Abstract
The amyloid-β (Aβ) protein forms fibrils and higher-order plaque aggegrates in Alzheimer's disease (AD) brain. The copper ion, Cu2+, is found at high concentrations in plaques, but its role in AD etiology is unclear. We use high-resolution pulsed-electron paramagnetic resonance (EPR) spectroscopy to characterize the coordination structure of Cu2+ in the fibrillar form of full-length Aβ(1-40). The results reveal a bis-cis-histidine (His) equatorial Cu2+ coordination geometry, and participation of all three N-terminal His residues in Cu2+ binding. A model is proposed, in which Cu2+–His6/His13 and Cu2+–His6/His14 sites alternate along the fibril axis, on opposite sides of the β-sheet fibril structure. The local intra-β-strand coordination structure is not conducive to Cu2+/Cu1+ redox-linked coordination changes, and the global arrangement of Cu sites precludes facile multi-electron and bridged-metal site reactivity. This indicates that the fibrillar form of Aβ suppresses Cu redox cycling and reactive oxygen species (ROS) production. The insulator configuration suggests application of Cu2+-Aβ fibrils as an amyloid architecture for switchable electron charge/spin coupling and redox reactivity.
INTRODUCTION
Alzheimer's disease (AD) is a neurodegenerative disorder of present and acute future human impact.1 The AD brain is characterized by extracellular histopathological lesions, or plaques.2 The primary component of the plaques is the amyloid-β (Aβ) peptide. The Aβ peptides are n=39-42 amino acids in length, and are denoted Aβ(1-n). Aβ in the plaques is predominantly in the form of amyloid fibrils, which aggregate to form higher-order structures. High-resolution solid-state nuclear magnetic resonance (SS-NMR) studies have led to molecular models for the structure of residues 9-40 of the de-metallated Aβ(1-40) fibril prepared under different conditions,3-5 and the Aβ(1-42) fibril,6 in which β-strand structure in central and C-terminal regions of the peptide promotes stacking of the β-turn β-fold in a parallel, in-register β-sheet arrangement. The β-sheet structure can extend along the fibril axis to micrometer lengths. The metal ion, Cu2+, is found at high concentrations of up to 400 μM in plaques, and it is directly coordinated by the Aβ peptide.7-10 Reactive oxygen species (ROS) generation by Cu2+-Aβ, and ensuing oxidative damage to cellular components, has been proposed to contribute to the etiology of AD,8,11,12 but a specific role of fibril-bound Cu2+ (promoting or ameliorating) has remained unclear.7,8,11,13,14 Mounting refined evidence that relatively low molecular mass oligomers are cytotoxic forms of Aβ,6,15,16 possibly abetted by enhanced Cu2+-promoted ROS generation,14,17 re-invigorates the question: What is the role of fibrillar Cu2+-Aβ in AD?
General features of Cu2+ coordination in the soluble and fibrillar forms of Aβ(1-40) or Aβ(1-42) at pH 7.2-7.4 have been addressed by using continuous-wave EPR18,19 and SS-NMR10 spectroscopies. The EPR parameters, g⊥, g∥, and the copper hyperfine coupling constant, A∥, of Cu2+ complexed with soluble Aβ(1-16), Aβ(1-28), and monomeric Aβ(1-40) and Aβ(1-42) are the same to within experimental uncertainty.18,19 The EPR line shape of the initially monomeric Cu2+-Aβ(1-40) complex did not change18 during fibrillization in the presence of Cu2+. Transmission electron microscopy (TEM) and SS-NMR showed that the presence of Cu2+ during Aβ(1-40) fibril formation did not influence the fibril morphology or reorganize the parallel, in-register β-sheet structure in Aβ(1-40) fibrils.10 These results show that the Cu2+ sites in soluble monomeric and fibrillar forms of Aβ are geometrically comparable, may involve identical or chemically-similar ligands, and that the structure of the N-terminal domain that binds Cu2+, and the distal β-sheet-forming domains of full-length Aβ, are independent.
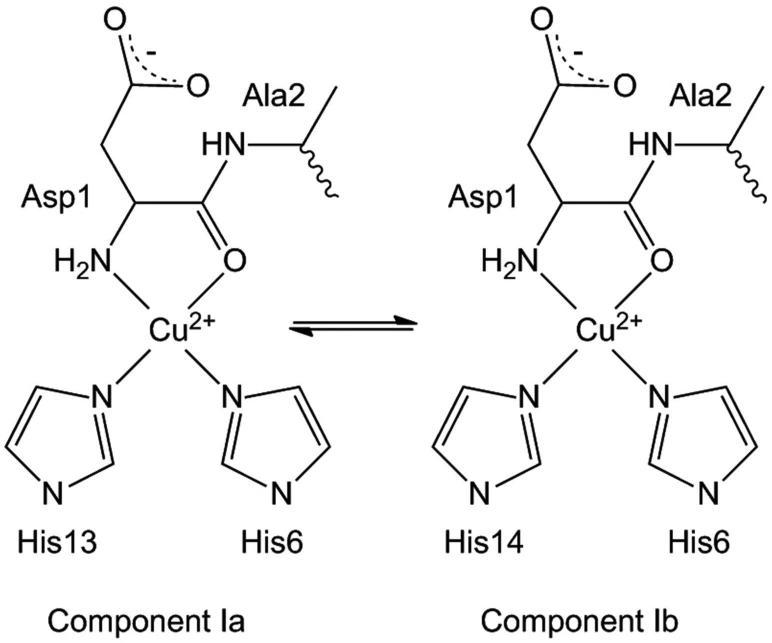
The detailed coordination structure of Cu2+ in the soluble (non-aggregating, non-fibril forming), monomeric complexes of Cu2+ with the truncated Aβ-peptides, Aβ(1-16)20-25 and Aβ (1-28),19,26 or both,27 has been addressed by using electron spin echo envelope modulation (ESEEM) and hyperfine sublevel correlation (HYSCORE) spectroscopies, in conjunction with isotopic labeling, His-to-Ala mutations, and chemical modification. The N-terminal metal binding domain in the Aβ peptide contains three histidine (His) residues, His6, His13, and His14, all of which have been implicated in Cu2+ coordination in soluble Aβ(1-16) and Aβ(1-28). A model of a two-component (Component Ia, Ib; Scheme 1), approximately equimolar mixture of His6/His13- and His6/His14-Cu2+ coordination in soluble Aβ(1-16) was proposed,22 and later revised and extended by additional spectroscopic studies.23-25,28
Scheme 1.
We have used high-resolution pulsed-electron paramagnetic resonance (EPR) spectroscopy to probe histidine (His) coordination of Cu2+ (electron spin, S=1/2; nuclear spin, I=3/2) in frozen solution samples of fibrillar Aβ(1-40), to determine the local coordination structure and global patterning of Cu2+ binding at the N-terminus of fibrillar Aβ(1-40). The results provide molecular-level insights into the chemistry and physiological impact of Cu2+ binding by Aβ fibrils, and suggest applications of Cu2+-Aβ(1-40) as an amyloid architecture29 for designed, switchable electron charge/spin conduction and redox reactivity.
