Abstract
Summary: DMEAS is the first user-friendly tool dedicated to analyze the distribution of DNA methylation patterns for the quantification of epigenetic heterogeneity. It supports the analysis of both locus-specific and genome-wide bisulfite sequencing data. DMEAS progressively scans the mapping results of bisulfite sequencing reads to extract DNA methylation patterns for contiguous CpG dinucleotides. It determines the DNA methylation level and calculates methylation entropy for genomic segments to enable the quantitative assessment of DNA methylation variations observed in cell populations.
Availability and implementation: DMEAS program, user guide and all the testing data are freely available from http://sourceforge.net/projects/dmeas/files/
Contact: davidxie@vt.edu
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
DNA methylation is a crucial epigenetic modification involved in many biological processes, from normal cellular differentiation to disease genesis and progression. Traditionally, DNA methylation analysis is limited to the determination and comparison of DNA methylation levels. A number of computational tools including BISMA, QDMR, BiQ Analyzer HT and CpG_MPs have been developed to analyze DNA methylation data derived from illumina Beadchip or bisulfite sequence reads (Lutsik et al., 2011; Rohde et al., 2010; Su et al., 2013; Zhang et al., 2011). Recently, great interest has been aroused in decoding DNA methylation patterning to understand the generation of cell diversity. In addition to tracing the cell lineage (Shibata, 2012; Tsai et al., 2012), DNA methylation patterns can be used as a measure of the epigenetic heterogeneity in cell populations (Xie et al., 2011). In particular, with the emergence of next-generation sequencing techniques, rapidly accumulating deep bisulfite sequencing data allow the securitization of DNA methylation patterns on genome-wide scale. However, existing DNA methylation analysis tools mainly focus on the bisulfite sequencing data mapping and the comparison at DNA methylation level. No software has been developed to assess DNA methylation variations embedded in bisulfite sequencing data. Here, we present DMEAS, a C# implementation of the algorithm for DNA methylation entropy calculation (Xie et al., 2011) as an interactive tool to evaluate the variation in DNA methylation patterns.
2 OVERVIEW OF DMEAS
2.1 Input data
DMEAS offers user-friendly interfaces for researchers to import the high-throughput methylation sequencing data analyzed with Bismark software (Krueger and Andrews, 2011). For each sequence read, Bismark provides one line annotation for mapping information including read ID, chromosome ID, genome start position and methylation calls for cytosines identified. With such sequence annotation, DMEAS identifies all possible genomic segments with at least four contiguous CpG dinucleotides and records the combination of their methylation statuses. Based on the mapping result for each segment, the sequence reads covering the corresponding genomic region will be identified and clustered. The DNA methylation pattern will be extracted and the methylation level/entropy will be determined for visualization and comparison. DMEAS also takes user-defined locus-specific methylation data. For each genomic region, the default input file should consist of sample information, locus information and multiple-line numerical data representing DNA methylation patterns. More specifically, DNA methylation statuses are represented with 0, 1 or 2 for unmethylated, methylated or unknown methylation status, respectively. DMEAS processes lines from input stream to extract DNA methylation patterns for all possible genomic segments with four contiguous CpG dinucleotides.
2.2 DNA methylation level and entropy analysis
DNA methylation entropy is calculated as described previously (Xie et al., 2011). Briefly, for a given genomic segment, the frequency of each distinct DNA methylation pattern observed is calculated based on all sequence reads mapped to the locus. Providing the number of CpG sites, the frequencies of all patterns observed and the total number of sequence reads generated for a given genomic locus, methylation entropy could be determined with a modified version of Shannon entropy equation (Xie et al., 2011). To ensure all 16 possible methylation patterns would be considered for a given 4-CpG segment, only genomic segments with ≥16× coverage will be included in the further analysis. DNA methylation level of a genomic region with multiple CpG sites is defined as the percentage of methylated CpG dinucleotides observed. For each high-throughput bisulfite sequencing dataset, DMEAS provides a descriptive statistical summary and the distribution plots for methylation level and entropy. Statistical analyses, including Pearson correlation, etc., are provided for pairwise comparisons and/or multi-sample comparisons. Similar functions have been implemented for the analysis of locus-specific methylation data as well.
