Abstract
Autophagy is an evolutionarily conserved cellular process through which long-lived proteins and damaged organelles are recycled to maintain energy homeostasis. These proteins and organelles are sequestered into a double-membrane structure, or autophagosome, which subsequently fuses with a lysosome in order to degrade the cargo. Although originally classified as a type of programmed cell death, autophagy is more widely viewed as a basic cell survival mechanism to combat environmental stressors. Autophagy genes were initially identified in yeast and were found to be necessary to circumvent nutrient stress and starvation. Subsequent elucidation of mammalian gene counterparts has highlighted the importance of this process to normal development. This review provides an overview of autophagy, the types of autophagy, its regulation and its known impact on development gleaned primarily from murine models.
Keywords: autophagy, regulation, development, knockout models
Overview
Mammalian autophagy refers primarily to three cellular processes: chaperone-mediated autophagy, microautophagy and macroautophagy. This review will focus on the latter, macroautophagy, hereafter referred to simply as “autophagy.” To form a mature autophagosome, autophagy proceeds in successive stages, which include: (1) de novo formation of a double-membrane-bound structure or phagophore, (2) elongation of this lipid-based membrane and (3) encapsulation of intracellular cargo to form the mature autophagosome. Autolysosomes, formed from the fusion of autophagosomes and lysosomes, then degrade and recycle the macromolecule components in order to maintain energetic homeostasis in the cell. Autophagy is an ancient process that is highly conserved among eukaryotes. The initial characterization of the autophagy pathway was worked out in yeast1-3 with the identification of approximately 30 autophagy-related (ATG) genes, and many mammalian genetic homologs have been identified. Autophagy has been implicated in longevity/life-span extension, disease prevention and promotion, as well as mammalian development (reviewed in refs. 4–7). This review will concentrate on the process and regulation of autophagy, as well as the role for autophagy in murine development.
Core Machinery Involved in Autophagosome Formation
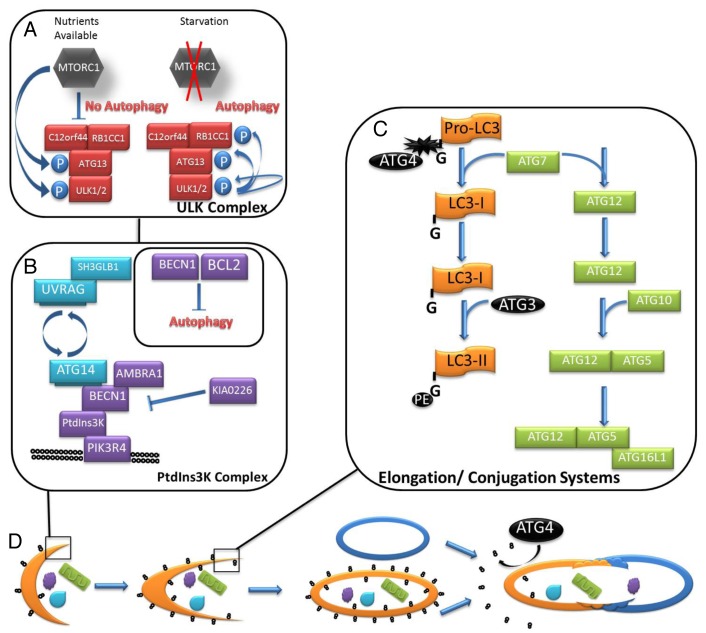
The mechanistic target of rapamycin (MTOR), upstream of autophagy induction, senses cellular nutritional levels, regulates cell growth and survival and directly inhibits autophagy. MTOR is part of a cellular survival pathway that is the crux of many interconnected cellular pathways and integrates information to modulate cellular growth, metabolism and survival. MTOR is active in nutrient-rich conditions, inhibiting autophagy and protein degradation. In nutrient-poor conditions MTOR is inactive and autophagy is induced by alleviating MTOR phosphorylation of ULK1 and ATG13. When MTOR is inactivated by starvation conditions, dephosphorylated ULK1 dissociates from the MTOR complex and phosphorylates ATG13 and RB1CC1/FIP200 (RB1-inducible coiled-coil 1) to induce the nucleation phase (Fig. 1A).8 Autophagosome nucleation is a highly orchestrated process relying on the PtdIns3K (class III phosphatidylinositol 3-kinase) complex that serves to phosphorylate phosphatidylinositol to phosphatidylinositol 3-phosphate (PtdIns3P) (Fig. 1B). This modified lipid recruits complexes and lipids to expand the autophagosome membrane. The PtdIns3K complex, anchored by the interaction between core members PIK3C3/VPS34 and BECN1, exhibits different functions depending upon the composition of ancillary proteins in the complex. UVRAG (UV irradiation resistance associated) and ATG14 (proposed mammalian homolog of yeast Atg14, or Barkor) are found in BECN1 complexes in a mutually exclusive manner. For example, the complex of ATG14, PIK3R4/VPS15, PIK3C3 and BECN1 positively regulate autophagosome formation at the nucleation step. In contrast, a complex including UVRAG, SH3GLB1/BIF1, PIK3C3, PIK3R4/VPS15 and BECN1 reportedly controls autophagosome maturation and RAB7 (GTPase)-dependent lysosomal fusion.9 Autophagy regulation may be influenced through additional protein–protein interactions to either promote or inhibit autophagy. SH3GLB1 directly associates with UVRAG and is a positive regulator of autophagy; similarly, AMBRA1 (autophagy/Beclin 1 regulator 1) directly binds BECN1 to stimulate autophagy. In contrast, KIAA0226/Rubicon also directly binds BECN1 but acts as a negative regulator of autophagy.10
Figure 1. The core machinery of autophagy. Autophagy is a complex degradation process in which general cytoplasm or organelles are engulfed by a double-membrane bound structure and degraded and recycled following fusion with a lysosome. (A) Dephosphorylated ULK1 dissociates from the MTOR complex and phosphorylates itself, ATG13 and RB1CC1 to induce the nucleation phase. (B) The PtdIns3K complex is assembled at the site of the nascent autophagosomal membrane. UVRAG and ATG14 are found in BECN1 complexes in a mutually exclusive manner. BECN1 is inhibited when bound by anti-apoptotic BCL2, which results in downregulated autophagy. (C) The two ubiquitin-like conjugation systems essential for membrane elongation are outlined schematically. (D) The autophagosomal membrane (orange crescent) is studded with LC3–PE (stylized in black). The membrane elongation is dependent on the ATG12–ATG5-ATG16L1 conjugation system. During fusion with the lysosome (blue oval) LC3–PE associated with the outer membrane is cleaved and recycled by ATG4 while LC3–PE associated with the inner-membrane is degraded by lysosomal proteases along with the cargo of the autophagosome.
Elongation of the phagophore membranes is accomplished by two ubiquitin-like conjugation systems (Fig. 1C). An ATG12–ATG5-ATG16L1 complex associates on the forming membrane. ATG12 is first activated in an ATP-dependent reaction with ATG7, an E1-like enzyme. ATG12 is then conjugated to ATG5 by ATG10, an E2-like enzyme. ATG16L1 then interacts with the ATG12–ATG5 conjugate, forming a dimeric complex. Components of the complex dissociate from the autophagosome and return to the cytoplasm when elongation is complete. The second ubiquitin-like conjugation that contributes to membrane elongation modifies MAP1LC3, the mammalian homolog of yeast protein Atg8. ATG4B, one of the four mammalian ATG4 homologs, cleaves the C-terminal 22 residues of precursor LC3 (proLC3) producing LC3-I. Cytoplasmic LC3-I is then conjugated with phosphatidylethanolamine (PE) by ATG7 and ATG3, an E2-like enzyme. Lipidated LC3 (LC3-II) is selectively incorporated into the forming autophagosomal membrane. LC3-II remains associated with the autophagosome until, or just prior to, fusion with the lysosome, when LC3-II associated with the outer membrane dissociates and LC3-II associated with the inner membrane is degraded by lysosomal proteases along with the autophagosomal cargo (Fig. 1D). This specific association of LC3-II makes it an attractive autophagy marker and will be discussed in further detail in later sections. The autophagosome fuses with a lysosome, forming the autolysosome. Fusion with the lysosome results in breakdown of the inner autophagosomal membrane and cargo by lysosomal proteases and recycling of macromolecules. Soluble NSF attachment protein receptors (SNAREs), RAB7 and the homotypic fusion and vacuole protein sorting (HOPS) complex likely are primary regulators of autophagosome/lysosome fusion.11,12
Sources of Autophagosomal Membranes
The main lipid sources that are recruited in the formation of autophagosomes are thought to emanate from the endoplasmic reticulum (ER), mitochondria, Golgi apparatus or the plasma membrane (PM). Integral membrane proteins normally found in the rough ER have been localized to the inner and outer autophagosome membranes by immunostaining.13 This was also supported by electron microscopy (EM)-3D tomograms that identified points of contact between the rough ER and the nascent phagophore, which indicated that the growing phagophore membrane might be a subdomain of the rough ER.14,15 Moreover, ER regions enriched in PtdIns3P are sites of emerging omegasomes. The ER protein ZFYVE1 can be found in omegasomes extending out from the ER by EM-3D tomography, and the PtdIns3K-complex protein ATG14 is localized at the ER surface.16 Contrary to the ER-based models, Hailey et al.17 have proposed a novel model suggesting that the mitochondrial outer membrane may provide a membrane source for the biogenesis of autophagosomes. Following amino acid starvation, investigators observed by fluorescence microscopy that fluorescently labeled LC3 and ATG5 transiently localize to the outer mitochondrial membrane (OMM) and NBD–PE (7-nitro-2-1,3-benzoxadiazol-4-yl-phosphoserine, loaded into cells as NBD–PS and converted to NBD–PE in the mitochondria), can be detected in autophagosomes. Furthermore, loss of the mitochondrial protein MFN2 causes a dramatic depletion in starvation-induced autophagy. Other recent observations have suggested that cytoplasmic vesicles derived from the Golgi apparatus may act as a membrane source for forming autophagosomes in an ATG9-dependent manner. In yeast overexpressing Atg9, EM confirmed that under starvation conditions, post-Golgi ATG9-positive vesicles are transported to the vacuole and can be seen forming large tubulo-vesicular structures.18,19 Recent reports have described findings indicating that exocytic and endosomal SNARE proteins may mediate fusion of post-Golgi Atg9-positive vesicles and remodeling of tubulo-vesicle clusters in phagophore assembly.20 Exocytic Q/t-SNAREs Sso1/2 and Sec9 were shown to be essential for homotypic fusion of Atg9-positive vesicles in starvation-induced autophagy in yeast. As well, the endosomal Q/t-SNARE Tlg2 and R/v-SNAREs Ykt6 and Sec22 were found to interact with Q/t-SNAREs Sso1/2 and Sec9 and are essential for trafficking of post-Golgi Atg9-positive vesicles to the vacuole and during phagophore assembly. Finally, a number of studies have also indicated that the PM is capable of directly contributing membrane to forming phagophores. An analysis by Ravikumar et al.21 described interaction of the clathrin heavy chain (associated with endocytic vesicles) with ATG16L1 at the extending phagophore and later confirmed that disruption of the interaction results in decreased autophagosome formation.22 Recently, experimental data have also suggested that the action of PM SNAREs in complexes with tethering proteins may be essential for fusion of membranes in the phagophore. In HeLa cells, it was shown that the membrane protein LC3 is sufficient to tether PM-integral PE, potentially facilitating the activity of PM SNAREs.23
Cellular Stress Responses
Although the core machinery for autophagy was initially identified in yeast, seminal experiments defining the induction of autophagy were performed in a mammalian cell, the hepatocyte. These studies defined the importance of hormonal regulation, energy status and nutrient levels as key modulators of autophagy. Initially, the presence of double-membrane organelles was found to be induced in rat hepatocytes exposed to glucagon;24 years later, these organelles were eventually demonstrated to be autophagosomes.25 Conversely, treatment of hepatocytes with insulin gave the opposite result, which invariably defined autophagy as a catabolic, energy-generating mechanism for the cell.26 The dependence of the cell on autophagy for energy homeostasis was confirmed the same year by showing that amino acid supplementation inhibits autophagy in the rat liver.26 A final, important mechanistic link between autophagy and nutrient-energy sensing was established when rapamycin, an inhibitor of the energy sensor MTOR, was found to induce autophagy in hepatocytes.27 Thus, cellular stress became a focal point upon which to understand how autophagy could be regulated.
In addition to nutrient status and hormonal regulation, environmental stressors such as hypoxia,28 heat stress,29 and reactive oxygen species (ROS) accumulation can also induce autophagy (reviewed in ref. 30). Additionally, ER stress is also a potent inducer. The ER is a highly active and tightly regulated organelle responsible for protein folding fidelity, biogenesis of membrane structures, metabolism and a veritable menagerie of cellular processes. If the delicate microenvironment of the ER is disrupted, for instance by accumulation of unfolded (or misfolded) proteins, the unfolded protein response (UPR) is activated. There are three main pathways activated by the UPR, involving ATF6 (activating transcription factor 6), ERN1/IRE1 (endoplasmic reticulum to nucleus signaling 1) and EIF2AK3/PERK (eukaryotic translation initiation factor 2-α kinase 3). All of these proteins are typically bound to an inactivating chaperone molecule, HSPA5/BiP/GRP78, and released/activated in response to unfolded proteins. Autophagy is activated in response to ER stress and UPR by both EIF2AK3 and ATF6 pathways, perhaps in an effort to ameliorate the accumulation and aggregation of misfolded proteins. Interestingly, ERN1 activation inhibits autophagy in some systems and is required for autophagy induction in others; however, more systematic work is needed to define these pathways in mammalian systems.31 Although evidence is supportive of ER stress inhibiting MTOR- and AKT-mediated cell survival pathways, which would alleviate autophagy inhibition independently of the UPR, additional studies are needed to elucidate this process (reviewed in ref. 32).
