Abstract
Plants produce a variety of secondary metabolites that possess strong physiological activities. Unfortunately, however, their production can suffer from a variety of serious problems, including low levels of productivity and heterogeneous quality, as well as difficulty in raw material supply. In contrast, microorganisms can be used to produce their primary and some of their secondary metabolites in a controlled environment, thus assuring high levels of efficiency and uniform quality. In an attempt to overcome the problems associated with secondary metabolite production in plants, we developed a microbial platform for the production of plant isoquinoline alkaloids involving the unification of the microbial and plant metabolic pathways into a single system. The potential applications of this system have also been discussed.
Keywords: aromatic amino acid metabolism, isoquinoline alkaloid biosynthesis, metabolic engineering, microbial production
Introduction
Higher plants produce a wide range of chemicals, with more than 25,000 terpenoids, 12,000 alkaloids and 8,000 phenolic substances having been identified and reported in the literature to date.1) These chemicals serve a variety of different functions in plants, where they defend against herbivores and pathogens, aid in inter-plant competition and attract beneficial organisms such as pollinators. They can also have protective effects with regard to abiotic stresses such as UV exposure, and changes in the temperature, water status and mineral nutrient composition. In addition, many secondary metabolites produced in plants have been used by humans in a variety of different applications, including spices, dyes, fragrances, flavoring agents and pharmaceuticals. Many of these chemicals have also been reported to promote human health and can therefore enrich our lives in a variety of different ways.2)
The use of plant metabolites as natural medicines has a long history that can be traced back over more than 3,500 years.3) In the more recent years, research has focused predominantly on the identification of the active ingredients. Following the first identification of morphine as the principal active ingredient in opium (1804), many compounds have been identified from plant materials and used as medicines, including the pain-killing and fever-reducing salicylic acid from willow bark extract (1826), the anticancer agent paclitaxel from the Pacific yew tree (1971), and the anti-malarial drug artemisinin from Artemisia annua (1972). Japanese scientists have also provided a significant contribution to the isolation and identification of natural products from medicinal plants. The isolations of ephedrine from Ephedra vulgaris and matrine from Sophora flavescens (1885) by Dr. Nagayoshi Nagai highlight the early success of natural product research in Japan. Following on from this early success, many excellent works have been produced, including the structural determination studies of sinomenine in Sinomenium acutum and its synthesis by Dr. Kakuji Goto.4)
The use of biochemical and molecular characterization techniques in natural product research, however, has been largely limited, because of the low levels of enzymatic activity often encountered in the secondary metabolism of plants, especially towards the production of alkaloids. In the late 1970s and early 1980s, several groups developed cell culture systems for the production of large quantities of secondary metabolites.5,6) Although the levels of productivity encountered were often not sufficient for the industrial production of the desired metabolites, because of limitations regarding the market size and difficulties associated with the use of these materials as crude medicines, these materials have provided useful information pertaining to the study of the biosynthetic pathways associated with their production. Zenk et al. in particular studied a variety of different alkaloid pathways, including those of the isoquinoline alkaloids (IQAs).5)
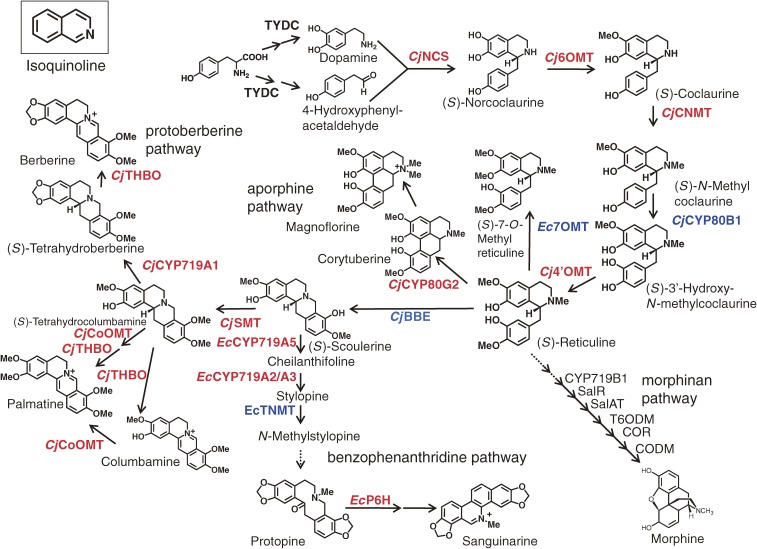
IQAs are a large and diverse group of alkaloids that are derived from tyrosine with approximately 2,500 defined structures, which all contain a heterocyclic isoquinoline backbone (Fig. 1).6) Some significant examples from this group of natural products include the analgesic morphine from Papaver somniferum L., the anti-gout colchicine from Colchicum autumnale L., the emetic and antiamoebic emetine from Cephaelis ipecacuanha (Brot.) A. Rich., the skeletal muscle relaxant tubocurarine from Strychnos toxifera Bentham, and the antimicrobial compounds berberine and sanguinarine from divergent plant species including Berberis spp. and Sanguinaria spp. It is noteworthy that many of these compounds and their derivatives have been used as pharmaceutical agents. Sato et al. independently selected a cultured cell line with a high level of IQA productivity for a particular medicinal herb known as the Japanese goldthread (Coptis japonica Makino) and characterized the molecular basis of the IQA pathway to consolidate the results of their biochemical characterization.7,8)
Figure 1.
Outline of isoquinoline alkaloid biosynthesis pathway. The cDNAs of the enzymes shown in red were first cloned and/or characterized in Sato’s laboratory, and those in blue were cloned as described in the literature, from cultured Coptis japonica (Cj) and California poppy (Eschscholzia californica: Es). Genes of the following enzymes have been also cloned from Papaver somniferum by other several groups;83) TYDC, CYP719B1, SalR (salutaridine reductase), SalAT (salutaridinol 7-O-acetyltransferase), T6ODM (thebaine 6-O-demethylase), CODM (codeinone O-demethylase) and COR (codeinone reductase).
In this review, we have provided a summary of the progress that has been made towards the biochemical and molecular biological characterization of the IQA biosynthetic pathways using cultured C. japonica and California poppy (Eschscholzia californica) cells. The application of this progress to the metabolic engineering, and microbial production of plant IQAs will also be discussed.
Clarification of the molecular basis of the IQA biosynthetic pathways and metabolic engineering
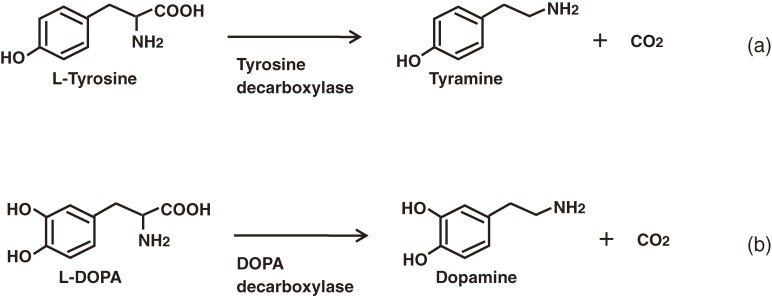
IQA biosynthesis is generally considered to begin with the conversion of tyrosine to both dopamine and 4-hydroxyphenylacetaldehyde via a sequential decarboxylation, ortho-hydroxylation and deamination pathway (Fig. 1).6,9) From these early steps, only the tyrosine/DOPA decarboxylase (an aromatic L-amino acid decarboxylase; TYDC), which converts tyrosine and DOPA to their corresponding amines, and the aminotransferase have been purified and characterized.10,11) The general lack of a detailed understanding of the other early step enzymes has led to the use of alternative techniques for the supply of the substrate via reconstituted pathways in microbial cells (this will be discussed in a later section).
Dopamine and 4-hydroxyphenylacetaldehyde are condensed by norcoclaurine synthase (NCS; EC 4.2.1.78) to yield (S)-norcoclaurine, which is the central precursor for the construction of all of the benzylisoquinoline alkaloids. NCS has been purified and characterized from cultured Thalictrum flavum spp., and a TfNCS cDNA belonging to the pathogenesis-related protein (PR)10 family was cloned.12,13) It is noteworthy that a novel dioxygenase (CjNCS) from cultured C. japonica cells was also shown to catalyze this NCS reaction.14)
(S)-Norcoclaurine is converted to coclaurine by S-adenosyl methionine (SAM)-dependent norcoclaurine 6-O-methyltransferase (6OMT; EC 2.1.1.128),15) which is in turn converted to N-methylcoclaurine by SAM-dependent coclaurine N-methyltransferase (CNMT; EC 2.1.1.140),16) before being converted to 3′-hydroxy-N-methyl coclaurine by P450 hydroxylase (CYP80B1; EC 1.14.13.71),17) and then finally converted to (S)-reticuline by SAM-dependent 3′-hydroxy N-methylcoclaurine 4′-O-methyltransferase (4′OMT; EC 2.1.1.116).15) Both of the enzymes were co-purified by SDS-gel electrophoresis, because of the high level of similarity between the proteinaceous properties of 6OMT to those of 4′OMT, to give a final preparation containing homologous 40- and 41-kD polypeptides. To effectively discriminate the two enzymes, the screening of a cDNA library of cultured C. japonica cells was carried out with oligonucleotide probes designed on the basis of the internal amino acid sequences of the two polypeptides. The two kinds of cDNA obtained in this way were expressed in Escherichia coli, and characterization of the recombinant proteins enabled us to confirm that the 40-kDa polypeptide corresponded to the 6OMT whereas the 41-kD corresponded to the 4′OMT. Similarly, the cloning of the cDNA of CNMT was carried out based on the amino acid sequence of the purified polypeptide.16)
Detailed biochemical studies using recombinant enzymes have demonstrated the strict reaction specificities of these enzymes, with the enzymes themselves regulating the biosyntheses sequentially and in a coordinated manner. For example, CNMT prefers coclaurine to 6-O-methylnorlaudanosoline, whereas 4′OMT prefers an N-methylated substrate,15,16) which suggested that the pathway shown in Fig. 1 was preferred to a sequence consisting of N-methylation, hydroxylation and 4′-O-methylation. Current data also indicate that all of the enzymes involved in the early steps of (S)-reticuline biosynthesis from norcoclaurine, except for the membrane-bound P450 CYP80B1, are located in the cytosol. (S)-Reticuline, which is produced by reaction with 4′OMT, is the central intermediate in the branch pathways leading to many divergent IQAs, including the aporphine alkaloids (e.g., magnoflorine), benzophenanthridine alkaloids (e.g., sanguinarine), morphinan alkaloids (e.g., morphine and thebaine) and protoberberine alkaloids (e.g., berberine) (Fig. 1).