MATERIALS AND METHODS
Sample Preparation
Wild type Aβ(1–16) and Aβ(1–40) peptides were purchased from rPeptide (Athens, GA) or Bachem (King of Prussia, PA). Glycerol, sodium phosphate (NaPi), sodium chloride, phosphotungstic acid (PTA), and 1,1,1,3,3,3 hexafluro-2-proponal (HFIP) were purchased from Fisher Scientific (Pittsburgh, PA). Cu2+-Aβ samples were prepared as described previously.18 Briefly, Aβ(1–40) peptides were monomerized with HFIP and stored at −80 °C. An aliquot of the stock solution was removed for determination of the concentration, by using a bovine serum albumin calibration curve. Fibrillar Aβ(1-40) were prepared by using 100 μM Aβ(1-40) peptide in 50 mM NaPi and 75 mM NaCl (pH 7.2), which was incubated at 37 °C for 7-14 days under quiescent conditions in the presence of equimolar Cu2+. Progress of fibril formation was assayed by TEM. Samples for EPR experiments were separated from the supernatant and concentrated by centrifugation of the pooled volumes in a microfuge (60 min, 16000 rcf), resuspended and washed with buffer, centrifuged a second time (30 min, 16000 rcf) and resuspended in a buffer of 50 mM NaPi and 75 mM NaCl (pH 7.2) containing 50% glycerol (v/v). Samples of the soluble Aβ(1-16) were prepared by resuspending dried peptide in buffer containing 50 mM NaPi, 75 mM NaCl (pH 7.2), and 50% glycerol (v/v). An equimolar amount of Cu2+ was added to the sample prior to freezing in liquid nitrogen.
EPR Spectroscopy and Simulations
CW-EPR spectroscopic measurements were made by using a Bruker ELEXSYS E500 EPR spectrometer with an ER 4123SHQE X-band cavity resonator and a Bruker ER 4131VT liquid nitrogen flow temperature control system. Spectra were acquired at 120 K under non-saturating conditions.
CW-EPR simulations were performed by using the SpinCount software package (M. Hendrich, Carnegie Mellon University: www.chem.cmu.edu/groups/hendrich/facilities/index.html) by diagonalization of the following spin Hamiltonian:
| (1) |
with S= 1/2 and I = 3/2 for Cu2+. In Eq. 1, S and I are the electron and nuclear spin operators, g and A are the electron g and Cu2+ hyperfine tensors, βe is the Bohr magneton, B is the applied magnetic field, and h is Planck's constant.
ESEEM Spectroscopy and Simulations
ESEEM was collected at 6 K on a home-designed and constructed pulsed EPR spectrometer using the three-pulse (π/2-τ-π/2-T-π/2-τ-echo) microwave pulse sequence.30 ESEEM waveforms were cosine Fourier transformed to generate ESEEM frequency spectra. Data processing and analysis was performed by using codes written in Matlab (Mathworks, Natick, MA). All spectra were recorded under the same conditions, with B0 = 303.0 mT and τ = 310 ns. The τ value of 310 ns was chosen to suppress the undesired modulation from 1H shf coupling, and to enhance the contribution of the 14N ΔmI = ±2 (2νdq) combination modulation.
Numerical simulations of the ESEEM were performed by using the OPTESIM software package.31 The spin Hamiltonian for the Cu2+–14N interaction is:
| (2) |
where A is the shf coupling tensor, Q is the nqi tensor, gn is the nuclear g factor, βn is the nuclear magneton, B is the applied magnetic field, and h is Planck's constant. The traceless tensor Q is related to the nuclear quadrupole coupling constant, e2qQ/h, and the electric field gradient asymmetry parameter, η, as follows:
| (3) |
| (4) |
where e is the elementary charge, q is the electric field gradient, and Q is the nuclear quadrupole moment. The relative orientation of the PAS of the nqi and shf tensors is defined by the Euler angles [αQ, βQ, γQ]. For systems with >1 coupled 14N nucleus, the orientation of the shf tensors is defined by the Euler angles, [αA, βA, γA]. Hybrid optimization in the simulations was performed by sequential application of genetic and Nelder-Mead simplex algorithms.31
RESULTS AND DISCUSSION
EPR spectroscopy of natural isotopic abundance fibrillar Cu2+-Aβ(1-40) and soluble Cu2+-Aβ(1-16)
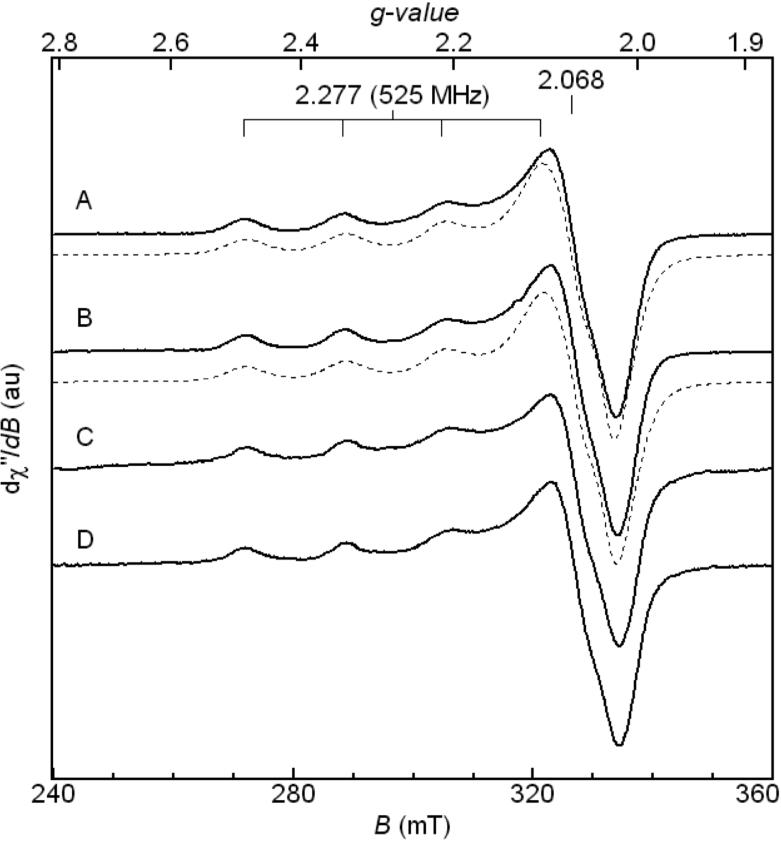
Continuous-wave (CW)-EPR spectra of the soluble Cu2+-Aβ(1-16) and fibrillar Cu2+-Aβ(1-40) complexes are shown in Figure 1. The EPR spectra of the two complexes are closely similar, as quantified by the EPR simulations (Figure 1, A and B; dashed lines), and simulation parameters, which are presented in Table 1. The single set of g⊥ , g∥ and A⊥, A∥ values required in the simulations are the same for the Aβ(1-16) and Aβ(1-40) complexes, to within one standard deviation. The g∥ and A∥ parameters in Table 1 are typical of a tetragonal, type-2 Cu2+ center.32 A three-nitrogen/one-oxygen (3N1O) equatorial coordination of Cu2+ is suggested by the Peisach-Blumberg correlation of g∥ and A∥.33
Figure 1.
