Abstract
We exploited the amenability of the fungus Aspergillus nidulans to genetics and live-cell microscopy to investigate autophagy. Upon nitrogen starvation, GFP-Atg8-containing pre-autophagosomal puncta give rise to cup-shaped phagophores and circular (0.9-μm diameter) autophagosomes that disappear in the vicinity of the vacuoles after their shape becomes irregular and their GFP-Atg8 fluorescence decays. This ‘autophagosome cycle’ gives rise to characteristic cone-shaped traces in kymographs. Autophagy does not require endosome maturation or ESCRTs, as autophagosomes fuse with vacuoles directly in a RabS (homolog of Saccharomyces cerevisiae Ypt7 and mammalian RAB7; written hereafter as RabSRAB7)-HOPS-(homotypic fusion and vacuole protein sorting complex)-dependent manner. However, by removing RabSRAB7 or Vps41 (a component of the HOPS complex), we show that autophagosomes may still fuse, albeit inefficiently, with the endovacuolar system in a process almost certainly mediated by RabARAB5/RabBRAB5 (yeast Vps21 homologs)-CORVET (class C core vacuole/endosome tethering complex), because acute inactivation of HbrA/Vps33, a key component of HOPS and CORVET, completely precludes access of GFP-Atg8 to vacuoles without affecting autophagosome biogenesis. Using a FYVE2-GFP probe and endosomal PtdIns3P-depleted cells, we imaged PtdIns3P on autophagic membranes. PtdIns3P present on autophagosomes decays at late stages of the cycle, preceding fusion with the vacuole. Autophagy does not require Golgi traffic, but it is crucially dependent on RabORAB1. TRAPPIII-specific factor AN7311 (yeast Trs85) localizes to the phagophore assembly site (PAS) and RabORAB1 localizes to phagophores and autophagosomes. The Golgi and autophagy roles of RabORAB1 are dissociable by mutation: rabOA136D hyphae show relatively normal secretion at 28°C but are completely blocked in autophagy. This finding and the lack of Golgi traffic involvement pointed to the ER as one potential source of membranes for autophagy. In agreement, autophagosomes form in close association with ring-shaped omegasome-like ER structures resembling those described in mammalian cells.
Keywords: autophagosome, phagophore, intracellular traffic, nitrogen starvation, Rab, SNARE, Atg8
Introduction
Macroautophagy (hereafter ‘autophagy’) is a nutrient-starvation response conserved across eukaryotes. During autophagy, single-membrane-bound structures (known as phagophores) form, undergo expansion by addition of lipid membrane, and ultimately give rise to spherical, double-membrane-bound autophagosomes that fuse with the endolysosomal system. Studies with S. cerevisiae, including seminal genetic screens that resulted in the identification of 36 ATG (autophagy-related) genes,1,2 have been instrumental in our current understanding of autophagy.3
The biogenesis of autophagosomes requires delivery of lipid membrane to the phagophores. The source(s) of this membrane has been intensively debated. In S. cerevisiae electron microscopy studies identified Atg9-containing tubulovesicular clusters denoted “Atg9 reservoirs.”4 Because Atg9 is an integral membrane protein that cycles between these reservoirs and the PAS, Atg9 reservoirs must contribute membrane to the phagophore. The source(s) of autophagosome membrane has also been investigated using panels of mutations in different membrane trafficking steps. These experiments led to conclusions that are not easy to reconcile with data obtained with mammalian cells, which strongly suggest that the endoplasmic reticulum (ER) is an important contributor to autophagosomal membranes.5 For example, it has been reported that yeast post-Golgi proteins such as Sec4, plasma membrane SNAREs, such as Sso1, or late Golgi proteins such as Sec7 are required for autophagy.6-8 Autophagy also requires Ypt1 (mammalian RAB1), which is recruited to the PAS in a TRAPP (transport protein particle)-III-dependent manner,9 as well as subunits of the COG (conserved oligomeric Golgi) complex.10 A recent study concluded that delivery of membranes to the autophagosome is only prevented by double-null mutations in pathways connecting the Golgi and the endosomal system, a finding attributable to “the ability of the yeast Golgi-endosomal system to compensate for single gene deletions.”11 Aspergillus nidulans is a genetically amenable filamentous ascomycete that lacks this plasticity, offering a potentially useful alternative to budding yeast.
A protein playing a central role in autophagy in yeast is the ubiquitin-like Atg8, which localizes to the PAS, phagophore and autophagosome,12,13 thereby being widely accepted as a prototypical protein marker for autophagy. The Atg4 cysteine protease removes the C-terminal residue of the yeast Atg8 primary translation product to expose a glycine residue that is subsequently conjugated to phosphatidylethanolamine (PE) through a ubiquitin-like conjugation reaction, mediated by the E1-like Atg7 and E2-like Atg3.14 Lipidation anchors Atg8 to membranes and plays a key role in autophagosome biogenenesis.6,15 Upon autophagosome closure, Atg4 deconjugates Atg8p from PE, releasing the former from membranes.16 Deconjugation precedes autophagosome-vacuole fusion and is required for efficient autophagosome biogenesis.17
While genetic, biochemical and immunoelectron microscopy studies have successfully exploited the full potential of S. cerevisiae, live-microscopy studies have been useful, in combination with autophagy mutants, to establish a hierarchy of action of the different Atg products.18 However, conventional microscopy is arguably more limited to correlating genetic and morphological observations, due to the small volume of the yeast cell and the relatively small size of yeast autophagosomes. Yeast GFP-Atg8 localizes to the PAS, which are thought to give rise to autophagosomes.19,20 Xie et al. performed live studies on autophagosome biogenesis using GFP-Atg8. Although these studies could not resolve the different intermediates beyond punctate structures, they provided a blueprint of the autophagosome cycle, showing that Atg8 is released from the autophagosomal structures at late stages preceding autophagosome-vacuole fusion.21 Quantitative studies using fluorescence microscopy also revealed stoichiometric information on Atg proteins at the PAS.22
The genetic model A. nidulans, whose multinucleated hyphal cells are notably larger than those of S. cerevisiae, is ideally suited for in vivo microscopy and intracellular trafficking studies.23 Atg8 had been characterized in A. oryzae, an asexual industrial species in which Atg8 is delivered to the vacuole by autophagy.24 However, the full genetic potential of A. nidulans to investigate autophagy remained unexplored. In this work we set up basic tools to investigate autophagy, including state-of-the-art time-lapse microscopy that we used to resolve the different stages of autophagosome biogenesis that precede fusion with the vacuoles. We used these tools to track in vivo the PtdIns3P cycle on autophagic membranes. We further showed that neither post-Golgi steps nor traffic across the Golgi or endosomes is required for autophagy. By using a ts mutation in rabO to dissociate the secretory and autophagic functions of RabORAB1, we demonstrated that autophagy is exquisitely dependent on this Rab protein. These findings drove our attention to the ER as a potential source of autophagic membranes, leading us to detect the presence of omegasome-like structures, akin to those reported in mammalian cells, associated with fungal autophagosomes.
Results
Basic methodology for monitoring autophagy in A. nidulans
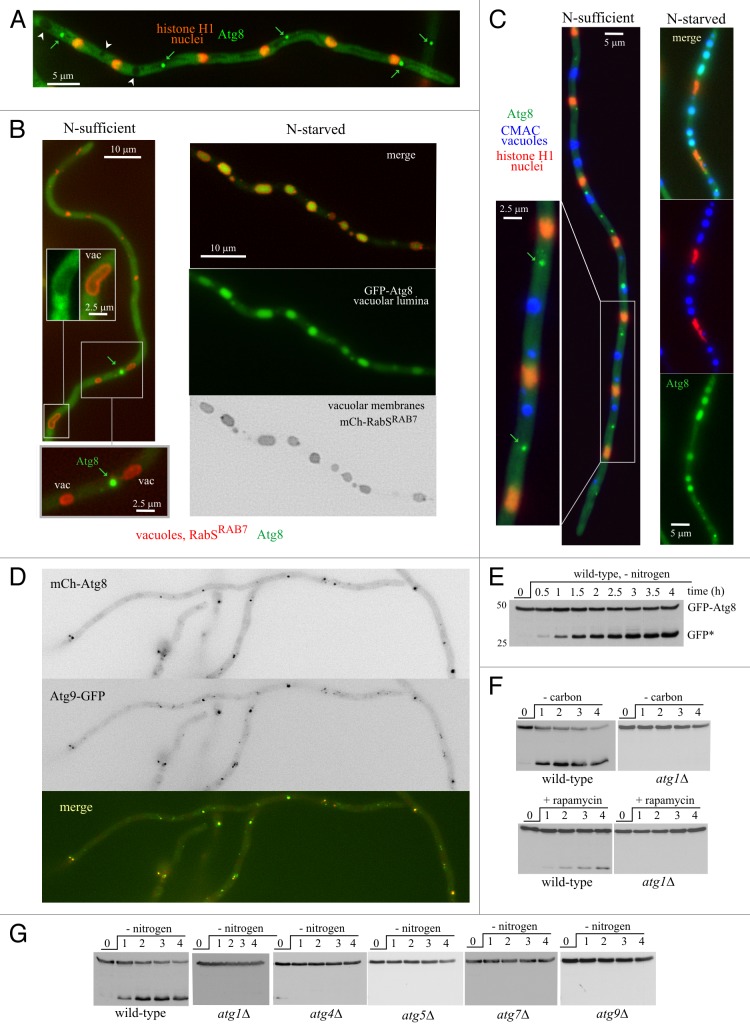
We first developed basic assays and technology to investigate autophagy in A. nidulans, including a description of the pathway largely based on live microscopy and proteolysis of GFP-Atg8. In A. nidulans hyphae cultured in nitrogen-sufficient conditions (ammonium as nitrogen source) GFP-Atg8 localizes to a cytosolic haze and to bright punctate structures (Fig. 1A–C; Movie S1). Atg8 puncta correspond to the PAS, as determined by co-imaging with the well-characterized PAS marker Atg9, endogenously tagged with GFP, which also localizes to puncta (Fig. 1D; and to fainter, diffuse structures possibly representing ‘Atg9 reservoirs’).4 Colocalization of mCherry-Atg8 puncta and Atg9-GFP puncta was essentially total (Fig. 1D) (85% of n = 358 Atg8 puncta colocalized with Atg9 dots).
Figure 1. Autophagy in A. nidulans, monitored with GFP-Atg8. (A) Maximal intensity projection of a z-series stack of images depicting a cell cultured in nitrogen-sufficient conditions that expresses GFP-Atg8 and mCherry-histone H1. The latter labels evenly spaced nuclei (spacing 15.6 μm ± 1.3 SD). Examples of bright GFP-Atg8 puncta are marked with green arrows. Note the ‘empty’ vacuoles (white arrowheads). (B) In nitrogen-sufficient conditions, bright GFP-Atg8 puncta (arrows) do not associate with GFP-negative vacuoles, whose membranes were visualized with mCherry-RabSRAB7. Insets are shown at double magnification. After 4 h of incubation in nitrogen starvation conditions, the lumen of the vacuoles becomes GFP-positive. (C) GFP-Atg8 localization in hyphal cells in which nuclei had been stained with mCherry-histone H1 and the vacuolar lumen with CMAC (colored in blue). Examples of bright Atg8 puncta seen under nitrogen-sufficient conditions are marked with arrows. Note that GFP is in the lumen of the vacuoles following the shift to nitrogen starvation conditions. (D) Nitrogen-sufficient conditions: Atg9-GFP, expressed at physiological levels after gene replacement, colocalizes with mCherry-Atg8 puncta. (E) GFP-Atg8 proteolysis assays. Wild-type GFP-Atg8 mycelia were cultured overnight with ammonium and shifted to medium without nitrogen. Samples were taken before (0 time point) and at the indicated times after the transfer (in h) and total cell extracts were analyzed by anti-GFP western blotting. The bands corresponding to full-length GFP-Atg8 and the proteolysis-recalcitrant GFP* moiety (“free GFP”) are indicated. (F) GFP-Atg8 proteolysis experiments in wild type and atg1∆ cells. Autophagy was induced after starving for carbon (cells shifted from 1% glucose medium to medium without any carbon source) or by addition of rapamycin (0.2 μg/ml), as indicated. (G) GFP-Atg8 processing experiments in atg∆ mutants starved for nitrogen. atg1∆, atg4∆, atg5∆, atg7∆ and atg9∆ are completely deficient in GFP-Atg8 processing.
