Abstract
Autophagy is a process in which a eukaryotic (but not prokaryotic) cell destroys its own components through the lysosomal machinery. This tightly regulated process is essential for normal cell growth, development, and homeostasis, serving to maintain a balance between synthesis and degradation, resulting in the recycling of cellular products. Here we try to expand the concept of autophagy and define it as a general mechanism of regulation encompassing various levels of the biosphere. Interestingly, one of the consequences of such an approach is that we must presume an existence of the autophagic processes in the prokaryotic domain.
Keywords: autophagy, biosystem, prokaryotes, bacteria, multicellular organization, self-regulation, protophagy
Autophagy Overview
Autophagy (from the Greek for self-eating) is a cellular mechanism describing the chaperone- or vesicle-mediated recycling of excessive or damaged proteins, protein complexes and organelles, conducted by enzymes originating from the same cell.1-3 Such recycling serves several essential functions including nutrient acquisition,2 maintenance of cellular homeostasis,1,4 adaptivity,3 immunity and differentiation.5 In this article we do not touch on the molecular basis and functions of autophagy, as they have already been described in a number of excellent reviews.1-3,6 Herein we will focus on autophagy from a purely conceptual point of view.
Typically, the term autophagy is applied to cellular processes. Meanwhile, analogous processes are observed in various self-regulated communities at different levels of the biosphere (Table 1). These autophagy-like processes occur at the level of a single eukaryotic cell (as a community of organelles), in organisms (as a community of cells and tissues), in ecosystems (as a community of living organisms) and finally in the entire biosphere (as a community of ecosystems). For example, at the organismal level, one of the manifestations of an autophagy-like process is fat consumption during starvation, when the organism as a system consumes part of its own structure and redistributes the energy freed from adipose tissue.7 This process compensates for energy influx oscillation and is vitally important for the organism. Other phenomena, such as placentophagy (consuming of the placenta after delivery in mammals),9 exuviae eating (eating the old skin after molting in amphibians and insects),10 or cannibalism in animals,11 at first glance also seem reminiscent of autophagy. However, classifying these latter phenomena as true autophagy is in fact arguable since they are not regular and/or absolutely essential for survival of animals, and thus might be viewed as episodic manifestations of the autophagic principle. At the level of ecosystems autophagy-like mechanisms are also present. Stability of an entire ecosystem as a self-regulated system is maintained by permanent component redistribution (known as “trophic chains” or “trophic webs”),12 which can also be described in terms of autophagy. Indeed, like mitochondria being consumed to provide energy for a cell,13 weak herbivores being hunted by carnivores to redistribute energy for more viable components of an ecosystem. Indeed, like mitochondria being consumed to provide energy for a cell13 weak herbivores being hunted by carnivores to redistribute energy for more viable components of an ecosystem.14
Table 1. Analogs of autophagy at different levels of animate matter.
| Community | Process | Examples |
|---|---|---|
| Cell |
Autophagy |
Mitophagy (degradation of mitochondria)2 Pexophagy (degradation of peroxisomes)2 Ribophagy (degradation of ribosomes)2 Reticulophagy (degradation of ER)2 |
| Organism |
Hibernation, fasting or starvation |
Consumption by organism of its own tissues (e.g., adipose tissue)7 Oophagy (e.g., stronger embryos of sharks consume less-developed siblings in utero)8 |
| Ecosystem and Population | Trophic chain |
Predator-prey relationships12 Herbivore-plant relationships12 |
Thus, in various biosystems autophagy-like processes are essential for self-regulation and survival. These processes carry out synonymous functions at all levels: providing energy during starvation, supporting homeostasis and differentiation, promoting development, etc. A key example of the autophagic principle is observed in energy recycling, which is an important characteristic of living matter. Since every biosystem (from cell to biosphere) requires constant energy influx from outside, which is inherently unreliable, the system prudently recycles any damaged or excessive constituents rather than waste them. Starvation is usually provoked by two factors: food deficit and/or malfunctioning of energy-producing elements. In both cases, autophagy-like processes are able to restore energy flow through dismantling of expendable components down to elementary blocks for their consequent reusage. In such a way, biosystems obtain additional energy for restoration, adaptation, transformation or even migration. The aforementioned analogy between cellular autophagy, organismal fat consumption and trophic chains illustrates global employment of the autophagy principle in energy recycling.1
Another example of autophagy function is maintaining homeostasis. Supporting a stable state of internal environment is an important property of animate systems, and is implemented through a number of mechanisms. One of them is constant updating of the biosystem’s elements, mediated by autophagy-like processes. Indeed, a time of existence of any differentiated community (be it an association of organelles, cells or organisms) is many times longer than an average life span of its components. This is achieved through constant removal of old or damaged constituents by the system, which in essence is autophagy. Removing and subsequent recycling of old components protects a system from hazardous consequences of their malfunctioning. Akin to the way cellular autophagy (e.g., mitophagy)2 updates organelle content during cell life time, trophic chains (e.g., predator-prey interactions)12 regulate organismal content of an ecosystem by removing weak and sick animals.
