Abstract
The N-end rule pathway is a cellular proteolytic system that utilizes specific N-terminal residues as degradation determinants, called N-degrons. N-degrons are recognized and bound by specific recognition components (N-recognins) that mediate polyubiquitination of low-abundance regulators and selective proteolysis through the proteasome. Our earlier work identified UBR4/p600 as one of the N-recognins that promotes N-degron-dependent proteasomal degradation. In this study, we show that UBR4 is associated with cellular cargoes destined to autophagic vacuoles and is degraded by the lysosome. UBR4 loss causes multiple misregulations in autophagic pathways, including an increased formation of LC3 puncta. UBR4-deficient mice die during embryogenesis primarily due to defective vascular development in the yolk sac (YS), wherein UBR4 is associated with a bulk lysosomal degradation system that absorbs maternal proteins from the YS cavity and digests them into amino acids. Our results suggest that UBR4 plays a role not only in selective proteolysis of short-lived regulators through the proteasome, but also bulk degradation through the lysosome. Here, we discuss a possible mechanism of UBR4 as a regulatory component in the delivery of cargoes destined to interact with the autophagic core machinery.
Keywords: N-recognin, UBR box, UBR4, angiogenesis, ubiquitin ligase, yolk sac
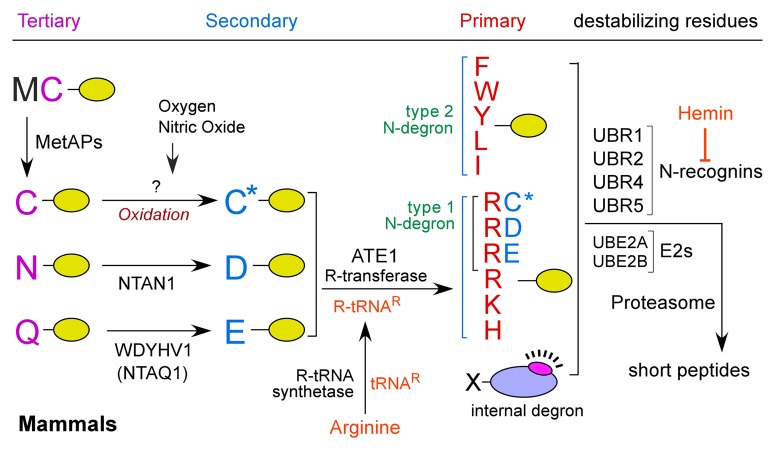
In mammals, N-end rule degradation determinants include type 1 (Arg, Lys and His; positively charged) and type 2 (Phe, Tyr, Trp, Leu and Ile; bulky hydrophobic) N-terminal residues (Fig. 1). N-degrons can be generated when internally embedded residues are exposed by proteolytic cleavage or through an enzymatic cascade that mediates post-translational modifications of precursors (pro-N-degrons), including N-terminal Asn, Gln, Cys, Asp and Glu. In this cascade, the tertiary residues Asn and Gln are respectively deamidated by the N-terminal asparagine amidase NTAN1 and the N-terminal glutamine-specific amidase WDYHV1/NTAQ1 (encoded by the gene WDYHV motif containing 1), generating the secondary residues Asp and Glu (Fig. 1). N-terminal Asp and Glu are conjugated with the amino acid L-Arg by ATE1-encoded R-transferases. In the mammalian N-degron code, N-terminal Cys is oxidized into CysO2(H) or CysO3(H) to generate an arginylation-permissive secondary residue. The resulting Arg and other degrons are recognized by N-recognins that mediate polyubiquitination, a signal for proteasomal degradation. In mammals, at least four N-recognins (UBR1, UBR2, UBR4 and UBR5/EDD) bind to type 1 or type 2 residues. Canonical N-recognins, UBR1 and UBR2 and their sequelogs, have been implicated in selective degradation of short-lived proteins through N-degron-dependent polyubiquitination. In contrast, the function of UBR4, a newly identified 570-kDa N-recognin, in the N-end rule pathway remains unclear. Different from canonical N-recognins carrying a RING finger ubiquitination domain, UBR4 does not have such a signature unique to substrate recognition components of the ubiquitin-proteasome system, raising a possibility that its action mechanism could be different from those of canonical N-recognins.