CW-EPR spectra of Cu2+-Aβ complexes. (A) Soluble Cu2+-Aβ(1-16), experiment and simulation (dashed line), (B) Fibrillar Cu2+-Aβ(1-40), experiment and simulation (dashed line), (C) 13C,15N-His13-Cu2+-Aβ(1-40), (D) 13C,15N-His14-Cu2+-Aβ(1-40). Acquisition parameters: microwave frequency = 9.45 GHz; microwave power = 2 mW; modulation amplitude = 1 mT; modulation frequency = 100 kHz; time constant = 10.24 ms; conversion time = 81.92 ms; T = 120 K. Spectra A and B are an average of 10 scans, and spectra C and D are an average of 25 scans. Simulation parameters for spectra A and B are presented in Table 1.
Table 1.
CW-EPR simulation parameters for the soluble, monomelic Cu2+-Aβ(1-16) and fibrillar Cu2+-Aβ(1-40) complexes.
| Parametera | Cu2+-Aβ(1-16) | Cu2+-Aβ(1-40) |
|---|---|---|
| g ⊥ | 2.066 ±0.003 | 2.065 ±0.003 |
| g ∥ | 2.277 ±0.003 | 2.277 ±0.003 |
| σg ⊥ | 0.030 | 0.031 |
| σg ∥ | 0.025 | 0.026 |
| A∥ (MHz)b | 524 ±5 | 527 ±5 |
Simulations are for S = 1/2, I = 3/2, and have a Gaussian line width of 1.0 mT. Standard deviations are given.
The copper hyperfine parameter (A∥) is assumed to be from the 65Cu isotope.
The EPR simulation parameters (g∥=2.272, g⊥=2.056, A∥=527 MHz) reported here for the fibrillar Cu2+-Aβ(1-40) complex are comparable with values reported previously for Cu2+ in soluble Aβ(1-40) (g∥=2.268, g⊥=2.061, A∥=534 MHz)18 and in fibrillar Aβ(1-40/42) (g∥=2.268, g⊥=2.061, A∥=534 MHz).18,19 These parameters are also comparable to those for Component I of Cu2+-Aβ(1-16). Our results, in agreement with the previous EPR studies,18,19 thus indicate that the Cu2+ site in fibrillar Aβ(1-40) has a comparable geometry and set of equatorial ligand atoms, as for Component I in Cu2+-Aβ(1-16).
The CW-EPR spectra of the fibrillar 13C,15N-His13-Cu2+-Aβ(1-40) and 13C,15N-His14-Cu2+-Aβ(1-40) complexes are shown in Figure 1, C and D. The EPR line shapes of the isotope-labeled complexes are comparable to the line shapes for the natural isotopic abundance spectra. Although hyperfine coupling with the I=3/2 copper nucleus is displayed at g∥, an influence of the change from 12C (I=0) to 13C (I=1/2), and from 14N (I=1) to 15N (I=1/2), on the EPR line shape is not apparent, owing to inhomogeneous broadening. High resolution techniques of EPR spectroscopy, such as ESEEM, are necessary to resolve the ligand superhyperfine coupling.
14N ESEEM spectroscopy of natural isotopic abundance fibrillar Cu2+-Aβ(1-40) and soluble Cu2+-Aβ(1-16)
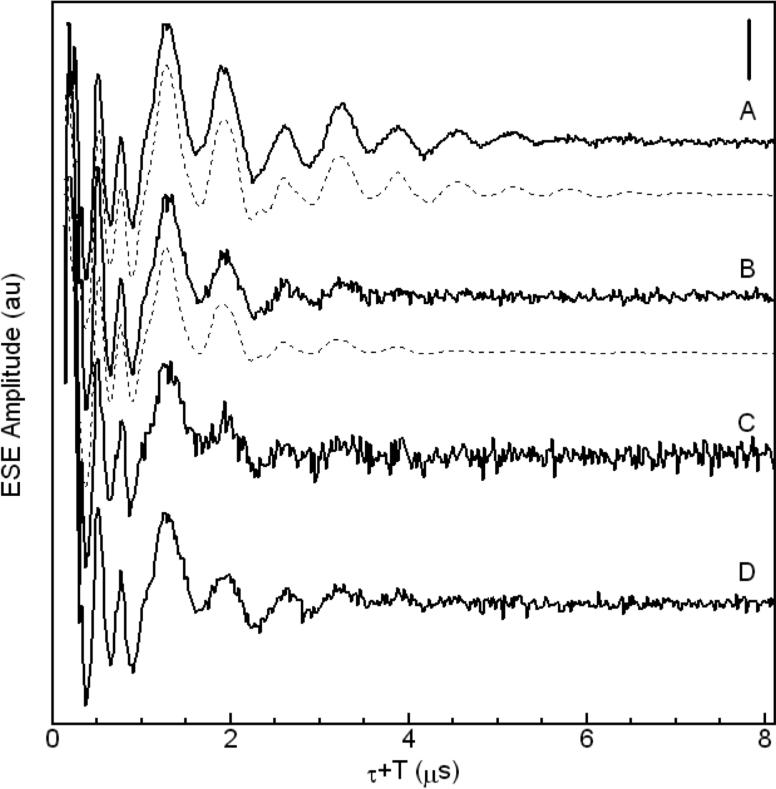
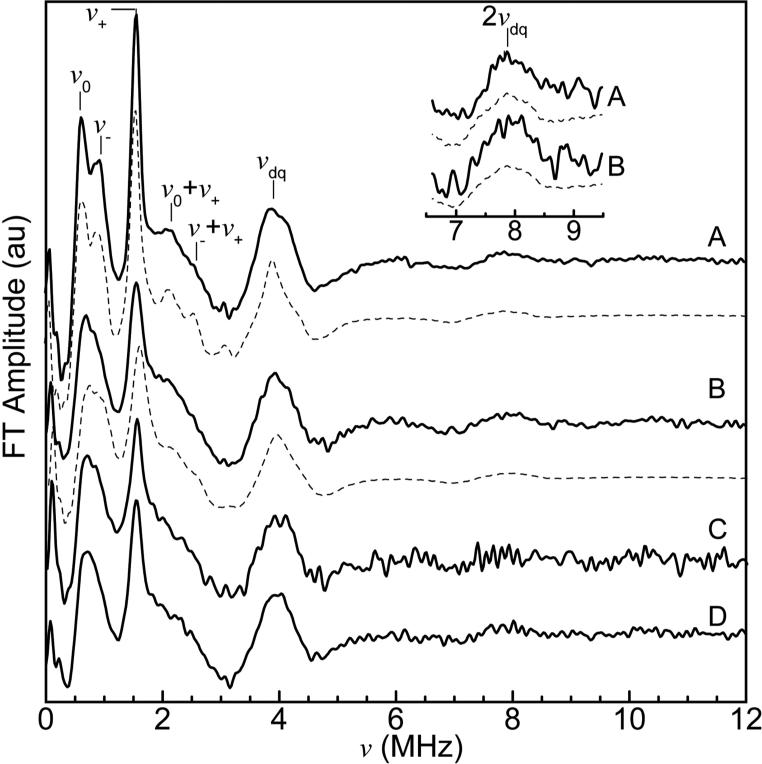
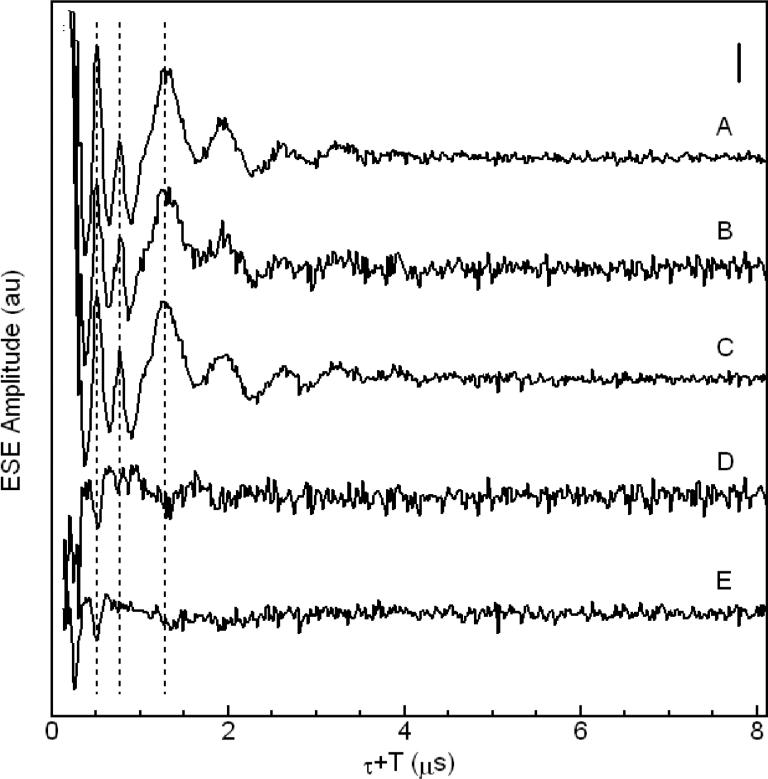
Three-pulse ESEEM waveforms and corresponding cosine Fourier transform (FT) spectra of four Cu2+-Aβ complexes, are shown in Figures 2 and 3, respectively. The spectral features are from the “remote,” non-coordinated 14N of His imidazole, and are characteristic of the “exact cancellation” condition34 for coupling of the unpaired electron spin from Cu2+ with 14N. The ν0, ν− and ν+ “pure” nuclear quadrupole interaction (nqi) frequencies, and the frequencies of the ΔmI = ±1, and ΔmI = ±2 (double