Autophagy assays exploited the nitrogen starvation-mediated delivery of GFP-Atg8 to the vacuolar lumen. In nitrogen-sufficient conditions, GFP-Atg8 is excluded from the vacuoles (Fig. 1A, arrowheads; Fig. 1B, left inset). In contrast, the lumen of the vacuoles becomes conspicuously fluorescent 2–4 h after nitrogen starvation (Fig. 1B and C). As expected for an autophagy-mediated process, null atg1Δ, atg4∆, atg5∆, atg7Δ and atg9∆ mutations, which result in deficient conidiation at 42°C, prevented the vacuolar GFP accumulation (Fig. S1). Thus, visual inspection of vacuolar GFP fluorescence provided a simple way of assessing autophagy. [Of note, none of the atg∆ mutations delocalized GFP-Atg8 from puncta (Fig. S1)]. For a more systematic monitoring, we used the GFP-Atg8 partial proteolysis assay developed for S. cerevisiae,25 which exploits the resistance of the GFP moiety to vacuolar proteases, such that activation of autophagy leads to the accumulation of “free” GFP in the vacuolar lumen. In nitrogen-sufficient conditions, anti-GFP western blotting detected a 47-kDa band corresponding to full-length GFP-Atg8. Shifting cells to nitrogen-starvation conditions led to the detection of an additional band with the mobility of “free” GFP, whose levels increased with time, apparently at the expense of full-length GFP-Atg8 (Fig. 1E). GFP-Atg8 proteolysis was also induced, somewhat less efficiently, by adding rapamycin (0.2–0.4 µg/ml) to nitrogen-sufficient cultures, or by shifting cells to medium lacking a carbon source (Fig. 1F) and was completely prevented by atg1∆, atg4∆, atg5∆, atg7Δ and atg9∆ mutations (Fig. 1G). Thus, GFP accumulation following vacuolar GFP-Atg8 proteolysis is a bona fide indicator of autophagy.
A direct way of monitoring autophagy consisted of the visualization of autophagic structures by time-lapse microscopy (see also below). When we filmed, with a time resolution of 30 frames/h, the fate of GFP-Atg8 fluorescent puncta over a 3–4 h period after starving hyphae for nitrogen, Atg8 puncta became ‘unstable,’ eventually ‘developing’ into hollow autophagic structures consisting of Atg8-containing membranes enclosing a portion of cytoplasm. The maturation of Atg8 puncta into autophagic structures peaked at ~1.5–3 h after starving cells for nitrogen and was a relatively synchronous process within the cell population. Following this ‘wave’ of autophagosome biogenesis, the vacuolar lumina, previously devoid of GFP, became markedly fluorescent (Movie S2). Nitrogen starvation also resulted in a large increase in the number and size of vacuoles, such that after ~4 h vacuoles occupied a large fraction of the cells (Fig. 1B and C; Movie S3).
In wild-type hyphae undergoing autophagy, essentially every GFP-Atg8 punctum developed into an autophagosome: Of n = 105 puncta tracked over 10–20 min periods, 100 matured into autophagosomes, whereas only five remained stable over the time of observation. This contrasted sharply with the marked stability of GFP-Atg8 puncta in nitrogen-sufficient medium (none out of n = 127 puncta from 99 hyphae cultured with ammonium gave rise to autophagosomes). Moreover, no conversion of puncta into autophagosomes was detectable in atg1∆ (n = 163 puncta analyzed in 116 cells), atg5∆ (n = 49 puncta in 47 cells) and atg9∆ hyphae (n = 46 puncta in 39 cells) starved for nitrogen. These data established that A. nidulans PAS are, sensu stricto, autophagosomal precursors and provide strong evidence that the hollow GFP-Atg8 structures induced by nitrogen starvation are autophagosomes captured at different stages of maturation.
The different stages of the autophagosome cycle visualized by time-lapse microscopy
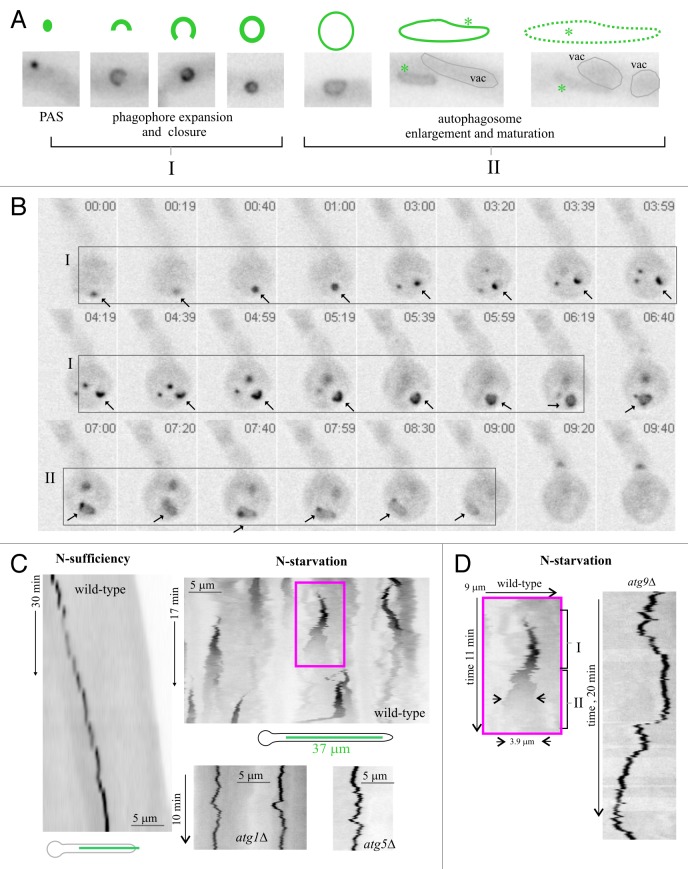
To study the different stages of the maturation of GFP-Atg8 puncta into autophagic structures we used time-lapse microscopy with a greater time resolution (12 frames/min over 10–20 min periods; i.e., 120–240 frames per sequence). Movie S4 shows a cell undergoing autophagy, with vacuoles seen as oblong structures with faintly fluorescent lumina. GFP-Atg8 PAS appear de novo at different times of the sequence and mature into autophagosomes that gradually lose fluorescence before finally disappearing. This maturation follows a highly reproducible chain of events (Fig. 2A and B). Initially, GFP-Atg8 puncta give rise to expanding phagophores engulfing a portion of cytoplasm. These phagophores are eventually sealed, giving rise to brightly fluorescent spherical autophagosomes (0.9 ± 0.1-μm diameter; n = 84). This ‘stage I’ of ‘phagophore closure’ leading to ‘primary’ autophagosomes is followed by an autophagosome enlargement ‘stage II’ during which 0.9-µm spheres become less fluorescent and increase in size as they acquire irregular (usually oblong) shapes (Fig. 2A and B; Movie S5 show several examples). At a later stage, oblong organelles disappear in the vicinity of vacuoles. Co-imaging of RabSRAB7-labeled vacuoles and Atg8-labeled autophagosomes strongly indicated that faint oblong structures ‘disappear’ as they fuse with the vacuoles (Fig. S2; see below).
Figure 2. The autophagosome cycle. (A) Different intermediates of a prototypic autophagosome cycle, based on observations of tens of individual cycles. Images (GFP fluorescence in inverted contrast) show the different intermediates described in the text: PAS, cup-shaped phagophores, closed autophagosomes and oblong structures. The two different stages (see also text) are indicated. (B) Selected frames extracted from a time-lapse sequence illustrating a complete GFP-Atg8 autophagosome cycle in a basal conidium (arrow). Time is in min:sec. I and II indicate the two stages of the autophagosome cycle as shown in (A). (C) The autophagosome cycle studied with kymographs. Left, a GFP-Atg8-labeled PAS in a hyphal tip cell undergoing apical extension in nitrogen-sufficient conditions was filmed over a 30 min period. The corresponding time stack was used to draw a kymograph across the line indicated in the scheme. The PAS is stable under these conditions, giving rise to a vertical line, parallel to the time dimension, if the PAS is static or, as in the example, to a diagonal line if it moves. Top right, autophagosome cycles in the wild type, nitrogen-starvation conditions: A time-lapse sequence (204 frames; 1 frame every 5 sec) was used to draw a kymograph across a 37 μm-long linear region of a hyphal cell, as indicated in the scheme below. Autophagosome cycles give rise to characteristic conical (triangular in the two-dimensional representation) shapes. Bottom right: examples of stable PASs in kymographs drawn from time-lapse sequences of GFP-Atg8 in atg1∆ and atg5∆ cells cultured in nitrogen starvation conditions. Note that the time and distance dimensions are shown at the same scales and thus the different kymographs can be compared directly. (D) Left, enlarged view of the wild type autophagosome cycle boxed in (C); I and II indicate the two stages of the autophagosome cycle; right, example of stable atg9∆ PAS. Time and distance scales are directly comparable.
The whole autophagosome cycle involving changes in the size and brightness of the different intermediates over time can be conveniently monitored using kymographs. In these plots, stable GFP-Atg8 puncta (wild type in nitrogen-sufficient conditions or atg∆ mutants under nitrogen-sufficient or nitrogen-starved conditions) give rise to lines that are parallel to, or if they move, diagonal to, the (vertical) time dimension (Fig. 2C and D). In sharp contrast, a PAS maturing into an autophagosome gives rise to a characteristic triangular profile whose base, height and vertex correspond to the width of the ‘terminal’ autophagosome, the duration of the complete cycle and the position of the ‘parental’ PAS, respectively (Fig. 2C; enlarged example in Fig. 2D). Kymographs showed that the transition between the spherical and the oblong structures was marked by a sharp reduction in fluorescence (Fig. 2C and D), unattributable to photobleaching because a similar reduction was never observed in vertical traces of nonmaturing atg∆ structures (Fig. 2C and D). The fluorescence of the oblong structures further decreases with time, such that the end of the ‘cycle’ corresponds to the time-point at which the autophagosome becomes indistinguishable from the background, a stage almost certainly corresponding to the fusion of the autophagosome with the vacuole. The whole process takes, on average, 500 sec ± 100 SD (n = 21 maturation events) at 28°C.