These examples show that autophagy is not limited by the eukaryotic cell membrane but is a general mechanism encompassing various levels of the biosphere. Literally, almost any differentiated animate community is using autophagy as a potent mechanism of surviving and self-regulation. We use the word “almost” because autophagy has never been described in prokaryotes.15 Without lysosomes and membrane transport, it seems prokaryotes are unable to realize autophagy. Nevertheless, the absence in prokaryotes of such an important principle looks strange, and herein we try to show some evidence of autophagy existing in the prokaryotic domain.
Prokaryotes as Multicellular Organisms
The presence of autophagy has never been reported in a prokaryotic cell. Nevertheless, prokaryotes have several recycling mechanisms, such as different types of proteolysis16 and proteasomal degradation.15,17 Recently, ubiquitin-like tags, which mark proteins for proteasomal degradation, were found in several bacteria (e.g., Pup in Mycobacterium tuberculosis,18 and Samp proteins in Haloferax volcanii18,19). In addition, prokaryotes also have an analog of deubiquitinases: a recent discovery of a deaminase of Pup (Dop),20 which removes the Pup-tag from bacterial proteins, suggests that prokaryote proteolysis systems are as flexible as those found in eukaryotes.
Several bacteria, especially the large bacteria,21 have a primitive vesicular traffic that provides sorting of multiple factors (e.g., quorum sensing molecules),22 toxin secretion23 and DNA transfer24 (for more examples see refs. 22, 23 and ref. therein). Even though functions of these vesicles are far from the complexity of eukaryotic endosomal pathways, they can mediate precise cargo transport in a bacterial cell. Furthermore, Podar et al.25 have discovered the presence of vesicle-tethering proteins (critical regulators of endosomal traffic) in prokaryotes. They have shown that a V4R domain (predicted hydrocarbon-binding domain, COG1719) in bacteria and archaea is highly homologous to the Bet3 protein—a component of the TRAPPI complex, a conserved eukaryotic vesicle-tethering complex, involved in ER-Golgi vesicular exchange.
In sum, bacteria have vesicles, vesicle-tethering proteins and ubiquitin-like tags, which in theory may constitute a platform for a bona fide autophagic mechanism. Nevertheless, autophagy has never been identified in bacterial cells, leading to a safe conclusion that it is absent from this domain of life.
Convergent pieces of evidence indicate that in nature prokaryotes exist as differentiated multicellular forms rather than isolated cells.26-29 Considering that autophagy-like processes are a feature of differentiated communities, we presume that analogs of autophagy might exist in prokaryotic multicellular formations. Indeed, multicellular forms of prokaryotes such as colonies or biofilms display multiple characteristics of differentiated multicellular organisms. Among these characteristics are quorum sensing,28 collective digesting,30 collective prey hunting31 and resistance to antibiotics32 (Table 2).