Figure 1. The N-end rule pathway. In the mammalian N-end rule pathway, the tertiary destabilizing N-terminal residues Asn and Gln are deamidated by two distinct enzymes, NTAN1 and WDYHV1/NTAQ1 into Asp and Glu, respectively. The secondary destabilizing N-terminal residues Asp and Glu are arginylated by ATE1 R-transferase isoforms produced through alternative splicing of ATE1. N-terminal Cys is a tertiary destabilizing residue in mammals but not in S. cerevisiae. The destabilizing activity of N-terminal Cys requires its oxidation prior to arginylation by ATE1. C* denotes the oxidized N-terminal Cys residue, either Cys-sulfinic acid [CysO2(H)] or Cys-sulfonic acid [CysO3(H)]. N-terminal Arg together with other type 1 and type 2 residues are directly bound by a set of N-recognins characterized by the UBR box, including UBR1, UBR2, UBR4 and UBR5. UBR box proteins can target substrates through the recognition of internal degrons as well as N-degrons.
In this study, we found that UBR4 stably expressed in HEK293 cells forms UBR4-positive cytosolic puncta that correspond to ATG12–ATG5- and LC3-positive phagophores and LC3-positive autophagosomes. Lysosomal degradation of a subpopulation of UBR4, likely together with associated cargoes, is accelerated when autophagy is stimulated by starvation, and is slowed down by pharmacological inhibition of autophagy. Immortalized embryonic fibroblast cells and the YSs from ubr4−/− mouse embryos as well as siRNA-based UBR4-knockdown HEK293 cells exhibit multiple misregulations in autophagic pathways, including an increased formation of autophagic vacuoles. The overall autophagic flux, as determined by the level and degradation of the selective autophagy substrate SQSTM1/p62, is accelerated in ubr4−/− mouse embryonic fibroblasts. Transient expression of recombinant UBR4 in UBR4-knockdown HEK293 cells reverts the induced LC3 puncta back to the basal level. Our results collectively suggest that UBR4 is associated with (unidentified) autophagic cargoes and regulates their delivery to autophagic vacuoles. How does UBR4 loss induce autophagy? UBR4 loss for a prolonged period of time may cause misregulation in the degradation of autophagic cargoes, leading to cellular stress (e.g., nutrient insufficiency and/or accumulation of damaged organelles). A mutually nonexclusive possibility is that UBR4 has a direct function as a regulator of autophagic flux.
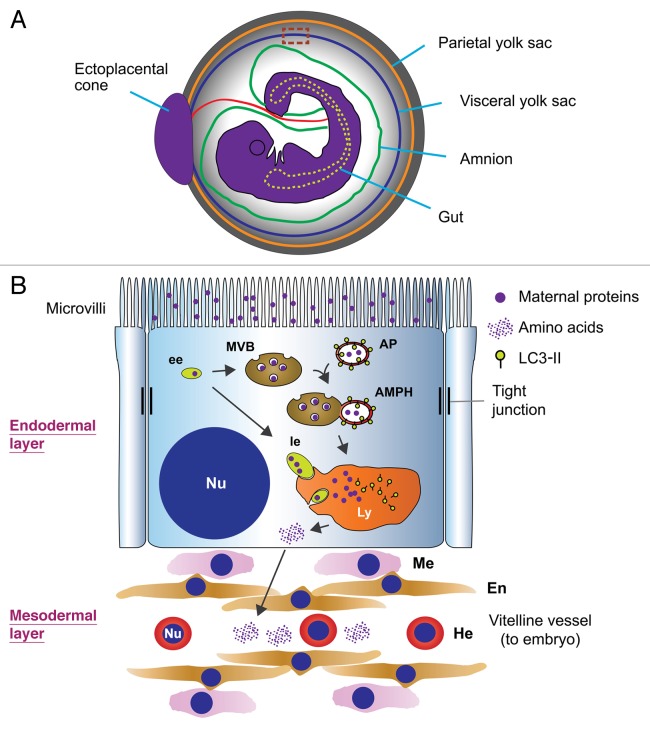
We also found that UBR4-deficient mice die at embryonic day (E) 9.5–10.5 primarily due to defects in vascular development of the YS. Differentiation of UBR4-deficient endothelial cells in the YS normally advances to form a network of primitive vessels, but subsequent remodeling of vascular smooth muscle cells is arrested, leading to vascular failure and embryonic death. How can the YS vascular defect be explained by UBR4 function in autophagy? Mammalian embryos at early stage (e.g., E9.5 in mice) do not have functional placenta yet and, thus, cannot take up circulating amino acids and other nutrients from the mother’s blood. Through whole embryo ex vivo culture studies, embryologists in the late '70s already noticed that amino acids essential for protein synthesis in early murine embryo are not synthesized de novo, but must be supplied through lysosomal degradation of maternal proteins absorbed in the YS. For the past more than three decades, however, this lysosomal degradation system, which is “bulk” (as opposed to “selective”) and “constitutive” (as opposed to “inducible”) in nature, has not received attention from the fields of both the autophagy-lysosome system and the ubiquitin-proteasome system. As illustrated in Figure 2, the visceral YS is composed of two distinct layers in which vascular development occurs in the inner layer derived from the mesoderm, whereas the endoderm-derived outer layer absorbs maternal proteins from the YS cavity and digests this “foreign material” (as opposed to the cells’ own constituents in autophagy) into amino acids using lysosomal hydrolases. Although little is known about components and mechanisms underlying this bulk degradation system, recent findings from cultured cells suggest that the delivery of endosomes carrying maternal proteins to apical vacuoles/lysosomes in the YS endoderm may involve fusion with late