Abstract
Atg13 is a subunit of the Atg1 complex that is involved in autophagy. The middle and C-terminal regions of Atg13 are intrinsically disordered and rich in regulatory phosphorylation sites. Thus far, there have been no structural data for any part of Atg13, and no function assigned to its N-terminal domain. We crystallized this domain, and found that it has a HORMA (Hop1, Rev7, Mad2) fold. We showed that the Atg13 HORMA domain is required for autophagy and for recruitment of the phosphatidylinositol (PtdIns) 3-kinase subunit Atg14, but is not required for Atg1 interaction or Atg13 recruitment to the PAS. The HORMA domain of Atg13 is similar to the closed conformation of the spindle checkpoint protein Mad2. A pair of conserved arginines was identified in the structure, and tested functionally in yeast. These residues are important for autophagy, as mutations abrogate autophagy and block Atg14 recruitment. The location of these Arg residues in the structure suggests that the Atg13 HORMA domain could act as a phosphorylation-dependent conformational switch.
Keywords: Atg13, HORMA, Atg14, protein structure, protein crystallography, yeast genetics, protein degradation
Autophagy is a cellular process that provides metabolic precursors through cellular self-digestion when cells are starved. It is mediated by more than 30 autophagy-related (Atg) proteins, together with certain SNAREs, Rab GTPases, and other general purpose membrane-trafficking proteins.
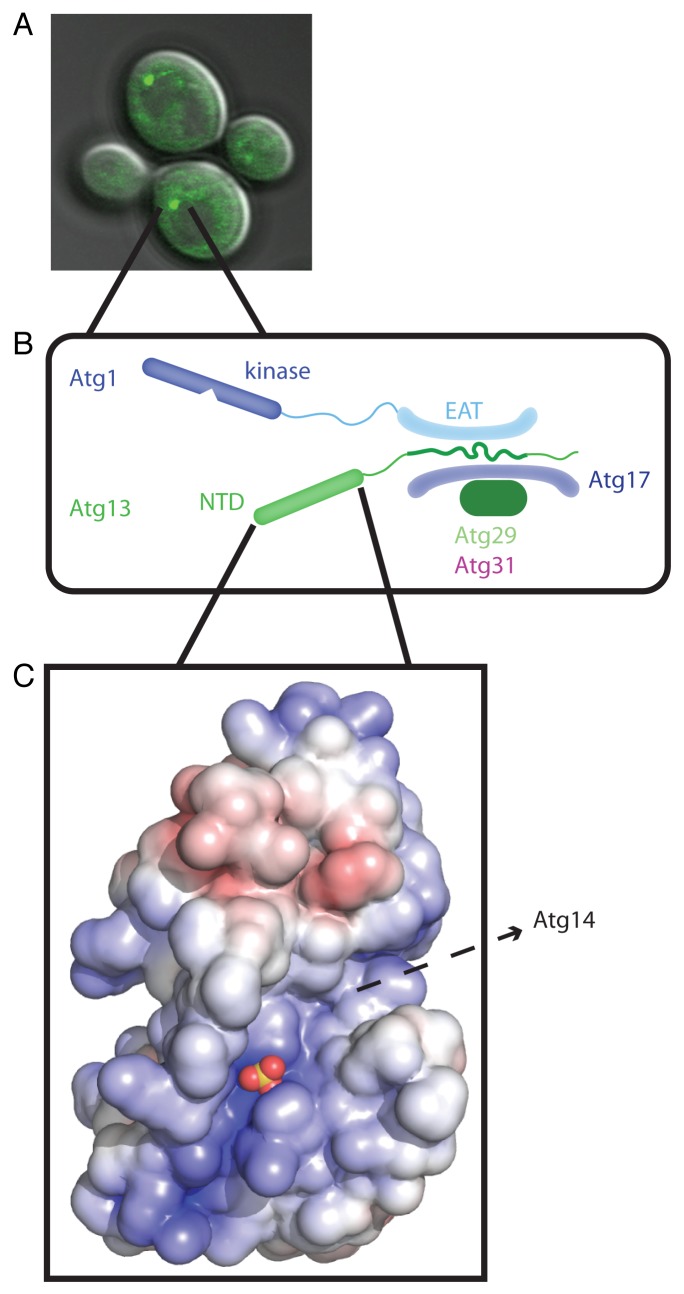
When autophagy is initiated in yeast, autophagy components are recruited to the phagophore assembly site (PAS). The PAS is visualized by fluorescence microscopy as bright puncta next to the vacuole, which also contain Atg proteins (Fig. 1A). The complex containing the proteins Atg17-Atg31-Atg29 is the first to localize to the PAS. TOR (target of rapamycin) becomes inactive in starvation, resulting in the hypophosphorylation of Atg13, enabling it to bind to Atg17, and leading it to translocate to the PAS. The protein kinase and vesicle tether Atg1 is subsequently recruited to the PAS by binding to Atg13.