Table 2. Examples of multicellular organization and cell-cell interactions of some prokaryotes.
| Taxon | Example of multicellular organization and cell-cell interactions |
|---|---|
| Cyanobacteria |
Photosynthetic bacteria that can differentiate into specialized cells (heterocysts) which lack chlorophyll but can convert nitrogen gas into a usable form for photosynthetic neighbors (vegetative cells).33 Form intercellular channel system for exchange of fixed nitrogen and photosynthetic products between these two types of cells. Often grow as connected chains of cells or as a mat, and in many ways resemble multicellular algae (were first classified as members of the plant kingdom).27 |
| Deltaproteobacteria |
Myxococcus xanthus uses cell-cell interactions to behave cooperatively when hunting for food. Predation involves the release of lytic substances that degrades prey organisms, thereby creating a public pool of growth substances. When starved for nutrients, the group of M. xanthus cells undergo a change in which the cells form a fruiting body containing spores that can disperse and rejuvenate into motile cells when they sense that prey are present.34 |
| Gammaproteobacteria | A small number of antibiotic-resistant mutants of E.coli can provide protection to other sensitive cells, enhancing the survival capacity of the overall population in stressful environments.32 |
From this point of view, autophagy should appear as another characteristic of multicellular form of prokaryote. Indeed, a single bacterium is similar to eukaryotic organelles like mitochondria,6 and can be considered as a membrane-bordered organelle-like element of a multicellular bacterial community. This presumption leads to an interesting conclusion: that autophagy is indeed present in the prokaryotic world, but it is an attribute of a prokaryotic community and not of a single bacterium. Actually, autophagy-like processes are well described in prokaryotic colonies but in different terms—cannibalism,35,36 altruism,37 autolysis38 or programmed cell death.38 Below are several examples.
Typical autophagy-like patterns have been described in bacteria during starvation.35 This pattern is widespread in bacterial species and known as toxin-antitoxin systems.37,39 One of the functions of these two-gene modules are regulation of colony density in response to different stimuli such as amino acid starvation or by antibiotics.39 Briefly, under nutrient limitation, bacteria secrete a toxic peptide (e.g., SdpC in Bacillus subtilis or mazF in Escherichia coli) that induces death of part of a colony lacking the antitoxin (protein SdpI or mazE, respectively).35 Dead cells are lysed and consumed by their neighbors, providing them with enough energy for sporulation.35,36 Another example of an autophagy-like process is inducible autolysis, such as that seen in colonies of Streptococcus pneumoniae.38,40 Under overcrowded conditions S. pneumoniae cells secrete the pheromone CSP that activates two-component signaling transduction kinases ComD and ComE, which activate expression of the lytA gene, responsible for autolysis.40 LytA requires activation by another kinase called VncS. Expression of the latter is regulated by different stress signals and is usually activated in defective or old cells, which are undergoing autolysis. After autolysis, DNA from destroyed cells is absorbed by the healthy neighboring cells.38 It is logical to extrapolate to the hypothesis that other biomolecules from destroyed cells can be acquired along with DNA, providing a recycling process within the colony. Moreover, similar two-gene altruistic models have been created experimentally.41
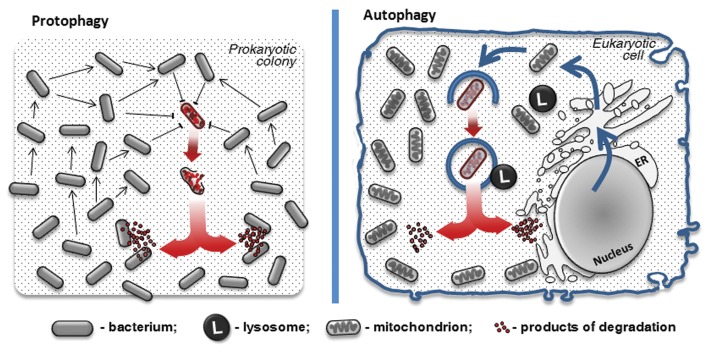
Such self-destructive cooperation can be seen as an extreme form of the division of labor between at least two phenotypes, in which one does not survive. For simplicity, here we will introduce the term “protophagy” as a synonym of bacterial cannibalism, autolysis, programmed cell death and other self-destructing patterns within bacterial colonies. From a bird’s-eye view, protophagy processes abide by the same rules as, and share a set of similarities with, eukaryotic autophagy (Fig. 1). Both operate with a similar cargo size (in most cases the size of a single lysed bacterium is approximately equal to a mitochondrion6,42 or a peroxisome43); have the same triggers (starvation or nonfavorable conditions);3,39 have the same principal mechanism (regulated partial self-consumption of constituents by the biosystem);1,22 and also achieve the same final goal (survival of a biosystem under stress conditions and maintaining homeostasis).3,22 In protophagy, the role of cargo vesicles is assumed by a prokaryotic cell, while a prokaryotic community is a biosystem, which recycles parts of digested bacteria to maintain self-stability.39
Figure 1. Key similarities between protophagy (left) and autophagy (right).