endosomes/multivesicular bodies (MVBs), which, in turn, are fused with autophagosomes to form amphisomes. In either case, the lysosome-derived amino acids with a maternal origin are collected by blood vessels in the YS mesoderm and subsequently supplied to the embryo via the vitelline circulation (Fig. 2). Interestingly, we found that UBR4 expression in the YS is not readily detected in the layer of vascular development but, is prominent in an outer layer that supplies lysosome-generated catabolites and secretes paracrines, such as growth factors, to orchestrate vascular development in the underlying layer. UBR4 in endodermal cells is associated with cytosolic puncta that colocalize with LC3-positive autophagic vacuoles enriched in a specific subcellular cytosolic compartment between the plasma membrane and lysosomes/vacuoles. Moreover, UBR4-deficient YSs exhibit a markedly increased formation of LC3 puncta. These findings, together with our more recent results using cultured cells, collectively suggest that UBR4 in the YS endoderm plays a role in the delivery of cargo-loaded endosomes to lysosomes, a process essential for YS vascular development. It would be of interest in the field of proteolysis to elucidate the components and regulation of bulk lysosomal degradation of maternal materials in the visceral YS.
Figure 2. The structure and function of the visceral YS of murine embryos. (A) A cross-section of a mouse embryo at E9.5 with the YS intact. (B) An enlarged view of a dotted rectangle in (A). The visceral YS of early mouse embryos at E9.5 is composed of two developmentally distinct layers: an outer layer derived from the endoderm and an underlying layer derived from the mesoderm. The YS at E9.5 contains a constitutive, bulk protein degradation system that absorbs maternal proteins from the YS cavity and digests these foreign materials (as opposed to its own cellular constituents) using lysosomal hydrolases. The model illustrates that endocytosed maternal proteins are delivered to and digested by the lysosome through early endosomes (ee) and late endosomes (le). Lysosome-derived amino acids in the YS endoderm are supplied to the embryo via the vitelline circulation or used to synthesize paracrines that control vascular development in the mesodermal layer. AP, autophagosome; AMPH, amphisome; En, endothelial cells; Ly, lysosome; Nu, nucleus; Me, mesenchymal cell; He, hematopoietic cell.
Our earlier work identified UBR4 as an adaptor that recognizes N-degrons of short-lived regulators in the ubiquitin-proteasome system. The current study implicates UBR4 as a putative adaptor in nonselective proteolysis through autophagic machinery. What is unique to UBR4 as a regulatory component in both proteolytic systems? In the N-end rule pathway, UBR4 recognizes substrates through the UBR box, an ~70-residue zinc finger domain essential for binding type 1 and type 2 residues. Moreover, the binding to the UBR box of canonical N-recognins induces allosteric conformational change, resulting in accelerated degradation of N-end rule substrates. This raises an interesting possibility that the binding of the destabilizing N-terminal residues of proteins or their structural homologs (small molecule ligands) to the UBR box of UBR4 induces an allosteric conformational change to regulate the delivery of UBR4 substrates to autophagic vacuoles.
Studies for the past decade have revealed the role of UBR4 in a variety of biological processes in mammals as well as the plant Arabidopsis thaliana and the fly Drosophila melanogaster. Given an emerging role of UBR4 in autophagy, these functions in and outside the N-end rule pathway should be reevaluated in light of bulk degradation by the lysosome.
Acknowledgments
We are grateful to the Kwon and Kim research team members at University of Pittsburgh, Seoul National University, and Korean Research Institute of Bioscience and Biotechnology for helpful discussions. This work was supported by NIH grant HL083365 (to Y.T.K.), World Class University (R31-2008-000-10103-0 to Y.T.K.) and World Class Institute (WCI 2009-002 to B.Y.K.) through the National Research Foundation funded by the Ministry of Science, ICT and Future Planning, Korea, and Promoted Research from Kanazawa Medical University (S2012-7 to T.T.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24643