quantum, νdq) transitions, that originate from the non-cancellation electron spin manifold, and that include both nqi and superhyperfine (shf) contributions, are identified in Figure 3. Multiple 14N couplings result in the presence of weak modulation at sums of the fundamental modulation periods.34,35 The “combination lines” are denoted in the FT spectra in Figure 3. In addition to the combination lines at the nuclear quadrupole frequencies, a broad feature centered at 7.8 MHz is observed, that corresponds to the 2νdq, double quantum combination line.31 Combination features of order >2 are not observed. The spectra in Figure 3 thus indicate that two histidines coordinate Cu2+ in fibrillar Aβ(1-40).
Figure 2.
Three-pulse ESEEM waveforms of Cu2+-Aβ complexes. (A) Soluble Cu2+-Aβ(1-16), experiment and simulation (dashed line), (B) Fibrillar Cu2+-Aβ(1-40), experiment and simulation (dashed line), (C) 13C,15N-His13-Cu2+-Aβ(1-40), (D) 13C,15N-His14-Cu2+-Aβ(1-40). The vertical scale bar corresponds to 25% of the echo amplitude at τ + T = 8 μs. Acquisition Parameters: microwave frequency, 8.750 GHz, B0 = 303.0 mT, T = 6 K, τ = 310 ns. Spectrum A is an average of 10 scans, spectrum B is an average of 16 scans, and spectra C and D are an average of 25 scans. Simulation parameters for waveforms A and B are presented in Table 2.
Figure 3.
FT spectra of the three-pulse ESEEM waveforms of Cu2+-Aβ complexes. (A) Soluble Cu2+-Aβ(1-16), experiment and simulation (dashed line), (B) Fibrillar Cu2+-Aβ(1-40), experiment and simulation (dashed line), (C) 13C,15N-His13-Cu2+-Aβ(1-40), (D) 13C,15N-His14-Cu2+-Aβ(1-40). Inset: Expanded view of double quantum harmonic region, for (A) soluble Cu2+-Aβ(1-16) and (B) fibrillar Cu2+-Aβ(1-40). Acquisition Parameters: microwave frequency, 8.750 GHz, B0 = 303 mT, T = 6 K, τ = 310 ns. Spectrum A is an average of 10 scans, spectrum B is an average of 16 scans, and spectra C and D are an average of 25 scans. Simulation parameters for spectra A and B are presented in Table 2.
14N ESEEM simulations for the fibrillar Cu2+-Aβ(1-40) and soluble Cu2+-Aβ(1-16) complexes are shown as dashed lines in Figure 3. Simulations were obtained by using the hybrid optimization protocol in the OPTESIM software.31 Simulation parameters are presented in Table 2. Simulation of the ESEEM was optimized by using a minimal set of two coupled 14N. The simulation parameters for the fibrillar Cu2+-Aβ(1-40) and soluble Cu2+-Aβ(1-16) complexes are the same, to within the 99% confidence interval, with the exception of the Axx, parameter, which deviates from overlap of the confidence regions by 0.015 MHz, or 1.3%. The simulations indicate that interactions of the coordinating bis-His-imidazole side chains with Cu2+ in the fibrillar Aβ(1-40) and soluble Aβ(1-16) complexes are comparable, and suggest that the physical-chemical properties of the other ligands involved in Cu2+ coordination are also comparable.
Table 2.
ESEEM simulation parameters for the Cu2+ coupling to the histidine imidazole remote 14N nuclei in the soluble Aβ(1-16) and fibrillar Aβ(1-40) complexes.