The next step in the biosynthesis of berberine from (S)-reticuline involves the conversion of the N-methyl group of (S)-reticuline to the methylene bridge moiety linked to (S)-scoulerine by the berberine bridge enzyme (reticuline oxidase, BBE; EC 1.21.3.3).18) This unique enzyme is soluble and localized in the vesicles.19) (S)-Scoulerine is converted to (S)-tetrahydrocolumbamine (THC) by the SAM-dependent scoulerine 9-O-methyltransferase (SMT; EC 2.1.1.117),20) and then to tetrahydroberberine (THB; canadine) by a P450-dependent canadine synthase (CYP719A1; EC 1.14.21.5).21) The characterization of these enzymes expressed from the corresponding cDNAs effectively confirmed that the biosynthesis of berberine proceeds via canadine in Coptis cells and not via columbamine. Once again, the enzyme substrate specificity shows a clear preference for this pathway. Although the hydrophobic N-terminal region of SMT suggests that this enzyme may be targeted to the membrane fraction, its localization in the cytosol as well as within the lumen of alkaloid-specific vesicles has been reported.20,22)
(S)-Tetrahydroberberine oxidase (THBO or STOX; EC 1.3.3.8) is the enzyme involved in the final step of the berberine biosynthesis.23–25) A cDNA of THBO was cloned based on the partial amino acid sequence of the purified enzyme and its deduced amino acid sequence suggested that CjTHBO belongs to the FAD-containing BBE oxidoreductase family. As a consequence of the difficulties encountered during the attempted production of active recombinant CjTHBO in E. coli and Saccharomyces cerevisiae cells, recombinant CjTHBO was heterologously expressed in E. californica cells and its metabolic activities subsequently characterized.26)
As well as the biosynthesis of berberine, the cDNAs of several other biosynthetic enzymes involved in the biosynthesis of related protoberberine alkaloids have been also cloned. For example, SAM-dependent columbamine O-methyltransferase (CoOMT; EC 2.1.1.118), which is involved in the biosynthesis of palmatine, was cloned from cultured C. japonica cells using expressed sequence tag information. Recombinant CjCoOMT clearly indicated that THC could be a used as a substrate for the formation of tetrahydropalmatine (Fig. 1),27) whereas columbamine was thought to be the exclusive substrate of CoOMT in Berberis plants based on the substrate specificity of Berberis CoOMT.28) Differences in substrate specificity have also been reported for the CNMT of Coptis and Berberis, in that the Coptis enzyme could N-methylate norlaudanosoline, whereas the Berberis enzyme could not.16) These results suggest that the pathways associated with the secondary metabolism may vary considerably depending on the enzyme(s) that each plant has acquired during its evolution.
The cDNA of a novel P450 enzyme involved in the biosynthesis of aporphine-type alkaloids from (S)-reticuline was also cloned from cultured C. japonica cells.29) A P450 corytuberine synthase (CYP80G2; EC 1.14.21.-) was found to have a significant level of amino acid sequence similarity to that of berbamunine synthase (CYP80A1; EC 1.14.21.3) in the biosynthesis of bisbenzylisoquinoline alkaloids via a C–O phenol-coupling reaction.30) Recombinant CYP80G2 produced from yeast showed intramolecular C–C phenol coupling activity to produce the aporphine alkaloid corytuberine from (S)-reticuline (Fig. 1). In contrast, the cDNA of a native N-methyltransferase capable of producing magnoflorine from corytuberine has not yet been cloned, although the potential catalytic activity of CNMT towards corytuberine could be used for the production of magnoflorine from (S)-reticuline in a microbial system.
The benzophenanthridine alkaloid sanguinarine is also produced from (S)-reticuline via (S)-scoulerine (Fig. 1). This conversion begins with the action of the P450-dependent oxidases, (S)-chelanthifoline synthase (CYP719A5; EC 1.14.21.2) and (S)-stylopine synthase (CYP719A2/A3; EC 1.14.21.1), which catalyze the formation of a methylenedioxy-bridge to produce (S)-stylopine from (S)-scoulerine. Their cDNAs were also cloned from cultured E. californica cells.31,32) (S)-Stylopine was then converted to cis-N-methylstylopine by (S)-tetrahydroprotoberberine N-methyltransferase (TNMT; EC 2.1.1.122),33) before being converted to protopine by N-methylstylopine hydroxylase (MSH; EC 1.14.13.37), and then to dihydrosanguinarine by protopine 6-hydroxylase (P6H; EC 1.14.13.55).34) Although the cDNA of MSH was not yet been cloned, the cDNA of P6H (CYP82N2v2) has been cloned from E. californica cells, based on an integrated analysis of metabolite accumulation and the mRNA expression profile of transgenic cells.35) The identification of CYP82N2v2 as a protopine 6-hydroxylase for the production of dihydrosanguinarine from protopine, as well as its broad range of reaction specificity in the conversion of corycavine to corynoloxine, indicate that CYP82N2v2 plays an important role in the biosynthesis of a diverse range of benzophenanthridine alkaloids.36)
In total, Sato et al. successfully cloned the cDNAs of 12 enzymes belonging to the IQA biosynthetic pathways to provide a better understanding of the molecular basis of these particular pathways. Furthermore, seven of these cDNAs were subsequently used in the later work to construct a microbial platform for the production of plant IQAs.
Unique properties of the biosynthetic enzymes of the IQA pathways.
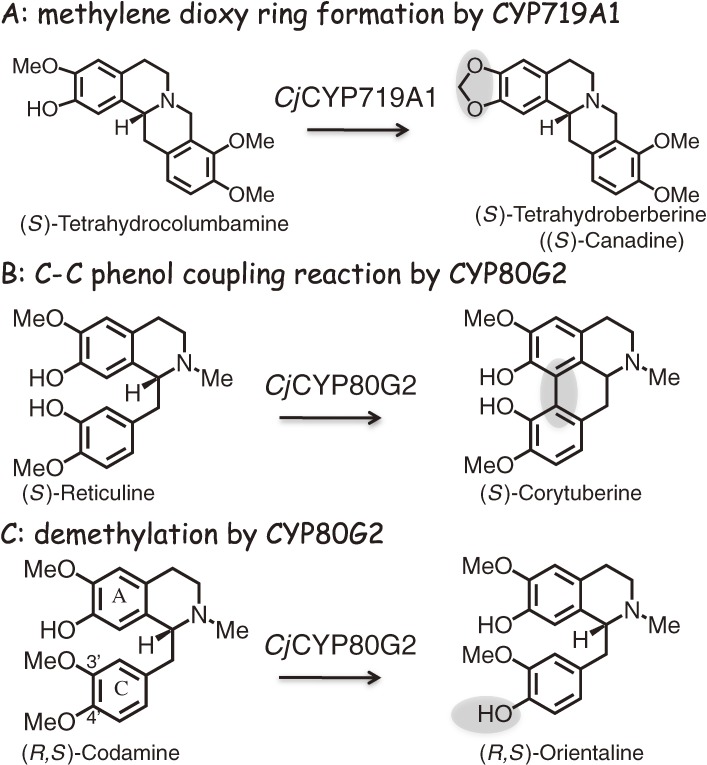
The cloning of the cDNAs of the biosynthetic enzymes from the IQA biosynthetic pathways enabled the detailed characterization of their enzymatic properties. Of the enzymes characterized from the IQA biosynthetic pathway, CYP719A1 and CYP80G2 provided the first molecular basis for methylenedioxy ring formation and C-C phenol coupling reactions in eukaryotic cells.21,29) Interestingly, the CYP719 family of enzymes was not found in Arabidopsis and rice, with these members of the cytochrome P450 super-family being unique to benzylisoquinoline alkaloid biosynthesis.37)
Although the oxidative cyclization of an ortho-hydroxymethoxy-substituted aromatic ring (i.e. methylenedioxy bridge formation) is a P450-dependent reaction, this reaction is not a simple monooxygenation reaction as demonstrated by the lack of oxygen insertion into the products. The P450-dependent hydroxylation of the methoxy group yields an intermediate corresponding to the hemiacetal of formaldehyde that can cyclize to form a methylenedioxy bridge via an ionic mechanism involving the methylene oxonium ion intermediate.38) All of the methylenedioxy bridge-forming reactions involved in IQA biosynthesis have been found to be catalyzed by members of the CYP719A subfamily, which exhibit rather strict levels of substrate specificity. CYP719A1 from C. japonica has been identified as a canadine synthase and can only convert (S)-THC to (S)-THB (Fig. 2).21) E. californica CYP719A5 can convert (S)-scoulerine to (S)-cheilanthifoline, whereas CYP719A2 and CYP719A3 have been identified as stylopine synthases responsible for catalyzing the conversion of (S)-cheilanthifoline to (S)-stylopine. CYP719A9 from E. californica can convert (R,S)-reticuline to the corresponding products containing a methylenedioxy ring.31,32)
Figure 2.