Autophagosomes normally fuse directly with vacuoles
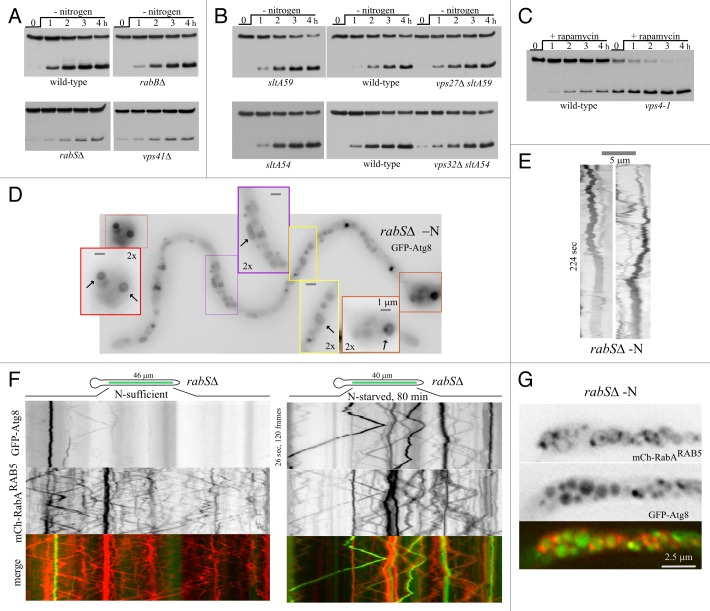
We next ‘mapped’ the step(s) of the endocytic pathway at which autophagosomes fuse with the endovacuolar system. rabB/srgH encodes the major A. nidulans RAB5, such that rabB∆ results in a severe defect in the maturation of early endosomes into late endosomes.26 However, rabB∆ does not affect autophagy (Fig. 3A). Autophagy is also unaffected by the absence of ESCRTs. Null A. nidulans ESCRT mutations are very severely debilitating, but can be rescued by loss-of-function mutations in the regulatory gene sltA.28 We tested vps27∆ and vps32∆ (removing the “gatekeeper” of the multivesicular body pathway, and the major structural component of ESCRT-III,29 respectively) in combination with sltA- mutations for autophagy activity. Single sltA- controls and double vps27∆ sltA- and vps32∆ sltA- mutants showed wild-type GFP-Atg8 processing (Fig. 3B). We also monitored GFP-Atg8 proteolysis in vps4–1 cells, using rapamycin to induce autophagy, as vps4–1 is a conditional expression allele of vps4/AN3061 that is repressible by ammonium.30 Ammonium downregulation of Vps4 to levels insufficient to support normal growth did not prevent GFP-Atg8 processing (Fig. 3C). The finding that neither impaired endosome maturation nor inability to form multivesicular endosomes prevented the autophagic delivery of GFP-Atg8 to the vacuoles strongly indicated that autophagosomes fuse directly with the latter. In contrast, rabSΔ removing the A. nidulans vacuolar RAB7/Ypt7 ortholog27 that is required for autophagosome-lysosome/vacuole fusion markedly reduced GFP-Atg8 proteolysis, to an extent comparable to that obtained in the absence of Vps41, a key subunit of the HOPS oligomeric tethering complex, the effector by which RabSRAB7 exerts its fusogenic role27 (Fig. 3A). Despite being markedly inefficient in proteolyzing GFP-Atg8 by autophagy, rabSΔ cells were capable of forming abundant autophagosomes, indicating that autophagosome biogenesis was unaffected (Fig. 3D and E). Therefore autophagosomes fuse with vacuoles in a RabSRAB7-HOPS-dependent manner.
Figure 3. The endocytic pathway intersects with autophagy mostly at the level of late endosomes and vacuoles. (A) GFP-Atg8 proteolysis assays in the indicated endovacuolar mutants. (B) GFP-Atg8 processing assays in vps27∆ and vps32∆ strains (in double mutants with the sltA− loss-of-function mutations sltA59 and sltA54, respectively) compared with the corresponding single sltA− mutant controls and the wild type. (C) GFP-Atg8 processing assays in a strain carrying vps4–1, a conditional expression allele in the gene encoding Vps4. Levels of Vps4 are virtually nil on ammonium. Therefore, to test the role of Vps4 in autophagy, strains were cultured for 15 h in liquid minimal medium containing 20 mM (NH4)2SO4 before inducing autophagy by addition of 400 ng/ml of rapamycin (0 h time point). (D) rabS∆ hypha incubated in nitrogen-starvation conditions showing the marked abundance of autophagosomes. The insets are shown at double magnification and improved contrast to visualize autophagosomes in different stages of maturation. (E) Two examples of autophagosome cycles in rabS∆ (note the relatively short time coverage of this sequence). (F) Kymographs (regions-of-interest as indicated in the schemes) of time-lapse sequences. The red and green channels were simultaneously acquired with a Dual-Viewer, using the stream acquisition function of Metamorph software, which results in a high density of information per time unit, allowing trajectories of moving endosomes (diagonal lines) to be followed unambiguously. (G) Image of mini-vacuoles of a rabS∆ GFP-Atg8 cell expressing the early endosome marker mCherry-RabARAB5, after incubation for 4 h under autophagy conditions. Note the faint labeling of the vacuolar membranes with early endosomal RabARAB5 that is seen in the rabS∆ background.27
The fact that rabS∆ does not abolish GFP-Atg8 proteolysis indicates that the protein is able to gain access to the mutant vacuoles, albeit inefficiently. rabS∆ results in mini-vacuoles retaining some degree of endosomal identity: They contain the RAB5 paralogs, RabA and RabB, and a proportion of them show long-distance movement characteristic of early endosomes.27,31 In nitrogen-starved rabS∆ hyphae GFP-Atg8 localized, in addition to autophagosomes, to motile structures (Fig. S3); so we hypothesized that autophagosomes might fuse with endosomes. Therefore, we co-imaged GFP-Atg8 with the endosomal marker mCherry-RabARAB5. In nitrogen-fed rabS∆ cells, hardly any moving GFP-labeled structures were detectable by kymographs, contrasting with the abundant moving mCherry-labeled endosomes (Fig. 3F, left). In sharp contrast, moving GFP-Atg8 structures colocalizing with mCherry-RabARAB5 were very conspicuous in autophagy conditions (Fig. 3F, right) establishing that GFP-Atg8 traffics to the rabS∆ population of motile endosomes/mini-vacuoles. Indeed, after > 4 h in nitrogen-starved conditions (allowing autophagy to proceed to completion), mini-vacuoles containing GFP-Atg8 fluorescence rimmed by faint mCherry-RabARAB5 fluorescence were clearly observed (Fig. 3G).
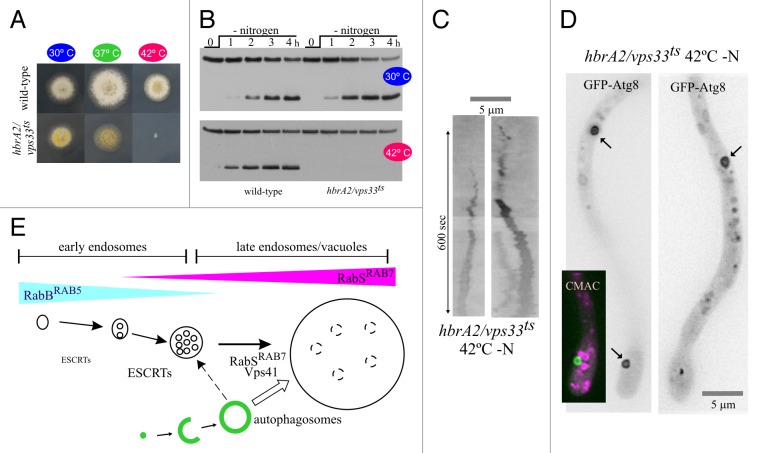
We hypothesized that residual HOPS-independent fusion of autophagosomes with the endovacuolar system of rabS∆ and vps41∆ mutants would be mediated by RabA/RabB and their effector CORVET, implying that the simultaneous inactivation of HOPS and CORVET should block autophagy completely. This prediction cannot be tested with null mutants because a single vps8∆ mutation inactivating CORVET and a double rabA∆ rabB∆ deletion abolishing CORVET recruitment to endosomes are virtually lethal. Therefore we used hbrA2, a ts mutation in the gene encoding the Sec1/STXBP1/Munc-18 protein HbrA/Vps33, a key component of both HOPS and CORVET.32 hbrA2 prevents growth at 42°C (Fig. 4A), as expected for the simultaneous inactivation of both complexes.26,27 GFP-Atg8 proteolysis assays of hbrA2 (hbrA/vps33ts) cells revealed that autophagy proceeds normally at 30°C but is completely blocked at 42°C (Fig. 4B), even though hbrA2 cells are, at 42°C, capable of forming autophagosomes (Fig. 4C and D; Fig. S3). Thus, the block in GFP-Atg8 proteolysis must result from the complete inability of hbrA2 autophagosomes to fuse with the endovacuolar system, not from an intrinsic defect in their biogenesis. We conclude that autophagosomes fuse with vacuoles in a HOPS-dependent manner, although they have the capability of fusing with endosomes inefficiently, in a CORVET-dependent manner. This second pathway has little physiological relevance, as removal of RabBRAB5, the main recruiter of CORVET, has no detectable effect on GFP-Atg8 proteolysis (Fig. 4E).
Figure 4. Simultaneous inactivation of CORVET and HOPS prevents GFP-Atg8 proteolysis but not the formation of autophagosomes. (A) Growth tests of a wild-type strain compared with hbrA2/ vps33ts. (B) GFP-Atg8 processing assays in hbrA2/vps33ts. When indicated, strains were shifted to 42°C for 45 min before inducing autophagy. (C) Two examples of autophagosome cycles in hbrA2 (vps33ts) cells in which autophagy had been induced after shifting cells to 42°C in an incubator. To prevent the (minor) decrease in the incubation temperature that might occur during image acquisition from permitting recovery of functional HbrA2/Vps33, the culture was photographed within 10 min after the incubation chamber was transferred from the 42°C incubator to the microscope set-up adjusted to 37°C. (D) Examples of GFP-Atg8 autophagosomes (marked with arrows) in a hbrA2/vps33ts cell shifted to nitrogen-starvation conditions at 42°C. (Note that, upon shifting cells to the restrictive temperature, the mutation arrests apical extension, leading to tip swelling.) One cell was counterstained with CMAC (magenta pseudocolor) to show that none of the numerous mini-vacuoles seen under these conditions overlaps with the autophagosome. (E) Our experiments show that autophagosomes fuse with late endosomes/vacuoles in a RabSRAB7-dependent manner, although they can also fuse with early endosomes inefficiently (discontinuous line).