Autophagy may also be induced as a survival mechanism in response to hypoxic conditions in normal and tumor cells.33 Induction of autophagy can proceed through different pathways depending on the severity of the hypoxia and cell type.34 The best-characterized means of hypoxia-induced autophagy is by activation of the transcription factor HIF1A (hypoxia inducible factor 1, α subunit).35 HIF1A is capable of transcriptional activation of a variety of target genes involved in offsetting the damaging effects of hypoxia including erythropoiesis, angiogenesis and autophagy. The HIF1A-target gene Bnip3 (BCL2/adenovirus E1B 19 kDa interacting protein 3) encodes a putative BH3-only (BCL2 homology domain 3-only) protein that is necessary and sufficient to induce autophagy by competitively binding BCL2 (B-cell CLL/lymphoma 2) and disrupting the BCL2–BECN1 interaction.36,37 Bnip3 has also been identified as a target gene of the transcription factor E2F, which can be activated by inhibition of the RB1 (retinoblastoma 1) protein via severe hypoxia.38 This suggests that BNIP3 can trigger autophagy by HIF1A-dependent and HIF1A-independent mechanisms. Severe hypoxia/anoxia (< 0.1% oxygen) can also induce autophagy by AMP-activated protein kinase (AMPK)-mediated MTOR inhibition as well as protein kinase C activation of MAPK8 (mitogen-activated protein kinase 8) and BECN1.39-41
The ubiquitin-proteasome system (UPS) and autophagy act as the major pathways for cellular catabolism, and were initially thought to function independently of one another. However, new observations suggest that the two degradation pathways function in a highly coordinated manner to maintain cellular homeostasis. The UPS pathway specifically targets soluble proteins in the nucleus or cytoplasm, which are labeled for proteasomal degradation by the addition of the small peptide ubiquitin at various lysine residues. Through the action of three classes of enzymes, E1 ubiquitin-activating, E2 ubiquitin-conjugating and E3 ubiquitin-ligase, the UPS pathway can ensure high levels of specificity in labeling protein targets for degradation. The most well-known limitations of the UPS pathway result from the small size and cylindrical structure of the 26S proteasome to which polyubiquitinated proteins must enter for degradation by a series of peptidases. It appears that proteins may need to be partially denatured and monomeric in order to be degraded within the proteasome, greatly limiting the UPS pathway for clearance of aggregated proteins or large multimeric complexes.42 In contrast, autophagy is limited in its activity to the cytoplasm, where it can efficiently degrade soluble proteins, protein aggregates and organelles by sequestration within autophagosomes followed by fusion with the lysosome. Previously, autophagy was viewed as a nonspecific degradation pathway for cellular recycling; however, it is clear that several soluble proteins and aggregates are selectively degraded in the autolysosome, rather than the proteasome.43-45 In addition, ubiquitinated proteins can be selectively degraded by autophagy.45 Suppression of the UPS pathway by siRNA is offset by an increase in autophagy.46 However, inhibition of autophagy results in inhibited degradation of UPS substrates.47,48 As a result, current models for the autophagy and UPS pathways suggest not only overlapping roles for the two systems, but a more dynamic and coordinated approach than previously described (reviewed in ref. 49).
SQSTM1 (sequestosome 1) is a scaffolding protein with several known functions in various tissues and is the most well-known target of selective autophagy.43 As SQSTM1 is constitutively expressed, it shows a consistent turnover and normally forms aggregates in the cytoplasm, which are selectively degraded by autophagy. These characteristics are easily observed by immunohistochemistry and have been used to monitor successful autophagy in vitro. Tissue-specific ablation of autophagy results in accumulation of SQSTM1 aggregates and proteins carrying polyubiquitinated Lys63 residues.50 The SQSTM1 protein contains both a ubiquitin-binding domain as well as an LC3-interacting domain, which has led to the conclusion that it acts as a central link between autophagy and the UPS pathway. Current models describe SQSTM1 as a cargo receptor for autophagic degradation of various proteins.51,52
Autophagy can also be pharmacologically stimulated or inhibited in a number of ways; each method is accompanied by its own set of advantages and disadvantages (Table 1). Autophagy can be stimulated indirectly through inhibiting the UPS pathway, inducing an ER stress response, reducing intracellular calcium levels, or modifying the acetylproteome. Use of proteasomal inhibitors MG132 or bortezomib in human cancer cell lines induces both the accumulation of the lipidated form of LC3B and the localization of a green fluorescent protein (GFP)-tagged reporter (GFP-LC3) to autophagosomes.53 Tunicamycin, an inhibitor of N-acetylglucosamine phosphotransferase, is used widely in the literature to induce ER stress.54 Tunicamycin treatment in vitro increases autophagosomes and autolysosomes as detected by EM and GFP-LC3 puncta formation, a fluorescent marker of autophagy. Although thapsigargin acts as an ER stressor to inhibit the ATP2A1/SERCA1 ATPase (ATPase, Ca2+ transporting, cardiac muscle, fast twitch 1), it can also act directly by blocking the fusion of autophagosomes with lysosomes by preventing recruitment of RAB7.55 Intracellular calcium can activate CAPN1 [calpain 1, (mu/I) large subunit], a protease that targets ATG5 for degradation, thus inhibiting autophagy.56 Fluspirilene induces autophagy through reducing calcium stores, which leads to a deactivation of CAPN1 and stabilization of the ATG12–ATG5 conjugate. Lastly, resveratrol and spermidine are being studied for their ability to induce autophagy in vivo in order to promote longevity.57,58 The apparent mechanism though which they act is via modification of the acetylproteome, principally through cytoplasmic deacetylation reactions mediated by the sirtuins.59
Table 1. Pharmacological inducers and inhibitors of autophagy.
| Autophagy inducer | Action | Ref. | |
|---|---|---|---|
|
MG132 |
Inhibit 26S proteasome |
53 |
|
|
Bortezomib |
Inhibit 26S proteasome |
53 |
|
|
Tunicamycin |
Induce ER stress by inhibiting N-acetylglucosamine phosphotransferase |
54 |
|
|
Thapsigargin |
Induce ER stress via ATP2A1 inhibition Prevent autophagosome/lysosomal fusion by inhibiting RAB7 recruitment |
55 |
|
|
Fluspirilene |
Stabilize ATG12–ATG5 by preventing Ca2+-mediated CAPN1 activation |
56 |
|
|
Resveratrol |
Sirtuin-mediated deacetylation of cytoplasmic proteins |
57, 59 |
|
|
Spermidine |
Sirtuin-mediated deacetylation of cytoplasmic proteins |
58, 59 |
|
|
Rapamycin |
Inhibit MTOR |
60–62 |
|
|
Lithium chloride |
Increase PtdIns3P levels by inhibition of IMPAD1 |
63 |
|
|
L-690,330 |
Increase PtdIns3P levels by inhibition of IMPAD1 |
64 |
|
|
Carbamazepine |
Increase PtdIns3P levels by inhibition of IMPAD1 |
65,66 |
|
|
Xestospongin B |
Antagonize ITPR1 (and ITPR1-mediated BCL2–BECN1 interaction) |
69–71 |
|
|
Xestospongin C |
Inhibit ITPR1 and ER Ca2+ receptors |
68 |
|
|
Autophagy inhibitor |
Action |
Ref. |
|
|
3-methyladenine |
Inhibit PtdIns3K and PtdIns3K (PIK3CB stably, and PIK3C3 transiently) |
72 |
|
|
Wortmannin |
Inhibit PtdIns3K and PtdIns3K (PIK3CB and PIK3C3) |
73 |
|
|
Bafilomycin A1 |
Inhibit vacuolar ATPase |
75 |
|
| Spautin-1 | Inhibit USP10 and USP13 deubiquitinase activity | 74 | |
Rapamycin is a highly specific inhibitor of MTOR and is commonly used both in vitro and in vivo.60-62 In contrast, lithium chloride is an MTOR-independent inducer of autophagy that inhibits IMPAD1 (inositol monophosphatase domain containing 1), leading to a decrease in inositol availability. IMPAD1 inhibition results in increasing cellular levels of PtdIns3P, thus inducing autophagy.63 L-690,330 also inhibits IMPAD1 and is more potent, but has a reduced ability to permeate the PM and blood-brain barrier.64 Carbamazepine also works by a similar mechanism to lithium, and has been used in vivo to ameliorate proteinopathies in mice either suffering from mutant SERPINA1 (α1-antitrypsin) Z or TAR DNA binding protein (TARDBP) accumulation.65,66 A broader range of pharmacological agents used to induce autophagy in the treatment of proteinopathies has been previously reported.67 The xestospongin family of natural compounds purified from marine sponges, xestospongin B (XeB) and xestospongin C (XeC), are inducers of autophagy. XeC inhibits both the inositol 1,4,5-trisphosphate receptor 1 (ITPR1) and ER Ca2+ receptors indiscriminately, thus altering Ca2+ flux.68 XeB was originally identified as an inhibitor of inositol 1,4,5-trisphosphate-mediated Ca2+ signaling69 and has more recently been shown to act as an antagonist of ITPR1 on the ER membrane. Evidence supports a mechanism wherein ITPR1 facilitates BCL2-mediated sequestration of BECN1 to inhibit autophagy. XeB would rapidly induce autophagy by interfering with these protein–protein interactions, freeing BECN1 in an organelle-specific manner.70 The induction of autophagy by XeB appears to be independent of steady-state Ca2+ levels in the ER or cytoplasm.71
For autophagy inhibition, 3-methyladenine (3-MA) reduces autophagy in nutrient-poor conditions by blocking PtdIns3K and autophagosome formation. However, 3-MA inhibits both PIK3CB/PIK3C1 and PI3CK3 indiscriminantly, which could lead to pleiotropic cellular effects. Interestingly, in complete media, 3-MA is pro-autophagic, leading to an accumulation of autophagic markers and increased conversion of LC3-I to LC3-II. Wu and colleagues were able to show that the increase in autophagic markers is a result of increased autophagic flux rather than an effect of autophagosome accumulation, and seems to be cell-line independent. Therefore, use of 3-MA as an autophagic inhibitor is well supported and substantiated only when used in starvation conditions and at relevant concentrations.72 Another PtdIns3K inhibitor, wortmannin, functions in a similar way but does not show pro-autophagic effects in complete media. This difference is attributed to wortmannin more stably inhibiting PtdIns3K compared with 3-MA, which has a transient effect on PtdIns3K but a stable effect on PIK3CB.73 The second group of inhibitors either targets PtdIns3K stabilization or autophagosome–lysosome interaction. A recently identified inhibitor, spautin-1 (specific and potent autophagy inhibitor-1), blocks the deubiquitinase activities of USP10 and USP13, leading to the UPS-mediated degradation of the kinase component of the PtdIns3K complex.74 Vacuolar ATPase is an enzyme that resides on the lysosomal membrane and regulates lysosomal acidification. Bafilomycin A1 (BafA1) is a vacuolar-type ATPase inhibitor. BafA1 inhibits fusion of autophagosomes with lysosomes and leads to an accumulation of autophagosomes in the cell as well as a reduction of protein degradation, another indication that autophagy is impaired.75 The fusion blockage seems to be an effect secondary to the reduced acidification of the lysosome.76 Conflicting reports in the literature imply that both cell type and length of BafA1 treatment affect the degree of inhibition as well as more general effects. It is important to note here that interpretation of commonly used autophagic activity and flux assays may be altered when using drugs affecting lysosomal acidification. In a short communication, Klionsky et al.,77 discuss the complications that may arise such as reduced LC3-II degradation at early time points and the difficulty in discriminating autophagosomes from autolysosomes in this system due to GFP persistence in the less acidified lysosomes.
Types of Autophagy
Aggrephagy
Aggrephagy refers to the autophagic process of degrading proteins that are assembled into large protein aggregates, which are less toxic to the cell than more numerous small protein aggregates. Although this is seen as a companion system to the UPS to promote protein degradation, there are subtle distinctions as to the intertwining of these pathways. For example, proteins targeted for degradation by this means can be ubiquitinated, but do not necessarily have to be. Histone deacetylase 6 (HDAC6)-mediated dynein transport of proteins along microtubules occurs preferentially with proteins that have Lys63 (K63) linked polyubiquitin chains. Conversely, BAG3-mediated aggresome formation, which also uses dynein transport along microtubules, does not require such protein modifications. The forming aggresome requires K63 linked polyubiquitination to recruit autophagy receptors SQSTM1, NBR1 and WDFY3. These receptors provide a physical link to ATG8, found in the developing phagophore, to ultimately envelope the aggresomes.78
Allophagy, crinophagy and zymophagy
Allophagy refers to the autophagic degradation of paternally-derived mitochondria upon fertilization in the zygote. As such, this mechanism is a developmental-specific form of mitophagy. Sperm mitochondria, located in the mid-piece region, are initially tagged with K63-linked ubiquitin prior to fertilization. This labeling increases immediately after fertilization, presumably to insure their quick and successful degradation prior to fusion of the male and female pronuclei.79 Historically, crinophagy refers to the process by which secretory granules containing hormones are directly routed to lysosomes without contribution from autophagy. However, a parallel mechanism involving encapsulating these granules within autophagosomes does occur, as demonstrated within Paneth cells and pancreatic β-cells.80,81 This provides a turnover mechanism to regulate appropriate granule numbers in these cells. Autophagy is also responsible for the regulation of neurotransmitter vesicle levels, as dopamine responses are substantially increased in the dorsal striatum of Atg7-deficient mice.82 Conversely, induction of autophagy through rapamycin administration attenuates dopamine responses. Although it has not been demonstrated, these processes are most likely ubiquitin-dependent. For example, the closely related process of zymophagy allows for the autophagic degradation of activated zymogen granules,83 which is dependent on VMP1 (vacuole membrane protein 1), SQSTM1 and the ubiquitin protease USP9X.84 This mechanism prevents acute pancreatitis from occurring by removing potentially harmful activated zymogen granules.