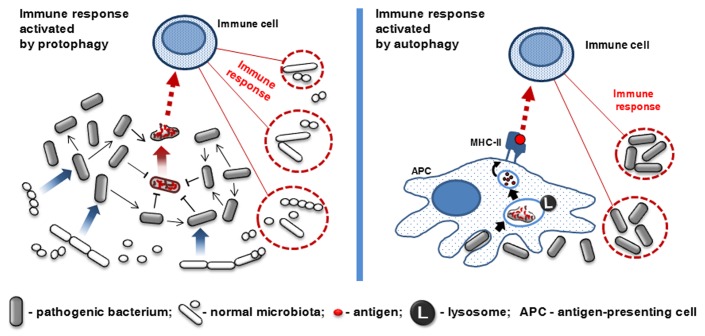
Similar to eukaryotic autophagy, protophagy functions are not limited to energy homeostasis and quality control only. For example, protophagy is employed by pathogenic bacteria for host invasion (Fig. 2). In order to eliminate competition for resources within commensal microbiota, some pathogenic bacteria use protophagy to manipulate the host’s immune system. To remove competing microbiota, a part of the bacterial population dies and releases intracellular toxins to boost inflammation. Activation of the host immune response kills or restricts commensal bacteria and allows the pathogen to take advantage of reduced competition to invade host tissues.44 This mechanism is employed by the enteropathogenic bacteria Salmonella enterica serovar Typhimurium,41 and Clostridium difficile.35,44 A similar protophagic strategy is also used by Streptococcus pneumoniae during lung colonization,45 colicinogenic strains of E. coli,46 Staphylococcus aureus,47 and Pseudomonas aeruginosa.48 Interestingly, in the absence of competing microbiota, such as germ-free mice, S. typhimurium colonizes the intestine efficiently without causing inflammation.44 This suggests that protophagy is used by a number of bacteria as an advanced mechanism of survival in a dense microbial community.
Figure 2. Analogous role of protophagy and autophagy in induction of inflammation (see the text for an explanation). MHC-II, major histocompatibility complex class II.
Some Applications of the Protophagy Concept
Introduction of the concept of protophagy not only has theoretical value, but may also be useful in practical applications. Classifying the processes listed above as related to autophagy might serve as a basis for new insights into prokaryotic life. In bioinformatics, the concept of protophagy may uncover new patterns of the evolution of recycling processes. We did not find evolutionary connections between protophagy and autophagy genes using BLAST and PSI-BLAST (using standard protocols),49 which may suggest that protophagy is not a direct evolutionary predecessor of eukaryotic autophagy, but rather an independent parallel realization of the recycling principle. However, more sophisticated professional analysis may find some homologies between the autophagy and protophagy genes in a similar way to the discovery that many apoptotic genes are conserved between pro- and eukaryotes.50
Moreover, understanding of general patterns that govern bacterial life may bring a great practical benefit. Industry widely uses bacteria as biofactories, and manipulation through protophagy could help tackle some hurdles associated with growing large-scale bacterial cultures. For example, when large biomass production is required, modulators of protophagy may improve the yield by means of enhancing natural mechanisms of eliminating impaired or damaged microorganisms. Protophagy may also be a beneficial concept in prediction and modeling of the behavior of multicellular (natural) forms of bacteria in biodegradation or bioremediation fields, where microorganisms applied over large areas can spontaneously differentiate into their natural multicellular forms.