Some unique reactions catalyzed by P450 enzymes found in the IQA pathway. CYP719A1 from C. japonica catalyzes the oxidative cyclization of an ortho-hydroxymethoxy-substituted aromatic ring (i.e. methylenedioxy bridge formation) and converts only tetrahydrocolumbamine to (S)-tetrahydroberberine. CjCYP80G2 not only catalyzes the intramolecular C-C phenol coupling reaction and converts (S)-reticuline to (S)-corytuberine, but also 4-O-demethylates codamine to form the demethylated product. The reaction mechanism has been discussed in the text.
Detailed characterization of CjCYP80G2, which catalyzes the intramolecular C-C phenol coupling reaction for the conversion of (S)-reticuline to (S)-corytuberine,29) indicated that CjCYP80G2 reacts initially with the C-ring and not the A-ring during the 4-O-demethylation of codamine. The product obtained from the reaction of codamine with CjCYP80G2 suggested that hydrogen abstraction occurred as the first step from the 4-methoxy group of the C-ring, followed by hydroxy radical rebound to the same position of the substrate, with the eventual release of formaldehyde to form the demethylated product (Fig. 2). Similarly, when CYP80G2 was reacted with (S)-reticuline, the initial hydrogen atom abstraction step occurred at the 3-hydroxy group of the C-ring to generate a phenoxy radical, followed by subsequent oxidation at the 7-hydroxy group of the A-ring to generate a second radical. The terminal reaction was then achieved by the coupling of the diradical species (i.e. bond formation between the C8 and C2 positions of (S)-reticuline). The reaction mechanism of CjCYP80G2 is therefore best explained by the diradical process.
Metabolic engineering.
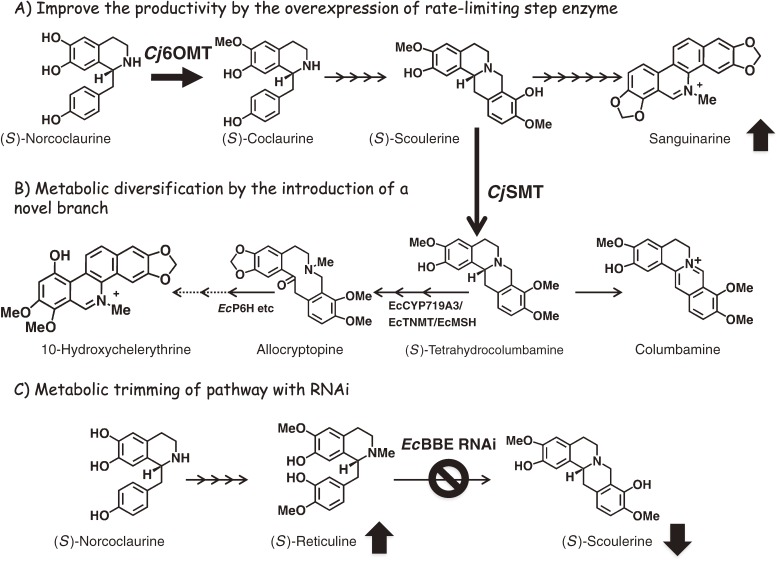
The isolation of the genes of biosynthetic enzymes has enabled the molecular design of the metabolic flow of plant cells (Fig. 3). Using the Agrobacterium-mediated transformation technique, Sato et al.8) reported the molecular engineering of IQA biosynthesis in C. japonica and E. californica cells with C. japonica genes, and demonstrated the high level of flexibility of the IQA biosynthetic pathway towards its genetic alteration.
Figure 3.
Outline of metabolic engineering in transgenic E. californica cells. (A) Overexpression of the rate-limiting enzyme, norcoclaurine 6-O-methyltransferase (6OMT), using C. japonica cDNA in California poppy cells led to an increase in the accumulation of the main alkaloid of the cells, sanguinarine, as shown by the upward arrow. (B) Introduction of a branch pathway with scoulerine 9-O-methyltransferase (SMT) also induced a marked shift in the metabolic flow, as shown by the arrows. SMT overexpression led to a significant reduction in the accumulation of sanguinarine-type alkaloids with two methylenedioxy ring structures, but induced the production of protoberberine-type alkaloids, such as berberine and columbamine, and of allocryptopine- and chelerythrine-type alkaloids with one methylenedioxy ring and two methoxy groups. (C) The shutdown of the berberine bridge enzyme (BBE) expression with an RNAi vector clearly downregulated the accumulation of sanguinarine and the intermediate scoulerine, as shown by the downward arrow. In contrast, RNAi of BBE induced a marked accumulation of reticuline, which is a substrate of BBE, as shown by the upward arrow.
Attempts have been made to overexpress the genes responsible for producing the enzymes involved in the rate-limiting steps of these processes with the aim of improving the yields of the associated IQAs. For example, the overexpression of C. japonica SMT and 4′OMT using a strong constitutive promoter, Cauliflower mosaic virus 35S, led to an increase in berberine production in C. japonica cells and plants, respectively.39,40) Similarly, heterologous overexpression of C. japonica 6OMT improved the level of alkaloid productivity in E. californica cells.41) It is important to note, however, that the overexpression of a biosynthetic enzyme does not always result in a successful increase in the levels of alkaloid production. For example, the overexpression of Cj4′OMT in E. californica cells only provided a marginal increase in alkaloid production.41) The bottleneck step(s) associated with the IQA biosynthetic pathways can vary across different plant species and isolated cell lines because of variations in the expression levels of the genes required to produce the enzymes. Furthermore, changes in the bottleneck step(s) may also occur as a consequence of modifications to the enzyme expression level resulting from metabolic engineering.
Given that there may be multiple bottleneck steps involved in a particular biosynthetic process, the overall regulation of the expression levels of biosynthetic genes in the pathway would provide a more effective means of control, and further studies are currently underway to isolate general transcription factors. The recent identification of the transcription factors CjWRKY1 and CjbHLH1 as possible general regulators in IQA biosynthesis using a transient RNAi screening system could represent a significant discovery in the field and provide significant improvements in the yields of IQAs by enabling the regulation of the transcription processes associated with their production.42–44)
As well as providing improvements in the yield, improvements in the quality of the IQAs are perhaps more desirable, in that although increases in metabolic diversity would be useful for drug discovery, the reduction of the metabolic diversity is critical for the facile preparation of pure compounds. The introduction of a novel branch pathway to a biosynthetic pathway represents one of the most useful approaches for increasing the level of metabolic diversity resulting from the pathway. We have ectopically expressed CjSMT in E. californica cells (Fig. 3).39) CjSMT catalyzes the O-methylation of scoulerine to produce THC in the berberine biosynthesis and is not involved in the benzophenanthridine alkaloid biosynthesis. These transgenic E. californica cells with CjSMT expression not only produced columbamine (i.e. the oxidized product of THC),39) but also provided novel products, including allocryptopine and 10-hydroxychelerythrine, which were produced from the CjSMT reaction products following their reaction with other endogenous enzymes in the benzophenanthridine alkaloid biosynthetic pathway, effectively demonstrating the enhanced diversification of the alkaloid profile in transgenic cells.35)
Modifications to the metabolic flow leading to a reduction in the levels of enzymes associated with undesired pathways represents another promising approach to increase the product levels of the desired metabolites, while reducing unwanted byproducts. The combination of this approach with the introduction of a new pathway allows for the production of novel products from transgenic cells. Of the methods used to reduce gene expression (e.g., antisense, RNA interference (RNAi), knockout with T-DNA insertion and mutation), RNA silencing with double-stranded RNA (i.e. RNAi with siRNA or miRNA) is the most powerful and reliable of these techniques.45) The versatility of RNAi for controlling the multiple genes responsible for metabolite production across a variety of different tissues and developmental steps has been well recognized. For example, in 2007, Fujii et al.46) reported the successful accumulation of reticuline, which is a direct substrate of BBE, by the use of an RNAi of BBE in E. californica cells (Fig. 3), whereas little accumulation of reticuline was reported by Park et al.47) using the antisense method.
Characterization of aromatic amino acid metabolizing enzymes in microorganisms
Kumagai et al. have been working on microbial enzymes capable of metabolizing aromatic amino acids and aromatic amines, such as L-tyrosine, L-DOPA and their corresponding amines. The enzymes used in their work include amino acid-lyases,48–57) aromatic amino acid decarboxylases,58–64) and amine oxidases.65–72) When they started their work, enzymes were not routinely purified or particularly well characterized. Consequently, the bacterial strains which provided the highest levels of enzyme activity from a number of microbial strains were screened first. The culture conditions were then investigated extensively, with particular emphasis on the induction conditions of the enzyme biosynthesis to obtain the highest enzyme activity, with the investigation ultimately contributing to the isolation of a pure enzyme fraction as crystals. The characterization of these aromatic amino acid metabolizing enzymes and the elucidation of the control mechanisms associated with the enzyme biosynthetic pathways in the microorganisms were indispensable for the construction of the microbial IQA production platform. This paragraph describes the synthesis of L-DOPA by tyrosine phenol-lyase from Erwinia herbicola, aromatic amino acid decarboxylase from Pseudomonas putida, and monoamine oxidase (tyramine oxidase) from Micrococcus luteus.
L-DOPA synthesis by tyrosine phenol-lyase and tyrosinase.
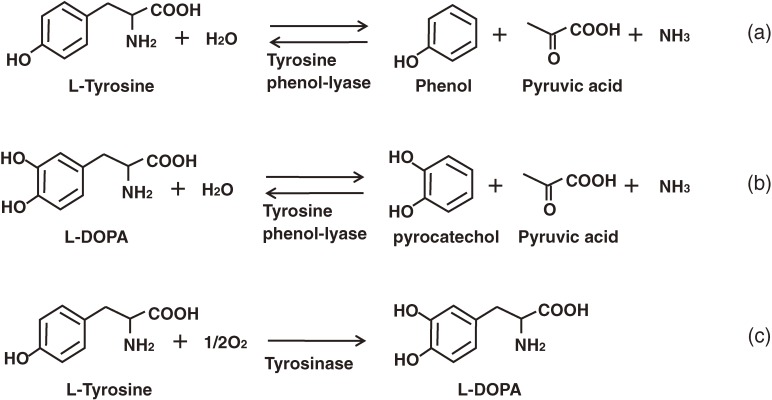
L-DOPA, which is otherwise known as 3,4-dihydroxy-L-phenylalanine, and dopamine are the starting materials required of IQA production in microorganisms. Although tyrosine phenol-lyase (TPL; EC 4.1.99.2) is not directly used in the microbial IQA production platform, work focused on the enzymatic properties of TPL and the control mechanisms associated with the biosynthesis of TPL has contributed greatly to the industrial production of L-DOPA.