Exophagy
Autophagy is also associated with nondegradative processes involved in protein secretion known as exophagy. Deretic et al.85 review the roles of autophagy in conventional (regulated and constitutive) and unconventional secretion. Conventional secretion pathways normally route through the Golgi complex or occasionally directly from the ER; however, the TOR-autophagy spatial coupling compartment86 is a newly identified region that is responsible for constitutive secretion of IL6 and IL8. Regulated secretion examples include lysozyme release by Paneth cells and CTSK/cathepsin K by osteoclasts. Autophagy-based unconventional secretion, or autosecretion, involves omegasome formation at the ER to secrete proinflammatory factors IL1B and HMGB1 in mammalian cells.
Heterophagy and endosomal microautophagy
Heterophagy is distinguished from autophagy in the sense that it is a process devoted to degrade extracellular material that has been internalized within the cell, in contrast to the degradation of pre-existing intracellular material. Upon endocytosis, proteins are routed into early endosomes and late endosomes/multivesicular bodies (MVBs) for fusion with lysosomes. However, there is synergy with the endosomal system and autophagy, as early endosomes and MVBs can both fuse with autophagosomes to form amphisomes, which in turn fuse with lysosomes. These amphisomes carry the protein markers early endosome antigen 1 (EEA1) and mannose-6-phosphate receptor (cation dependent; M6PR), present in early and late endosomes, respectively.87 These fusion events are driven by GTPases, as MVB-autophagosome fusion is RAB11-dependent, and amphisome-lysosome fusion is RAB7-dependent.88 In contrast to internalized materials, cytosolic proteins can be routed into MVBs through endosomal microautophagy that is either HSPA8-mediated or through a nonspecific mechanism. This process occurs during MVB formation and requires the ESCRT I and ESCRT III protein machinery.89
Immunophagy
More broadly, autophagy plays a larger role in both innate and adaptive immunity in a process termed immunophagy.90 As recently reviewed, immunophagy is subdivided into three types.91,92 Type I immunophagy involves the processing of foreign or endogenous immunologically active molecules. This would include xenophagy, the autophagic activation of macrophages, pattern recognition receptor activation, MHC class II endogenous antigen presentation, and thymic selection. Type II immunophagy regulates cell viability and immune cell function. Specific roles for this type include: T/B cell homeostasis, T cell maturation and Paneth cell maintenance. Type III immunophagy utilizes specific ATG proteins but does not require the entire process of autophagy to occur. Examples in this class include inhibition of RIG-I-like receptor signaling by ATG12–ATG5, and the negative regulation of TBK1 signaling for type I interferon secretion by ATG9.
Lipophagy
Lipophagy involves the metabolic regulation of lipids through degradation of lipid droplets (LDs) by autophagy. Ultimately, fusion with lysosomes contributes to lipolysis of LDs, or the breakdown of triglycerides into free fatty acids. In addition to lipophagy, cytosolic lipases generate free fatty acids from lipolysis in an autophagy-independent fashion. Although these cytosolic lipases are well characterized, the autophagy proteins involved in the detection and mobilization of LDs are not known. This field has recently expanded due to the pathologies associated with autophagy-deficient mice. Liver-specific ablation of Atg7 results in LD accumulation known as liver steatosis (fatty liver).93 Similar studies have shown that lipophagy is a sensor for appetite regulation in hypothalamic cells,94 whereas reduced lipolysis in macrophages causes their premature conversion into foam cells, which promotes atherosclerosis.95
Mitophagy
Mitophagy is the selective degradation of mitochondria through autophagy, although the process may be cell specific within mammals. For example, reticulocytes lose their mitochondria as they mature into red blood cells, an interaction-dependent process driven by BNIP3L (BCl2/adenovirus E1B 19 kDa interacting protein 3-like) located on mitochondria, and LC3 found on phagophores.96 Within other cells, a PINK1 (PTEN-induced putative kinase 1)-PARK2/Parkin system seems to regulate mitophagy.97,98 PINK1 localization at the OMM of damaged mitochondria recruits the E3 ubiquitin ligase PARK2 to cause K63-ubiquitination of three known mammalian proteins: mitofusin1, mitofusin2 and voltage-dependent anion selective channel protein 1 (VDAC1). Autophagy can subsequently be directed by SQSTM1- and HDAC6-dependent mechanisms. Given the enrichment of PARK2 within skeletal muscle, brain, heart and liver, it is possible that other tissues may use distinct E3 ubiquitin ligases for selective removal of mitochondria. A recent genomic mammalian screen revealed 96 proteins required for PARK2-mediated mitophagy, underscoring the complexity and uncertainty of this mechanism.99
Nucleophagy
Although yeast undergo piecemeal microautophagy of the nucleus, where portions of the yeast nuclear membrane and nucleoplasm are invaginated into a vacuole for degradation,100 mammalian cells can exhibit complete encapsulation of the nucleus known as nucleophagy. This was initially demonstrated in murine models that exhibit nuclear envelopathies from mutations in nuclear membrane-associated proteins such as LMNA/lamin A and EMD/emerin.101 Murine embryonic fibroblasts (MEFs) isolated from these mutant mice demonstrate decreased cell viability and increased nuclear abnormalities when pharmacological inhibitors of autophagy are used. Wild-type MEFs also exhibit signs of nucleophagy, although at lower levels than mutant MEFs, suggesting a control mechanism when nuclear damage occurs. This notion is strengthened by findings that anticancer drugs that elicit DNA damage can trigger nucleophagy.102 Topoisomerase inhibitors (e.g., camptothecin, etoposide), DNA intercalating agents (e.g., cisplatin) and oxidative stress damage [e.g., vanadyl (IV)] can induce nucleophagy in cancer cells. It is possible that basal piecemeal nucleophagy occurs to maintain nuclear architecture, energy production and nucleotide stores for DNA repair enzymes.
Pexophagy
Pexophagy, the selective degradation of peroxisomes through autophagy, is probably the most utilized of the three known mechanisms, which also includes LON protease-mediated and 15-LOX-mediated turnover, to eliminate damaged or superfluous peroxisomes. Analysis of liver-specific Atg7-deficient mice showed pexophagy contributes to about 70–80% of the turnover, compared with the remainder that is linked to both a LON protease-mediated mechanism and 15-LOX-mediated autolysis.103 The only known mammalian peroxisome receptor involved in pexophagy is PEX14, which can associate with LC3 on the phagophore membrane.104 An LC3-RAB7-FYCO1-kinesin complex is responsible for the transport of engulfed peroxisomes along microtubules to the lysosome. Although ubiquitination of a distinct membrane protein of the peroxisome allows for SQSTM1-mediated autophagy to occur, the identity of this protein is currently not known.105
Reticulophagy and ribophagy
Degradation of the ER through reticulophagy occurs in response to ER stress, and is seen as an additional stress coping mechanism like the UPR and ER-associated degradation.106,107 The trigger for all three mechanisms lies in the accumulation of unfolded protein aggregates within the ER lumen. Activation of the resident ER membrane protein EIF2AK3 occurs when chaperones dissociate from the luminal side of EIF2AK3 to assist in protein folding. This causes dimerization of EIF2AK3, phosphorylation of EIF2A, and activation of ATG12, thus triggering the autophagic response via ATG12–ATG5-ATG16L1 complex formation. Ribophagy, the selective elimination of free ribosomes in the cytosol, is also linked to the ER stress response similar to reticulophagy. ER stress leads to reduced translation levels, via phosphorylated EIF2A, to avoid the additional burdening of chaperone recruitment to nascent peptides.108 In modeling neurodegenerative disorders in Purkinje cells, polyribosomes are disassembled into nontranslational monosomes, which become associated with autophagosomes.109 However, it is not known which ATG proteins mediate the recognition and sequestration of ribosomes and ER fragments into autophagosomes.
Xenophagy
Viruses, bacteria and parasites can be eliminated in an autophagic process involved in innate immunity defense termed xenophagy, which has been previously reviewed.92,110 Invading bacteria can generally be classified as vacuolar (e.g., Salmonella) or cytosolic (e.g., Listeria, Shigella). Cytosolic bacteria can undergo ubiquitin-dependent and ubiquitin-independent mechanisms for autophagosomal envelopment followed by translocation to lysosomes. Vacuolar bacteria can be routed into autophagosomes, or in the instance of Mycobacteria, autophagy proteins can resume the maturation of the vacuole and promote fusion with the lysosome.111 The main recognition receptors that link detection and autophagy induction include the membrane TLRs (toll-like receptors) and the cytoplasmic nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs). The receptors can recognize the lipopolysaccharides and peptidylglycans of gram-negative bacteria. Microbial interference with autophagy can occur due to the adaptive nature of bacteria. For example, Shigella flexneri secretes the protein lcsB, which prevents ATG5-induced autophagy at the bacterial surface.112 Yersinia pseudotuberculosis resides within arrested autophagosomes in macrophages, since it can inhibit the fusion process with lysosomes.113
Transcriptional Regulation
A multitude of studies have focused on discovering the transcription factors responsible for controlling ATG genes. Thus far these studies have culminated in nine transcription factors that orchestrate the expression of autophagy-related genes in mammals. We have learned that autophagy is under the regulatory control of circadian, metabolic, inflammatory and cell death factors.
Circadian Regulation
As might be expected, autophagy genes are transcribed with a circadian rhythm under control of the biological clock due to metabolic demands. A circadian rhythm refers to the oscillation of a biochemical process (e.g., transcription, translation, phosphorylation, etc.) that goes through a cycle roughly every 24 h. The circadian clock is a transcription/translation feedback loop between the positive transcription factors CLOCK and ARNTL/BMAL1 and the negative transcription factors PER1 and PER2. These signals originate in two clusters of hypothalamic neurons of the suprachaismatic nucleus (SCN), which is reset by a light-dark cycle. The oscillation in the SCN is considered the “master pacemaker” and is responsible for coordinating “subordinate pacemakers” throughout an organism, giving rise to circadian rhythms in every tissue. Autophagy is under circadian control likely because metabolism demands change throughout the day, an idea recently reviewed.114 While only ~10% of transcripts are globally rhythmically expressed,115-118 research using mice has shown that the mRNA expression of many autophagy genes such as Becn1, and Map1lc3b are rhythmic in the distal colon,119 liver, kidney, heart and skeletal muscle.120 In addition, the conversion of LC3-I to LC3-II is rhythmic in the liver of mice.120 The expression of the transcription factor CEBPB/CEBPβ, known to be controlled by the clock, was found to coordinate autophagy gene rhythms by directly binding to the promoters of autophagy genes, such as, Bnip3, cathepsin L (Ctsl) and GABA(A) receptor-associated protein (Gabarap). Additionally, CEBPB has been identified as the transcription factor responsible for rhythmic autophagy gene expression in the mouse liver.120 Furthermore, this rhythm is abolished in Arntl knockout livers, proving that autophagy rhythms from CEBPB are dependent on the clock and thus, are circadian.120 It remains untested if CEBPB or another component of the clock controls autophagy rhythmicity in other tissues such as the distal colon, but logic would dictate this to be the case. While there have not been any direct links to disease caused by interrupting the link between autophagy and the circadian clock, several lines of research suggest this link exists. Metabolic disorders such as diabetes are similarly exacerbated independently by both autophagy and CLOCK deficiencies.121,122 It is sure to be found that the link between autophagy and the biological clock is stronger than we currently understand.
FOXO Family
The FOXO (forkhead box, class O) family is comprised of four proteins in mammals: FOXO1, FOXO3, FOXO4 and FOXO6. FOXO proteins are transcription factors involved in metabolism, longevity, oxidative-stress resistance, apoptosis and autophagy. Once FOXO proteins are activated in the cytosol, FOXO proteins are shuttled into the nucleus where they bind to promoters to initiate transcription. Their role in controlling autophagy gene transcription comes from studies on skeletal muscle,123-125 neurons,126 ischemic insults on the heart127 and human cancer cell lines.128 Upon starvation or denervation of skeletal or heart muscles, both autophagy and FOXO activity are increased, suggesting a transcriptional control link. These studies have shown that FOXO1 and FOXO3 can induce the expression of Gabarapl1 [GABA(A) receptor-associated protein like 1), Lc3b, Atg12l, Atg4b, Pik3c3, Ulk2, Becn1, Bnip3 and Bnip3l. Further analysis by ChIP has found FOXO proteins bound to the consensus sequence (T/C/G) (G/A/T)AAA(C/A)A found in the promoters of several autophagy genes including Becn1, Gabarapl1, Lc3b, Atg12 and Bnip3.127 The activation of autophagy by FOXO is one major arm of autophagy activation as FOXO is independent of MTORC1 signaling. FOXO1 can be activated in neurons through loss of Mapk8.126 Studying mapk8−/− mouse brains and neurons it is clear that MAPK8 is a negative regulator of autophagy by inhibiting FOXO1. mapk8−/− neurons have increased levels of dephosphorylated, nuclear-localized, activated FOXO1 and greater autophagic flux. In cardiomyocytes, sirtuin 1 (SIRT1) and nuclear protein, transcription regulator, 1 (NUPR1 also known as p8) play crucial roles in regulating autophagy through modification of FOXO1 and FOXO3.129,130 SIRT1 regulates the activation of autophagy by directly deacetylating and activating FOXO1 during glucose deprivation (GD).131 Knockdown of either Sirt1 or Foxo1 leads to no increase in expression of Atg genes and failure of autophagy induction during GD.130 Utilizing a mutated FOXO1 (3A/LXXAA), which cannot be recognized by SIRT1, leads to increased acetylation of FOXO1 and failure to induce autophagy after GD.130 Additionally, Foxo1 conditional KO or Foxo1 (3A/LXXAA) overexpression in cardiomyocytes of mice significantly deteriorates cardiac function after GD, and is proposed to be a direct cause of the significant decrease in autophagic flux. Knockdown of Nupr1, a transcriptional corepressor of Foxo3, leads to activation and nuclear localization of FOXO3 where it associates more with target gene promoters such as Bnip3, which in turn have increased mRNA and protein expression. Subsequently, Nupr1 knockout mice have increased Bnip3 expression and increased autophagic flux in cardiac tissue, likely due to the increased FOXO3 activity.129 In primary skeletal muscle myotubules, AMPK is a potent activator of autophagy through both activation of FOXO3 and inhibition of MTORC1.125 While more regulators of FOXO proteins are likely to be found, this research shows a clear link between FOXO activation and autophagy induction, through the increased expression of Atg genes in several cell types.