Live imaging of the PtdIns3P cycle in autophagic structures
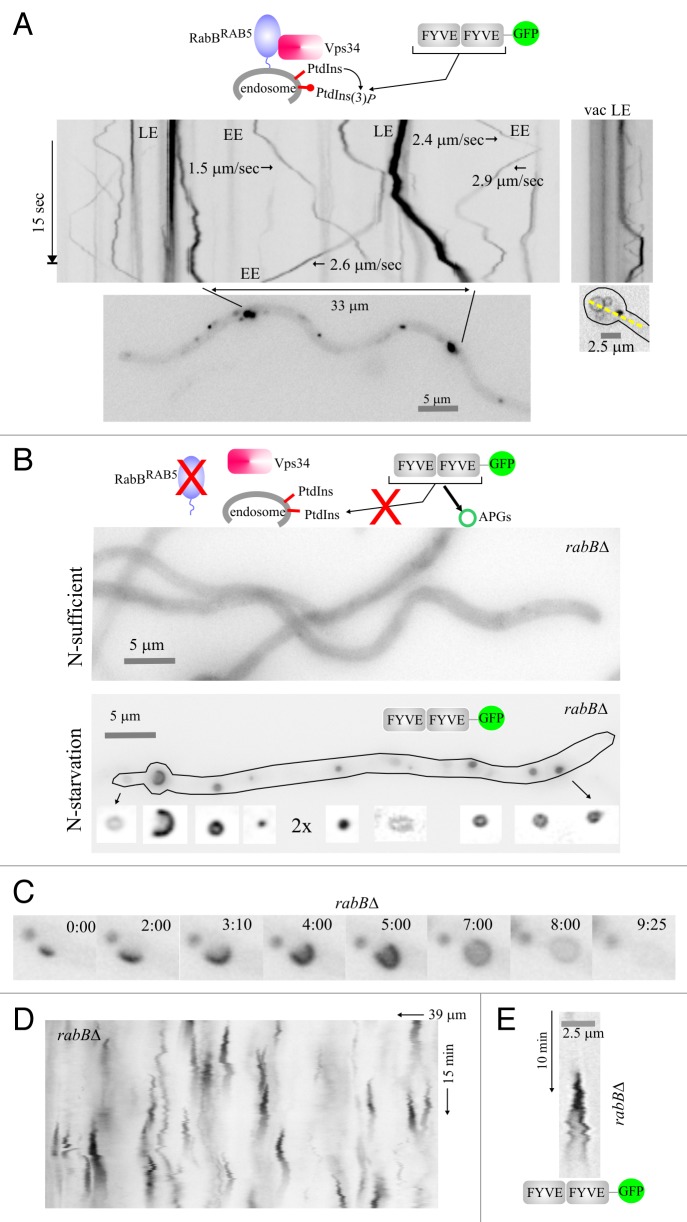
The above data provided the basic background to investigate autophagy in A. nidulans. In a second set of experiments, we exploited the RabBRAB5-independency of autophagy for tracking in vivo PtdIns3P specifically on autophagic structures, using a probe consisting of two copies of the A. nidulans Vps27 FYVE domain fused to GFP (FYVE2-GFP). Under nitrogen-sufficient conditions FYVE2-GFP labels endosomes in a strictly RabBRAB5-dependent manner (Fig. 5A and B; RabBRAB5 is the sole recruiter of the Vps34 PtdIns3-kinase to endosomal membranes).26 In autophagy conditions FYVE2-GFP localizes to autophagosomes in addition to endosomes (data not shown), as PtdIns3P plays a well-documented role in autophagy.33,34 However, the motility and abundance of early endosomes obstructed visualization of autophagic structures. We reasoned that depletion of the endosomal PtdIns3P pool by means of a rabB∆ mutation would restrict FYVE2-GFP to autophagic structures. This was indeed the case (Fig. 5B–E). FYVE2-GFP was essentially cytosolic in rabB∆ cells cultured in nitrogen-sufficient conditions. However, the reporter was recruited to the different autophagic structures (puncta, phagophores and autophagosomes) upon induction of autophagy, showing that all these structures contain PtdIns3P (Fig. 5B). Time-lapse imaging (Fig. 5C; Movie S6) and kymograph analyses (Fig. 5D) demonstrated that FYVE2-GFP puncta were short-lived, rapidly maturing into autophagosomes. Importantly, while PtdIns3P was present in autophagosomes through the complete cycle, the reporter signal sharply declined toward its end, roughly paralleling the behavior of Atg8. Figure 5E depicts a kymograph representation of the cycle of an autophagosome that stayed in sharp focus throughout a 10 min sequence. These in vivo observations complement recent genetic and cell biology data demonstrating that PtdIns3P clearance from the autophagosome is an essential step for the completion of the autophagosome cycle.35
Figure 5. Visualization of PtdIns3P across the autophagosome cycle. (A) In the wild type, cultured in nitrogen-sufficient conditions, RabBRAB5 recruits the Vps34 PtdIns 3-kinase to endosomes. Thus, PtdIns3P-binding FYVE2-GFP localizes to motile endosomes, larger and more static late endosomes and, faintly, to the vacuolar membranes (fluorescence shown in inverted contrast). Stream-acquired time-lapse sequences of the PtdIns3P reporter were used to derive kymographs. The top left image kymograph corresponds to the hyphal region indicated in the bottom image. The rate at which early endosomes move is indicated for some examples. The top right image is a kymograph representation, drawn across the yellow line on the bottom right image, of a basal conidiospore region showing a vacuole and a late endosome. (B) In the rabB∆ mutant, endosomes are depleted of PtdIns3P. Therefore, in nitrogen-sufficient conditions FYVE2-GFP is cytosolic. In contrast, in nitrogen-starved conditions Vps34 synthesizes PtdIns3P on autophagic structures to which FYVE2-GFP is very efficiently recruited. The hypha displayed contains numerous autophagic structures labeled with FYVE2-GFP (shown below the cell at double magnification and improved contrast). (C) Images of a rabB∆ time-lapse sequence showing the different intermediates of an autophagosome cycle, labeled with FYVE2-GFP. Time is in min::sec. (D) Kymograph of a rabB∆ cell expressing FYVE2-GFP, cultured in nitrogen-starved conditions. Fluorescent puncta give rise to cone-shaped figures corresponding to autophagosome cycles. (E) Example of a rabB∆ autophagosome cycle as labeled with FYVE2-GFP. Note that fluorescence gradually declines as the autophagosome enlarges its size.
A. nidulans autophagy does not require Golgi or post-Golgi traffic
A key issue in the field, previously investigated in mammalian cells and in yeast, is the source of membrane used to build up autophagic structures.5 We exploited the hyphal mode of growth of A. nidulans to investigate whether autophagy requires Golgi or post-Golgi traffic. A. nidulans hyphae grow by apical extension, which is critically dependent on the optimal functioning of the secretory pathway to deliver cell wall-modifying enzymes/materials to the apex. Therefore, acute impairment of secretion by drugs or ts mutations sharply arrests apical extension, often leading to tip swelling.23 The presence/absence of a morphogenetic defect permits the observer to assess, by microscopy, if any given secretory protein function has become limiting for growth in any particular cell.
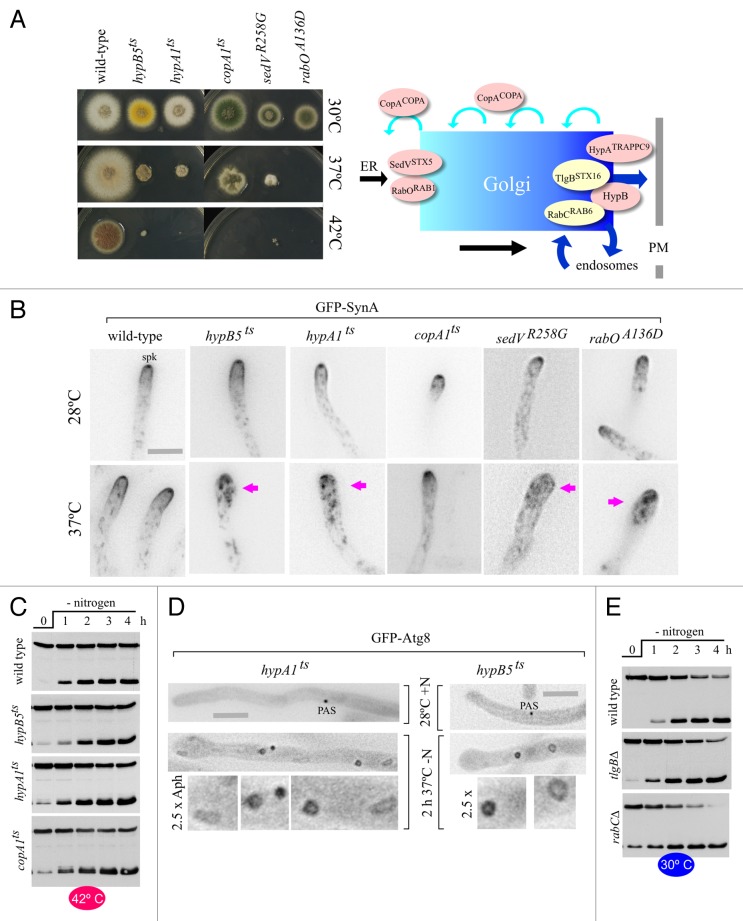
We tested the effects of mutations affecting the Golgi at different levels (Figs. 6 and 7). Experiments with hypA1ts and hypB5ts alleles showed that autophagy does not require post-Golgi traffic to the plasma membrane or endosomes. hypA1ts is a mutation in the gene encoding the TRAPPII-specific component that corresponds to yeast Trs120 (A. nidulans HypATRAPPC9).36 TRAPPII appears to be the GEF for yeast Ypt31/3237 and of its A. nidulans ortholog AN0347/RabE (Pinar M, Arst HN, Pantazopoulou A, Peñalva MA, unpublished). hypA1ts strains grow well at 30°C. However, they show severely reduced growth at 37°C and no growth at 42°C, indicating that the essential function of HypATRAPPC9 is severely impaired and abolished, respectively, at these temperatures (Fig. 6A). Then we used the synaptobrevin homolog SynA (Snc1 in S. cerevisiae), the v-SNARE of plasma membrane-bound carriers, as a well-characterized reporter of secretion. SynA localizes to a plasma membrane apical crescent and to the Spitzenkörper (‘spk’ in Fig. 6B), an apical cluster of secretory vesicles awaiting fusion with the plasma membrane.31,38-40 To monitor the effects of hypA1ts on secretion, we cultured mutant hyphae at 28°C and examined SynA localization by microscopy before and after shifting cells to 37°C (actual temperature measured within the incubation chamber medium, see Materials and Methods; higher temperatures would damage the objective). In the wild type, temperature shift-up did not alter SynA localization, which was also completely normal in hypA1ts cells cultured at 28°C (Fig. 6B). In contrast, SynA relocalized to internal structures within 30–45 min after shifting hypA1ts cells to 37°C (Fig. 6B). This relocalization correlated with swelling of the hyphal tips. Together with growth tests, these observations strongly indicated that hypA1ts largely impairs secretion at 37°C (and abolishes it at 42°C). Despite this, hypA1ts cells showed essentially normal kinetics of GFP-Atg8 proteolysis at 42°C (Fig. 6C) and, by microscopy, were able to form autophagosomes well beyond the time at which secretion had arrested (Fig. 6D; Movie S7); (note that the main ‘wave’ of autophagosome formation occurred 1.5 h after the shift to 37°C).
Figure 6. Intra-Golgi traffic and post-Golgi steps are not required for autophagy. (A) Growth tests of strains carrying the different ts alleles used in Golgi experiments, cultured at the indicated temperatures. The scheme on the right indicates the location at which the gene products affected by the different mutations play their roles. (B) Tip swelling and redistribution of secretion marker GFP-SynA to internal structures in cells carrying ts mutations affecting key Golgi regulatory proteins, following a shift from 28°C to 37°C. Hyphae were either incubated at 28°C and photographed (top row) or incubated at 28°C and then shifted to 37°C on the stage before acquiring images (bottom row). The actual temperature in the medium took 15 min to reach the 37°C target. All bottom row pictures were taken within 45 min after the shift. The Spitzenkörper, indicated in the 28°C wild-type example with ‘spk’, is visible in the wild type and in the copA1ts mutant at both temperatures, and in all other mutants at the permissive temperature only. Magenta arrows indicate mutant tips that had clearly swollen after the temperature shift, correlating with GFP-SynA relocalization to internal structures. Scale bar: 5 µm (for all panels). (C) GFP-Atg8 processing assays in cells carrying the indicated heat-sensitive (ts) mutations. Cells were cultured at 30°C in nitrogen-sufficient medium, shifted to 42°C for 45 min and then transferred to medium without a nitrogen source (0 h time point). The lower and faster mobility bands correspond to GFP-Atg8 and GFP* in previous figures. (D) GFP-Atg8 autophagosomes photographed in hypA1ts and hypB5ts hyphal cells cultured in the indicated medium and temperature conditions. Note the numerous autophagic structures seen in mutant cells in which the function affected by the respective ts mutation became limiting, resulting in conspicuous tip swelling. Scale bars: 5 µm. Autophagic structures are enlarged 2.5 times below the corresponding hyphae. (E) GFP-Atg8 processing assays in cells carrying the indicated null alleles. Cells were cultured at 30°C in nitrogen-sufficient medium and shifted to medium without a nitrogen source at the 0 h time point.