L-DOPA, is used in the treatment of Parkinson’s diseases. Approximately 250 tons of L-DOPA is produced around the world every year. Nearly half of this material is synthesized according to an enzymatic method involving TPL.48–50) TPL normally catalyzes the α,β-elimination reaction of L-tyrosine to produces pyruvate, ammonia and phenol (Fig. 4a). This reaction is reversible, and when catechol is substituted for phenol, the process produces L-DOPA (Fig. 4b).51) TPL is inducible by tyrosine and, from a practically perspective, E. herbicola cells with a high level of TPL activity are prepared by cultivation in a medium supplemented with L-tyrosine before being harvested and transferred to the reactor together with the substrates, pyruvate, ammonia and catechol. Bai et al.73) and Kumagai et al.52,53) elucidated the regulatory mechanism underlying the L-tyrosine-mediated induction of TPL, and identified TyrR as the transcriptional activator of tpl. The tyrR gene was subsequently cloned from E. herbicola cells and randomly mutagenized to obtain a mutant protein with an enhanced ability to activate tpl. One such mutant, TyrRV67A Y72C E201G, was found to effectively increase TPL activity expression.54) Furthermore, when the mutant gene was introduced to E. herbicola, the resulting strain showed improved levels of L-DOPA production.55) Kumagai et al.56) subsequently identified a second-site suppressor mutations (N324D or A503T) that rescued the inability of TyrRE275Q to activate the tpl promoter. N324 is located in the central domain of TyrR and is thought to be involved in the formation of a TyR oligomer, which is required for tpl-activation. Amino acid replacement of the residue (N324D) compensated for the impaired ability of TyrRE275Q to form an oligomer.56) The amino acid substitutions that lead to the enhanced TPL expression were scattered into three domains, including: (1) V67A, Y72C and E201G in the N-terminal domain (NT); (2) N324D in the central domain (Cen); and (3) A503T in the C-terminal domain (CT). These mutations were genetically combined by the domains, and the resulting tyrR alleles were inserted into a low-copy-number plasmid. These plasmids were then introduced to tyrR-deficient E. herbicola (ΔtyrR::kan+) cells, and the TPL activities of the cell-free extracts of the recombinant strains were measured. The strain carrying TyrRNT Cen CT exhibited its highest level of L-DOPA productivity when cultivated under the non-induced conditions, and its level of productivity was 30-fold greater than that of the tyrosine-induced wild-type cells that are currently utilized in the industrial production of L-DOPA (11.1 g/L/h vs. 0.375 g/L/h).57)
Figure 4.
Reaction catalyzed by tyrosine phenol-lyase and tyrosinase.
TPL requires high concentrations of it pyruvate, ammonia and catechol substrates to carry out an L-DOPA producing reaction, and the concentrations of these substrates can be so high that they can become toxic to bacterial cells. The construction of an IQA producing bacterial platform therefore requires the use of Ralstonia solanacearum tyrosinase (TYR; EC 1.14.18.1, monophenol, L-DOPA: oxygen oxidoreductase) to produce L-DOPA from L-tyrosine. TYR is a copper containing monophenol monooxygenase that catalyzes the oxidation of the 3′-position of the L-tyrosine phenol-ring (Fig. 4c). This particular reaction mechanism was developed on the basis of an X-ray crystallographic investigation conducted by Matoba et al.74) R. solanacearum TYR has low o-diphenolase activity,75) and its introduction to the IQA producing bacterial platform was considered to increase the amount of accumulation of L-tyrosine in the bacterial cells,76) as will be described in a later section.
Aromatic amino acid decarboxylases from bacteria.
Aromatic amino acid decarboxylases (AADCs; EC 4.1.1.28) catalyze the decarboxylation reactions of aromatic amino acids (Fig. 5) and are found in a variety of different organisms, where they play distinct physiological roles. AADCs from higher eukaryotes have been well studied because they are involved in the synthesis of biologically important molecules such as neurotransmitters and alkaloids. In contrast, bacterial AADCs have received less attention because of the simplicities associated with their physiology and target substrate (tyrosine) (Fig. 5a). Although the existence of strongly active AADCs has been reported in some lactic acid bacteria,77) these AADCs have never been purified and characterized as pure preparations because of their instability.
Figure 5.
Reactions catalyzed by aromatic amino acid decarboxylase.
The formation of aromatic L-amino acid decarboxylase in bacteria was studied with intact cells in a reaction mixture containing the aromatic amino acid. The activity was found to be widely distributed in such genera as Achromobacter, Micrococcus, Staphylococcus and Sarcina. Strains belonging to Micrococcus showed especially high levels of decarboxylase activity towards L-tryptophan, 5-hydroxy-L-tryptophan and phenylalanine. Micrococcus percitreus AJ1065 was selected as a promising source of aromatic L-amino acid decarboxylase. M. percitreus was found to constitutively produce an enzyme that exhibited decarboxylase activity towards L-tryptophan, and cultivation conditions to produce high levels of decarboxylase activity were subsequently examined.58)
The AADC from M. percitreus was purified and successfully crystallized for the first time, allowing for its complete characterization.59,60) The enzyme that found to require pyridoxal phosphate (PLP) as a cofactor and to be deactivated by the addition of L-DOPA to the reaction mixture as a substrate.61) The mechanism of the inactivation was investigated by enzyme- and organic-chemical methods, which revealed that L-DOPA reacted slowly with the PLP cofactor resulting in the formation of the so-called tetrahydroisoquinoline adduct compound during the reaction.62) Inhibition of the enzyme was also found to occur when α-methyl aromatic amino acids were used as substrates, and analysis of this inhibition mechanism resulted in the proposal of a reaction mechanism for the AADC from M. percitreus.63)
A eukaryotic-type AADC exhibiting a high level of specificity for L-DOPA (Fig. 5b) has recently been found and its genetic and enzymatic characteristics determined.64) The purified enzyme converted L-DOPA to dopamine with Km and Kcat values of 0.092 mM and 1.8 s−1, respectively. The enzyme was inactive towards other aromatic amino acids such as 5-hydroxy-L-tryptophan (HTP), L-phenylalanine, L-tryptophan and L-tyrosine (Table 1). The observed strict substrate specificity was distinct from that of any other AADC characterized to date. The name DOPA decarboxylase (DDC) was subsequently proposed for the enzyme. Quantitative RT-PCR experiments revealed that the expression of this gene was induced by L-DOPA. DDC was found to be encoded in a cluster together with a LYSR-type (LysineR-type) transcriptional regulator and a major facilitator superfamily transporter. This genetic organization was found to be conserved among all of the sequenced P. putida strains that inhabit the rhizosphere environment, where DOPA acts as a strong allelochemical. These findings suggest the possible involvement of this enzyme in the detoxification of the allelochemical in the rhizosphere, and potential occurrence of a horizontal gene transfer event between the pseudomonad and its host organism.64)
Table 1.
Substrate specificity of P. putida DDC
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| DOPA | 9.2 (±1.9) × 10−2 | 1.8 (±0.1) × 100 | 2.1 (±0.4) × 101 |
| 5-HTP | 9.3 (±2.3) × 10−1 | 9.5 (±0.6) × 10−2 | 1.0 (±0.2) × 10−1 |
| Phe | 8.8 (±1.9) × 100 | 2.3 (±0.2) × 10−1 | 2.7 (±0.4) × 10−2 |
| Trp | 8.8 (±2.1) × 100 | 4.4 (±0.4) × 10−3 | 5.1 (±0.8) × 10−4 |
| Tyr | 1.1 (±0.1) × 10−3 | ||
| 3-Methoxy-Tyr | 6.3 (±0.3) × 10−4 |
The kcat values were calculated based on the reported Vmax values. The estimated molecular mass was 54 kDa.
DDC from P. putida is particularly interesting because its physiological role is related to that of the host plant. In addition, its strict specificity for L-DOPA could be useful for the construction of a microbial platform for plant IQA production because it actually plays an important role in this context, and this will be discussed in more detail in a later section.76)
Aromatic amine oxidases from bacteria.
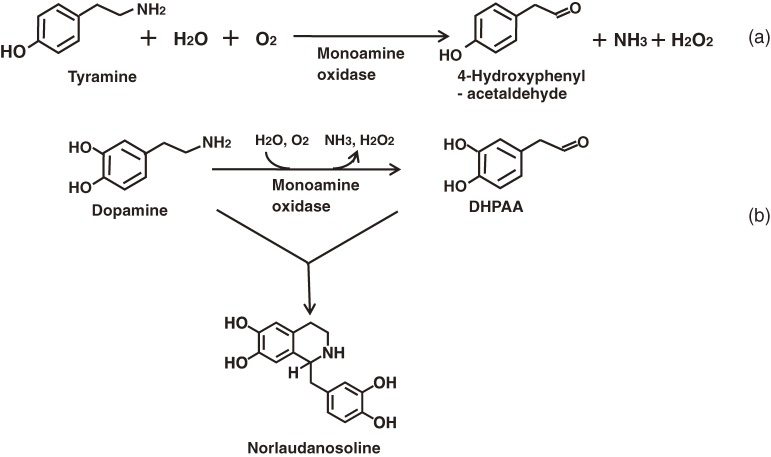
Amine oxidases catalyze the oxidative deamination of amines to produce the corresponding aldehydes, together with ammonia and hydrogen peroxide (Fig. 6a). The Gram-positive bacterium Sarcina lutea (M. luteus, later) contains an amine oxidase specific for tyramine when it grows on nutrient medium. It was purified as a crystalline preparation and characterized. This enzyme is a monoamine oxidase (MAO; EC 1.4.3.4) and uses FAD as a cofactor.65–67) When M. luteus was grown on synthetic medium containing tyramine as a sole nitrogen source, its level of MAO increased extensively to give 20- to 30-fold more crystalline MAO than that obtained under the former cultivation conditions in a nutrient medium.68) The mechanism of the reaction catalyzed by the MAO was studied based on the overall reaction kinetics as well as by the stopped flow.69)
Figure 6.