TRP53
Transformation-related protein 53/tumor protein p53 (TRP53/TP53/p53) is a tumor-suppressor protein with well-known roles in regulating both cell cycle progression and the cell survival/death axis. Recently, it has also been shown to provide both positive and negative regulation of autophagy. In normal cells, TRP53 remains in the cytoplasm at low levels, which are maintained by the E3 ubiquitin ligase MDM2 and the UPS. Upon activation by post-translational modifications, TRP53 translocates to the nucleus where it functions to transactivate a number of target genes involved in cell survival, death and cell cycle regulation. Initial observations that TRP53 inhibition results in increased autophagy were complicated by the discovery that nuclear TRP53 is able to induce autophagy by transactivation of specific target genes.132 The current model for TRP53 regulation of autophagy describes distinct, independent roles for cytoplasmic TRP53 and nuclear TRP53. Analysis in vitro in a variety of cell types and in vivo in various organisms has shown that cytoplasmic TRP53 functions as a repressor of autophagy. Trp53 knockout (in enucleated cells) as well as inhibition or inactivation of cytoplasmic TRP53 results in increased levels of autophagy, similar to that of cells following induction by starvation or treatment with rapamycin.133 As well, induction of autophagy by starvation or rapamycin results in rapid depletion of cytoplasmic TRP53 by UPS degradation. Mutant TRP53, lacking a ubiquitination site, strongly inhibits the induction of autophagy in vitro.133,134 In contrast, nuclear TRP53 transactivates several genes known to stimulate autophagy following genotoxic stress. The TRP53 target gene, Dram1, encodes a highly conserved lysosomal protein whose function is unclear, but strongly induces autophagy in a variety of cell types.132 Other nuclear TRP53 transactivation targets include Sesn2, Tsc2 and genes encoding AMPK α/β subunits, which function together to stimulate autophagy during nutrient deprivation. AMPK can activate the TSC1–TSC2 complex, which functions as a potent and sensitive inhibitor of MTOR, thereby derepressing autophagy. The AMPK activator SESN2 functions to amplify the TRP53-directed autophagic response.135,136
While it is widely accepted that TRP53 levels in normal cells are maintained by MDM2 ubiquitination and proteasomal degradation,137 it is also well demonstrated that tumor cells commonly carry various TRP53 mutants capable of escaping the regulation by MDM2 and proteolytic degradation.138-140 Accumulation of mutant TRP53/TP53 in tumor cells is associated with metastases and resistance to chemotherapeutic agents. It has also previously been shown that various post-translational modifications can stabilize and destabilize cytoplasmic TRP53.141 Recent analyses have shown that glucose restriction of tumor cells results in deacetylation and subsequent autophagy-dependent degradation of mutant TRP53, followed by apoptotic cell death. Inhibition of autophagy prevents degradation of the mutant TRP53 and promotes tumor cell survival during glucose restriction.142 This finding presents an interesting link in regulation of TRP53 and the cell death-survival axis by selective autophagy because cytoplasmic TRP53 has classically been characterized as a negative regulator of autophagy.
RB1–E2F1
The Rb1–E2F pathway is a well-characterized regulator of the G1/S cell cycle transition. Under normal conditions, RB1 directly inhibits E2F so that the cell remains in G1. When RB1 is phosphorylated by CCND1−CDK4, RB1 dissociates from E2F proteins, which allows them to bind to promoters of several genes including Ccne1 and virtually all initiators of the pre-replication complex necessary for entrance into S-phase. As this pathway is the major pathway augmented to increase proliferation in many cancer models, Rb1 and E2F have become targets of many therapies. E2F1, a member of the E2F family of eukaryotic transcription factors, is unique in that when DNA damage is sensed E2F1 can directly promote apoptosis through expression of TRP73 and indirectly through TRP53 accumulation by expression of CDKN2A/p19arf (reviewed in ref. 143). Evidence for how RB1-E2F regulates autophagy is confusing, as both RB1 and E2F1 can induce autophagy separately and often in an inverse relationship to each other. E2F1 indirectly increases the expression of autophagy-related genes after E2F activation in U2-OS cells.144 A comprehensive analysis of Atg gene promoters in the presence of E2F1 reveals that E2F1 promotes the expression of Ulk2, Atg4b, Atg4d, Atg7, Gabarap2, Lc3a, Lc3b, Atg9, Atg10, Atg12 and Dram1.145 E2F1 overexpression in SK-MEL2 and MEFs induces autophagic flux by LC3-II conversion and GFP-LC3, irrespective of a functional transactivation domain.146 While the E2F2 transactivation domain contains the RB1 binding site, autophagy induction cannot be linked to E2F1-RB1 interaction since the expression of RB1 compared with E2F1 was not investigated. These data also do not address whether E2F1tr (deletion of transactivation domain) can still induce expression of Atg genes. As might be expected, removal of RB1 a negative regulator of E2F1 increases autophagic flux and expression of Atg genes in myoblasts.147 Together these data suggest that regulation of autophagy by RB1-E2F is dependent on active E2F to promote transcription. However, when Rb1 is overexpressed in several human cancer cell lines (U2-O2, Saos-2, Hep3b and U-87MG) autophagic flux is dramatically increased by detection through LC3-II conversion, GFP-LC3 localization and transmission EM.148 Furthermore, when mutant RB1 that cannot bind to E2F family members is overexpressed in these same cell lines, autophagy is not induced, which suggests that the repression of E2F by RB1 is necessary for autophagy induction. To corroborate these findings, knockdown of E2F1 in the same cells increases autophagy, suggesting that the regulation of autophagy by RB1-E2F is dependent on RB1 inhibiting E2F. To best determine if the control of autophagy by RB1-E2F is physiological, more studies conducted in noncancer cell lines are needed since discrepancies could be due to the models used. Either way, RB1-E2F is clearly a transcriptional regulator of Atg genes and could be involved in post-transcriptional regulation.
TFEB
The transcription factor EB (TFEB) is a well-characterized master regulator of lysosome biogenesis. TFEB directly promotes the transcription of several lysosomal-specific genes. Recent work in several labs has shown that when TFEB is phosphorylated, it remains inactive at the lysosome membrane and does not translocate to the nucleus. Along these lines, it has been elucidated that MTORC1 at the lysosomal membrane is responsible for the inhibiting phosphorylation at Ser142 of TFEB.149-151 Furthermore, the inhibiting activity of MTORC1 is blocked during nutrient starvation, which is strikingly similar to MTORC1’s inhibition of ULK1. When TFEB is overexpressed in HeLa cells, Atg genes UVRAG, WIPI1, LC3B, SQSTM1, VPS11, VPS18 and ATG9B are upregulated along with lysosomal genes.152 Analysis of these same cells showed that autophagic flux is increased as well, indicating a parallel pathway to induce autophagic flux due to starvation. It is interesting that TFEB, a lysosomal master switch, is implicated in the induction of Atg genes post-starvation and that it is regulated through the starvation sensor MTORC1.
Autophagy-Apoptosis Crosstalk
BECN1–BCL2/BCL2L1 interaction
Classically, crosstalk between autophagy and apoptosis has focused primarily on the interaction of the pro-autophagic protein BECN1 and the anti-apoptotic protein BCL2.153,154 While the mechanisms behind the regulation of autophagy remain elusive and highly debated, the most widely accepted theory describes BCL2 as a BECN1 antagonist, through direct interaction with the BH3 domain found on BECN1, preventing activation of the PtdIns3K complex and induction of autophagy.155-157 Further analyses have identified additional components of regulation in that only ER-localized BCL2 or BCL2L1 (BCL2-like 1/BCLX) in complex with the ITPR1 complex and the NAF1 protein can sufficiently inhibit the induction of autophagy under starvation conditions.158 It has been stated that BCL2 and BCL2L1 in complex with BECN1 retain their anti-apoptotic capabilities; however, these data can be misleading as the interaction with BECN1 is dependent upon ER localization of BCL2 and BCL2L1 while the regulation of intrinsic apoptosis occurs primarily at the mitochondria.159 This may suggest multiple independent roles of the BCL2 and BCL2L1 proteins, dependent upon subcellular localization; however, it is unknown what drives the specific distribution of ER- and mitochondrial-BCL2/BCL2L1 proteins and whether or not redistribution may occur under starvation conditions as a functional link between apoptosis and autophagy. Recent data suggest that BCL2 may indirectly regulate BECN1 by interaction with AMBRA1, a positive regulator of BECN1 and autophagy. BCL2 interaction with AMBRA1 is disrupted upon autophagic stimuli, at which point AMBRA1 competes with ER-localized BCL2 for interaction with BECN1. Interestingly, AMBRA1 is preferentially bound by the mitochondrial fraction of cellular BCL2, rather than ER-localized BCL2, further linking the regulation of apoptosis and autophagy.160
BECN1–BCL2L11 interaction
In contrast to the previous model, a novel interaction has been observed that suggests that the pro-apoptotic protein BCL2L11 (also known as BIM) can inhibit autophagy by recruitment of BECN1. BCL2L11 is a BH3-only BCL2 family protein that has been previously shown to function in an active, phosphorylated state or an inactive, dephosphorylated state. Phosphorylated BCL2L11 induces mitochondrial pore formation and apoptosis by activating pro-apoptotic BAX-BAK1 proteins, while the dephosphorylated BCL2L11 is found in a complex with the dynein, light chain, LC8-type 1 protein (DYNLL1). Luo et al.161 recently described the complex formed by BCL2L11 bridging the BECN1–DYNLL1 interaction, which appears to sequester and inhibit BECN1 at the dynein motor complex. Upon knockdown or knockout of Bcl2l11, cells in culture exhibit an increase in autophagosome formation, which is reversible by overexpression of Bcl2l11 (in mutants unable to induce apoptosis). While this interaction appears sufficient to reduce levels of autophagy in nutrient-rich conditions, starvation-induced phosphorylation of BCL2L11 eliminates its interaction with BECN1, suggesting that inactive BCL2L11 may act to repress BECN1 until autophagy is induced by nutrient deprivation. This model also proposes an interesting link between autophagy and apoptosis in that BCL2L11 phosphorylation potentially enables BECN1-dependent autophagosome formation and also activates pro-apoptotic BAX-BAK1 proteins, inducing mitochondrial apoptosis.
Extrinsic apoptosis and autophagosomes
While the models mentioned above primarily involve interactions between BECN1 and the BCL2 family proteins, a recent study has described autophagosomes as potential platforms for formation of the death-inducing signaling complex (DISC) for the activation of caspase 8 (CASP8). Classically, the DISC forms near the PM off of adaptor proteins bound to the cytoplasmic domain of various death receptors, activated by extracellular apoptotic signaling. CASP8 is recruited to the DISC as a monomer, which then self-associates and cross activates by cleavage of the prodomain. Active CASP8 cleaves and activates caspase 3 (CASP3) triggering the apoptotic cascade. It has been previously reported that CASP8 requires SQSTM1 for efficient self-association; however, Young et al. have described complex formation between ATG5 and CASP8, as well as ATG5 and the adaptor protein FADD at the autophagosomal membrane. Atg5 knockdown is associated with reduced CASP8 activation upon induction of apoptosis.162 While other amplification mechanisms have been described in the apoptotic machinery, this appears to be the first that is dependent upon autophagy/autophagosome formation.
Transgenic Models for Autophagy Detection
GFP-LC3
The best-characterized and most widely used detection model is the GFP-LC3 transgenic mouse generated by Mizushima and colleagues. This robustly expressing transgenic mouse, in which LC3 is driven by a constitutive CAG promoter, displays punctate GFP fluorescence that corresponds to LC3-positive phagophores and autophagosomes.163 With this transgenic model quantification of autophagosomes and phagophores is feasible using a high-resolution fluorescence microscope. This reporter line has been crossed into many of the knockout and floxed autophagy models generated in the field. For example, atg5−/− mice are autophagy deficient and atg5−/−; GFP-LC3 mice do not exhibit the punctate fluorescence indicative of autophagosome formation.164 (Protocols for use are widely available and published references are helpful; for detailed information see ref. 165). This model is limited in that only phagophore and autophagosome number, not autophagic flux can be evaluated. In basal conditions, lysosomal degradation clears the autophagosome and contents from the cell, to maintain a “balance” of autophagosome formation and degradation. An accumulation of LC3-positive structures may represent either an increase in formation or a decrease in fusion events. Ferreting out these differences is relevant for proper data interpretation, especially when using chemical autophagy inhibitors and inducers. Measuring autophagic flux in vivo has been problematic to date and the field is in need of an appropriate reporter model; currently, tandem fluorescent-tagged autophagy proteins are a valuable in vitro tool.166 Due to its chemical nature, GFP is quenched by the low pH of the autolysosome; however, red fluorescent proteins are more pH stable, and thus will retain their fluorescence in the lower pH of the autolysosome. The need for better detection mechanisms with regard to cardiac autophagy has led to the generation of a double-transgenic reporter. A cardiomyocyte-specific myosin, heavy chain 6, cardiac muscle, α (MYH6) promoter was used to drive expression of a mCherry-LC3 construct. These mice were crossed with the GFP-LC3 model previously described to produce a double label, which allows for the detection and evaluation of autophagic flux.164 GFP-LC3 will still function for visualization of LC3-positive phagophores and autophagosomes, while mCherry-LC3 puncta will mark phagophores, autophagosomes and autolysosomes. Any double-labeled puncta are indicative of phagophores or autophagosomes, while red-only puncta correspond to autolysosomes. The comparison of double-labeled structures to red fluorescent-only labeled structures is informative to distinguish an increase in formation and a decrease of fusion events. Although this model is cardiac-specific, a similar strategy could be used to target other tissues or to generate a global transgenic model.