Figure 7.A. nidulans RabORAB1 plays a key role in autophagy (A) GFP-Atg8 processing assays of rabOA136D and sedVR258G strains, cultured at 30° or 42°C. The slower and faster mobility bands correspond to GFP-Atg8 and GFP* in previous figures. (B) Autophagy proceeds normally upon major secretion impairment by the sedVR258G mutation. sedVR258G hyphal tip cell shifted to 37°C and autophagy-inducing conditions on the microscopy stage. Note the characteristic swelling of the tip (encircled by dotted line), indicative of apical extension arrest and thus deficient secretion. Three autophagosomes are indicated with arrows. (C) Example of a complete autophagosome cycle in a nitrogen-starved sedVR258G cell incubated at 37°C for more than 1 h. Time is in min:sec. (D) Kymographs derived from time-lapse sequences of sedVR258G cells shifted to 37°C showed multiple examples of cone-shaped autophagosome cycles when cells were starved for nitrogen (bottom panel), but not when cells were cultivated in the presence of a nitrogen source (top panel). (E) GFP-Atg8 is delivered to the lumina of vacuoles (vac) in sedVR258G cells that had been cultured in nitrogen-starved conditions at 37°C. (F) At 28°C, GFP-Atg8 PAS are brighter in the rabOA136D strain than in the wild type. Maximal intensity projections of z-stack images taken under identical conditions were used to determine the average intensity of n = 147 GFP-Atg8 puncta for each genetic background. Scale bars indicate SD. The difference was clearly detectable by bare eye. (G) mCherry-Atg8 puncta in the rabOA136D mutant contain Atg9-GFP. Although not every Atg9 structure contains Atg8, every Atg8 punctate structure contains Atg9. Red arrows indicate 21 mCherry-Atg8 puncta visible in the image. Green arrows indicate the colocalizing Atg9-GFP puncta. (H) Top image, control kymograph of a time-lapse sequence showing a wild-type control cultured in nitrogen-starved conditions: Atg8 puncta give rise to typical autophagosome cones at 28°C. Bottom image, a kymograph derived from a 28°C rabOA136D time series showed that the mutant’s Atg8 puncta were completely unable to produce autophagosomes, giving rise to vertical lines. (I) GFP-Atg8 rabOA136D cells co-expressing mCherry-RabSRAB7 to label the vacuolar membranes were cultured at 28°C in nitrogen-sufficient conditions and shifted to nitrogen-starved conditions for 4 h. The volume of the vacuolar compartment increases notably in autophagy conditions, but the vacuoles rimmed by mCherry-RabSRAB7 are clearly devoid of GFP fluorescence, unlike the situation in the wild type (Fig. 1).
Yeast Sec7, the late Golgi Arf1 GEF, is needed by traffic exiting the Golgi toward the plasma membrane and endosomes. We studied the autophagy phenotype of hypB5ts, having a mutation in the gene encoding the A. nidulans Sec7 ortholog.41,42 Like hypA1ts strains, hypB5ts strains did not grow at all at 42°C and showed markedly impaired growth at 37°C, indicating a major block in secretion at the restrictive or semi-restrictive temperatures (Fig. 6A). In agreement, hypB5ts resulted in SynA mislocalization and tip swelling within 45 min after shifting cells to 37°C (Fig. 6B). Importantly, GFP-Atg8 proteolysis at 42°C (a fully restrictive temperature for growth) was normal (Fig. 6C) and autophagy-mediated, as shown with a hypB5ts atg1∆ double mutant strain (Fig. S4). By microscopy, autophagosomes formed normally in hypB5ts cells shifted to 37°C, far beyond the time point at which SynA became delocalized and tips swelled (Fig. 6D; Movie S8). Therefore, the absence of a hypA1ts effect indicates that AN0347/RabE activity is dispensable for autophagy, whereas the absence of a hypB5ts effect shows that the late Golgi activity of AN1126/ArfAARF1 is not required either. These data strongly indicate that exit from the Golgi is not required for autophagy. The absence of HypB involvement pointed at an important difference between A. nidulans and S. cerevisiae, as Sec7 is required for yeast autophagy.7
We also analyzed mutations affecting known Golgi resident proteins. One was rabC∆. RabCRAB6 maintains the structure of the Golgi, its absence resulting in significantly smaller Golgi bodies and impaired secretion.39 However rabC∆ did not impair GFP-Atg8 proteolysis (Fig. 6E). Another mutation that we examined was tlgB∆, removing the late Golgi syntaxin TlgBSTX16. tlgB∆ is virtually lethal when combined with rabC∆.39 Similar to rabC∆, tlgB∆ caused no autophagy defect (Fig. 6E). Last, we analyzed copA1, a ts mutation affecting the A. nidulans COPI α-coatomer subunit.43 GFP-CopACOPA localizes to the Golgi,44 in agreement with the view that COPI carriers are involved in intra-Golgi retrograde traffic.45 copA1, which allows substantial growth at 37°C (Fig. 6A) and affects SynA localization only very weakly at this temperature (Fig. 6B), is unsuitable for microscopy shift-up experiments. However, copA1 cells do not grow at 42°C, a temperature at which they do not show any autophagy defect as determined by GFP-Atg8 proteolysis (Fig. 6C, bottom). Taken together, these data indicate that autophagy does not require Golgi traffic, pointing at the ER-Golgi interface as a potential source of membranes for autophagy.
Thus, we investigated the roles of the early Golgi syntaxin SedVSTX5/Sed5 and the A. nidulans Ypt1 ortholog RabORAB1 using gene-replaced A. nidulans rabOA136D and sedVR258G ts alleles mimicking the S. cerevisiae ypt1-A136D46 and sed5–1 (R258G)47 ts mutations, respectively (M. Pinar, A. Pantazopoulou, Herbert N. Arst and Miguel A Peñalva, to be described elsewhere). At 25–30°C, rabOA136D and sedVR258G strains grew slightly more slowly than the wild type (Fig. 6A) and displayed a minor defect in SynA localization (Fig. 6B), indicating that at permissive temperatures the mutations impaired secretion only weakly. In contrast, sedVR258G strains grew marginally, whereas rabOA136D strains did not grow at all at 37°C (Fig. 6A), making these mutations ideally suited for temperature shift-up experiments. Both mutations completely prevented growth at 42°C (Fig. 6A), giving strong weight to conclusions drawn from GFP-Atg8 proteolysis assays performed after shifting cells to this temperature.
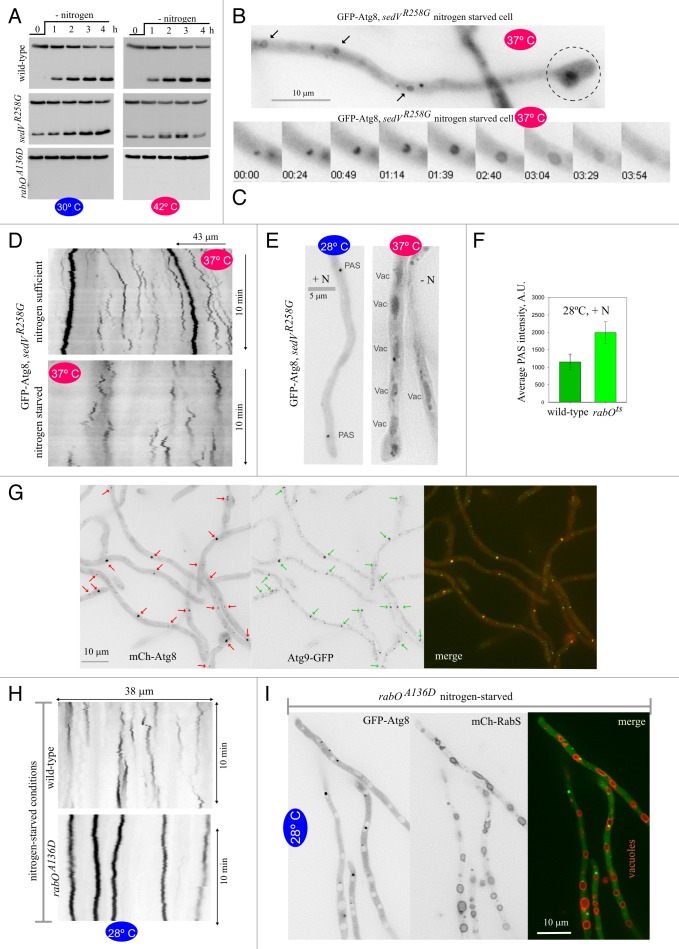
Orthologs of the early Golgi-localizing SedVSTX5 syntaxin39 play a well-demonstrated role at the level of Golgi entry. sedVR258G did not prevent GFP-Atg8 proteolysis at 30°C and, more importantly, it did not prevent proteolysis at 42°C either (Fig. 7A), indicating that SedVSTX5 is not required for autophagy. Shifting sedVR258G cells to 37°C led to SynA mislocalization and conspicuous tip swelling within 30 min (Fig. 6B), indicating that secretion was largely impaired if not completely blocked by the temperature up-shift. However, sedVR258G hyphae with swollen hyphal tips were able to form autophagosomes (Fig. 7B and C; Movie S9), with kymographs derived from GFP-Atg8 time-lapse series revealing the characteristic triangular traces of autophagosome cycles (Fig. 7D). Moreover, after incubation for 3–4 h under autophagy conditions at 37°C, the vacuoles became GFP-positive (Fig. 7E). These data strongly indicate that autophagy proceeds normally under conditions in which sedVR258G largely impairs secretion and, together with the above experiments, strongly supports the conclusion that autophagy does not require traffic across the Golgi.
RabORAB1 is required for autophagy and localizes to expanding phagophores and autophagosomes
Recent work has revealed that yeast Ypt1 plays a key role in autophagy, independent of its role in secretion.9,48 At 42°C, rabOA136D hyphae were completely unable to proteolyze GFP-Atg8 (Fig. 7A), indicating that RabORAB1 is also crucial for autophagy. Unexpectedly, rabOA136D cells were equally unable to proteolyze GFP-Atg8 at the permissive temperature (Fig. 7A), essentially behaving as an atg∆ mutant. This occurred in spite of the fact that the mutant hyphae grew at the permissive temperature (Fig. 6A), underwent apical extension (Movie S10) and displayed relatively normal SynA localization (Fig. 6B), showing that at permissive temperatures RabORAB1 (A136D) was able to maintain secretion sustainably.
We confirmed the 28°C autophagy-deficient phenotype of rabOA136D cells by microscopy. In nitrogen-sufficient conditions rabOA136D GFP-Atg8 puncta were significantly brighter than in the wild type (Fig. 7F). These puncta represented PAS because essentially every Atg8 punctum contained Atg9 (Fig. 7G). At 28°C, rabOA136D GFP-Atg8 puncta never gave rise to autophagic structures upon autophagy conditions: None of the 64 Atg8 puncta (n = 22 cells) that remained in focus over 10 min-long time-lapse sequences of hyphae incubated in autophagy conditions developed into an autophagosome as determined by kymographs (vertical lines in Fig. 7H); moreover no autophagosomes were ever observed by direct inspection either. Last, we confirmed that GFP-Atg8 was not delivered to rabOA136D vacuoles. In the rabOA136D mutant, nitrogen starvation led to a marked increase in vacuolar size/number, like in the wild type (Fig. 7I; Fig. 1B and C). However, in contrast to the wild type, the rabOA136D vacuolar lumina were completely devoid of GFP fluorescence (Fig. 7I). [No GFP staining of the vacuoles was observed at 37°C either (Fig. S5)]. In summary, we established by three different criteria that rabOA136D blocks autophagy at 28°C, a permissive temperature for apical extension and secretion. Thus, RabORAB1 plays two roles, one in Golgi traffic and a second in the biogenesis of autophagosomes. At 28°C, the A136D substitution selectively impairs the RabORAB1 role in autophagy, either directly or by impairing function to a degree that is sufficient to maintain secretion but insufficient to promote autophagy. At 37°C, however, RabOA136D is completely inactivated for both roles.