Figure 1. Structure and function of the Atg13 HORMA domain. (A) The Atg1 complex initiates autophagy at the PAS, shown here as marked by Atg13-GFP. (B) Schematic of the Atg1 complex highlighting the context of the HORMA domain. The Atg1 complex is a dimer of pentamers, with just one pentamer shown here for simplicity. (C) Space filling model of the Atg13 HORMA domain, with the bound sulfate ion shown as a marker for a likely biological phosphate binding site. Note the prominent peptide-sized groove running southwest to northeast. EAT, early autophagy targeting/tethering; NTD, N-terminal domain. Adapted from Jao CC, Ragusa MJ, Stanley RE, Hurley JH. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci U S A 2013; 110:5486–91; PMID:23509291; 10.1073/pnas.1220306110.
To understand the molecular mechanisms of autophagy initiation at the PAS, we analyzed the structure and function of Atg13. Atg13 is well conserved from yeast to humans, and is predicted to contain a structured N-terminal region, and intrinsically disordered middle and C-terminal regions. Previous reports showed that the unstructured region in the middle of Atg13 mediates binding to both Atg1 and Atg17 (Fig. 1B). In this study, we sought to determine the structure and function of the N-terminal region, which we found to comprise a HORMA domain.
Using yeast autophagy assays, we began to deconstruct the function of Atg13. Expression of neither the N- nor C-terminal regions alone rescues autophagy in cells lacking Atg13. We next tested which region of Atg13 is required for PAS localization. We expressed different GFP-tagged constructs of Atg13 in atg13Δ yeast cells. After rapamycin treatment, which mimics starvation, Atg13-GFP recruitment to the PAS was assessed. Both full-length- and C-terminal region-expressing cells contained fluorescent puncta corresponding to an Atg13-positive PAS, but the HORMA domain-expressing cells did not, indicating that the HORMA domain does not autonomously localize to the PAS.
The generation of PtdIns3-phosphate by the autophagy-specific class III PtdIns 3-kinase complex is a key event in autophagy. Recruitment of PtdIns 3-kinase subunits occurs immediately downstream of Atg13 recruitment. To test whether the Atg13 HORMA domain plays a role in recruitment of the autophagy-specific PtdIns 3-kinase subunit Atg14, we looked for Atg14 recruitment to the PAS by fluorescence microscopy. After rapamycin treatment, cells that express full-length Atg13 recruit Atg14 to the PAS, but cells that express Atg13 lacking the HORMA domain do not. This demonstrated that the Atg13 HORMA domain is required for Atg14 recruitment, even though it is not required for Atg13 recruitment to the PAS.
To understand the role of the Atg13 N-terminal domain in PAS initiation, we crystallized the first 260 residues of Atg13 from the thermotolerant budding yeast L. thermotolerans, and solved its crystal structure to a resolution of 2.3 Å (Fig. 1C). Surprisingly, our structure is very similar to the closed conformation of Mad2 (mitotic arrest-deficient 2), a protein that contains the HORMA fold. Bound sulfate ions were seen in the structure located along an electropositive groove. Sulfate ions are often markers for biological phosphate binding sites. This electropositive groove is of the correct dimensions to accommodate a phosphopeptide. The sulfate ion is coordinated by two conserved arginine residues, and mutation of these residues to aspartic acid residues results in the absence of autophagy after rapamycin treatment or starvation. We hypothesize that these residues could act as a phosphate sensor, although the identity of the phosphorylated ligand is unknown.
It is important to emphasize that we have not been able to identify a direct interaction between the Atg13 HORMA domain and Atg14. There could be many reasons for this. For example, the interaction may, in fact, be indirect, mediated by some third bridging factor. Alternatively, the Atg13-Atg14 interaction might be direct, but only occurs after conformational switching of the HORMA domain by some unidentified factors, or the interaction might be direct, but only occurs in the context of having Atg14 incorporated into the intact, assembled PtdIns 3-kinase complex. Candidates for the putative phophorylated ligand for the Atg13 HORMA domain include the C-terminal part of Atg13, Atg14 or some other subunit of the autophagy-specific PtdIns 3-kinase complex, or some other factor important in bridging between Atg13 and Atg14.
Acknowledgments
This work was supported by the NIH/NIDDK Intramural Program (J.H.H.), fellowships from the NIH/NIGMS (C.C.J., M.J.R.), and a Damon Runyon Cancer Research Fellowship (R.E.S.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24896