GFP-GABARAP
GFP-GABARAP transgenic mice were originally generated to address the question of the role of GABARAP in podocytes. Since GABARAP was reported to be highly expressed in podocytes, a pCAG-GFP-GABARAP transgenic mouse was produced in order to examine subcellular localization in this specialized cell type.167 The expression level of GFP-GABARAP is low, yet visible, ameliorating many of the potential effects of highly expressing fluorescent proteins. In podocytes, GFP-GABARAP merges with SQSTM1 aggregates, but not LC3-II. Although it was shown that GABARAP is not the preferred Atg8 ortholog for conjugation in podocytes this is a valuable reporter model for use across the field as differences in Atg8 orthologs are ferreted out.
Autophagy and Development
Since the seminal genetic studies conducted in yeast, there has been an explosion in global and tissue-specific mouse knockout models produced to ascertain the role of autophagy in mammalian development and disease. Although this section stresses development over disease, the pathologies that arise from induced mutations frequently lead to diseased states. More commonly, mouse models are generated to profile and to uncover treatment regimens to address human diseases. The models that we present are segregated into one of four distinct complexes that the encoded protein primarily functions in rather than by resultant phenotype, these being the: (1) ULK1 complex, (2) PtdIns3K complex, (3) ATG9 complex and (4) ATG12 conjugation and LC3-lipidation system. Lastly, we include a section on induced mutations that affect autophagy more indirectly beyond these complexes.
ULK1 complex
ULK1 is the namesake member of the ULK kinase complex, and was initially identified as the mammalian homolog of yeast Atg1. ULK1 kinase complex functions early in the autophagy pathway, during the induction phase, and consists of ULK1, ATG13, RB1CC1 and C12orf44/ATG101. ULK1 and ATG13 are phosphorylated by activated MTORC1 in nutrient-rich conditions; however, during stress, MTORC1 is inactivated and the ULK1 complex is liberated to induce autophagy. In this sense, ULK1 is a direct link to nutrient sensing in the cell. Contrary to expectations based on other autophagy component knockouts, the ULK1 conventional knockout mice are viable and have no overt developmental defects. It is noted by the authors that Ulk1 knockout mice exhibit LC3 conversion at the same rate as wild-type controls, indicating that starvation-induced autophagy is not impaired in these mutants. Under closer inspection Ulk1 knockout mice exhibit specific red blood cell lineage populations retaining mitochondria and ribosomes, as well as delayed mitochondria elimination in reticulocytes and increased reticulocyte numbers.168 It is evident from these initial studies that ULK1 is important for organelle clearance during erythrocyte maturation but not necessarily for starvation-induced autophagy. Ulk2 knockout mice, like Ulk1 knockouts, are phenotypically normal and viable, which is not surprising since there is most likely some degree of functional redundancy in the mammalian system.169 To investigate this possibility, the authors generated Ulk1 Ulk2 double-knockout mice, which die shortly after birth. This result suggests that elimination of ULK function yields a similar phenotype to the other autophagy deficient models. ulk1−/−ulk2−/− MEFs are specifically impaired in response to amino acid deprivation, though they have a similar response to glucose deprivation as wild-type MEFs.169
RB1CC1, the mammalian functional counterpart of yeast Atg17, is a direct interacting partner of ULK1 and a member of the ULK kinase induction phase complex.170,171 Rb1cc1 knockout mice die in utero at approximately E14.5-E15.5, with the lethality attributed to massive liver and cardiac cell death from increased levels of apoptosis. At E14.5, Rb1cc1 knockout embryos exhibit ventricular abnormalities: the left ventricular wall is lacking in trabeculation, contains fewer cells, and is thinner than control littermates.172 Autophagy may also have an impact on the differentiation of hematopoietic stem cells (HSCs) as revealed in the conditional knockout of Rb1cc1. Tek/Tie2-Cre; Rb1cc1fl/fl mice die perinatally from severe erythroblastic anemia and have 6-fold fewer HSCs than control mice. There is not a proliferation defect, as apoptosis rates are similar between groups and proliferation is actually slightly higher in the conditional knockout (35% compared with 25%). However, a 4-fold increase in myeloid cells is found in the livers of CKO fetuses at E14.5, possibly explaining the depletion of fetal HSCs.173,174 Eight proteins have been identified as binding partners of RB1CC1, including TSC1, a tumor suppressor gene, which is also a negative regulator of MTOR. This interaction mediates cell size and growth. As a compliment to the Rb1cc1 KO studies, Tsc1 knockout embryos also show defects in the heart and liver; however, the heart defects consist of thickened, rather than thinned, ventricular walls.175
As part of the ULK1 kinase complex ATG13 functions during the induction phase. ATG13, the mammalian homolog of yeast Atg13, is directly phosphorylated by both MTORC1 and ULK1/2 and has been characterized as an adaptor protein. At this time there is not an Atg13 knockout mouse, though intriguing in vitro studies indicate that ATG13 is essential for autophagy induction. Interestingly, when researchers simultaneously knock out both Ulk1 and Ulk2, RB1CC1 and ATG13 are able to induce autophagy, suggesting that ATG13 and RB1CC1 are functioning independently of MTORC1 input to ULK1/2.176 This higher order autophagy regulation network is deserving of further attention.
PtdIns3K complex
AMBRA1 promotes the positive association of PtdIns3K and BECN1 to form the core multi-protein autophagy induction complex. Cecconi and colleagues initially reported that Ambra1 knockout mice, generated by using a gene trap strategy, are nonviable.177 The majority of Ambra1 null embryos exhibit severe neural tube defects and spina bifida at E10 through E14.5. Additionally, a myriad of autophagy-deficient effects are seen including cell cycle abnormalities, accumulation of ubiquitinated proteins and increased apoptotic cell death. AMBRA1 is expressed specifically in the developing nervous system and appears to serve a role in controlling neuron survival. Like the Becn1 knockout model, the phenotype of the Ambra1 knockout is particularly severe and strongly affects the developing embryo. Interestingly, new evidence suggests that AMBRA1, like its binding partner BECN1, is a regulator of the autophagy-apoptosis crosstalk. Pagliarini et al. report that AMBRA1 is selectively and irreversibly degraded by caspases and calpains.178 This would help shift the cell from a survival program into an apoptotic program by destabilizing the autophagy promotion complex (BECN1-AMBRA1-PIK3C3). In support of this hypothesis, a noncleavable mutant of Ambra1 introduced into a 2F cell line shows a delay and partial prevention of apoptosis by extending the pro-survival effect of activated autophagy.177 It has been widely reported in the literature that PARK2, an E3 ubiquitin ligase involved in the pathogenicity of Parkinson disease, translocates from the cytosol preferentially to depolarized mitochondria and prompts their selective degradation via mitophagy.179 Emerging evidence shows that AMBRA1 directly interacts with PARK2, although it is not a target for ubiquination. Additionally, Ambra1 overexpression leads to an increase of mitophagy but only in the presence of PARK2. These findings suggest a novel mechanism whereby recruitment of AMBRA1, by PARK2, to the area of depolarized mitochondria elicits the nucleation of a phagophore.
BECN1 is a core protein component of the PtdIns3K complex needed for the nucleation phase of autophagy, and serves as a scaffolding protein whereby a dynamic grouping of autophagy-related proteins hub. BECN1 is unique among the autophagy-related proteins in that it contains an N-terminal BH3 domain, which can be bound and inhibited by BCL2 and family member BCL2L1.180 This inhibition is primarily disrupted by phosphorylation of BCL2/BCL2L1.181 BECN1 contains a central coiled-coil interaction domain, which affords it the ability to oligomerize.182 Interestingly, the other domains of BECN1 are nonessential for self-oligomerization, and self-oligomerization is not affected by starvation, rapamycin or overexpression of Bcl2l1, Uvrag or Pik3c3. Higher-level understanding of this property has yet to be elucidated. The evolutionarily conserved domain is responsible for autophagic function. BECN1 interacts with PIK3C3 directly to induce autophagosome nucleation and promote elongation. Perhaps attributed to their unique juxtaposition between two programmed cell death pathways or autophagy-independent functions, Becn1 knockout mice have a more severe phenotype than other autophagy-related genes. becn1−/− mice die in utero around E7.5 d, with the embryonic lethality ascribed to a developmental failure to close the pro-amniotic canal.183 As a classical tumor suppressor, heterozygous disruption of Becn1 results in an increase in tumor incidence.183,184 Additional induced mutant models for Becn1 demonstrate a developmental-specific role for autophagy in blood cell lineages. Becn1-deficient rag1−/− chimeras have a dramatic reduction in early marrow-derived thymocytes and B cells, while displaying normal levels of peripheral B and T cells. Thus, autophagy is required for undifferentiated lymphocyte progenitor maintenance and is not needed for the peripheral T and B cell compartments.185 Naive T-cell homeostasis is crucial for protective immunity against infection and it appears that autophagy is necessary for this process. A T cell-specific deletion of Pik3c3 shows that T cell development does not require autophagy; however, naïve T cell survival is contingent upon autophagy to effectively remove damaged mitochondria via mitophagy.186
The class III PtdIns3K is a highly conserved ancient kinase, and the only PtdIns3K identified in yeast. PIK3C3 phosphorylates phosphatidylinositol to generate phosphatidylinositol 3-phosphate, which is used in the elongating the phagophore membrane. A global knockout of Pik3c3 was generated by crossing a conditional Pik3c3 allele with a Meox1-Cre transgenic strain, for conversion to a null allele early in development. As with several other autophagy knockout models, the hemizygous mouse is viable and exhibits no obvious phenotype. In stark contrast to the hemizygous state, the homozygous Pik3c3 knockout is lethal in the early embryonic stages (E7.5). The embryos fail to form a recognizable mesoderm and both the endosomal trafficking and autophagosomal pathways are disrupted.187 In a liver-specific knockout of Pik3c3 (Alb-Cre; Pik3c3fl/fl), mice are smaller and have enlarged, pale livers. At the molecular level, hepatocytes have reduced levels of the autophagy nucleation complex and an increase in intracellular lipid droplet formation. Mitochondria are smaller, despite fed conditions, indicating a potential mitochondrial fusion defect. Additionally, when challenged with a 24-h starvation, there are no observable GFP-LC3 puncta formed in the mutants, indicating a cessation of autophagic flux in the Pik3c3 liver-specific knockout mice.188 A cardiomyocyte-specific knockout (Ckm/Mck-Cre; Pik3c3fl/fl) results in mice with cardiomegaly, with an increased left ventricular wall thickness, decreased cardiac contractility and reduced cardiac output.188 Although these mutants appear to be healthy at birth, they die between 5 and 13 weeks of age. Consistent with the liver-specific knockout, small and deformed mitochondria are observed, and autophagic flux is disrupted. It is evident that PtdIns3K is essential for autophagy regulation as well as autophagy-independent functions such as endosomal trafficking.
UVRAG is a coiled-coil, BECN1 interacting protein. It has been characterized as a positive regulator of autophagy acting in a complex with requisite autophagy proteins BECN1 and PIK3C3. UVRAG has tumor suppressor activity and a negative effect on tumor cell proliferation, as UVRAG is recurrently monoallelically mutated in human colon cancers.189 Although BECN1 serves a host of autophagy-independent functions, UVRAG may have autophagy-independent roles as well. Liang et al. elegantly showed that a UVRAG-PtdIns3K complex coordinates both late endosome fusion and trafficking, independently of its BECN1-dependent role in mediating autophagosomal formation and maturation.9 Although neither UVRAG knockout nor conditional knockout mice currently exist, the generation of a model would be of great value to the field, aiding in ascribing autophagy-dependent and independent functions to each member of the induction complex.
Atg9 complex
ATG9 is the only multimembrane spanning autophagy-related protein involved in autophagosome formation identified to date. Proposed to function in membrane trafficking, it dynamically shuttles between organelles during starvation conditions. There are two mammalian homologs: Atg9a, with a global expression profile, and Atg9b, with a restricted expression in the placenta and pituitary gland.190 Not surprisingly, atg9a−/− mice, like atg5−/− and atg7−/− mice, die as neonates. Thus, several autophagy-deficient mouse strains are unable to survive the transition from placenta-derived nutrition without an intact autophagic response. ATG9A is necessary for LC3 to conjugate PE, which is essential for autophagy function. This study reveals that Atg9a is an essential autophagy gene. In addition to autophagy-related functions, Saitoh and colleagues show that ATG9A regulates double-stranded DNA innate immune response.191
ATG12 conjugation and LC3 lipidation
ATG3 functions as the E2-like enzyme acting with LC3 in the LC3 ubiquitin-like conjugation system. Similar to atg5−/−, atg7−/− and atg9a−/− mice, atg3−/− neonates die within 1 d of parturition, as amino acid levels are reduced and the overall energy homeostasis is disrupted after termination of the placental nutrient supply. The ATG12–ATG5 conjugate system is affected as well as the LC3 conjugation system,192 suggesting cooperation between the two distinct conjugation complexes during elongation. atg3−/− cells contain small autophagosome-like, ATG16L1-positive structures, disorganized phagophores, and altered levels of GABARAP and GABARAPL2/GATE-16.