Another critical area that may benefit is medicine. A growing problem of contemporary medicine is development of bacterial resistance to antibiotics. It is still a challenge to develop more effective and less toxic agents to treat chronic bacterial infections. Acceptance of multicellular organization of bacteria suggests a path for the development of a new type of drug: instead of killing each individual bacterium (as antibiotics do) it may be possible to target the global properties of infection by the disorganization of bacterial regulatory systems.41 Indeed, many infectious diseases are associated with bacterial biofilms — microbial accretions covered with a mucus shield that adhere to biological or nonbiological surfaces. Bacterial biofilms are implicated in a number of chronic infections including gastrointestinal and urinary tract infections, coronary heart disease, pulmonary diseases and others.51 Antibiotic resistance and immune evasion that is conveyed by biofilms are serious factors that may impair the effectiveness of traditional antibacterial therapies. A promising way to treat such infections is disorganization or blocking of bacterial communication networks.51 Some experimental therapies that employ this strategy, such as phage therapy52 or quorum-sensing quenchers,53 have already been proposed. In this sense, activation of protophagy might impair the protective barrier of biofilms and enhance biofilm dispersal and breakdown as well as activation of the immune system. In other words, activation of protophagy within bacterial multicellular formation may present new paths for drugs that are able to make biofilms vulnerable and detectable for host defense.
In conclusion, here we propose the protophagy concept for further evaluation and discussion. From our point of view, protophagy is a manifestation of the global autophagy mechanism in the bacterial world. We think that this concept might be useful to provide new insights into prokaryotic life and the evolution of autophagy.
Acknowledgments
The authors wish to express their appreciation to Drs. Lev Starokadomskyi, Oleg Volkov, Fiona J. McDonald, Andriy Nemchenko, and Ilya Bezprozvanny for reading the manuscript and valuable discussions. This study was partially supported by Poland-Belarus-Ukraine Cross-border Cooperation Programme 2007–2013 (Reference number PBU/0452/11).
Disclosure of Potential Conflicts of Interest
The authors declare that they do not have any conflicts of interest in connection to this work.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24544
References
- 1.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–55. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 4.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–5. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deretic V. Autophagy of intracellular microbes and mitochondria: two sides of the same coin? F1000 Biol Rep. 2010;2:1–4. doi: 10.3410/B2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein JL, Zhao TJ, Li RL, Sherbet DP, Liang G, Brown MS. Surviving starvation: essential role of the ghrelin-growth hormone axis. Cold Spring Harb Symp Quant Biol. 2011;76:121–7. doi: 10.1101/sqb.2011.76.010447. [DOI] [PubMed] [Google Scholar]
- 8.Lund R. Viviparity and intrauterine feeding in a new holocephalan fish from the lower carboniferous of montana. Science. 1980;209:697–9. doi: 10.1126/science.209.4457.697. [DOI] [PubMed] [Google Scholar]
- 9.Young SM, Benyshek DC. In search of human placentophagy: a cross-cultural survey of human placenta consumption, disposal practices, and cultural beliefs. Ecol Food Nutr. 2010;49:467–84. doi: 10.1080/03670244.2010.524106. [DOI] [PubMed] [Google Scholar]
- 10.Mira A. Exuviae eating: a nitrogen meal? J Insect Physiol. 2000;46:605–10. doi: 10.1016/S0022-1910(99)00146-8. [DOI] [PubMed] [Google Scholar]
- 11.Rudolf VH. Impact of cannibalism on predator-prey dynamics: size-structured interactions and apparent mutualism. Ecology. 2008;89:1650–60. doi: 10.1890/07-0709.1. [DOI] [PubMed] [Google Scholar]
- 12.Estes JA, Terborgh J, Brashares JS, Power ME, Berger J, Bond WJ, et al. Trophic downgrading of planet Earth. Science. 2011;333:301–6. doi: 10.1126/science.1205106. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):R169–76. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweiger O, Biesmeijer JC, Bommarco R, Hickler T, Hulme PE, Klotz S, et al. Multiple stressors on biotic interactions: how climate change and alien species interact to affect pollination. Biol Rev Camb Philos Soc. 2010;85:777–95. doi: 10.1111/j.1469-185X.2010.00125.x. [DOI] [PubMed] [Google Scholar]
- 15.Hughes T, Rusten TE. Origin and evolution of self-consumption: autophagy. Adv Exp Med Biol. 2007;607:111–8. doi: 10.1007/978-0-387-74021-8_9. [DOI] [PubMed] [Google Scholar]