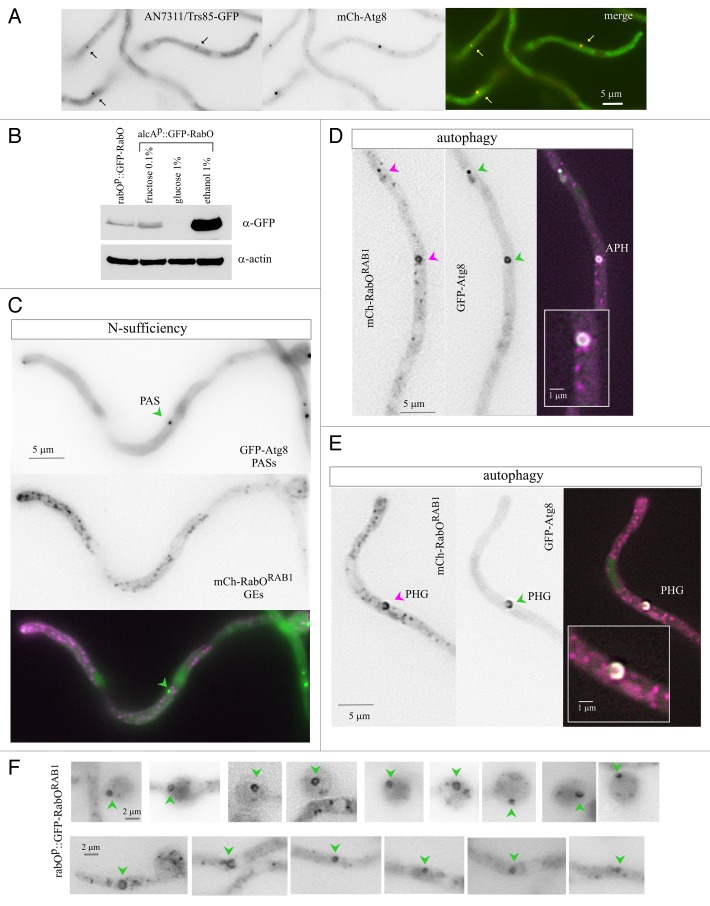
Work with S. cerevisiae showed that a proportion of Ypt1 (RAB1) localizes to the PAS,9,48 where TRAPPIII rather than the Golgi-specific GEF TRAPPI49 mediates Ypt1 nucleotide exchange.9 AN7311/Trs85, the specificity subunit targeting TRAPPIII to the PAS,9,48 was tagged endogenously with GFP. The AN7311/Trs85-GFP signal was weak, precluding dynamic analyses, but sufficient to show that under nitrogen-sufficient conditions AN7311/Trs85-GFP localized to discrete puncta that colocalized with mCherry-Atg8 PAS (Fig. 8A; Atg8 colocalization detected for 92% of n = 152 AN7311/Trs85 puncta). Next we examined whether RabORAB1 was also present in autophagosomes, using two different constructs, single copy-integrated into known chromosomal locations. One construct directs mCherry-RabORAB1 expression under the control of the alcohol dehydrogenase promoter (alcAp). The other drives GFP-RabORAB1 expression under the control of the native rabO promoter (rabOp). Using anti-GFP western blots to monitor relative expression levels, we determined that full induction (on ethanol) of the alcAp construct led to RabORAB1 overexpression, whereas noninduced (on fructose) levels were similar to those driven by the native promoter, rabOp (Fig. 8B).
Figure 8. Localization of RabORAB1 to autophagosomes. (A) AN7311/Trs85-GFP, expressed at physiological levels from an endogenously tagged allele, localizes to cytosolic puncta that also contain mCherry-Atg8. (B) Western blot analysis of GFP-RabORAB1 levels driven by the rabOp and the alcAp promoters. In the latter case, glucose represses the promoter, ethanol at 1% induces it, and fructose at 0.1% leads to basal levels of expression, which are similar to those driven by rabOp. Anti-actin western blotting of the same samples was used as a loading control. (C) Strain co-expressing mCherry-RabORAB1 and GFP-Atg8. In nitrogen-fed cells cultured with 0.1% fructose as carbon source, mCherry-RabORAB1, expressed at approximately physiological levels under the control of the alcAp driver, localizes to polarized Golgi cisternae. Images in (C–E) display the mCherry and GFP single channel images in inverted contrast, as indicated, and the “merge” of the two channels in color. In the “merge” images, the mCherry signal is shown in magenta, whereas the GFP signal is shown in green. Regions of signal overlap are white. (D) Localization of mCherry-RabORAB1 to cytosolic puncta and autophagosomes (APH) containing GFP-Atg8 in cells cultured under autophagy-inducing conditions. (E) Example of colocalization of mCherry-RabORAB1 and GFP-Atg8 in an expanding cup-shaped phagophore (PHG). For (D and E) the insets shown are at double magnification. (F) Several examples of autophagic structures labeled with GFP-RabORAB1, expressed at physiological levels under the control of the rabO promoter; cells were incubated in autophagy conditions. Autophagosomes are marked with a green arrowhead.
Complementation experiments showed that, at 37°C, alcAp::mCherry-RabORAB1 rescued rabOA136D growth to an extent similar to that of the wild-type allele in a rabOA136D/rabO+ heterozygous diploid (Fig. S6; note that rabOA136D is co-dominant), indicating that the fusion protein is functional. We investigated the localization of mCherry-RabORAB1 on fructose (at approximately physiological levels of expression), in cells co-expressing GFP-Atg8. In nitrogen-sufficient conditions, mCherry-RabORAB1 localized to punctate Golgi structures (Fig. 8C; to be described elsewhere). In nitrogen-starved conditions the punctate Golgi pattern persisted, but GFP-Atg8 facilitated the visualization of hollow autophagosomes (Fig. 8D) and expanding phagophores (Fig. 8E) containing mCherry-RabORAB1. Next we examined a strain expressing GFP-RabORAB1 under the native rabOp promoter, without any autophagosomal reporter, asking whether despite the relatively weak GFP signal we would be capable of detecting GFP-RabORAB1 autophagosomes by themselves, using their characteristic morphology to distinguish them from Golgi puncta. Indeed, using this “blind” procedure we could detect with relative ease, but only in nitrogen-starved conditions, numerous examples of GFP-RabORAB1 phagophores and autophagosomes (Fig. 8F). These data strongly support the conclusion that RabORAB1 is present in the expanding phagophores, remaining in them until the closed autophagosome is formed. In addition, they further support the conclusion that RabORAB1 plays a role in autophagy independent from its secretory role.
ER-connected omegasome-like structures associate with autophagosomes
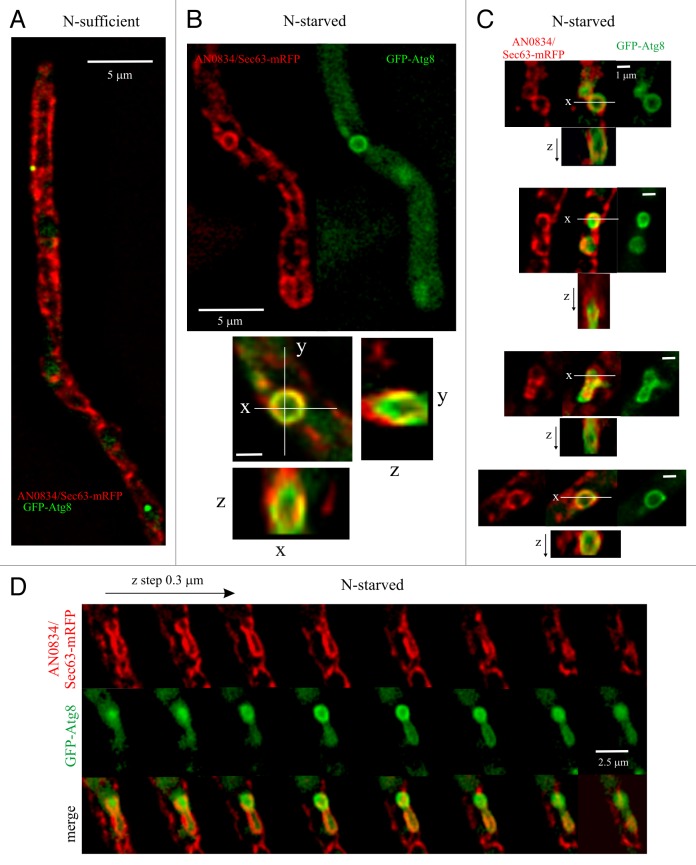
The crucial importance of RabORAB1 for autophagy in conjunction with the lack of involvement of several proteins regulating Golgi traffic suggested that the ER might be a source of autophagosomal membranes. In mammalian cells the ER forms cup-shaped structures, denoted omegasomes, that cradle autophagosome biogenesis.50-52 We imaged the A. nidulans ER, labeled with endogenously tagged AN0834/Sec63-mRFP, in cells expressing GFP-Atg8 (a detailed description of the ER will be published elsewhere). Figure 9A provides an overall view of the network of ER strands. In nitrogen-sufficient conditions a few scattered Atg8 puncta were intermingled with these strands. Next, we shifted hyphae to autophagy conditions and acquired z-stacks in the red and green channels, which were subsequently deconvolved to improve sharpness. Even though the overall organization of the ER essentially remained unchanged, we observed that many GFP-Atg8-containing autophagic structures were associated with discrete ER structures resembling omegasomes. These structures were frequently circular, as they generally followed the shape of an autophagosome (Fig. 9B and C). In x,y projections, many ‘omegasome-like’ structures apparently colocalized with GFP-Atg8 autophagosomes. However, optical z-sections clearly showed that ‘omegasome-like’ structures and autophagic structures were distinctive entities, located in different focal planes (Fig. 9B–D). In x,z and y,z projections the ER was frequently seen as a cup surrounding the autophagic structure (Fig. 9B and C). Of n = 111 autophagic structures, 55 (50%) were associated with underlying ER “cradles,” indicating that autophagosomes form in their close proximity. Due to the resolution limit, we could not discriminate if regions of signal overlap (Fig. 9B–D) actually represented continuities between ER and autophagic membranes. Movie S11 displays, as animated dual-channel serial z-sections, eight different examples that summarize our observations on ER structures associated with autophagic structures. In conclusion, our data strongly suggest that omegasome-like structures derived from the ER may be involved in autophagosome biogenesis in an ascomycete fungus.
Figure 9. Autophagic structures are closely associated with similarly shaped structures formed by ER strands. (A) General view of a GFP-Atg8 hyphal cell also expressing AN0834/Sec63-mRFP from an endogenously tagged allele, cultured in nitrogen-sufficient conditions. The image is a maximal intensity projection of a z-series stack showing the overall structure of the ER and the position of two GFP-Atg8 puncta. (B) Example of a circular ER structure associated with a GFP-Atg8-labeled autophagosome. The merge shown below (x, y) is a combination of the maximal intensity projections of deconvolved z-stacks acquired in each of two channels. Also shown are optical sections in the z-axis across the indicated x and y lines, showing that, despite their very similar shapes, the ER and Atg8 structures are in different z-planes. (C) Further examples of differently shaped ER structures associated with autophagic structures. Red, merge and green images of maximal intensity projections are shown and, below each composite, optical sections in the z dimension across the indicated lines (note that due to the approximately round shapes of the structures, y,z sections—not shown—are similar to x, z sections). (D) Series of optical sections taken every 0.3 µm in the z-axis showing that ER (red) and autophagic structures (green) are not in the same focal planes.