| Parameters | Cu2+-Aβ(1–16) | Cu2+-Aβ(1–40) | ||
|---|---|---|---|---|
| Values | 99% Confidence Interval | Values | 99% Confidence Interval | |
| Axx (MHz) | 1.14 | 1.09-1.19 | 1.30 | 1.21-1.39 |
| Ayy (MHz) | 1.74 | 1.70-1.76 | 1.76 | 1.70-1.80 |
| Azz (MHz) | 2.36 | 2.35-2.39 | 2.40 | 2.36-2.45 |
| e2Qq/h (MHz) | 1.59 | 1.57-1.59 | 1.57 | 1.56-1.59 |
| η | 0.71 | 0.70-0.71 | 0.71 | 0.70-0.72 |
| αQ (°) | 357 | 354-359 | 0 | −5.40-16.6 |
| βQ (°) | 31.4 | 30.9-33.9 | 30.7 | 26.8-34.8 |
| γQ (°) | 246 | 243-251 | 250 | 245-256 |
| αA (°) | 71.0 | 61.7-80.1 | 71.7 | 57.1-83.7 |
| βA (°) | 98.2 | 87.7-106 | 89.1 | 77.9-101.0 |
| γA (°) | 1.70 | −6.1-6.2 | 0 | −24.0-13.8 |
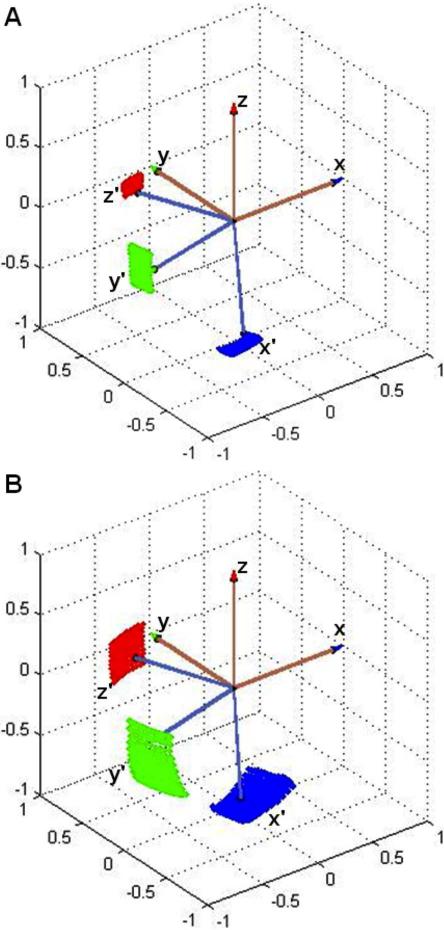
The line shape of the 2νdq feature depends sensitively on the coordination geometry and relative orientation of the His imidazole rings.31 The Euler angles, [αA βA γA], for the relative orientation of the two 14N shf principal axis systems (PAS), specify cis-coordination of Cu2+ by the two His imidazole in fibrillar Cu2+-Aβ(1-40), and also directly confirm the bis-cis-His coordination proposed for soluble Cu2+-Aβ(1-16) Component I.21,22,24 This is illustrated in Figure 4 by the approximately orthogonal x-axes. The x-axis of the dipolar shf PAS lies in the imidazole ring plane, and is directed at an angle of 13°, relative to the imidazole N-N axis.36 The x-axis thus points roughly toward the coordinated Cu2+. The z-axis is directed approximately perpendicular to the imidazole ring plane, at an angle of 4° with respect to the ring plane normal.36 Thus, Figure 4 also indicates that the imidazole rings are tilted by about 90°, with respect to each other. The robustness of the bis-cis-His assignment is shown by the dramatically different waveforms and line shapes of the 2νdq feature in different geometries (Supporting Information, Figures S1 and S2).
Figure 4.
Model for the mutual orientation of the histidine imidazole remote 14N dipolar superhyperfine principal axis systems in (A) soluble Cu2+-Aβ(1-16) and (B) fibrillar Cu2+-Aβ(1-40). Figure axes represent the direction cosines, relative to the reference x, y, z-axis system (brown). The dotted surfaces show the 99% confidence interval, that corresponds to the Euler angles, [αA, βA, γA].
Influence of Selective 15N-Substitution of Histidine Residues on 14N ESEEM from Cu2+ Bound to Fibrillar Aβ(1-40)
Figures 2 and 3 show the ESEEM waveforms and cosine FT spectra for the Cu2+-Aβ(1-40) complexes with 15N-imidazole ring-labeled His13 and His14. Under the exact cancellation condition [νN=A/2, where νN is the nuclear Larmor frequency and A is the shf coupling constant], substitution of 14N (I=1; nqi present) with 15N (I=1/2; no nqi) results in a dramatic change in the ESEEM from the coupled nitrogen.34 The dominant isotropic contribution to the shf coupling of the remote imidazole nitrogen is expected to lead to a doublet line shape in the FT spectrum upon 15N-substitution, with dipolar shf-broadened features near vN−A/2≈0 and vN+A/2≈2vN, that replace the narrow, large-amplitude ν0, ν− and ν+ nqi lines, and distinctive νdq line, that are characteristic of the 14N shf coupling. In addition, ESEEM amplitudes are dependent on the degree of nuclear state mixing,37 which is strong for 14N coupling, because of the combined nuclear quadrupole and shf interactions, with generally non-coincident PAS.34 In contrast, mixing is relatively weaker for 15N coupling. Thus, the 15N ESEEM may not be resolved at modest signal-to-noise ratios, in the presence of exact cancellation ESEEM from other coupled, 14N-nitrogen nuclei.38 This is the case in Figures 2 and 3. The predominant effect of 15N-substitution is, therefore, to eliminate the strong exact cancellation ESEEM from the 14N that is replaced.38 Thus, the qualitatively comparable ESEEM for the 15N-imidazole-labeled and natural abundance fibrillar Cu2+-Aβ(1-40) complexes eliminates a single equatorial bis-His coordination mode for Cu2+, because all 14N combination features would be lost upon 15N-substitution on His13 or His14. This is exemplified by simulations of single- and bis-cis- imidazole coordinated 14N in Supporting Information, Figures S3 and S4. Therefore, a single bis-His coordination mode by the following combinations is ruled out: His6/His13, His6/His14, or His13/His14.
The effects of 15N-substitution are revealed by division of the waveform from the 15N-labeled samples by the all-14N-His waveform. As shown in Figure 5, weak modulation in the quotient waveform is present, with peaks and troughs at positions that correspond to the features in the individual ESEEM waveforms, but with opposite phase. This quotient waveform is consistent with the 15N-substitution, because the waveform in the numerator of the envelope division includes reduced contributions from coupled 14N, relative to the waveform in the denominator. The shf and nqi simulation parameters, obtained for the natural abundance Cu2+-Aβ(1-40) complex in Table 2, were used to perform an ESEEM simulation analysis of the effect of 15N-substitution on a minimal model of two equimolar pairs of bis-cis-His-Cu2+ sites. The simulated quotient waveform in Supporting Information, Figure S5, which is obtained from the division of a 1:1 mixture of [15N, 14N] and [14N, 14N] sites by the all-14N model, shows weak, negative-phase modulation, with opposite-phase peaks and troughs at positions that correspond to features in the individual simulated ESEEM waveforms. The quotient simulation thus agrees with the experimental quotient ESEEM in Figure 5.
Figure 5.
Three-pulse ESEEM waveforms of (A) Fibrillar Cu2+-Aβ(1-40), (B) 13C,15N-His13-Cu2+-Aβ(1-40), (C) 13C,15N-His14-Cu2+-Aβ(1-40), (D) envelope division; B divided by A, (E) envelope division; C divided by A. The vertical scale bar corresponds to 25% of the echo amplitude at τ + T = 8 μs. Aquisition Parameters: microwave frequency, 8.750 GHz, B0 = 303 mT, T = 6 K, τ = 310 ns.
