin and the Protamine-Cre mice, Katrina Waymire for assistance setting up the RAB7 mouse colony, and Eigen Peralta, Garret Guenther, Hikmat Assi, Pinal Patel, Mary Anne Nguyen, and Ronika Sitapara Leang for assistance with animal husbandry and genotyping.
Glossary
Abbreviations:
- atg5
autophagy-related 5
- CD3
CD3-T-cell receptor complex
- CD4
T cell surface glycoprotein CD4
- CD8
T cell surface glycoprotein CD8
- CD28
T cell surface glycoprotein CD28
- CD44
CD44 molecule (Indian blood group)
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- CQ
chloroquine
- DAPI
4',6-diamidino-2-phenylindole
- DCFDA
5-(and-6)-carboxy-2',7'-dichlorofluorescein diacetate
- IL2RA
interleukin 2 receptor, alpha (CD25)
- MAP1LC3B
microtubule-associated protein 1 light chain 3 beta (LC3B)
- MEF
murine embryonic fibroblast
- MLR
mixed lymphocyte reaction
- MTG
MitoTracker Green
- rab7
RAB7A, member RAS oncogene family
- ROS
reactive oxygen species
- SELL
selectin L (CD62L)
- SLC3A2
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 (4F2 heavy chain/CD98)
- TCR
T cell receptor
- TFRC
transferrin receptor (CD71)
- TKO
T cell knockout
- TOMM20
translocase of outer mitochondrial membrane 20 homolog (yeast)
Disclosure of Potential Conflicts of Interest
The authors have no conflicts of interest to declare.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24468
References
- 1.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–80. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–52. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang T, Ming Z, Xiaochun W, Hong W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal. 2011;23:516–21. doi: 10.1016/j.cellsig.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–91. doi: 10.1016/S0962-8924(01)02147-X. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–97. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 6.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–96. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–57. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 11.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 13.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 17.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–6. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 20.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006;175:121–33. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunol Rev. 2010;236:95–109. doi: 10.1111/j.1600-065X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–55. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson LM, Miller BC, Ng A, Eisenberg J, Zhao Z, Cadwell K, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–35. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–57. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A. 1997;94:14602–7. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol. 2001;167:6869–76. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 31.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 32.Edinger AL, Cinalli RM, Thompson CB. Rab7 prevents growth factor-independent survival by inhibiting cell-autonomous nutrient transporter expression. Dev Cell. 2003;5:571–82. doi: 10.1016/S1534-5807(03)00291-0. [DOI] [PubMed] [Google Scholar]
- 33.Gottesdiener KM, Karpinski BA, Lindsten T, Strominger JL, Jones NH, Thompson CB, et al. Isolation and structural characterization of the human 4F2 heavy-chain gene, an inducible gene involved in T-lymphocyte activation. Mol Cell Biol. 1988;8:3809–19. doi: 10.1128/mcb.8.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nii T, Segawa H, Taketani Y, Tani Y, Ohkido M, Kishida S, et al. Molecular events involved in up-regulating human Na+-independent neutral amino acid transporter LAT1 during T-cell activation. Biochem J. 2001;358:693–704. doi: 10.1042/0264-6021:3580693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacLeod CL. Regulation of cationic amino acid transporter (CAT) gene expression. Biochem Soc Trans. 1996;24:846–52. doi: 10.1042/bst0240846. [DOI] [PubMed] [Google Scholar]
- 36.Richardson DR, Lane DJ, Becker EM, Huang ML, Whitnall M, Suryo Rahmanto Y, et al. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci U S A. 2010;107:10775–82. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 38.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–30. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suraweera A, Münch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48:242–53. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol Cell. 2000;6:683–92. doi: 10.1016/S1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 41.Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. 2002;13:2276–88. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200–12. doi: 10.4161/auto.8.2.18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Zvi I, Kivity S, Langevitz P, Shoenfeld Y. Hydroxychloroquine: from malaria to autoimmunity. Clin Rev Allergy Immunol. 2012;42:145–53. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123:1107–17. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellman I, Fuchs R, Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 46.Codogno P, Meijer AJ. Atg5: more than an autophagy factor. Nat Cell Biol. 2006;8:1045–7. doi: 10.1038/ncb1006-1045. [DOI] [PubMed] [Google Scholar]
- 47.Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–30. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–39. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 50.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]