Reaction catalyzed by monoamine oxidase (a) and formation of the IQA norlaudanosoline from the reaction of MAO with dopamine (b).
The enzyme was found to oxidize tyramine and dopamine at a similar rate, but other monoamines, diamines and polyamines were not oxidized at all. The aldehyde formed during the reaction with dopamine was found to condense with unchanged dopamine to yield the IQA compound norlaudanosoline (i.e. tetrahydropapaveroline) (Fig. 6b).70) This finding suggested the idea of the microbial production of isoquinoline compounds, with MAO and its gene playing important roles in the construction of microbial platform for the production of IQAs.
Kumagai et al.71) have completed the gene cloning and three-dimensional structure prediction experiments for M. luteus MAO. The amine oxidase of the fungi Aspergillus niger has also been reported to be inactivated during the reaction with an analog of the substrate (bromo-ethylamine), and its reaction mechanism was elucidated by analysis of the materials resulting from its suicide inactivation.72)
Construction of the microbial platform for plant IQA production
First-generation microbial platforms for plant IQA production.
Following an increase in the availability of molecular biological tools, the reconstitution of an entire metabolic pathway became feasible in microorganisms.78–80) Microorganisms have a simpler internal structure than plants because of their smaller genome size, and this effectively simplifies metabolite transport between the enzymatic steps.81) The reconstitution of a secondary metabolite pathway in an engineered microbial host offers several advantages over other methods directed towards native plant hosts including the specific production of key intermediate molecules, such as reticuline, the rapid accumulation and facile purification of the target IQA, and the availability of genetic tools for further engineering and pathway optimization of the IQA production.82) A combination of microbial and plant-derived genes offers some advantages for the establishment of efficient and productive systems for the entry compound.79) Unfortunately, however, microbial systems also possess disadvantages, such as a limited substrate availability for plant metabolites, although this limitation is currently being challenged (see below).
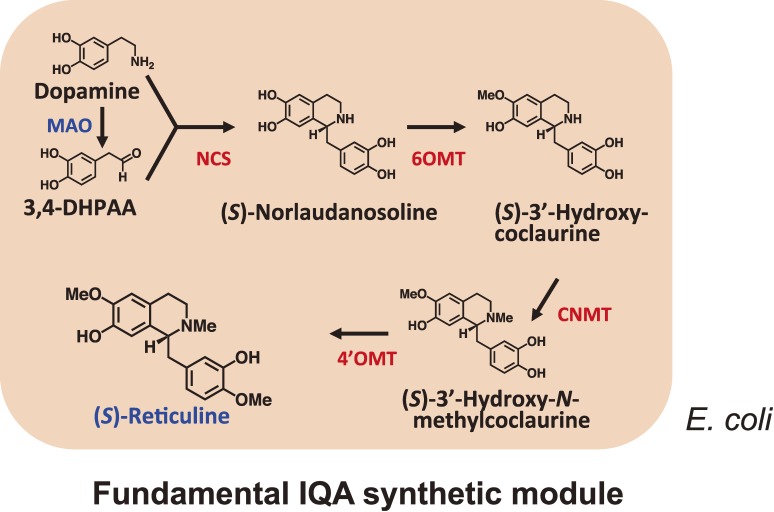
The first reported example of a reconstitution was performed in yeast cells for the production of artemisinic acid according to an engineered isoprenoid pathway.80) The microbial production of alkaloids, however, was not reported in the literature until the pathway from dopamine to reticuline was reconstituted in E. coli. The first artificial pathway for the production of reticuline to be reported in the literature was assembled in E. coli via the introduction of genes for MAO together with C. japonica NCS, 6OMT, CNMT and 4′OMT to a plasmid-based expression systems (Fig. 7).79) The use of microbial MAO allowed for the incorporation of the hydroxy group early in the reticuline pathway via the synthesis of 3,4-dihydroxyphenylacetaldehyde (3,4-DHPAA) from dopamine, thereby avoiding the need to express the plant cytochrome P450 hydroxylase (CYP80B1) in the bacterium, which can often be problematic in a microbial expression. Following the induction of enzyme expression and dopamine supplementation, the final yield of (S)-reticuline synthesized in vitro by the crude enzymes from recombinant E. coli was 55 mg L−1 within 1 hour. In a separate report, Hawkins and Smolke82) used S. cerevisiae as the sole host organism for the assembly of an artificial IQA pathway. In this particular case, three enzymes were combined from plant sources (i.e. 6OMT, CNMT and 4′OMT derived from either T. flavum or P. somniferum) and a human P450 enzyme, and (R,S)-norlaudanosoline was used as a substrate.82) Two systems are therefore now available for the production of plant IQAs, with one of these systems requiring dopamine and the other requiring norlaudanosoline.
Figure 7.
Production of IQA from dopamine in E. coli. The “fundamental IQA synthetic module” for the production of (S)-reticuline, which is the key substance in the IQA synthetic pathway, was constructed in E. coli with the genes of four enzymes (NCS, 6OMT, CNMT and 4′OMT) cloned from C. japonica and a microbial enzyme (MAO).
Second-generation platform for microbial alkaloid production.
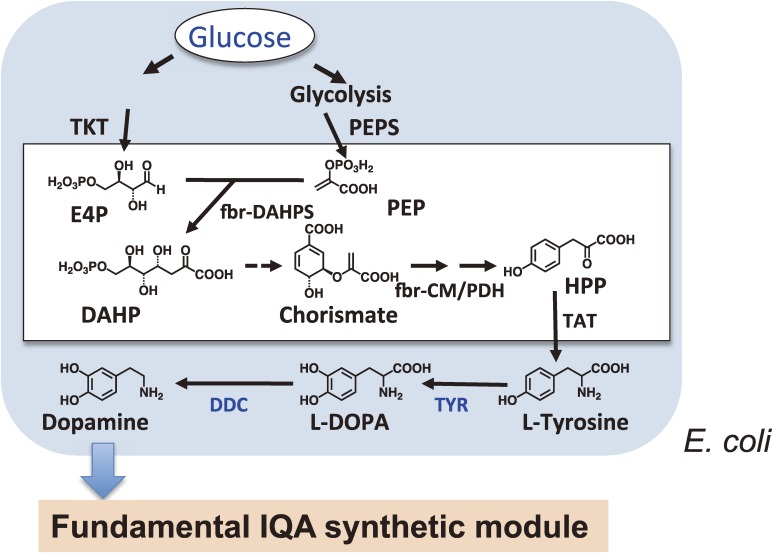
The reconstitution of alkaloid biosynthesis in microbial systems has shown promising results, as described above. Unfortunately, however, the existing need to exogenously supply expensive intermediate precursors has limited the commercial application of this technique. An incomplete understanding of the complex alkaloid biosynthetic networks, especially during the early steps, has effectively hindered progress towards the synthesis of alkaloids from simple precursors. In addition, the cytotoxicity of alkaloids in yeast may also pose a similar problem in other microbial systems. The development of practical and effective strategies to minimize toxicity is therefore needed to create alkaloid-overproducing microbes, such as the engineering of transcription factors to increase microbial tolerance towards toxic metabolites. Further attempts have enabled the successful production of (S)-reticuline from simple carbon sources without the need for an additional substrate in an E. coli fermentation system (Figs. 8 and 9). Nakagawa et al.76) produced tyrosine-overproducing E. coli via the overexpression of biosynthetic enzymes (TKT, transketolase; PEPS, phosphoenolpyruvate synthetase; fbr-DAHPS, feedback-inhibition-resistant 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase; fbr-CM/PD, feedback-inhibition-resistant chorismate mutase/prephenate dehydrogenase and deletion of the repressor of aromatic amino acid biosynthesis; tyrR), and then modified the pathway to produce dopamine from tyrosine with the introduction of pathway-specific tyrosinase (TYR) of R. solanacearum and DDC of Pseudomonas.62) The combination of these modifications with a previously developed alkaloid-production module enabled reticuline production from glucose or glycerol. This achievement could potential lead to the industrial-scale production of alkaloids in the near future.
Figure 8.
Construction of a platform for the production of IQA precursors from glucose in E. coli. The “universal aromatic amino acid synthetic module” was prepared by genetic modifications in E. coli to facilitate overproduction of L-tyrosine from glucose as the sole carbon source. The “tailor-made biosynthetic module” was also assembled and optimized for the production of the two IQA precursors, dopamine and 3, 4-DHPAA, from L-tyrosine. E4P, erythrose-4-phosphate; DAHP, 3-deoxy-d-arabino-heptulosonate-7-phosphate; HPP, para-hydroxyphenylpyruvate
Figure 9.