Discussion
Despite its amenability to genetic analysis, live-cell microscopy and studies on intracellular trafficking,23 the ascomycete A. nidulans had not been exploited to gain insight into autophagy. We provide here basic tools for studying this pathway and exploit them to explore the source of autophagosomal membranes. Our results imply that A. nidulans uses membranes for autophagy in a way that appears more similar to mammalian cells than to S. cerevisiae
Live microscopy of S. cerevisiae GFP-Atg8 has been useful to outline the pathway of autophagosome biogenesis.21 However important, these observations could not go beyond establishing that the pathway involves an ‘Atg8 cycle’ initiated with recruitment of Atg8 to the PAS and terminated with its release from the mature autophagosome, because these studies were limited by the fact that, by conventional microscopy, yeast autophagosomes are seen as puncta. A. nidulans GFP-Atg8 localizes to punctate cytosolic structures that also contain Atg9, indicating that they represent a PAS. We tracked the fate of these GFP-Atg8 puncta individually, concluding that they are remarkably stable in nitrogen-sufficient conditions or in autophagy-deficient mutants. However, upon nitrogen starvation, in virtually every instance GFP-Atg8 puncta developed into optically resolvable expanding phagophores and autophagosomes. Therefore, our data functionally establish that GFP-Atg8 puncta represent authentic phagophore assembly sites.
By visual inspection backed by kymograph analyses of time-lapse sequences we were able to resolve the biogenesis of phagophores and autophagosomes in vivo, tracking the latter until they fuse with the vacuoles. In the initial stage, GFP-Atg8 puncta give rise to cup-shaped phagophores that grow into closed rings, approximately 0.9 μm in diameter, representing primary autophagosomes. Up to this stage the fluorescence (and thus Atg8 content) of the enlarging structures increases, presumably reflecting the addition of membrane. In the second stage, these rings become notoriously irregular and undergo further expansion, but gradually lose fluorescence before becoming undetectable in the proximity of a vacuole. At least in part, this decrease in GFP-Atg8 fluorescence must result from delipidation of GFP-Atg8–PE by Atg4, a step required to complete autophagosome biogenesis.17 A. nidulans autophagosomes fuse with RabSRAB7-containing vacuoles27 in a HOPS tethering complex-dependent manner. However, inefficient fusion with the endovacuolar system can clearly occur without RabSRAB7 or Vps41, circumstances under which Atg8 reaches the lumen of motile structures containing the early endosomal marker RabARAB5. We hypothesized that the RAB5 paralogs, RabA and RabB, and their oligomeric tethering effector CORVET might mediate the residual fusion that takes place in HOPS-deficient conditions. The finding that inactivation of HbrA/Vps33 (a component shared by HOPS and CORVET) completely blocks autophagy strongly supports this interpretation.
PtdIns3P is a key player in autophagosome biogenesis and endosome maturation. PtdIns3P synthesis on endosomes is guided by RabBRAB5, which recruits Vps34 to this locale.26 As endosomal maturation is not required for autophagy, we used a rabB∆ mutation to deplete endosomal PtdIns3P, which allowed us to specifically monitor autophagic PtdIns3P in vivo. PtdIns3P is present in phagophores and autophagosomes throughout their complete cycle, resembling Atg8. A recent study concluded that PtdIns3P clearance is a prerequisite for the fusion of autophagosomes with vacuoles.35 Indeed, like Atg8, the FYVE2-GFP signal progressively decays toward the end of the cycle, confirming that the in vivo-observed changes in the levels of autophagosome PtdIns3P meet one of the predictions derived from this study.
A. nidulans autophagy does not require post-Golgi steps, traffic across early endosomes or the Golgi, or even entry of ER-derived traffic into the Golgi. For example, cells in which the function of the early Golgi t-SNARE SedVSTX5 is severely compromised are fully competent in autophagy. In contrast, A. nidulans autophagy absolutely requires RabORAB1, a protein that intimately cooperates with SedVSTX5/Sed5 at the ER-Golgi interface, regulating entry of anterograde traffic into the Golgi. This apparent conundrum is explained because RAB1 orthologs play Golgi-independent functions in autophagy. Thus, Ypt1 is recruited to PAS by the TRAPPIII oligomeric GEF, itself directed to the PAS by its Trs85 component.9,48 As in yeast, A. nidulans AN7311/Trs85 localizes to the PAS. Our data further extend the above reports by showing that RabORAB1 is also present in expanding phagophores and autophagosomes, suggesting that it might be needed throughout autophagosome biogenesis. We establish that the roles of RabORAB1 in Golgi traffic and autophagy are dissociable by mutation. rabOA136D impedes secretion at 37°C, but permits relatively normal secretion at 28°C. However, at 28°C, rabOA136D completely blocks autophagy. It is formally possible that rabOA136D preferentially affects the recruitment of RAB1 effectors needed for autophagy, such as Atg11.48 An alternative, yet speculative, interpretation is that autophagosome biogenesis necessitates higher levels of RAB1 function than does Golgi entry.
Autophagosome biogenesis requires the delivery of lipid bilayer to the expanding phagophore. Understanding the transport route(s) of these membranes represents a key issue.53 The integral membrane protein Atg9 follows one such route, which involves recently characterized Atg9 vesicles representing one source of autophagosome membrane.54 In S. cerevisiae, evidence that autophagy requires post-Golgi trafficking is compelling. Atg9 has to move beyond the ER and early Golgi to play its role, and the formation of autophagosomes requires the cycling of Atg9 between the forming autophagosome and “Atg9 reservoirs,” a compartment derived from the Golgi4 whose organization requires post-Golgi SNAREs6 as well as Sec7.7 A. nidulans autophagy does not require HypATRAPPC9 or HypB, and thus is independent of Golgi exit (Fig. 6). Therefore, our data collectively indicate that the delivery of A. nidulans Atg9 to “Atg9 reservoirs,” the normal functioning of these reservoirs exchanging membrane with autophagosomes and, more broadly speaking, the supply of membranes to the autophagosomes does not necessitate the Golgi. We note, however, that our conclusions are based on experiments with ts mutants (hypA1ts, hypB5ts, sedVR258G) shifted from permissive to restrictive conditions. Therefore, one alternative possibility would be that the reservoirs built up under permissive conditions had sufficient capacity to sustain autophagy during the course of our analysis once the cells are shifted to restrictive conditions.
The lack of Golgi involvement implicates the A. nidulans ER as one supplier of membrane for autophagy. In this context, the observation that autophagosomes are frequently associated with ER-derived “omegasome-like” structures is highly suggestive. Such ER structures are so closely associated, and are so similar in shape to autophagosomes that in x,y projections they often appear to overlap with them. If these ER structures were one source of autophagic membrane, they might not be the only source, as we detected them in only ~50% of the examples. It is well documented that, in mammalian cells, the ER is one source of autophagosomal membranes.50,52,55,56 Thus, a major outcome of this work is that ‘omegasome-like’ structures are also present in an ascomycete fungus related to S. cerevisiae. Given the large differences at the level of subcellular organization between S. cerevisiae and A. nidulans,23 it may come as no surprise that these two ascomycete fungi have evolved mechanisms to obtain their supply of autophagic membrane from different stores. As noted by Mari et al.,5 cells could be capable of deriving membrane from the most suitable or expendable reservoirs, and in a hyphal fungus the ER might be one of them.
Materials and Methods
Aspergillus strains, media and molecular genetics
Aspergillus complete medium (MCA) and synthetic complete medium (SC) containing 1% glucose and 5 mM ammonium tartrate (i.e., 10 mM NH4+) as carbon and nitrogen source, respectively, were used for growth tests and strain maintenance.57 Strains, which carried markers in standard use, are listed in Table S1. Cassettes for gene deletion and gene replacement were constructed by fusion PCR,58 using A. fumigatus pyrG (pyrGAf) as selective marker and strains carrying pyrG89 (resulting in pyrimidine auxotrophy) as recipients for transformation.59 Recipient strains also carried a nkuA∆ mutation to prevent nonhomologous recombination.60 The following entry codes correspond to the systematic designation of genes used in this work in the A. nidulans database AspGD (http://www.aspgd.org): atg1, AN1632; atg8, AN5131; atg7, AN7428; atg9, AN3734; hbrA/vps33, AN2418; atg4, AN3470; atg5, AN5174; rabO, AN4281; sec63, AN0834; trs85, AN7311; and sedVSTX5, AN9526.
Heat-sensitive mutations rabOA136D and sedVR258G were constructed by gene replacement. Their detailed characterization will be reported elsewhere (M. Pinar, A. Pantazopoulou, H. N. Arst and M. A. Peñalva). We determined by sequencing that the previously uncharacterized ts mutation hbrA2 in the gene encoding the A. nidulans Vps33 ortholog results in a C718G transversion leading to Arg240Gly substitution in HbrAVPS33.
GFP-Atg8 is a fusion protein in which GFP carrying a quintuple repeat of a Gly-Ala di-peptide attached to its C terminus was fused in frame to the N terminus of A. nidulans Atg8. The resulting GFP-(Gly-Ala)5-Atg8 protein (for brevity GFP-Atg8) was expressed under the control of the gpdAmini promoter, using a single-copy integration construct targeted to pyroA, as described previously.42 An equivalent transgene, assembled and integrated in the same manner, drove expression of mCherry-Atg8. mCherry-RabSRAB7 was expressed from a single copy transgene integrated at the argB locus, under the control of the alcA (alcohol dehydrogenenase gene) promoter (alcAp).27 Expression of mCherry-RabORAB1 was also driven by an alcAp construct, single-copy integrated at argB, using 0.1% fructose as carbon source to obtain noninduced levels of expression. GFP-RabORAB1 expression was driven by the rabOp promoter, using an integrative construct targeted to the pyroA locus. Expression of mCherry-tagged histone H1 (this histone is encoded by the A. nidulans hhoA gene), was expressed after endogenously tagging the gene using a hhoA::mCherry::pyroAAf cassete and a pyroA4 recipient strain.61 AN0834/Sec63 was endogenously tagged with mRFP at the C terminus, using an AN0834/sec63::mrfp::pyrGAf cassette for homologous recombination. Atg9 and AN7311/Trs85 were tagged at their C-termini, using atg9::gfp::pyrGAf and AN7311/Trs85::gfp::pyrGAf cassettes, respectively. In all three cases a pyrG89 nkuA∆ strain was used as recipient for transformation.
A. nidulans Rab nomenclature
We have updated the standard nomenclature of the A. nidulans Rabs at http://www.aspgd.org. In the former designation, Rab proteins were denoted as ‘Srg’ (secretion related GTPase) proteins. In the updated names, all 10 A. nidulans Rab GTPases are denoted as ‘Rab’, as follows [standard name/alias/systematic name]: RabA/SrgG/AN4915 and RabB/SrgH/AN3842 are the two RAB5 paralogs; RabC/SrgC/AN7602 is the RAB6 ortholog; RabE/SrgE/AN0347 is the RAB11 ortholog; RabD/srgA/AN6974 is the S. cerevisiae Sec4 ortholog; RabF/previously unnamed/AN9072 is the RAB4 ortholog; RabO/SrgB/AN4281 is the RAB1 ortholog; RabS/AvaA/AN0089 is the RAB7 ortholog; RabT/srgD/AN5106 is the RAB2 ortholog; and RabX/previously unnamed/AN2474 is an uncharacterized Rab.
GFP-Atg8 processing assays in flask cultures
Autophagy conditions were achieved by nitrogen starvation or, when indicated, by adding 0.4 μg/ml of rapamaycin (LC Laboratories, R-5000) to nitrogen-sufficient cultures. In the nitrogen-starvation procedure, cells were cultured for 14 h at 30°C in 1% glucose SC medium containing 40 mM ammonium sulfate as nitrogen source (nitrogen-sufficient medium) before being collected by filtration, washed and transferred to the same medium without nitrogen source. After the transfer, mycelia were incubated for a further 4 h under nitrogen-starved conditions. In experiments involving ts mutations, the incubation temperature for nitrogen-sufficient cultures was shifted to 42°C 45–60 min before transferring mycelia to medium without nitrogen (pre-heated at 42°C). The subsequent incubation under autophagy-inducing conditions was performed at 42°C. In all cases, mycelial samples were collected at the indicated time points, lyophilized and reduced to a fine powder by using a cell disruptor (FP120 Fast Prep; Qbiogene) and 0.5-mm ceramic beads in 2-ml screw-cap tubes. Total proteins in equal samples (by weight) were extracted by the alkaline lysis method described by Hervás-Aguilar and Peñalva (2010).62 Samples in Laemmli loading buffer were resolved in 12% SDS-polyacrylamide gels before being electro-transferred to nitrocellulose filters, which were reacted with α-GFP antibody (1:5000; Roche, 11 814 460 001). Peroxidase-coupled goat-anti mouse IgG (H+L)(Jackson ImmunoResearch; 115-035-003) was used as secondary antibody, also at 1:5000. Bands were detected using ECL (GE Pharmaceuticals, RPN2106).