The results and quotient ESEEM analysis are consistent with the approximately equimolar mixture of Cu2+-coordinating bis-His pairs, [His6/His13, His6/His14], in the fibril. This is consistent with the approximately equimolar His6/His13 and His6/H14 contributions in the Component Ia, Ib model proposed for soluble Cu2+-Aβ(1-16),22 and other soluble Cu2+-Aβ complexes.24,26
Model for Cu2+ coordination in fibrillar Aβ(1-40)
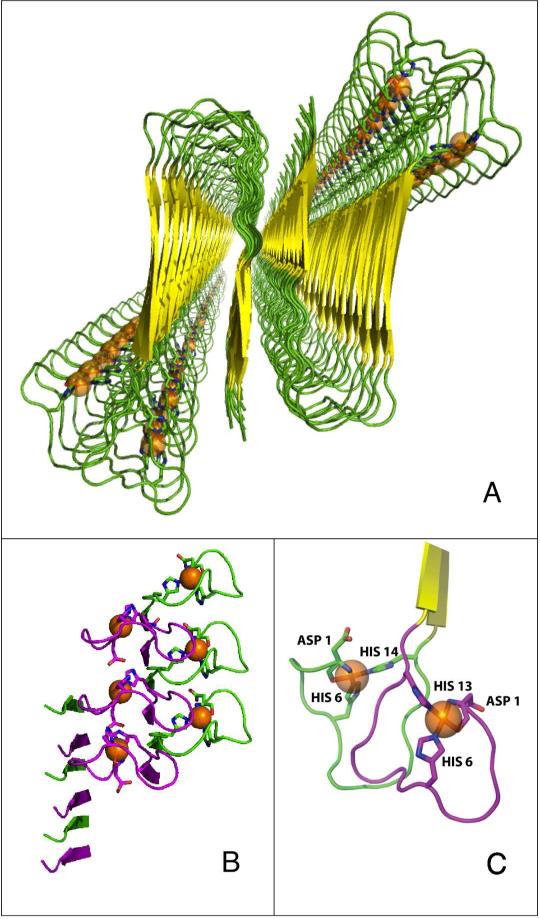
We propose a [His6/His13, His6/His14] model for His coordination of Cu2+ in fibrillar Aβ(1-40), as illustrated in Figure 6. The structure of residues 9-40 in the model is based on the Aβ(1-40) fibril structure determined by SS-NMR (Protein Data Bank ID: 2LMN).3 All atoms of residues 15-40 and backbone atoms of residues 9-14 were fixed in the model. Residues 1-8 are disordered (absent) in the 2LMN structure.3 Therefore, the dihedral angles of residues 1-8, and side chain dihedral angles of residues 1-14, were adjusted at the N-termini of the β-strands of 2LMN, according to the following considerations: (1) equatorial coordination by bis-cis-histidine, terminal-amine, and Ala2 carbonyl (as in Scheme 1), and (2) adherence to local steric constraints. The molecular visualization program, PyMOL (www.pymol.org), was used in model building. At the local level of the Cu2+ sites, the model is consistent with the paramagnetic quenching of the imidazole ring 13Cε and 13Cδ features in the CPMAS SS-NMR studies of either 13C-labeled- His13 or His14, which was reported as 30-60% (corresponding to 40-70% of narrow-line amplitude maintained).10 In addition, the model incorporates the symmetry of the imidazole ring 13C quenching determined by SS-NMR, which was Cε=Cδ>Cγ, and which indicated that His13 and His14 coordinate Cu2+ through the imidazole ring Nε.10 Although we cannot rigorously rule out Cu2+ coordination by a His13/His14 sub-population, as proposed for soluble Aβ(1-16),21 a His13–Cu2+–His14 complex would severely distort the β-strand structure of the liganding peptide around residues 13 and 14. This is because His13 and His14 are part of the β-strand region in the fibrils, and therefore, the imidazole rings extend from opposite sides of the β-sheet3. This is supported by the conclusions of the SS-NMR study,10 and TEM and CW-EPR studies,18 that Cu2+ binding in fibrils does not alter the parallel, in-register β-sheet structure.
Figure 6.
Model for the coordination of Cu2+ in the fibrillar Aβ(1-40) peptide. The model is based on the quaternary structure for the ordered residues 9-40 of Aβ(1-40), which was determined by SS-NMR3 (Protein Data Bank ID, 2LMN). (A) Protein secondary structure cartoon, showing global view down the fibril axis. The Cu2+ ions are represented as orange spheres. (B) Side-on view of the N-terminal region, showing the patterning of Cu2+ sites along the fibril axis. Purple and green loops represent His6/His13 and His6/His14 sites, respectively. (C) Local Cu2+ coordination site geometry.
The model in Figure 6 is consistent with previous studies,18,19 which concluded that the Cu2+-binding, N-terminal domain of Aβ was structurally independent from the β-sheet-forming central and C-terminal regions. Thus, the coordination geometry and atom-types of the non-His equatorial ligands of the Cu2+ site in the soluble complexes with Aβ(1-16) and Aβ(1-28), appear to be maintained in fibrils. We again note that the His13 and His14 side chains are constrained by the β-strand structure of the fibrils to protrude from opposite sides of the β-sheet.3-5 Therefore, the bis-cis-His6/His13-Cu2+ and bis-cis-His6/His14-Cu2+ sites are positioned on different β-sheet faces, and this pattern alternates along the fibril axis, as shown in Figure 6. The proposed alternating arrangement of sites separates Cu2+ beyond the ~7 Å limit of detectable dipolar broadening of the EPR spectrum.19 There is no evidence in the CW-EPR spectra for electron spin exchange-induced broadening. The shortest Cu2+–Cu2+ separation distance in the model is 11 Å, along the direction of the fibril axis. As depicted in Figure 6, this arrangement also suppresses the inter-β-strand coordination of Cu2+, which is consistent with earlier evidence against this binding mode,19 and with the general absence of Cu2+ binding effects on the structure of the demetallated fibril,18 Thus, in our view, the approximately isoenergetic Component Ia and Ib Cu2+ binding modes, that are identified in the soluble Aβ(1-16) peptide fragment,22,23 are a manifestation of the alternating site structure of Cu2+ binding in fibrillar full-length Aβ. Owing to the parallel, in-register stacking of the β-turn-β fold of Aβ(1-40) peptides in all reported high resolution fibril structures,3-5 the alternating pattern of N-terminal Cu2+ sites displayed in Figure 6 is expected to be a characteristic feature of Cu2+–Aβ(1-40) fibril polymorphs.