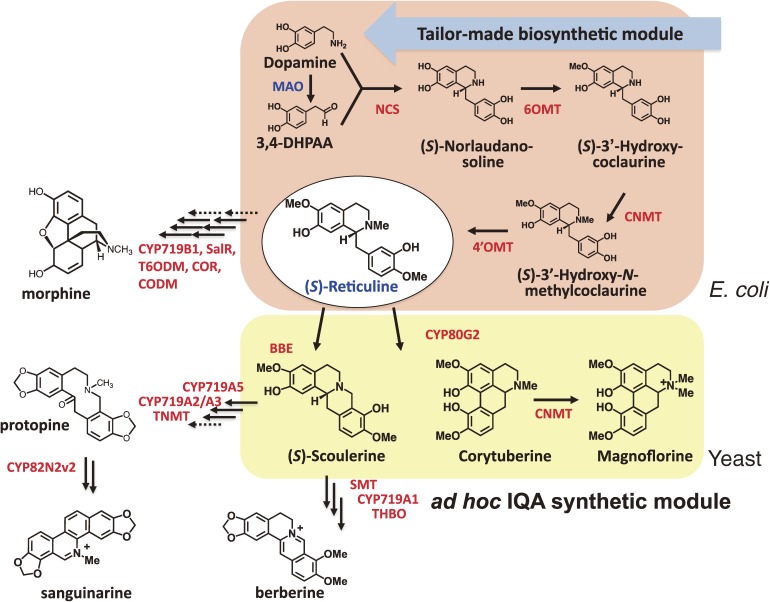
Platform to produce more complex IQAs. The “ad hoc IQA synthetic module” was prepared in S. cerevisiae to produce different IQAs from (S)-reticuline. Co-cultures of IQA producing E. coli and yeast cells efficiently produced (S)-reticuline and other IQAs. Further potential products are also shown with gene names. Broken lines indicate enzyme steps for which the genes have not yet been identified. The abbreviations used are as in Fig. 1.
Production of more complex alkaloids.
More sophisticated systems are required for the production of more complex alkaloids. Although synthetic biological approaches may in principle only require a single host system such as E. coli or S. cerevisiae, or a combination of the two, one of the main disadvantages of the E. coli system is that it lacks an internal membrane system. In 2008, Minami et al.79) examined a combination system of E. coli and S. cerevisiae to overcome these problems (Fig. 9). When reticuline-producing E. coli was co-cultured with S. cerevisiae expressing C. japonica BBE or CYP80G2 in the presence of dopamine, (S)-scoulerine and magnoflorine were detected at 8.3 and 7.2 mg L−1, respectively, following an incubation period of 48 to 72 hours. Although the use of two microbial systems for pathway construction should be useful for establishing different biosynthetic modules, as a strategy it could reduce the efficiency of alkaloid synthesis because of the need for metabolite transport between the cell membranes of the two microorganisms. As shown in Fig. 1, more genes are currently available. The possibility of producing a more divergent array of chemicals is therefore growing exponentially.83)
In summary, we have successfully constructed for the first time a microbial platform for plant IQA production. The platform consists of four modules, including (1) a “universal aromatic amino acid synthetic module” prepared through genetic modifications of the E. coli metabolic system to facilitate overproduction of L-tyrosine from glucose as a sole carbon source; (2) a “tailor-made biosynthetic module” that has been optimized for the production of the two IQA precursors dopamine and 3,4-DHPAA, from L-tyrosine using three microbial genes (Fig. 8); (3) a “fundamental IQA synthetic module” for the production of (S)-reticuline, which is the key substance in the IQA synthetic pathway (Fig. 7), from four genes cloned from C. japonica; and (4) an “ad hoc IQA synthetic module” consisting of three C. japonica genes that is capable of producing different IQAs from (S)-reticuline. The first three modules were installed in E. coli cells, whereas the last one was prepared with budding yeast. Co-cultures of these E. coli and yeast cells efficiently produced (S)-reticuline and other IQAs, demonstrating the overall utility of this constructed microbial platform for plant IQA production (Fig. 9).
The present investigation has opened up a new avenue for the microbial production of plant secondary metabolites, using IQAs as the model. The microbial platform that we have constructed will be useful for the production of phenylpropanoids and other phenylalanine- or tyrosine-derived plant metabolites and contribute to the development of new research areas in both synthetic biology and metabolic engineering.83)
Acknowledgments
The authors are deeply grateful for the continuous guidance and encouragement received from Prof. Emeritus Yasuyuki Yamada and Hideaki Yamada, as well as collaborations with previous and current colleagues and students at the Graduate School of Biostudies and the Faculty of Agriculture at Kyoto University, and at the Research Institute for Bioresources and Biotechnology at Ishikawa Prefectural University. The names of these people have been provided in the references. The authors also appreciate the financial support offered by the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Japan Society for the Promotion of Science, Future Program Grant JSPS-RFTF00L01606, New Energy and Industrial Technology Development Organization (NEDO), and the Program for the Promotion of Basic and Applied researches for Innovations in Bio-oriented Industry (Brain).
Note in proof
Very recently, cDNA of N-methylstylopine hydroxylase in sanguinarine biosynthesis was cloned from opium poppy.84)
Abbreviations
- AADC
aromatic amino acid decarboxylase
- BBE
berberine bridge enzyme
- bHLH
basic helix-loop-helix
- Cen
central domain
- CM/PD
chorismate mutase/prephenate dehydrogenase
- CNMT
coclaurine N-methyltransferase
- CoOMT
columbamine O-methyltransferase
- CT
C-terminal domain
- CYP80A1
berbamunine synthase
- CYP80B1
(S)-N-methylcoclaurine 3′-hydroxylase
- CYP719A1
canadine synthase
- CYP719A2/A3
stylopine synthase
- CYP719A5
cheilanthifoline synthase
- CYP719B1
salutaridine synthase
- CYP80G2
corytuberine synthase
- DAHPS
3-deoxy-d-arabino-heptulosonate-7-phosphate synthase
- 3,4-DHPAA
3,4-dihydroxyphenylacetaldehyde
- DDC
DOPA decarboxylase
- EST
expressed sequence tag
- fbr
feedback-inhibition-resistance
- IQAs
isoquinoline alkaloids
- MAO
monoamine oxidase
- MSH
N-methylstylopine hydroxylase
- NCS
norcoclaurine synthase
- NT
N-terminal domain
- 4′OMT
3′-hydroxy-N-methylcoclaurine-4′-O-methyltransferase
- 6OMT
norcoclaurine 6-O-methyltransferase
- 7OMT
(S)-reticuline 7-O-methyltransferase
- PEP
phosphoenolpyruvate
- PEPS
PEP synthetase
- P6H
protopine 6-hydroxylase
- PLP
pyridoxal phosphate
- PR10
pathogenesis-related protein 10
- RNAi
RNA interference
- SAM
S-adenosylmethionine
- SMT
(S)-scoulerine-9-O-methyltransferase
- TAT
tyrosine aminotransferase
- THB
tetrahydroberberine
- THBO
THB oxidase
- THC
tetrahydrocolumbamine
- TKT
transketolase
- TNMT
(S)-tetrahydroprotoberberine N-methyltransferase
- TPL
tyrosine phenol-lyase
- TYR
tyrosinase
- TYDC
tyrosine/DOPA decarboxylase
Biographies
Profile
Fumihiko Sato was born in 1953, graduated from Kyoto University, and started his research career in 1975 by studying the characterization of functional differentiation in plant cells at the Faculty of Agriculture at Kyoto University. He performed pioneering work on the selection of photoautotrophically cultured cells of higher plants in 1978 and of stable medicinal plant cell lines that produced desired natural products in high yield in 1984; this was the first demonstration of somatic heterogeneity in cultured plant cells. These cell cultures enabled extensive studies on biochemical reactions, especially in the pathways for plant isoquinoline alkaloids, as well as the discovery of many important enzymes and genes for the production of these alkaloids. Further metabolic engineering technologies provided a platform for the production of valuable alkaloids in high yield in transgenic medicinal plant cells or in a novel combinatorial microbial platform. He was promoted to Professor at the Faculty of Agriculture at Kyoto University in 1995, and moved to the Graduate School of Biostudies at Kyoto University in 1999, where he taught students about the cellular and molecular biology of plants. He was awarded the Scientific Contribution Award (Ichimura Foundation) in 2003, the Prize for Science and Technology (Research Category) from The Ministry of Education, Culture, Sports, Science and Technology, Japan in 2009, the Japan Society for Bioscience, Biotechnology and Agrochemistry Award in 2011, and the Japan Academy Prize in 2012.
Profile
Hidehiko Kumagai was born in 1940 and started his research career in 1964 under the guidance of Prof. Hideaki Yamada at the Institute for Food Science, Kyoto University where he worked on microbial enzymes capable of metabolizing amino acids and amines. He obtained his PhD degree for studies on a PLP-dependent multi-functional enzyme, tyrosine phenol-lyase in 1969. He was appointed as an Assistant Professor of the Institute for Food Science, Kyoto University in 1969. He moved to Faculty of Agriculture, Kyoto University in 1977 as an Associate Professor at the Department of Food Technology and was promoted to Professor in 1991. He moved to Graduate School of Biostudies, Kyoto University in 1999. After retirement of Kyoto University in 2004, he moved to Ishikawa Prefectural University as a Professor and now he is the President of the University. He served as a president of Japan Society for Bioscience, Biotechnology and Agrochemistry for 2003–2005. He is an emeritus member of Japan Society for Bioscience, Biotechnology and Agrochemistry and The Vitamin Society of Japan.
He received the Award of Vitamin Society of Japan in 1994 and the Japan Society for Bioscience, Biotechnology and Agrochemistry Award in 2001, and the Japan Academy Prize in 2012.