Microscopy of GFP-Atg8
For autophagy experiments, hyphae were pre-cultured in Lab-Tek chambers (Thermo Fischer Scientific, 115411; 0.3 ml of medium per well) at 25–28°C in pH 6.5 'watch minimal medium' (WMM) containing 0.1% glucose and 0.5 mM ammonium tartrate (i.e., 1 mM ammonium). In control experiments we established that these pre-culture conditions were nitrogen-sufficient (i.e., they did not result in any detectable autophagy in the wild type at 28°C, 37°C or 42°C, as observed by epifluorescence microscopy with GFP-Atg8). After 14–16 h of incubation, the medium was removed and the hyphae, which naturally adhered to the bottom coverslip, were washed four times and finally submerged in 0.3 ml of the same medium without a nitrogen source. These conditions were sufficient to induce autophagy at all tested temperatures and gave rise to all the observed intermediates in the autophagosome cycle. However, to achieve a more uniform response to nitrogen starvation, we added 0.5 μg/ml of rapamycin to the ‘autophagy medium,’ which ensured that all hyphae in the population underwent autophagy in a highly synchronous manner, starting after approximately 1.5 h after shifting them to nitrogen-starved conditions, thus enormously facilitating image acquisition and comparison between different strains. In experiments involving transgenes driven by the alcAp, which is regulable by the carbon source,31 we used 0.1% fructose (low expression levels) instead of 1% glucose (which represses the promoter).
The microscopy setup consisted of an inverted fluorescence microscope (Leica DMI6000B) driven by Metamorph® software (Molecular Dynamics) and coupled to a CCD camera (ORCA ER-II; Hamamatsu). The microscope is equipped with an external light source with metal halide lamp for epifluorescence excitation (Leica, EL6000), GFP (Semrock GFP-3035B), mCherry (Semrock TXRED-4040) and DAPI/CMAC) (Leica 513 875) filter sets, and a HCX 63x 1.4 NA objective (Leica 11506187). To minimize cell photodamage, the excitation light attenuator was set to positions 1–2. Exposure times were always equal to or less than 200 msec. These conditions permitted acquisition of time-lapse series of > 200 frames of live cells. When experiments ‘on the stage’ involved a temperature shift, the temperature of the room was set to 28°C. To set the actual temperature of the culture to 37°C we used a Heating Insert P (Leica 11531824) coupled to the microscope stage, in combination with an objective heater (PeCon 0280.010). Both devices were connected to a dual-channel temperature controller (PeCon 37-2). The incubation chamber temperature dial was set to 48°C and the objective heater was set to 38°C. With these settings, the actual temperature of the culture medium, as measured within the incubation chamber medium with a digital thermometer, reached 37°C within 15 min after switching on the temperature controls. This was the maximal temperature achievable with our set-up, as the microscope objective might be damaged at 39°C or above. Strains carrying rabOA136D and sedVR258G mutations did not grow at 37°C. However, hbrA2/vps33ts strains, although markedly inhibited, still showed some growth at 37°C. Thus, in this particular case (and in the corresponding wild-type controls) the LabTek chamber was placed into a 42°C external incubator for 1.5–4 h, then transferred to the stage incubation chamber pre-heated at 37°C, and imaged within 10 min after the transfer. Image acquisition, contrast adjustment, channel color combining, z-stack maximal intensity projections (contrasted, when indicated, with the “unsharp mask” filter) and assembly of kymographs from time-lapse series were made using Metamorph® software (Molecular Dynamics). Images were converted to 8-bit greyscale (and usually shown in inverted contrast) or to 24-bit RGB, and annotated with Corel Draw (Corel, version 14.0). Time-lapse sequences were converted to QuickTime using ImageJ 1.37 (http://rsb.info.nih.gov/ij/). Statistical analyses were performed using Prism (GraphPad, version 5.00 for Windows). In experiments involving the ER labeled with AN0834/Sec63-mRFP, images were deconvolved using a blind 3D deconvolution algorithm (MediaCybernetics, Autoquant X2 sofware), with standard settings. Last, when indicated, vacuoles were detected with CMAC (7-amino-4-chloromethylcoumarin, CellTracker Blue; Invitrogen C-2110).
Supplementary Material
Acknowledgments
Supported by grants BIO2012-30965 and IPT-2011-0752-900000 from the Ministerio de Economía y Competitividad (Spain) and S2010/BMD-2414 from Comunidad de Madrid. We thank Prof. Geoff Turner for generously providing hbrA2 strains, Dr. Antonio Galindo for help, Prof. Herbert N. Arst for critically reading the manuscript, Elena Reoyo for valuable technical assistance and three anonymous referees for useful suggestions.
Glossary
Abbreviations:
- CMAC
7-amino-4-chloromethyl-coumarin
- Cvt
cytoplasm-to-vacuole targeting
- EE
early endosomes
- CORVET
class C core vacuole/endosome tethering
- GEF
guanine nucleotide exchange factor
- HOPS
homotypic fusion and vacuole protein sorting
- PAS
phagophore assembly sites(s)
- PE
posphatidylethanolamine
- SD
standard deviation
- TRAPP
transport protein particle oligomeric complexes
Disclosure of Potential Conflicts of Interest
The authors declare that there are no potential conflicts of interest.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/24483
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24483
References
- 1.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 2.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–56. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–22. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep 2011; 3:25. [DOI] [PMC free article] [PubMed]
- 6.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen W-L, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–84. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–69. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–6. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yen W-L, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–14. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noda NN, Satoo K, Fujioka Y, Kumeta H, Ogura K, Nakatogawa H, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44:462–75. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 15.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair U, Yen W-L, Mari M, Cao Y, Xie Z, Baba M, et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8:780–93. doi: 10.4161/auto.19385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–6. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008;182:129–40. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peñalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, Jr., et al. Searching for gold beyond mitosis: Mining intracellular membrane traffic in Aspergillus nidulans. Cell Logist. 2012;2:2–14. doi: 10.4161/cl.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kikuma T, Ohneda M, Arioka M, Kitamoto K. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot Cell. 2006;5:1328–36. doi: 10.1128/EC.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–94. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abenza JF, Galindo A, Pantazopoulou A, Gil C, de los Ríos V, Peñalva MA. Aspergillus RabB Rab5 integrates acquisition of degradative identity with the long distance movement of early endosomes. Mol Biol Cell. 2010;21:2756–69. doi: 10.1091/mbc.E10-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abenza JF, Galindo A, Pinar M, Pantazopoulou A, de los Ríos V, Peñalva MA. Endosomal maturation by Rab conversion in Aspergillus nidulans is coupled to dynein-mediated basipetal movement. Mol Biol Cell. 2012;23:1889–901. doi: 10.1091/mbc.E11-11-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calcagno-Pizarelli AM, Hervás-Aguilar A, Galindo A, Abenza JF, Peñalva MA, Arst HN., Jr. Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT-0, I, II and III genes by inactivation of a salt-tolerance pathway allows examination of ESCRT gene roles in pH signalling. J Cell Sci. 2011;124:1–13. doi: 10.1242/jcs.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–89. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Galindo A, Calcagno-Pizarelli AM, Arst HN, Jr., Peñalva MA. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J Cell Sci. 2012;125:1784–95. doi: 10.1242/jcs.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abenza JF, Pantazopoulou A, Rodríguez JM, Galindo A, Peñalva MA. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 2009;10:57–75. doi: 10.1111/j.1600-0854.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- 32.Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–50. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–80. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–22. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 35.Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, et al. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012;22:1545–53. doi: 10.1016/j.cub.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi X, Sha Y, Kaminskyj S. Aspergillus nidulans hypA regulates morphogenesis through the secretion pathway. Fungal Genet Biol. 2004;41:75–88. doi: 10.1016/j.fgb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, et al. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat Cell Biol. 2006;8:1263–9. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- 38.Taheri-Talesh N, Horio T, Araujo-Bazán LD, Dou X, Espeso EA, Peñalva MA, et al. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008;19:1439–49. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pantazopoulou A, Peñalva MA. Characterization of Aspergillus nidulans RabC/Rab6. Traffic. 2011;12:386–406. doi: 10.1111/j.1600-0854.2011.01164.x. [DOI] [PubMed] [Google Scholar]
- 40.Taheri-Talesh N, Xiong Y, Oakley BR. The functions of myosin II and myosin V homologs in tip growth and septation in Aspergillus nidulans. PLoS One. 2012;7:e31218. doi: 10.1371/journal.pone.0031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, El-Ganiny AM, Bray GE, Sanders DAR, Kaminskyj SGW. Aspergillus nidulans hypB encodes a Sec7-domain protein important for hyphal morphogenesis. Fungal Genet Biol. 2008;45:749–59. doi: 10.1016/j.fgb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Pantazopoulou A, Peñalva MA. Organization and dynamics of the Aspergillus nidulans Golgi during apical extension and mitosis. Mol Biol Cell. 2009;20:4335–47. doi: 10.1091/mbc.E09-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittaker SL, Lunness P, Milward KJ, Doonan JH, Assinder SJ. sodVIC is an alpha-COP-related gene which is essential for establishing and maintaining polarized growth in Aspergillus nidulans. Fungal Genet Biol. 1999;26:236–52. doi: 10.1006/fgbi.1999.1117. [DOI] [PubMed] [Google Scholar]
- 44.Breakspear A, Langford KJ, Momany M, Assinder SJ. CopA:GFP localizes to putative Golgi equivalents in Aspergillus nidulans. FEMS Microbiol Lett. 2007;277:90–7. doi: 10.1111/j.1574-6968.2007.00945.x. [DOI] [PubMed] [Google Scholar]
- 45.Béthune J, Wieland F, Moelleken J. COPI-mediated transport. J Membr Biol. 2006;211:65–79. doi: 10.1007/s00232-006-0859-7. [DOI] [PubMed] [Google Scholar]
- 46.Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–90. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banfield DK, Lewis MJ, Pelham HRB. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–9. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- 48.Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci U S A. 2012;109:6981–6. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–13. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 52.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–5. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 53.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy. 2010;6:301–3. doi: 10.4161/auto.6.2.11134. [DOI] [PubMed] [Google Scholar]
- 56.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–61. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 57.Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–6. doi: 10.1016/S0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 58.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, et al. Fusion PCR and gene targeting in Aspergillus nidulans Nat Prot 2006; 1:3111-20. [DOI] [PubMed]
- 59.Tilburn J, Scazzocchio C, Taylor GG, Zabicky-Zissman JH, Lockington RA, Davies RW. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–21. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- 60.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, et al. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–66. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Etxebeste O, Markina-Iñarrairaegui A, Garzia A, Herrero-García E, Ugalde U, Espeso EA. KapI, a non-essential member of the Pse1p/Imp5 karyopherin family, controls colonial and asexual development in Aspergillus nidulans. Microbiology. 2009;155:3934–45. doi: 10.1099/mic.0.032615-0. [DOI] [PubMed] [Google Scholar]
- 62.Hervás-Aguilar A, Peñalva MA. Endocytic machinery protein SlaB is dispensable for polarity establishment but necessary for polarity maintenance in hyphal tip cells of Aspergillus nidulans. Eukaryot Cell. 2010;9:1504–18. doi: 10.1128/EC.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.