Implications of Cu2+-His coordination geometry and the patterning of Cu sites in fibrillar Aβ for dioxygen reactivity and ROS generation
Reported pro- and anti-oxidant properties of Aβ are dependent upon the specific measurement conditions and the peptide concentration.12,14 Any cytotoxicity of the Cu-Aβ complex, relative to the demetallated peptide, is thought to be caused by Cu1+-mediated reduction of dioxygen (O2), to produce hydrogen peroxide (H2O2) and other ROS, which is accompanied by Cu2+/Cu1+ redox cycling.8,12,14,39 The coordination geometry of Cu1+ is bis-trans-His in the soluble monomeric Aβ(1-16) and Aβ(1-40) complexes40 and in smaller soluble Aβ peptides.41 A His13–Cu1+–His14 dyad was assigned.40,41 The linear coordination mode is consistent with the preference for Cu1+ bis-trans-His geometry in the histidyl-histidine dipeptide42 and model imidazole complexes.43-45 The bis-cis-His Cu coordination by sequence-distant His pairs, in the model in Figure 6, precludes facile Cu1+ coordination by the sequence-adjacent His13-His14 dyad. Thus, Cu2+ reduction to linear Cu1+ can be achieved only at large inner sphere40,46 and outer sphere reorganization energies, which would substantially decrease the rate of reduction. The model in Figure 6 also spatially segregates the His imidazole side chains in each peptide into either [His6/His13, His14] or [His6/His14, His13]. This prevents facile achievement of the distorted-trigonal (or “pseudo-T-shaped”) interaction of a third imidazole ligand with the dyad.42,43 The distorted-trigonal interaction enhances the sluggish O2 reactivity of linear His-Cu1+-His in models45 and in monomeric Aβ.17 Thus, factors that would promote reoxidation of a linear His-Cu1+-His, if formed at equilibrium in fibrillar Aβ(1-40),47 are absent. As shown in Figure 6, the alternating pattern of Cu sites in fibrillar Aβ(1-40) also constrains formation of inter-peptide ligation of Cu, and maintains a minimum Cu-Cu distance of approximately 11 Å. This eliminates multi-electron redox reactivity and bridged-metal site chemistry.41 For example, a two-electron, two-proton process is proposed for ROS generation by Cu in soluble Aβ(1-16),14 and Cu-Cu distances in oxygen-activating enzymes are typically approximately 4.0-4.5 Å.48 Thus, in addition to the local Cu site constraints, the global arrangement of Cu sites also acts to suppress Cu2+/Cu1+ redox cycling and ROS production. In general, reactivity of the soluble monomer, or non-fibrillar oligomers, of Aβ appears to depend on the prevalent dynamics in these forms.39,49 Extensive configurational sampling39,50 will be suppressed in fibrillar forms of full-length Aβ. Based on the model in Figure 5, and the body of results considered above, we propose that the general physiological impact of fibrillar Aβ, at the levels of both the local Cu site coordination structure,17,40 and global Cu site arrangement, is suppression of Cu redox cycling and ROS production.
The amyloid protein structure provides a robust nanomaterial scaffold for templating ordered arrays of molecules to produce collective functions, including energy and charge transfer.29,51 Metal deposition on engineered amyloid frameworks has created nanowires with relatively high conductivity.52,53 The native Cu2+-Aβ(1-40) fibril accommodates site-specific, high-density metal ion loading, restricts electronic coupling between metal sites. Mutational and chemical modifications in residues 1-8 could be used to promote metal-metal interations, by introducting accessible alternative conformations, or to integrate additional metal ion binding sites. This suggests applications of Cu2+-Aβ fibrils as tunable, and potentially switchable (enabled, for example, by metal ion concentration or pH changes), electron charge/spin conductivity and redox reactivity architectures.
CONCLUSION
We have used 14N ESEEM spectroscopy to address His coordination of Cu2+ in frozen solution samples of fibrillar Aβ(1-40), by measuring the superhyperfine interaction of the unpaired electron spin with the remote imidazole nitrogen nucleus. Simulation analysis of the ligand geometry-dependent 2νdq combination line shape reveals a bis-cis-His coordination of Cu2+. The 14N ESEEM from native and selectively 15N-His-labeled Aβ(1-40) indicates an approximately equimolar mixture of Cu2+-coordinating bis-His pairs, [His6/His13, His6/His14], in the fibril. This is consistent with the approximately equimolar His6/His13 and His6/H14 contributions in the Component Ia, Ib model proposed for soluble Cu2+-Aβ(1-16),22 and other soluble Cu2+-Aβ complexes.24,26 The ESEEM results for fibrillar Cu2+-Aβ(1-40), and control Cu2+-Aβ(1-16), in agreement with the previous EPR studies,18,19 thus indicate that the Cu2+ site in fibrillar Aβ(1-40) has a comparable geometry and set of equatorial ligand atoms, as for Component I in Cu2+-Aβ(1-16). We propose a [His6/His13, His6/His14] model for His coordination of Cu2+ in fibrillar Aβ(1-40), in which residues 1-8 are configured in accord with the EPR and ESEEM-derived constraints, and residues 9-40 assume the β-sheet structure determined by SS-NMR.3 The fibril context dictates that the Cu2+-bis-cis-His6/His13 and Cu2+-bis-cis-His6/His14 sites are positioned on different β-sheet faces, and that this pattern alternates along the fibril axis. The alternating arrangement of sites separates Cu2+ beyond the ~7 Å limit of detectable dipolar broadening of the EPR spectrum.19 The local intra-β-strand coordination structure is not conducive to Cu2+/Cu1+ redox-linked coordination changes, and the global arrangement of Cu sites precludes facile multi-electron and bridged-metal site reactivity. It is therefore concluded that the fibrillar form of Aβ suppresses Cu redox cycling and reactive oxygen species (ROS) production.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Robert Tycko (Laboratory of Chemical Physics, National Institutes of Health) for the gift of the 15N-, 13C- labeled His13 and His14 Aβ(1-40) peptides. The purchase of the Bruker E500 EPR spectrometer was funded by NIH NCRR grant RR17767 and by Emory University.
Abbreviations
- Aβ
amyloid-β protein
- AD
Alzheimer's disease
- CPMAS
Carr-Purcell magic angle spinning
- CW
continuous-wave
- EPR
electron paramagnetic resonance
- ESEEM
electron spin echo envelope modulation
- EXAFS
Extended X-ray absorption fine structure
- FT
Fourier transform
- HYSCORE
hyperfine sublevel correlation
- nqi
nuclear quadrupole interaction
- OPTESIM
optimization of ESEEM simulation
- PAS
principal axis system
- shf
superhyperfine
- ROS
reactive oxygen species
- SS-NMR
solid-state nuclear magnetic resonance
- TEM
transmission electron microscopy
Footnotes