References
- 1).Croteau, R., Kutchan, T.M. and Lewis, N.G. (2000) Natural products (Secondary metabolites). In Biochemistry and Molecular Biology of Plants (eds. Buchanan, B.B., Gruissem, W. and Jones, R.L.). Am. Soc. Plant Physiol., Maryland, pp. 1250–1318. [Google Scholar]
- 2).Briskin D.P. (2000) Medicinal plants and phytomedicines. Linking plant biochemistry and physiology to human health. Plant Physiol. 124, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Raskin I., Ribnicky D.M., Komarnytsky S., Ilic N., Poulev A., Borisjuk N., Brinker A., Moreno D.A., Ripoll C., Yakoby N., O’Neal J.M., Cornwell T., Pastor I., Fridlender B. (2002) Plants and human health in the twenty-first century. Trends Biotechnol. 20, 522–531 [DOI] [PubMed] [Google Scholar]
- 4).Goto, K. (1964) Sinomenine: An Optical Antipode of Morphine Alkaloids. Kitasato Institute, Tokyo. [Google Scholar]
- 5).Zenk M.H. (1991) Chasing the enzymes of secondary metabolism: Plant cell cultures as a pot of gold. Phytochemistry 30, 3861–3863 [Google Scholar]
- 6).Sato, F. and Yamada, Y. (2008) Engineering formation of medicinal compounds in cell cultures. In Advances in Plant Biochemistry and Molecular Biology, Vol. 1. (eds. Bohnert, H.J., Nguyen, H. and Lewis, N.G.). Elsevier Ltd., Amsterdam, pp. 311–345. [Google Scholar]
- 7).Sato F., Yamada Y. (1984) High berberine-producing cultures of Coptis japonica cells. Phytochemistry 23, 281–285 [Google Scholar]
- 8).Sato, F., Inai, K. and Hashimoto, T. (2007) Metabolic engineering in alkaloid biosynthesis: case studies in tyrosine- and putrescine-derived alkaloids. In Applications of Plant Metabolic Engineering (eds. Verpoorte, R., Alfermann, A.W. and Johnson, T.S.). Springer, New York, pp. 145–173. [Google Scholar]
- 9).Facchini P. (2001) Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 29–66 [DOI] [PubMed] [Google Scholar]
- 10).Facchini P.J., Deluca V. (1994) Differential and tissue-specific expression of a gene family for tyrosine/dopa decarboxylase in opium poppy. J. Biol. Chem. 269, 26684–26690 [PubMed] [Google Scholar]
- 11).Lee E.J., Facchini P.J. (2011) Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol. 157, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Samanani N., Facchini P.J. (2002) Purification and characterization of norcoclaurine synthase. J. Biol. Chem. 277, 33878–33883 [DOI] [PubMed] [Google Scholar]
- 13).Samanani N., Liscombe D.K., Facchini P.J. (2004) Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 40, 302–313 [DOI] [PubMed] [Google Scholar]
- 14).Minami H., Dubouzet E., Iwasa K., Sato F. (2007) Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 282, 6274–6282 [DOI] [PubMed] [Google Scholar]
- 15).Morishige T., Tsujita T., Yamada Y., Sato F. (2000) Molecular characterization of the S-adenosyl-L-methionine: 3′-hydroxyl-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275, 23398–23405 [DOI] [PubMed] [Google Scholar]
- 16).Choi K.-B., Morishige T., Shitan N., Yazaki K., Sato F. (2002) Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 277, 830–835 [DOI] [PubMed] [Google Scholar]
- 17).Pauli H.H., Kutchan T.M. (1998) Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 13, 793–801 [DOI] [PubMed] [Google Scholar]
- 18).Dittrich H., Kutchan T.M. (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogen attack. Proc. Natl. Acad. Sci. U.S.A. 88, 9969–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Bock A., Wanner G., Zenk M.H. (2002) Immunocytological localization of two enzymes involved in berberine biosynthesis. Planta 216, 57–63 [DOI] [PubMed] [Google Scholar]
- 20).Takeshita N., Fujiwara H., Mimura H., Fitchen J.H., Yamada Y., Sato F. (1995) Molecular cloning and characterization of S-adenosyl-L-methionine: scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol. 36, 29–36 [PubMed] [Google Scholar]
- 21).Ikezawa N., Tanaka M., Nagayoshi M., Shinkyo R., Sasaki T., Inoue K., Sato F. (2003) Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 278, 38557–38565 [DOI] [PubMed] [Google Scholar]
- 22).Muemmler S., Rueffer M., Nagakura N., Zenk M.H. (1985) S-Adenosyl-L-methionine: (S)-scoulerine 9-O-methyltransferase, a highly stereo- and regio-specific enzyme in tertrahydroprotoberberine biosynthesis. Plant Cell Rep. 4, 36–39 [DOI] [PubMed] [Google Scholar]
- 23).Galneder E., Rueffer M., Wanner G., Tabata M., Zenk M.H. (1988) Alternative final steps in berberine biosynthesis in Coptis japonica cell cultures. Plant Cell Rep. 7, 1–4 [DOI] [PubMed] [Google Scholar]
- 24).Okada N., Shinmyo A., Okada H., Yamada Y. (1988) Purification and characterization of (S)-tetrahydroberberine oxidase from cultured Coptis japonica cells. Phytochemistry 27, 979–982 [Google Scholar]
- 25).Amman M., Nagakura N., Zenk M.H. (1988) Purification and properties of (S)-tetrahydroprotoberberine oxidase from suspension-cultured cells of Berberis wilsoniae. Eur. J. Biochem. 175, 17–25 [DOI] [PubMed] [Google Scholar]
- 26).Matsushima Y., Minami H., Hori K., Sato F. (2012) Pathway engineering of benzylisoquinoline alkaloid biosynthesis in transgenic California poppy cells with ectopic expression of tetrahydroberberine oxidase from Coptis japonica. Plant Biotechnol. 29, 473–481 [Google Scholar]
- 27).Morishige T., Dubouzet E., Choi K.B., Yazaki K., Sato F. (2002) Molecular cloning of columbamine O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 269, 5659–5667 [DOI] [PubMed] [Google Scholar]
- 28).Rueffer M., Amann M., Zenk M.H. (1986) S-Adenosyl-L-methionine: columbamine-O-methyltransferase, a compartmentalized enzyme in protoberberine biosynthesis. Plant Cell Rep. 3, 182–185 [DOI] [PubMed] [Google Scholar]
- 29).Ikezawa N., Iwasa K., Sato F. (2008) Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283, 8810–8821 [DOI] [PubMed] [Google Scholar]
- 30).Krause P.F.X., Kutchen T.M. (1995) Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C-O-phenol-coupling cytochrome-P450 from the higher-plant Berberis stolonifera. Proc. Natl. Acad. Sci. U.S.A. 92, 2071–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Ikezawa N., Iwasa K., Sato F. (2007) Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 274, 1019–1035 [DOI] [PubMed] [Google Scholar]
- 32).Ikezawa N., Iwasa K., Sato F. (2009) CYP719A subfamily of cytochrome P450 oxygenases and isoquinoline alkaloid biosynthesis in Eschscholzia californica. Plant Cell Rep. 28, 123–133 [DOI] [PubMed] [Google Scholar]
- 33).Liscombe D.K., Facchini P.J. (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 282, 14741–14751 [DOI] [PubMed] [Google Scholar]
- 34).Tanahashi T., Zenk M.H. (1990) Elicitor induction and characterization of microsomal protopine-6-hydroxylase, the central enzyme in benzophenanthridine alkaloid biosynthesis. Phytochemistry 29, 1113–1122 [Google Scholar]
- 35).Takemura T., Ikezawa N., Iwasa K., Sato F. (2010) Metabolic diversification of benzylisoquinoline alkaloid biosynthesis through the introduction of a branch pathway in Eschscholzia californica. Plant Cell Physiol. 51, 949–959 [DOI] [PubMed] [Google Scholar]
- 36).Takemura T., Ikezawa N., Iwasa K., Sato F. (2012) Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry (in press). doi:10.1016/j.phytochem.2012.02.013 [DOI] [PubMed] [Google Scholar]
- 37).Nelson D.R., Schuler M.A., Paquette S.M., Werck-Reichhart D., Bak S. (2004) Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135, 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Mizutani M., Sato F. (2011) Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 507, 194–203 [DOI] [PubMed] [Google Scholar]
- 39).Sato F., Hashimoto T., Hachiya A., Tamura K., Choi K.-B., Morishige T., Fujimoto H., Yamada Y. (2001) Metabolic engineering of plant alkaloid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 98, 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Inui T., Kawano N., Shitan N., Yazaki K., Kiuchi F., Kawahara N., Sato F., Yoshimatsu K. (2012) Improvement of benzylisoquinoline alkaloid productivity by overexpression of 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase in transgenic Coptis japonica plants. Biol. Pharm. Bull. 35, 650–659 [DOI] [PubMed] [Google Scholar]
- 41).Inui T., Tamura K., Fujii N., Morishige T., Sato F. (2007) Overexpression of Coptis japonica norcoclaurine 6-O-methyltransferase overcomes the rate-limiting step in benzylisoquinoline alkaloid biosynthesis in cultured Eschscholzia californica. Plant Cell Physiol. 48, 252–262 [DOI] [PubMed] [Google Scholar]
- 42).Dubouzet J.G., Morishige T., Fujii N., An C.I., Fukusaki E., Ifuku K., Sato F. (2005) Transient RNA silencing of scoulerine 9-O-methyltransferase expression by double stranded RNA in Coptis japonica protoplasts. Biosci. Biotechnol. Biochem. 69, 63–70 [DOI] [PubMed] [Google Scholar]
- 43).Kato N., Dubouzet E., Kokabu Y., Yoshida S., Dubouzet J., Yazaki K., Sato F. (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 48, 8–18 [DOI] [PubMed] [Google Scholar]