In yeast, a single Atg4 cysteine protease is responsible for cleaving Atg8 to expose an essential, terminal glycine residue. In humans and mice, four Atg4 homologs (also referred to as autophagins) have been identified: Atg4a, Atg4b, Atg4c and Atg4d. Of the Atg4 homologs, Atg4b and Atg4c knockout mouse models have been generated to date. Atg4c is the most widely expressed Atg4 homolog in human tissues, although Atg4b is the most efficient and broadly active family member in vitro.193 atg4b−/− mice are viable but have a depletion of ATG8-like proteins in normal and nutrient poor conditions. These mice have balance and coordination deficits resulting from defective development in the vestibular system. The otoconia within the inner ear do not develop normally, resulting in abnormal, giant or missing otoconia, a phenotype recapitulated in the atg5−/− model.194 In contrast, atg4c−/− mice have normal levels of basal autophagy, but are less responsive to starvation-induced autophagy in the diaphragm. This role in ameliorating cell stress is highlighted in this model’s increased susceptibility to methyl cholanthrene-induced fibrosarcoma formation.194 Given the functional redundancy of Atg4 homologs, double-knockout models should be highly informative.
ATG16L1 binds to ATG5 to regulate the localization of the ATG12–ATG5 conjugate to the nascent autophagosomal membrane. atg16L1−/− neonates do not survive the perinatal starvation period and have a disruption of LC3–PE conjugation.195 Crohn disease, a chronic inflammatory bowel disease, is strongly linked genetically to the autophagy gene ATG16L1 in humans,196 and hypomorphic Atg16l1 mice show altered Paneth cell morphology and decreased intestinal antimicrobial protein secretion.197 Immunological synapse dynamics between dendritic cells (DCs) and T cells seems to be autophagy-dependent, as RNAi knockdown of ATG16L1 in DCs stabilizes DC-T cell interaction and hyperstimulates the T cell response.198 This “hyperstability” is also found in DCs from Crohn disease patients with an ATG16L1 risk allele, which suggests that normally autophagy is activated at the synapse to negatively regulate T cell activation. In addition, atg16l1−/− mice may help to model an autophagy-inflammasome connection, since they have an induced expression of inflammatory cytokines including IL-1B.195
atg5−/− mice die perinatally, suggesting that autophagy is not essential for embryonic development;199 however, maternal mRNAs and proteins persist in the early embryo and may permit autophagic activity during the preimplantation period. Recent studies demonstrate that autophagy is essential for the oocyte-to-embryo transition. Analysis of GFP-LC3 embryos and oocytes show that autophagy is selectively upregulated in fertilized embryos from the one-cell to the four-cell stage.200 Very little evidence of autophagy is detectable in mature oocytes or ovulated, unfertilized oocytes, indicating that fertilization stimulates the induction of autophagy. Mating of oocyte-specific Atg5-deficient females (Zp3-Cre; Atg5fl/fl) with Atg5+/− males, leads to embryos devoid of maternally inherited autophagic protein that fail to develop.201 Autophagy may function to eliminate maternally inherited proteins or to catabolize cellular components for energy homeostasis. Since atg5−/− neonates die from starvation-induced amino acid deprivation,199 conditional gene deletion models have been developed to characterize autophagy later in development. From these models, autophagy has been shown to play a pivotal role in the prevention of neurodegeneration and the onset of age-related neurological diseases and cardiomyopathies. Nes/Nestin-Cre; Atg5fl/fl mice are viable and do not exhibit the suckling defect comparable to that found with Atg5- or Atg7-deficient neonates.202 After three weeks of age, progressive motor and behavioral defects become apparent including: growth retardation, ataxia, poor motor coordination and failed clasping reflex. High levels of neurodegeneration are found in the cerebellar Purkinje cells, and ubiquinated protein accumulations are observed across several anatomical regions of the brain.202 A Purkinje-cell specific deletion strategy (Pcp-Cre; Atg5fl/fl model) shows a later onset of motor defect around 10 mo of age compared with the more regional Nes-Cre.203 Autophagy-dependent cardiomyopathies may have a developmental component, as cardiac-specific Atg5 deletions in young and adult mice have different pathologies.204 Inducible deletion of Atg5 in adult mice leads to cardiac myopathy, contractile dysfunction, altered sarcomere structure and aberrant mitochondrial organization. Conversely, cardiac-specific Atg5 deletion during gestation does not result in these abnormalities, which points to adaptability of cardiomyocytes during early development.
Similar to Atg5, Atg7 is necessary for the survival of neonates through amino acid pool maintenance as shown in the traditional knockout model.199 Neuronal-specific atg7−/− mice share many of the Atg5 knockout phenotypes including: abnormal behavior, altered limb-clasping response and reduced motor coordination. The Nes-Cre; Atg7fl/fl model shows high levels of neurodegeneration in the cerebellar and cerebral cortex with death occurring by 28 weeks of age.205 Remaining neurons, which demonstrate normal proteasome function, accumulate polyubiquinated proteins and harbor inclusion bodies that increase with age. The Nes-Cre; Vps18fl/fl model exhibits a more severe neurodegeneration phenotype and results in lethality by postnatal day 12,206 perhaps due to VPS18 being part of a protein tethering complex at the lysosome needed for its fusion with autophagosomes or endosomes. Ablation of Atg7 within dopaminergic neurons of the substantia nigra has been performed to model Parkinson disease.207 Slc6a3/Dat-Cre; Atg7fl/fl and En1/engrailed-1-Cre; Atg7fl/fl adult mice have ubiquitinated protein aggregates and show a 40% to 60% reduction in dopaminergic neurons, respectively, and a corresponding decline in dopamine by 55% to 65%. Although Slc6a3-Cre; Atg7fl/fl mice do not display locomotive deficits, En1-Cre; Atg7fl/fl mice exhibit an ataxic gait, presumably from additional Atg7 ablation within the hindbrain, cerebellum and midbrain. Using a Th/tyrosine hydroxylase-specific Cre mouse (Thtm1(cre)Te/Th-IRES-Cre) with the Atg7fl/fl model, there is a 40% reduction in dopaminergic neurons compared with controls and an accumulation of SNCA/α-synuclein protein.208 Seizure activity, in human TSC patients, is thought to occur due to a hyperstimulated MTOR pathway.209 Inhibition of autophagy is demonstrated in Camkk2-Cre; Ptenfl/fl mice, Camkk2-Cre; Tsc1fl/fl mice, and brains of TSC patients.210 Moreover, Camkk2-Cre; Atg7fl/fl mice are prone to spontaneous tremors, as 75% of the mice develop symptoms by 6–7 weeks of age. A liver-specific Atg7 deletion results in hepatomegaly and an increase in hepatocyte size due to an accumulation of peroxisomes and deformed mitochondria.211 Insulin-producing pancreatic β-cells use autophagy to maintain homeostasis, as β-cell-specific Rip-Cre; Atg7fl/fl mice show β-cell mass reduction and exhibit hypoinsulinemia and hyperglycemia.212 Atg7-deficient islet cells have reduced numbers of insulin granules, increased mitochondrial swelling and a distended rough ER and Golgi complex. Interestingly, these mice do not become diabetic until bred with ob/ob mice to trigger an obesity-induced ER stress response. Obesity induces the UPR in β-cells, which in the absence of autophagy leads to an increase in β-cell death from an increase in reactive oxygen species.212,213
Since there are several Atg8 mammalian homologs (e.g., GABARAP, LC3s, GABARAPL1 and GABARAPL2), it is not surprising to find that lc3b−/− and gabarap−/− mice are viable, have normal life spans, and exhibit normal autophagy levels in fed and starved conditions.214,215 While the subcellular relocalization of LC3 is useful in monitoring autophagy, LC3 has been recently implicated in mitophagy. BNIP3L, an outer mitochondrial membrane-spanning protein, has a cytoplasmic LC3-interacting region through which it can recruit mitochondria to the autophagosome; ablation of this interaction severely affects mitochondrial clearance.96 Although SQSTM1 interacts with all of the LC3 orthologs, recent studies have revealed that only lipidated LC3 selectively binds to SQSTM1 and directs it to the autophagosome.163 Thus, closer examination of the lc3b−/− model may reveal defects in mitophagy or aggresome clearance in a cell-specific fashion.
Autophagy-modifying models
Activation of autophagy, through the ectopic overexpression of peroxisome proliferator-activated receptor gamma, coactivator 1 α (PPARGC1A, also known as PGC-1α), results in the degradation of HTT/huntingtin protein aggregates and ameliorates the phenotype of Huntington disease mice.216 This PPARGC1A-mediated proteolysis occurs by activation of TFEB, a known master regulator of autophagy. Induction of autophagy through rapamycin administration can reverse protein plaque formation in a murine model for Gerstmann–Sträussler–Scheinker disease, a prion disease that results in ataxia from extracellular PRNP amyloid plaques.217 Rapamycin treatment eliminates amyloid plaque formation, reduces symptoms and extends the survival time of these mice.218 Within the hippocampus, the region devoted to learning and memory, an increase in autophagosomes in aged mice has been correlated with short-term memory deficits.219 This suggests an impairment of productive autophagosome-lysosome fusion in aged hippocampal cells. A genetic affirmation of this mechanism is shown in a rescue of the TgCRND8 mouse model for Alzheimer disease.220 Ablation of cystatin B, a lysosomal cysteine protease inhibitor, reverses this defect through an increase in productive autophagy and reduction of amyloid protein deposition. The Sqstm1 knockout mouse displays an Alzheimer-like phenotype, characterized by accumulations of cytotoxic aggregates of ubiquitinated proteins in neurons and other tissues.221 Autophagy mitigates ROS-mediated cellular damage through productive mitophagy. The E3 ubiquitin ligase PARK2 initially was found to be necessary for mitophagy in neurons,222 and PARK2 mutations are associated with juvenile Parkinsonism.223
However, PARK2-mediated mitophagy is also important for increased myocardial survival after induced infarction. Fatalities from myocardial infarction increase from 20% in control mice to 60% in park2−/− mice during the first week after insult.224 Mitophagy may be a necessary process after injury to eliminate damaged mitochondria to avoid an inflammatory response. Lysosomal DNASE2 functions to degrade mitochondrial DNA upon autophagy-mediated trafficking of mitochondria to the lysosome. Upon pressure overload, mice with DNASE2-deficient hearts exhibit myocarditis from an inflammatory response and dilated cardiomyopathy.225 Autophagy could affect cell differentiation by restructuring the cell through the elimination of organelles, proteins, and structures while providing necessary “building block” metabolites. FGF signaling inhibits the transition from cardiac precursor cells to differentiated cardiomyocytes. Abrogation of FGF signaling through the conditional deletion of floxed Fgfr1/2 or Frs2 causes premature cardiomyocyte differentiation through precocious autophagy activation.226 A potential regulatory mechanism for autophagy induction comes from a recent study that examined potent miRNAs that are expressed upon hypertrophic stimuli.227 mir212−/− mir132−/− mice are protected from transaortic constriction-induced hypertrophy and exhibit increased basal levels of autophagy. Transgenic overexpression of Mir212-Mir132 causes a decrease in the pro-autophagic transcription factor FOXO3, and impairment of starvation-induced autophagy, and leads to hypertrophy and heart failure. Lastly, blocking the fusion of autophagosomes and lysosomes in cardiomyocytes through a cardiac-specific deletion of Mitofusin 2 (Mfn2) leads to increased sensitivity to ischemia and late onset cardiac dysfunction.228
Sonic hedgehog (SHH) is a crucial morphogen that promotes angiogenesis during development229 and after ischemia in adults.230 Ligation of mouse common carotid arteries induces neointimal lesion formation due to SHH-mediated, autophagy-dependent smooth muscle cell proliferation.231 Inhibition of autophagy with 3-MA or BafA1 inhibits cell proliferation and neointima formation. This potentially signifies an angiogenic role for autophagy in SHH-mediated smooth muscle cell proliferation during development as well. A reduced level of autophagy is associated with arterial endothelial cell aging and dysfunction in older humans, as autophagy markers are decreased by 50% and endothelium-dependent dilatation (EDD) is lowered by 30%.232 In addition, aged mice have a 40% decrease in autophagy markers and a 25% reduction in EDD due to suppressed nitric oxide generation and an increased inflammatory response. Induction of autophagy with trehalose reverses the EDD through normalizing both nitric oxide levels and inflammatory cytokine expression. The dependence of cell differentiation on autophagy is probably cell specific, since autophagy may inhibit skeletal muscle differentiation from myoblasts. TGFB1, an inhibitor of muscle differentiation, induces autophagy through phosphoprotein enriched in astrocytes 15 (PEA15/PED) activation.233 Ectopic overexpression of PEA15A in transgenic mice activates autophagy, leading to atrophic fiber accumulation in skeletal muscle.
Summary
Autophagy is a complex process with many constituent players and is highly regulated. Autophagic degradation of cellular contents and turnover of organelles is crucial for cell vitality and for the fidelity of organismal development. The concerted effort of specific types of autophagy is essential for cellular homeostasis and immune function. Transgenic mouse models of autophagic deficiency, both global and tissue/temporal specific, have revealed the importance of autophagy in the oocyte-to-embryo transition, postnatal survival, development, differentiation and aging. A synopsis of the induced genetic models discussed in this review is provided in Table 2. From the collection of phenotypes of these models, a common thread emerges to assert that autophagy may not be essential for embryogenesis. Six models (atg3−/−; atg5−/−; atg7−/−; atg9a−/−; atg16l1−/−; ulk1−/−ulk2−/−) exhibit a perinatal lethality partly based on an inability for the neonates to maintain a pool of amino acids after parturition. In contrast, a separate cluster of mutations, three within the PtdIns3K complex (ambra1−/−; becn1−/−; pik3c3−/−) and one in the ULK complex (rb1cc1−/−), demonstrate early to mid-gestation lethalities in the respective knockout models. Reconciliation of these phenotypic differences may be argued by functional redundancies or alternative mechanisms (e.g., cell-specific Atg5-Atg7-independent autophagy) found within the former group, or additional autophagy-independent roles for members of the latter group. With regard to the ULK complex, a recent study showed that both Atg13- and Rb1cc1-deficient avian DT40 cells have impaired autophagy, which is independent of Ulk1-Ulk2.176 Generation of an Atg13-deficient mouse model should help in discerning the developmental consequences of ULK1-ULK2-independent autophagy. In addition to this ULK1-ULK2-independent activation, ULK3 can also contribute to the induction of autophagy as well.234 This affirms Einstein’s notion that a problem, when looked at in the right context, becomes increasingly more complicated. It also illustrates that there is considerable work to be done in this area, especially as our methods of detection improve.