- 16.Visick JE, Clarke S. Repair, refold, recycle: how bacteria can deal with spontaneous and environmental damage to proteins. Mol Microbiol. 1995;16:835–45. doi: 10.1111/j.1365-2958.1995.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 17.Pouch MN, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete Frankia. Mol Microbiol. 2000;35:368–77. doi: 10.1046/j.1365-2958.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 18.Darwin KH. Prokaryotic ubiquitin-like protein (Pup), proteasomes and pathogenesis. Nat Rev Microbiol. 2009;7:485–91. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat Rev Microbiol. 2012;10:100–11. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun HY, Tamura N, Tamura T. Rhodococcus prokaryotic ubiquitin-like protein (Pup) is degraded by deaminase of pup (Dop) Biosci Biotechnol Biochem. 2012;76:1959–66. doi: 10.1271/bbb.120458. [DOI] [PubMed] [Google Scholar]
- 21.Schulz HN, Jorgensen BB. Big bacteria. Annu Rev Microbiol. 2001;55:105–37. doi: 10.1146/annurev.micro.55.1.105. [DOI] [PubMed] [Google Scholar]
- 22.Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61:839–46. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 23.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–5. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 24.Yaron S, Kolling GL, Simon L, Matthews KR. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol. 2000;66:4414–20. doi: 10.1128/AEM.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podar M, Wall MA, Makarova KS, Koonin EV. The prokaryotic V4R domain is the likely ancestor of a key component of the eukaryotic vesicle transport system. Biol Direct. 2008;3:2. doi: 10.1186/1745-6150-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordium VA, Moshynets EV, Tsapenko MV, Adamchuk-Chalaya NI, Irodov DM, Andrienko VI, et al. Microcosm of living: evidence of nonobviousness. Biopolymers Cell. 2008;24:412–25. doi: 10.7124/bc.0007BA. [DOI] [Google Scholar]
- 27.Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 28.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 29.Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–9. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirhonen M, Flego D, Heikinheimo R, Palva ET. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–76. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnham JC, Collart SA, Highison BW. Entrapment and lysis of the cyanobacterium Phormidium luridum by aqueous colonies of Myxococcus xanthus PCO2. Arch Microbiol. 1981;129:285–94. doi: 10.1007/BF00414699. [DOI] [Google Scholar]
- 32.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–5. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang CC, Laurent S, Sakr S, Peng L, Bédu S. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol Microbiol. 2006;59:367–75. doi: 10.1111/j.1365-2958.2005.04979.x. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser D, Robinson M, Kroos L. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb Perspect Biol. 2010;2:a000380. doi: 10.1101/cshperspect.a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–3. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 36.Engelberg-Kulka H, Hazan R. Microbiology. Cannibals defy starvation and avoid sporulation. Science. 2003;301:467–8. doi: 10.1126/science.1088051. [DOI] [PubMed] [Google Scholar]
- 37.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–14. doi: 10.1128/MMBR.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piñas GE, Cortes PR, Orio AG, Echenique J. Acidic stress induces autolysis by a CSP-independent ComE pathway in Streptococcus pneumoniae. Microbiology. 2008;154:1300–8. doi: 10.1099/mic.0.2007/015925-0. [DOI] [PubMed] [Google Scholar]
- 41.Tanouchi Y, Pai A, Buchler NE, You L. Programming stress-induced altruistic death in engineered bacteria. Mol Syst Biol. 2012;8:626. doi: 10.1038/msb.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poole AM, Penny D. Evaluating hypotheses for the origin of eukaryotes. Bioessays. 2007;29:74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- 43.Duhita N, Le HA, Satoshi S, Kazuo H, Daisuke M, Takao S. The origin of peroxisomes: The possibility of an actinobacterial symbiosis. Gene. 2010;450:18–24. doi: 10.1016/j.gene.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirst RA, Kadioglu A, O’callaghan C, Andrew PW. The role of pneumolysin in pneumococcal pneumonia and meningitis. Clin Exp Immunol. 2004;138:195–201. doi: 10.1111/j.1365-2249.2004.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–28. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402, h402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 51.André J-B, Godelle B. Multicellular organization in bacteria as a target for drug therapy. Ecol Lett. 2005;8:800–10. doi: 10.1111/j.1461-0248.2005.00783.x. [DOI] [Google Scholar]
- 52.Donlan RM. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009;17:66–72. doi: 10.1016/j.tim.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Hentzer M, Givskov M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J Clin Invest. 2003;112:1300–7. doi: 10.1172/JCI20074. [DOI] [PMC free article] [PubMed] [Google Scholar]