- 44).Yamada Y., Kokabu Y., Chaki K., Yoshimoto T., Ohgaki M., Yoshida S., Kato N., Koyama T., Sato F. (2011) Isoquinoline alkaloid biosynthesis is regulated by a unique bHLH-type transcription factor in Coptis japonica. Plant Cell Physiol. 52, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 45).Sato F. (2005) RNAi and functional genomics. Plant Biotechnol. 22, 431–442 [Google Scholar]
- 46).Fujii N., Inui T., Iwasa K., Morishige T., Sato F. (2007) Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res. 16, 363–375 [DOI] [PubMed] [Google Scholar]
- 47).Park S.U., Yu M., Facchini P.J. (2002) Antisense RNA-mediated suppression of benzophenanthridine alkaloid biosynthesis in transgenic cell cultures of California poppy. Plant Physiol. 128, 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Kumagai H., Yamada H., Matsui H., Ohkishi H., Ogata K. (1970) Tyrosine phenol-lyase. I. Purification, crystallization, and properties. J. Biol. Chem. 245, 1767–1772 [PubMed] [Google Scholar]
- 49).Katayama, T. and Kumagai, H. (2010) Tyrosine Phenol-Lyase. In The Encyclopedia of Industrial Biotechnology 7 (ed. Flickinger, M.C.). John Wiley & Sons, Inc., U.S.A., pp. 4752–4757. [Google Scholar]
- 50).Katayama, T. and Kumagai, H. (2010) L-DOPA, Microbial Production. In The Encyclopedia of Industrial Biotechnology 3 (ed. Flickinger, M.C.). John Wiley & Sons, Inc., U.S.A., pp. 1937–1940. [Google Scholar]
- 51).Yamada H., Kumagai H., Kashima N., Torii H., Enei H., Okumura S. (1972) Synthesis of L-tyrosine from pyruvate, ammonia and phenol by crystalline tyrosine phenol-lyase. Biochem. Biophys. Res. Commun. 46, 370–374 [DOI] [PubMed] [Google Scholar]
- 52).Katayama T., Suzuki H., Yamamoto K., Kumagai H. (1999) Transcriptional regulation of tyrosine phenol-lyase gene mediated through TyrR and cAMP receptor protein. Biosci. Biotechnol. Biochem. 63, 1823–1827 [DOI] [PubMed] [Google Scholar]
- 53).Suzuki H., Katayama T., Yamamoto K., Kumagai H. (1995) Transcriptional regulation of tyrosine phenol-lyase gene of Erwinia herbicola AJ2985. Biosci. Biotechnol. Biochem. 59, 2339–2341 [DOI] [PubMed] [Google Scholar]
- 54).Katayama T., Suzuki H., Koyanagi T., Kumagai H. (2000) Cloning and random mutagenesis of the Erwinia herbicola tyrR gene for high-level expression of tyrosine phenol-lyase. Appl. Environ. Microbiol. 66, 4764–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Koyanagi T., Katayama T., Suzuki H., Nakazawa H., Yokozeki K., Kumagai H. (2005) Effective production of 3,4-dihydroxyphenyl-L-alanine (L-DOPA) with Erwinia herbicola cells carrying a mutant transcriptional regulator TyrR. J. Biotechnol. 115, 303–306 [DOI] [PubMed] [Google Scholar]
- 56).Koyanagi T., Katayama T., Suzuki H., Kumagai H. (2008) Altered oligomerization properties of N316 mutants of Escherichia coli TyrR. J. Bacteriol. 190, 8238–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Koyanagi T., Katayama T., Suzuki H., Onishi A., Yokozeki K., Kumagai H. (2009) Hyperproduction of 3,4-dihydroxyphenyl-L-alanine (L-Dopa) using Erwinia herbicola cells carrying a mutant transcriptional regulator TyrR. Biosci. Biotechnol. Biochem. 73, 1221–1223 [DOI] [PubMed] [Google Scholar]
- 58).Nakazawa H., Sano K., Kumagai H., Yamada H. (1977) Distribution and formation of aromatic L-amino acid decarboxylase in bacteria. Agric. Biol. Chem. 41, 2241–2247 [Google Scholar]
- 59).Nakazawa H., Kumagai H., Yamada H. (1974) Constitutive aromatic L-amino acid decarboxylase from Micrococcus percitreus. Biochem. Biophys. Res. Commun. 61, 75–82 [DOI] [PubMed] [Google Scholar]
- 60).Nakazawa H., Kumagai H., Yamada H. (1981) Aromatic L-amino acid decarboxylase from Micrococcus percitreus. Purification, crystallization and properties. Agric. Biol. Chem. 45, 2543–2552 [Google Scholar]
- 61).Nakazawa H., Kumagai H., Yamada H. (1983) Cofacter requirement of constitutive aromatic L-amino acid decarboxylase from Micrococcus percitreus. Agric. Biol. Chem. 47, 1605–1609 [DOI] [PubMed] [Google Scholar]
- 62).Nakazawa H., Kumagai H., Yamada H. (1987) Inhibition of aromatic L-amino acid decarboxylase from Micrococcus percitreus by substrate analogs. Agric. Biol. Chem. 51, 2531–2537 [Google Scholar]
- 63).Nakazawa H., Kumagai H., Yamada H. (1985) Decarboxylation reaction of α-methyl amino acid catalyzed by aromatic L-amino acid decarboxylase from Micrococcus percitreus. Agric. Biol. Chem. 49, 159–165 [Google Scholar]
- 64).Koyanagi T., Nakagawa A., Sakurama H., Yamamoto K., Sakurai N., Takagi Y., Minami H., Katayama T., Kumagai H. (2012) Eukaryotic-type aromatic amino acid decarboxylase from the root colonizer Pseudomonas putida is highly specific for 3,4-dihydroxyphenyl-L-alanine, an allelochemical in the rhizosphere. Microbiology 158, 2965–2974 [DOI] [PubMed] [Google Scholar]
- 65).Yamada H., Uwajima T., Kumagai H., Watanabe M., Ogata K. (1967) Crystalline tyramine oxidase from Sarcina lutea. Biochem. Biophys. Res. Commun. 27, 350–355 [DOI] [PubMed] [Google Scholar]
- 66).Yamada H., Uwajima T., Kumagai H., Watanabe M., Ogata K. (1967) Bacterial amine oxidases Part I. Purification and crystallization of tyramine oxidase of Sarcina lutea. Agric. Biol. Chem. 31, 890–896 [Google Scholar]
- 67).Yamada H., Kumagai H., Uwajima T., Ogata K. (1967) Bacterial amine oxidases Part II. Substrate and inhibitor specificities of tyramine oxidase of Sarcina lutea. Agric. Biol. Chem. 31, 897–901 [Google Scholar]
- 68).Kumagai H., Matsui H., Ogata K., Yamada H. (1969) Properties of crystalline tyramine oxidase from Sarcina lutea. Biochim. Biophys. Acta 171, 1–8 [DOI] [PubMed] [Google Scholar]
- 69).Kumagai H., Yamada H., Suzuki H., Ogura Y. (1971) Action mechanism of tyramine oxidase from Sarcina lutea. J. Biochem. 69, 137–144 [DOI] [PubMed] [Google Scholar]
- 70).Kumagai H., Matsui H., Ogata K., Yamada H., Fukami H. (1968) Oxidation of dopamine by crystalline tyramine oxidase from Sarcina lutea. Memoirs of the Research Institute for Food Science, Kyoto University 29, 69–71 [Google Scholar]
- 71).Roh J.H., Wouters E.D., Yukawa H., Inui M., Minami H., Suzuki H., Kumagai H. (2000) Purification, cloning, and three dimensional structure prediction of Micrococcus luteus FAD-containing tyramine oxidase. Biochem. Biophys. Res. Commun. 268, 293–297 [DOI] [PubMed] [Google Scholar]
- 72).Kumagai H., Uchida H., Yamada H. (1979) Reaction of fungal amine oxidase with β-bromoethylamine. J. Biol. Chem. 254, 10913–10919 [PubMed] [Google Scholar]
- 73).Bai Q., Somerville R.L. (1998) Integration of host factor and cAMP receptor protein are required for TyrR-mediated activation of tpl in Citrobacter freundii. J. Bacteriol. 180, 6173–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Matoba Y., Kumagai T., Yamamoto A., Yoshitu H., Sugiyama M. (2006) Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J. Biol. Chem. 281, 8981–8990 [DOI] [PubMed] [Google Scholar]
- 75).Hernández-Romero D., Sanchez-Amat A., Solano F. (2006) A tyrosinase with an abnormally high tyrosine hydroxylase/dopa oxidase ratio. FEBS J. 273, 257–270 [DOI] [PubMed] [Google Scholar]
- 76).Nakagawa A., Minami H., Kim J.S., Koyanagi T., Katayama T., Sato F., Kumagai H. (2011) A bacterial platform for fermentative production of plant alkaloids. Nature Comm. 2, Article number: 326., doi:10.1038/ncomms1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Epps H.M. (1944) Studies on bacterial amino-acid decarboxylases. Biochem. J. 38, 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).Ajikumar P.K., Xiao W.-H., Tyo K.E.J., Wang Y., Simeon F., Leonard E., Mucha O., Phon T.H., Pfeifer B., Stephanopoulos G. (2010) Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330, 70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Minami H., Kim J.-S., Ikezawa N., Takemura T., Katayama T., Kumagai H., Sato F. (2008) Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. U.S.A. 105, 7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80).Ro D.K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M., Ho K.A., Eachus R.A., Ham T.S., Kirby J., Chang M.C.Y., Withers S.T., Shiba Y., Sarpong R., Keasling J.D. (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943 [DOI] [PubMed] [Google Scholar]
- 81).Leonard E., Runguphan W., O’Connor S., Prather K.J. (2009) Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat. Chem. Biol. 5, 291–300 [DOI] [PubMed] [Google Scholar]
- 82).Hawkins K., Smolke C. (2008) Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 4, 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Chow, Y.L. and Sato, F. (2012) Metabolic engineering and synthetic biology for the production of isoquinoline alkaloids. In Biotechnology for Medicinal Plants; Micropropagation and Improvement. (eds. Chandra, S., Lata, H. and Varma, A). Springer-Verlag, Berlin, pp. 327–343. [Google Scholar]
- 84).Beaudoin G.A., Facchini P.J. (2013) Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem. Biophys. Res. Commun. 431, 597–603 [DOI] [PubMed] [Google Scholar]