Table 2.Atg knockout and conditional knockout mouse phenotypes.
| Gene | Genotype | Phenotype | Ref. |
|---|---|---|---|
|
ULK1 complex | |||
|
Ulk1 |
ulk1-/- |
Viable; no developmental defects or impairment to starvation-induced autophagy; delayed mitochondria elimination from reticulocytes |
168 |
|
Ulk2 |
ulk2-/- |
Viable; no overt phenotype/fertility defects |
169 |
|
ulk1-/-; ulk2-/- |
Neonatal lethal; impaired response to amino acid deprivation |
||
|
Rb1cc1 |
rb1cc1-/- |
Embryonic lethal at E14.5-15.5; excessive liver and cardiac apoptosis; thin left ventricular wall, lacking trabeculation |
170-172 |
|
Tek-Cre; Rb1cc1fl/fl |
Perinatal lethal; severe erythroblastic anemia; 6-fold decrease in HSCs; increased number of myeloid cells in liver |
173-175 |
|
|
Atg13 |
|
Currently no KO mouse |
|
|
C12orf44 |
|
Currently no KO mouse |
|
|
PtdIns3K complex | |||
|
Ambra1 |
ambra1-/- |
Non-viable; embryos show neural tube defects, spina bifida at E10-E14.5, accumulation of ubiquitinated proteins, and increased apoptosis in various tissues |
177 |
|
Becn1 |
becn1-/- |
Embryonic lethal at E7.5; failure to close pro-amniotic canal |
183 |
|
Becn1+/- |
Increased tumor incidence in various tissues |
184 |
|
|
becn1-/-; rag1-/- |
Reduced levels of early marrow-derived thymocytes and B cells |
185 |
|
|
PtdIns3k |
Meox1-Cre; Pik3c3fl/fl |
Embryonic lethal at E7.5; embryos fail to form recognizable mesoderm |
187 |
|
Alb-Cre; Pik3c3fl/fl |
Smaller adult mice; enlarged, pale liver. In hepatocytes: Reduced levels of autophagy nucleation complex; arrested autophagic flux; increased intracellular lipid droplet formation; reduced size of mitochondria |
188 |
|
|
Ckm-Cre; Pik3c3fl/fl |
Lethal between 5-13 weeks; Cardiomegaly with increased thickness of left ventricular wall, decreased cardiac contractility/reduced cardiac output; arrested autophagic flux; Reduced size of mitochondria |
188 |
|
|
Uvrag |
|
Currently no KO mouse |
|
|
ATG9 complex | |||
|
Atg9a |
atg9a-/- |
Neonatal lethal during perinatal starvation |
191 |
|
Atg9b |
|
Currently no KO mouse |
|
|
ATG12 conjugation and LC3 lipidation | |||
|
Atg3 |
atg3-/- |
Neonatal lethal at P1; many cells contain small ATG16L1-positive structures and scattered phagophores |
192 |
|
Atg4a |
|
Currently no KO mouse |
|
|
Atg4b |
atg4b-/- |
Viable; inner ear developmental defects and associated defects in balance/coordination |
194 |
|
Atg4c |
atg4c-/- |
Viable; no overt phenotype/fertility defects; decreased starvation-induced autophagy in diaphragm |
194 |
|
Atg4d |
|
Currently no KO mouse |
|
|
Atg16l1 |
atg16l1-/- |
Neonatal lethal during perinatal starvation; induced expression of IL1B |
195 |
|
Atg5 |
atg5-/- |
Neonatal lethal during perinatal starvation |
199 |
|
Nes-Cre; Atg5fl/fl |
Viable; neurological defects appear after 3 weeks of age: growth retardation, ataxia, poor motor coordination, failed clasping response; increased accumulations of ubiquinated proteins; abundant neurodegeneration of cerebellar Purkinje cells |
202 |
|
|
Pcp-Cre; Atg5fl/fl |
Viable; neurological defects appear after 10 months of age |
203 |
|
|
Atg7 |
atg7-/- |
Neonatal lethal during perinatal starvation |
199 |
|
Nes-Cre; Atg7fl/fl |
Lethal by 28 weeks of age; abundant neurodegeneration /neuron death in cerebrum and cerebellum, ubiquitinated protein accumulations in remaining neurons; motor and clasping defects |
205 |
|
|
Slc6a3-Cre; Atg7fl/fl |
Viable; 40% reduction of dopaminergic neurons and 55% reduction in dopamine; ubiquitinated protein accumulations in remaining neurons |
207 |
|
|
En1-Cre; Atg7fl/fl |
Viable; 60% reduction of dopaminergic neurons and 65% reduction in dopamine; ubiquitinated protein accumulations in remaining neurons; taxic gait |
207 |
|
|
Thtm1(cre)Te-Cre; Atg7fl/fl |
Viable; 40% reduction of dopaminergic neurons; accumulation of SNCA protein |
208 |
|
|
Camkk2-Cre;Atg7fl/fl |
Viable; spontaneous tremors in 75% of mice by 6-7 weeks of age |
210 |
|
|
Alb-Cre; Atg7fl/fl |
Viable; enlarged hepatocytes; hepatomegaly; accumulation of peroxisomes and mitochondria |
211 |
|
|
Rip-Cre; Atg7fl/fl |
Viable; hypoinsulinemia and hyperglycemia; reduced number of insulin granules; swelling of mitochondria, rough ER, Golgi |
212 |
|
|
Lc3b |
lc3b-/- |
Viable; no overt phenotype/fertility defects |
214 |
|
Gabarap |
gabarap-/- |
Viable; no overt phenotype/fertility defects |
215 |
|
Autophagy-modifying models | |||
|
Sqstm1 |
sqstm1-/- |
Alzheimer-like phenotype; accumulation of ubiquitinated proteins in neurons and various tissues |
221 |
|
Park2 |
park2-/- |
40% increase in fatalities from induced myocardial infarction due to mitophagy defect |
224 |
| Mfn2 | mfn2-/- (cardiac specific) | Increased sensitivity to ischemia and late onset cardiac dysfunction | 228 |
Our understanding of autophagy has increased tremendously in the past decade and continues to do so as the research effort in this area continues to accelerate. More productive growth will greatly benefit from the advent of more specific pharmacological agents to induce or inhibit autophagy both in vitro and in vivo, as well as the generation of a global reporter model to monitor autophagic flux in currently existing induced mutant strains. Future autophagy-modifying agents will invariably be extended to novel, engineered peptides. Recently, a novel autophagy-inducing peptide, Tat-beclin 1, was engineered linking the HIV-1 Tat protein transduction domain to 18 amino acids derived from the BECN1 evolutionarily conserved domain. This peptide can induce autophagy in vitro and in vivo presumably by promoting the release of endogenous BECN1 from inhibitory complexes.235 As new tools and models are developed and refined over time, the tenuous nature of how autophagy integrates into multiple signaling pathways (e.g., AKT, MTOR, apoptosis) to orchestrate developmental programs will become more clear.
Glossary
Abbreviations:
- 3-MA
3-methyladenine
- ATG
autophagy-related
- BafA1
bafilomycin A1
- BH3
Bcl2 homology domain
- DC
dendritic cell
- DISC
death-inducing signaling complex
- EDD
endothelium-dependent dilatation
- EM
electron microscopy
- ER
endoplasmic reticulum
- ESCRTI-ESCRTIII
endosomal sorting complex required for transport complex I and III
- GD
glucose deprivation
- GFP
green fluorescent protein
- HOPS
homotypic vacuole fusion protein sorting complex
- HSC
hematopoietic stem cell
- ITPR1
inositol 1,4,5-trisphosphate receptor, type 1
- LD
lipid droplet
- MEF
murine embryonic fibroblast
- MVB
multivesicular body
- NBD-PS
7-nitro-2-1,3-benzoxadiazol-4-yl-phosphoserine OMM, outer mitochondrial membrane
- PE
phosphatidylethanolamine
- PM
plasma membrane
- PtdIns3P- phosphatidylinositol 3-phosphate
ROS, reactive oxygen species
- SNARES
soluble NSF attachment protein receptors
- SCN
suprachiasmatic nucleus
- UPR
unfolded protein response
- UPS
ubiquitin proteasome system
- XeB/XeC
xestospongin B/C
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24273
References
- 1.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–11. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–80. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 3.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 4.Jia K, Levine B. Autophagy and longevity: lessons from C. elegans. Adv Exp Med Biol. 2010;694:47–60. doi: 10.1007/978-1-4419-7002-2_5. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226:255–73. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–87. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fader CM, Sánchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901–16. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 13.Dunn WA., Jr. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–33. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 15.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–5. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 16.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–84. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 20.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen W-L, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravikumar B, Moreau K, Rubinsztein DC. Plasma membrane helps autophagosomes grow. Autophagy. 2010;6:1184–6. doi: 10.4161/auto.6.8.13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–17. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–6. doi: 10.1083/jcb.35.2.C11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–6. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- 27.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–6. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 28.David H, Ellermann J, Uerlings I. Primary phase of hepatocytic autophagocytosis under ischaemic conditions. Exp Toxicol Pathol. 1992;44:74–80. doi: 10.1016/S0940-2993(11)80191-0. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz AL, Brandt RA, Geuze H, Ciechanover A. Stress-induced alterations in autophagic pathway: relationship to ubiquitin system. Am J Physiol. 1992;262:C1031–8. doi: 10.1152/ajpcell.1992.262.4.C1031. [DOI] [PubMed] [Google Scholar]
- 30.Huang J, Lam GY, Brumell JH. Autophagy signaling through reactive oxygen species. Antioxid Redox Signal. 2011;14:2215–31. doi: 10.1089/ars.2010.3554. [DOI] [PubMed] [Google Scholar]
- 31.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dosenko VE, Nagibin VS, Tumanovska LV, Moibenko AA. Protective effect of autophagy in anoxia-reoxygenation of isolated cardiomyocyte? Autophagy. 2006;2:305–6. doi: 10.4161/auto.2946. [DOI] [PubMed] [Google Scholar]
- 34.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15:1572–81. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 35.Bohensky J, Shapiro IM, Leshinsky S, Terkhorn SP, Adams CS, Srinivas V. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207–14. doi: 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- 36.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazure NM, Pouysségur J. Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia. Autophagy. 2009;5:868–9. doi: 10.4161/auto.9042. [DOI] [PubMed] [Google Scholar]
- 38.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–42. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JL, Lin HH, Kim KJ, Lin A, Forman HJ, Ann DK. Novel roles for protein kinase Cdelta-dependent signaling pathways in acute hypoxic stress-induced autophagy. J Biol Chem. 2008;283:34432–44. doi: 10.1074/jbc.M804239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JL, Lin HH, Kim KJ, Lin A, Ou JH, Ann DK. PKC δ signaling: a dual role in regulating hypoxic stress-induced autophagy and apoptosis. Autophagy. 2009;5:244–6. doi: 10.4161/auto.5.2.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 43.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Onodera J, Ohsumi Y. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J Biol Chem. 2004;279:16071–6. doi: 10.1074/jbc.M312706200. [DOI] [PubMed] [Google Scholar]
- 45.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 46.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–92. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 47.Korolchuk VI, Menzies FM, Rubinsztein DC. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy. 2009;5:862–3. doi: 10.4161/auto.8840. [DOI] [PubMed] [Google Scholar]
- 48.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–27. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–8. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 50.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 51.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 52.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heifetz A, Keenan RW, Elbein AD. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry. 1979;18:2186–92. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- 55.Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–43. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M, et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010;6:61–6. doi: 10.4161/auto.6.1.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 58.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–29. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atack JR, Cook SM, Watt AP, Fletcher SR, Ragan CI. In vitro and in vivo inhibition of inositol monophosphatase by the bisphosphonate L-690,330. J Neurochem. 1993;60:652–8. doi: 10.1111/j.1471-4159.1993.tb03197.x. [DOI] [PubMed] [Google Scholar]
- 65.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–32. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 66.Wang IF, Guo BS, Liu YC, Wu CC, Yang CH, Tsai KJ, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A. 2012;109:15024–9. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010;285:11061–7. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Smet P, Parys JB, Callewaert G, Weidema AF, Hill E, De Smedt H, et al. Xestospongin C is an equally potent inhibitor of the inositol 1,4,5-trisphosphate receptor and the endoplasmic-reticulum Ca(2+) pumps. Cell Calcium. 1999;26:9–13. doi: 10.1054/ceca.1999.0047. [DOI] [PubMed] [Google Scholar]
- 69.Jaimovich E, Mattei C, Liberona JL, Cardenas C, Estrada M, Barbier J, et al. Xestospongin B, a competitive inhibitor of IP3-mediated Ca2+ signalling in cultured rat myotubes, isolated myonuclei, and neuroblastoma (NG108-15) cells. FEBS Lett. 2005;579:2051–7. doi: 10.1016/j.febslet.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 70.Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–17. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 71.Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14:1029–39. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 72.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–61. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–6. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 74.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 76.Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci U S A. 1978;75:3327–31. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–50. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 78.Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–7. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 80.Thachil E, Hugot JP, Arbeille B, Paris R, Grodet A, Peuchmaur M, et al. Abnormal activation of autophagy-induced crinophagy in Paneth cells from patients with Crohn’s disease. Gastroenterology. 2012;142:1097–9, e4. doi: 10.1053/j.gastro.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 81.Marsh BJ, Soden C, Alarcón C, Wicksteed BL, Yaekura K, Costin AJ, et al. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Mol Endocrinol. 2007;21:2255–69. doi: 10.1210/me.2007-0077. [DOI] [PubMed] [Google Scholar]
- 82.Hernandez D, Torres CA, Setlik W, Cebrián C, Mosharov EV, Tang G, et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. 2012;74:277–84. doi: 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vaccaro MI. Zymophagy: selective autophagy of secretory granules. Int J Cell Biol. 2012;2012:396705. doi: 10.1155/2012/396705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grasso D, Ropolo A, Lo Ré A, Boggio V, Molejón MI, Iovanna JL, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 2011;286:8308–24. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deretic V, Jiang S, Dupont N. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol. 2012;22:397–406. doi: 10.1016/j.tcb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–70. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berg TO, Fengsrud M, Strømhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–92. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 88.Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–50. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 89.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–9. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deretic V. Autophagy as an immune defense mechanism. Curr Opin Immunol. 2006;18:375–82. doi: 10.1016/j.coi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 91.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–83. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–67. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Orvedahl A, Sumpter R, Jr., Xiao G, Ng A, Zou Z, Tang Y, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–7. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–41. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- 102.Hughson LR, Poon VI, Spowart JE, Lum JJ. Implications of therapy-induced selective autophagy on tumor metabolism and survival. Int J Cell Biol. 2012;2012:872091. doi: 10.1155/2012/872091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yokota S, Dariush Fahimi H. Degradation of excess peroxisomes in mammalian liver cells by autophagy and other mechanisms. Histochem Cell Biol. 2009;131:455–8. doi: 10.1007/s00418-009-0564-6. [DOI] [PubMed] [Google Scholar]