Supporting Information. Simulated three-pulse ESEEM waveforms and FT spectra for representative bis-trans-His Cu2+ complexes; Simulated three-pulse ESEEM waveforms and FT spectra of 14N modulation for different numbers of coupled 14N; Simulated three-pulse ESEEM waveforms for fibrillar Cu2+-Aβ(1-40), which show the effect of 15N-substitution of the remote imidazole nitrogen for different coupling conditions. This information is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Selkoe DJ. Nature Med. 2011;17:1060. doi: 10.1038/nm.2460. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Ann. Intern. Med. 2004;140:627. doi: 10.7326/0003-4819-140-8-200404200-00047. [DOI] [PubMed] [Google Scholar]
- 3.Petkova AT, Yau W-M, Tycko R. Biochemistry. 2006;45:498. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paravastu AK, Leapman RD, Yau W-M, Tycko R. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18349. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi AJ. Am. Chem. Soc. 2011;133:16013. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. Nature Struct. Mol. Biol. 2010;17:561. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR. J. Neurol. Sci. 1998;158:47. doi: 10.1016/s0022-510x(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 8.Bush AI. Trends Neurosci. 2003;26:207. doi: 10.1016/S0166-2236(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 9.Miller LM, Wang Q, Telivala TP, Smith RJ, Lanzirotti A, Miklossy JJ. Struct. Biol. 2006;155:30. doi: 10.1016/j.jsb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Parthasarathy S, Long F, Miller Y, Xiao Y, McElheny D, Thurber K, Ma B, Nussinov R, Ishii YJ. Am. Chem. Soc. 2011;133:3390. doi: 10.1021/ja1072178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung YH, Bush AI, Cherny RA. J. Biol. Inorg. Chem. 2010;15:61. doi: 10.1007/s00775-009-0600-y. [DOI] [PubMed] [Google Scholar]
- 12.Smith DG, Cappai R, Barnham KJ. Biochim. Biophys. Acta. 2007;1768:1976. doi: 10.1016/j.bbamem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Barnham KJ, Bush AI. Curr. Op. Chem. Biol. 2008;12:222. doi: 10.1016/j.cbpa.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Hureau C, Faller P. Biochimie. 2009;91:1212. doi: 10.1016/j.biochi.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Ono K, Condron MM, Teplow DB. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14745. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh DM, Selkoe DJ. J. Neurochem. 2007;101:1172. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 17.Shearer J, Callan PE, Tran T, Szalai VA. Chem. Commun. 2010;46:9137. doi: 10.1039/c0cc02446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karr JW, Szalai VA. Biochemistry. 2008;47:5006. doi: 10.1021/bi702423h. [DOI] [PubMed] [Google Scholar]
- 19.Sarell CJ, Syme CD, Rigby SEJ, Viles JH. Biochemistry. 2009;48:4388. doi: 10.1021/bi900254n. [DOI] [PubMed] [Google Scholar]
- 20.Shin B.-k., Saxena S. Biochemistry. 2008;47:9117. doi: 10.1021/bi801014x. [DOI] [PubMed] [Google Scholar]
- 21.Shin B.-k., Saxena S. J. Phys. Chem. A. 2011;115:9590. doi: 10.1021/jp200379m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drew SC, Noble CJ, Masters CL, Hanson GR, Barnham KJ. J. Am. Chem. Soc. 2009;131:1195. doi: 10.1021/ja808073b. [DOI] [PubMed] [Google Scholar]
- 23.Drew SC, Barnham KJ. Acc. Chem. Res. 2011;44:1146. doi: 10.1021/ar200014u. [DOI] [PubMed] [Google Scholar]
- 24.Dorlet P, Gambarelli S, Faller P, Hureau C. Angew. Chem., Int. Ed. 2009;48:9273. doi: 10.1002/anie.200904567. [DOI] [PubMed] [Google Scholar]
- 25.Alies B, Eury H, Bijani C, Rechignat L, Faller P, Hureau C. Inorg. Chem. 2011;50:11192. doi: 10.1021/ic201739n. [DOI] [PubMed] [Google Scholar]
- 26.Syme CD, Nadal RC, Rigby SEJ, Viles JH. J. Biol. Chem. 2004;279:18169. doi: 10.1074/jbc.M313572200. [DOI] [PubMed] [Google Scholar]
- 27.Karr JW, Akintoye H, Kaupp LJ, Szalai VA. Biochemistry. 2005;44:5478. doi: 10.1021/bi047611e. [DOI] [PubMed] [Google Scholar]
- 28.El Khoury Y, Dorlet P, Faller P, Hellwig P. J. Chem. Phys. B. 2011;115:14812. doi: 10.1021/jp207328y. [DOI] [PubMed] [Google Scholar]
- 29.Knowles TPJ, Buehler MJ. Nature Nanotech. 2011;6:469. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 30.Schweiger A, Jeschke G. Principles of pulse electron paramagnetic resonance. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- 31.Sun L, Hernandez-Guzman J, Warncke K. J Magn Reson. 2009;200:21. doi: 10.1016/j.jmr.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wertz JE, Bolton JR. Electron Spin Resonance. Chapman and Hall; New York: 1986. [Google Scholar]
- 33.Peisach J, Blumberg WE. Arch. Biochem. Biophys. 1974;165:691. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]
- 34.Mims WB, Peisach J. J Chem Phys. 1978;69:4921. [Google Scholar]
- 35.McCracken J, Pember S, Benkovic SJ, Villafranca JJ, Miller RJ, Peisach J. J Am Chem Soc. 1988;110:1069. [Google Scholar]
- 36.Colaneri MJ, Peisach JJ. Am. Chem. Soc. 1995;117:6308. [Google Scholar]
- 37.Mims WB. Physical Review B. 1972;5:2409. [Google Scholar]
- 38.Warncke K, Brooks HB, Babcock GT, Davidson VL, Mccracken JJ. Am. Chem. Soc. 1993;115:6464. [Google Scholar]
- 39.Hureau C, Balland V, Coppel Y, Solari PL, Fonda E, Faller PJ. Biol. Inorg. Chem. 2009;14:995. doi: 10.1007/s00775-009-0570-0. [DOI] [PubMed] [Google Scholar]
- 40.Shearer J, Szalai VA. J. Am. Chem. Soc. 2008;130:17826. doi: 10.1021/ja805940m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Himes RA, Park GY, Siluvai GS, Blackburn NJ, Karlin KD. Angew. Chem., Int. Ed. 2008;120:9224. doi: 10.1002/anie.200803908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Himes RA, Park GY, Barry AN, Blackburn NJ, Karlin KD. J. Am. Chem. Soc. 2007;129:5352. doi: 10.1021/ja0708013. [DOI] [PubMed] [Google Scholar]
- 43.Sanyal I, Karlin KD, Strange JR, Blackburn NJ. J. Am. Chem. Soc. 1993;115:11259. [Google Scholar]
- 44.Sorrel TN, Jameson DL. J. Am. Chem. Soc. 1983;105:6013. [Google Scholar]
- 45.Le Clainche L, Giorgi M, Reinaud O. Eur. J. Inorg. Chem. 2000:1931. doi: 10.1021/ic000072r. [DOI] [PubMed] [Google Scholar]
- 46.Xie B, Elder T, Wilson LJ, Stanbury DM. Inorg. Chem. 1999;38:12. [Google Scholar]
- 47.Feaga HA, Maduka RC, Foster MN, Szalai VA. Inorg. Chem. 2011;50:1614. doi: 10.1021/ic100967s. [DOI] [PubMed] [Google Scholar]
- 48.Mirica LM, Ottenwaelder X, Stack TD. Chem. Rev. 2004;104:1013. doi: 10.1021/cr020632z. [DOI] [PubMed] [Google Scholar]
- 49.Hureau C, Coppel Y, Dorlet P, Solari PL, Sayen S, Guillon E, Sabater L, Faller P. Angew. Chem., Int. Ed. 2009;121:9686. doi: 10.1002/anie.200904512. [DOI] [PubMed] [Google Scholar]
- 50.Balland V, Hureau C, Saveant J-M. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17113. doi: 10.1073/pnas.1011315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang Y, Guo P, Pingali SV, Pabit S, Thiyagarajan P, Berland KM, Lynn DG. Chem. Commun. 2008:6522. doi: 10.1039/b814262a. [DOI] [PubMed] [Google Scholar]
- 52.Scheibel T, Parthasarathy R, Sawicki G, Lin X-M, Jaeger H, Lindquist SL. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4527. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carny O, Shalev DE, Gazit E. Nano Lett. 2006;6:1594. doi: 10.1021/nl060468l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.