- 104.Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res. 2008;314:3531–41. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 105.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–69. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deegan S, Saveljeva S, Gorman AM, Samali A. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1173-4. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. 2012;2012:182834. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–10. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 109.Baltanás FC, Casafont I, Weruaga E, Alonso JR, Berciano MT, Lafarga M. Nucleolar disruption and cajal body disassembly are nuclear hallmarks of DNA damage-induced neurodegeneration in purkinje cells. Brain Pathol. 2011;21:374–88. doi: 10.1111/j.1750-3639.2010.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Knodler LA, Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol. 2011;13:1319–27. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 113.Moreau K, Lacas-Gervais S, Fujita N, Sebbane F, Yoshimori T, Simonet M, et al. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol. 2010;12:1108–23. doi: 10.1111/j.1462-5822.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- 114.Ma D, Li S, Molusky MM, Lin JD. Circadian autophagy rhythm: a link between clock and metabolism? Trends Endocrinol Metab. 2012;23:319–25. doi: 10.1016/j.tem.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 117.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 118.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–9. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 119.Hoogerwerf WA, Sinha M, Conesa A, Luxon BA, Shahinian VB, Cornélissen G, et al. Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology. 2008;135:2019–29. doi: 10.1053/j.gastro.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011;30:4642–51. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gonzalez CD, Lee MS, Marchetti P, Pietropaolo M, Towns R, Vaccaro MI, et al. The emerging role of autophagy in the pathophysiology of diabetes mellitus. Autophagy. 2011;7:2–11. doi: 10.4161/auto.7.1.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stamenkovic JA, Olsson AH, Nagorny CL, Malmgren S, Dekker-Nitert M, Ling C, et al. Regulation of core clock genes in human islets. Metabolism. 2012;61:978–85. doi: 10.1016/j.metabol.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 123.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 124.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 125.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, et al. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 126.Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–22. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–31. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 129.Kong DK, Georgescu SP, Cano C, Aronovitz MJ, Iovanna JL, Patten RD, et al. Deficiency of the transcriptional regulator p8 results in increased autophagy and apoptosis, and causes impaired heart function. Mol Biol Cell. 2010;21:1335–49. doi: 10.1091/mbc.E09-09-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res. 2010;107:1470–82. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiong S, Salazar G, Patrushev N, Alexander RW. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. J Biol Chem. 2011;286:5289–99. doi: 10.1074/jbc.M110.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 133.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tasdemir E, Chiara Maiuri M, Morselli E, Criollo A, D’Amelio M, Djavaheri-Mergny M, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4:810–4. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 135.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, et al. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–6. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 136.Sanli T, Linher-Melville K, Tsakiridis T, Singh G. Sestrin2 modulates AMPK subunit expression and its response to ionizing radiation in breast cancer cells. PLoS One. 2012;7:e32035. doi: 10.1371/journal.pone.0032035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 138.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–3. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 139.Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 140.Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–89. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 141.Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol. 2006;173:533–44. doi: 10.1083/jcb.200512059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rodriguez OC, Choudhury S, Kolukula V, Vietsch EE, Catania J, Preet A, et al. Dietary downregulation of mutant p53 levels via glucose restriction: mechanisms and implications for tumor therapy. Cell Cycle. 2012;11:4436–46. doi: 10.4161/cc.22778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–48. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 144.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 145.Kusama Y, Sato K, Kimura N, Mitamura J, Ohdaira H, Yoshida K. Comprehensive analysis of expression pattern and promoter regulation of human autophagy-related genes. Apoptosis. 2009;14:1165–75. doi: 10.1007/s10495-009-0390-2. [DOI] [PubMed] [Google Scholar]
- 146.Garcia-Garcia A, Rodriguez-Rocha H, Tseng MT, Montes de Oca-Luna R, Zhou HS, McMasters KM, et al. E2F-1 lacking the transcriptional activity domain induces autophagy. Cancer Biol Ther. 2012;13:1091–101. doi: 10.4161/cbt.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ciavarra G, Zacksenhaus E. Rescue of myogenic defects in Rb-deficient cells by inhibition of autophagy or by hypoxia-induced glycolytic shift. J Cell Biol. 2010;191:291–301. doi: 10.1083/jcb.201005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, et al. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–93. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–14. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–9. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maiuri MC, Criollo A, Kroemer G. Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J. 2010;29:515–6. doi: 10.1038/emboj.2009.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 156.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–39. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–32. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 158.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29:606–18. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene. 2009;28:2128–41. doi: 10.1038/onc.2009.60. [DOI] [PubMed] [Google Scholar]
- 160.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O, et al. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell. 2012;47:359–70. doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455–68. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Terada M, Nobori K, Munehisa Y, Kakizaki M, Ohba T, Takahashi Y, et al. Double transgenic mice crossed GFP-LC3 transgenic mice with alphaMyHC-mCherry-LC3 transgenic mice are a new and useful tool to examine the role of autophagy in the heart. Circ J. 2010;74:203–6. doi: 10.1253/circj.CJ-09-0589. [DOI] [PubMed] [Google Scholar]
- 165.Deretic V. Autophagosome and phagosome. Methods Mol Biol. 2008;445:1–10. doi: 10.1007/978-1-59745-157-4_1. [DOI] [PubMed] [Google Scholar]
- 166.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 167.Takagi-Akiba M, Asanuma K, Tanida I, Tada N, Oliva Trejo JA, Nonaka K, et al. Doxorubicin-induced glomerulosclerosis with proteinuria in GFP-GABARAP transgenic mice. Am J Physiol Renal Physiol. 2012;302:F380–9. doi: 10.1152/ajprenal.00502.2010. [DOI] [PubMed] [Google Scholar]
- 168.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011;108:11121–6. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5:85–7. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 171.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006;175:121–33. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liu F, Guan JL. FIP200, an essential component of mammalian autophagy is indispensible for fetal hematopoiesis. Autophagy. 2011;7:229–30. doi: 10.4161/auto.7.2.14125. [DOI] [PubMed] [Google Scholar]
- 174.Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–14. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–34. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- 176.Alers S, Löffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, et al. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy. 2011;7:1423–33. doi: 10.4161/auto.7.12.18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 178.Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S, et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 2012;19:1495–504. doi: 10.1038/cdd.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, et al. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–61. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 181.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–48. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Adi-Harel S, Erlich S, Schmukler E, Cohen-Kedar S, Segev O, Mizrachy L, et al. Beclin 1 self-association is independent of autophagy induction by amino acid deprivation and rapamycin treatment. J Cell Biochem. 2010;110:1262–71. doi: 10.1002/jcb.22642. [DOI] [PubMed] [Google Scholar]
- 183.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Arsov I, Adebayo A, Kucerova-Levisohn M, Haye J, MacNeil M, Papavasiliou FN, et al. A role for autophagic protein beclin 1 early in lymphocyte development. J Immunol. 2011;186:2201–9. doi: 10.4049/jimmunol.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109:8670–5. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhou X, Takatoh J, Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One. 2011;6:e16358. doi: 10.1371/journal.pone.0016358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–8. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 190.Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, et al. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–90. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- 191.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–6. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–38. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mariño G, Fernández AF, Cabrera S, Lundberg YW, Cabanillas R, Rodríguez F, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 196.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wildenberg ME, Vos AC, Wolfkamp SC, Duijvestein M, Verhaar AP, Te Velde AA, et al. Autophagy attenuates the adaptive immune response by destabilizing the immunologic synapse. Gastroenterology. 2012;142:1493–503, e6. doi: 10.1053/j.gastro.2012.02.034. [DOI] [PubMed] [Google Scholar]
- 199.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4:1076–8. doi: 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- 201.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–20. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 202.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 203.Nishiyama J, Miura E, Mizushima N, Watanabe M, Yuzaki M. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy. 2007;3:591–6. doi: 10.4161/auto.4964. [DOI] [PubMed] [Google Scholar]
- 204.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 205.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 206.Peng C, Ye J, Yan S, Kong S, Shen Y, Li C, et al. Ablation of vacuole protein sorting 18 (Vps18) gene leads to neurodegeneration and impaired neuronal migration by disrupting multiple vesicle transport pathways to lysosomes. J Biol Chem. 2012;287:32861–73. doi: 10.1074/jbc.M112.384305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ahmed I, Liang Y, Schools S, Dawson VL, Dawson TM, Savitt JM. Development and characterization of a new Parkinson’s disease model resulting from impaired autophagy. J Neurosci. 2012;32:16503–9. doi: 10.1523/JNEUROSCI.0209-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, et al. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. J Neurosci. 2012;32:7585–93. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cao R, Li A, Cho HY. mTOR signaling in epileptogenesis: too much of a good thing? J Neurosci. 2009;29:12372–3. doi: 10.1523/JNEUROSCI.3486-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.McMahon J, Huang X, Yang J, Komatsu M, Yue Z, Qian J, et al. Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J Neurosci. 2012;32:15704–14. doi: 10.1523/JNEUROSCI.2392-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 212.Quan W, Lim YM, Lee MS. Role of autophagy in diabetes and endoplasmic reticulum stress of pancreatic β-cells. Exp Mol Med. 2012;44:81–8. doi: 10.3858/emm.2012.44.2.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Quan W, Hur KY, Lim Y, Oh SH, Lee JC, Kim KH, et al. Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia. 2012;55:392–403. doi: 10.1007/s00125-011-2350-y. [DOI] [PubMed] [Google Scholar]
- 214.Cann GM, Guignabert C, Ying L, Deshpande N, Bekker JM, Wang L, et al. Developmental expression of LC3α and β: absence of fibronectin or autophagy phenotype in LC3β knockout mice. Dev Dyn. 2008;237:187–95. doi: 10.1002/dvdy.21392. [DOI] [PubMed] [Google Scholar]
- 215.Le Grand JN, Chakrama FZ, Seguin-Py S, Fraichard A, Delage-Mourroux R, Jouvenot M, et al. GABARAPL1 (GEC1): original or copycat? Autophagy. 2011;7:1098–107. doi: 10.4161/auto.7.10.15904. [DOI] [PubMed] [Google Scholar]
- 216.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:42ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yang W, Cook J, Rassbach B, Lemus A, DeArmond SJ, Mastrianni JA. A New Transgenic Mouse Model of Gerstmann-Straussler-Scheinker Syndrome Caused by the A117V Mutation of PRNP. J Neurosci. 2009;29:10072–80. doi: 10.1523/JNEUROSCI.2542-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cortes CJ, Qin K, Cook J, Solanki A, Mastrianni JA. Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann-Sträussler-Scheinker disease. J Neurosci. 2012;32:12396–405. doi: 10.1523/JNEUROSCI.6189-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Soontornniyomkij V, Risbrough VB, Young JW, Soontornniyomkij B, Jeste DV, Achim CL. Increased hippocampal accumulation of autophagosomes predicts short-term recognition memory impairment in aged mice. Age (Dordr) 2012;34:305–16. doi: 10.1007/s11357-011-9234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, Ohno M, et al. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer’s disease ameliorates amyloid pathologies and memory deficits. Brain. 2011;134:258–77. doi: 10.1093/brain/awq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–20. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 222.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 224.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–26. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–5. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang J, Liu J, Huang Y, Chang JY, Liu L, McKeehan WL, et al. FRS2α-mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ Res. 2012;110:e29–39. doi: 10.1161/CIRCRESAHA.111.255950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, et al. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem. 2012;287:23615–25. doi: 10.1074/jbc.M112.379164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Byrd N, Grabel L. Hedgehog signaling in murine vasculogenesis and angiogenesis. Trends Cardiovasc Med. 2004;14:308–13. doi: 10.1016/j.tcm.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 230.Bijlsma MF, Spek CA. The Hedgehog morphogen in myocardial ischemia-reperfusion injury. Exp Biol Med (Maywood) 2010;235:447–54. doi: 10.1258/ebm.2009.009303. [DOI] [PubMed] [Google Scholar]
- 231.Li H, Li J, Li Y, Singh P, Cao L, Xu LJ, et al. Sonic hedgehog promotes autophagy of vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2012;303:H1319–31. doi: 10.1152/ajpheart.00160.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.LaRocca TJ, Henson GD, Thorburn A, Sindler AL, Pierce GL, Seals DR. Translational evidence that impaired autophagy contributes to arterial ageing. J Physiol. 2012;590:3305–16. doi: 10.1113/jphysiol.2012.229690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Iovino S, Oriente F, Botta G, Cabaro S, Iovane V, Paciello O, et al. PED/PEA-15 induces autophagy and mediates TGF-beta1 effect on muscle cell differentiation. Cell Death Differ. 2012;19:1127–38. doi: 10.1038/cdd.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–6. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]