2.3 Methylation pattern visualization and data output
DMEAS allows users to visualize methylation patterns at the genomic loci of interest. Heatmap representation is adopted for graphical displays of DNA methylation pattern. In heatmap style, red, blue and gray rectangles represent methylated, unmethylated and unknown methylation status, respectively. Owing to the large volume of high-throughput data, all sequence reads are sorted in ascending order according to their genomic coordinates to facilitate their retrieval later. To achieve a reasonable resolution, DNA methylation pattern is demonstrated in 1 kb window. If no methylation data are found for the 1 kb window targeted, DMEAS will automatically search the upstream and downstream for the most adjacent genomic region with methylation data and provide the corresponding genomic coordinates.
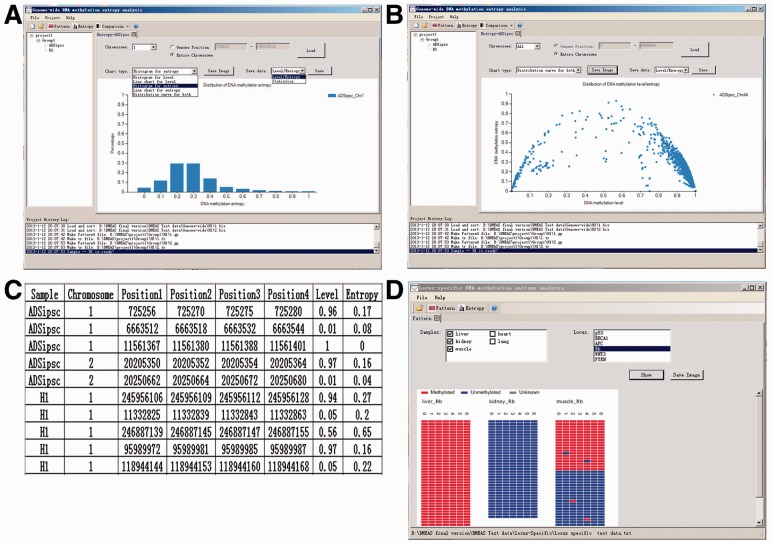
DMEAS also provides user-friendly interfaces to export the results in a wide variety of ways, including text file and image format. The distribution of DNA methylation level or DNA methylation entropy can be exhibited in either histogram (Fig. 1A) or line chart styles, and the correlation between methylation level and entropy is demonstrated in a scatter style (Fig. 1B). In addition, the methylation level and entropy results can also be exported directly to tables (Fig. 1C). Together with the heatmap representation (Fig. 1D), DMEAS generates the output file with basic statistical information for selected samples, such as the number of segments, total number of reads, total number of CpG sites and average methylation level and entropy. All the saved images and tables could be used for further analysis.
Fig. 1.
Methylation level/entropy analysis with DMEAS. (A) The histogram of DNA methylation level/entropy. (B) The scatter plot for the association between the methylation level and the methylation entropy. (C) The output table with DNA methylation entropy/level. (D) DNA methylation pattern heatmap for locus-specific data. The blue, red or gray represents for unmethylated, methylated or unknown methylation status, respectively
3 CONCLUSION
We have successfully developed DMEAS, the first software to enable the analysis of DNA methylation pattern and the quantification of epigenetic heterogeneity. For bisulfite sequencing data, locus-specific or genome-wide, DMEAS can automatically identify and determine the methylation levels and entropies for all possible 4-CpG segments. The visualization of DNA methylation pattern of each segment is implemented and the descriptive statistical summary on segments, including the distributions of methylation level and entropy, are provided. In addition, Pearson correlation was adopted to measure the correlation of the methylation levels and entropies across samples. We anticipate it will assist researchers to explore DNA methylation data in a new dimension.
Funding: This work was supported by grants from the Natural Science Foundation of China [31201002, 81270633]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsik P, et al. BiQ Analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing. Nucleic Acids Res. 2011;39:W551–W556. doi: 10.1093/nar/gkr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde C, et al. BISMA–fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata D. Cancer. Heterogeneity and tumor history. Science. 2012;336:304–305. doi: 10.1126/science.1222361. [DOI] [PubMed] [Google Scholar]
- Su J, et al. CpG_MPs: identification of CpG methylation patterns of genomic regions from high-throughput bisulfite sequencing data. Nucleic Acids Res. 2013;41:e4. doi: 10.1093/nar/gks829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, et al. Heterogeneity and randomness of DNA methylation patterns in human embryonic stem cells. DNA Cell Biol. 2012;31:893–907. doi: 10.1089/dna.2011.1477. [DOI] [PubMed] [Google Scholar]
- Xie H, et al. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 2011;39:4099–4108. doi: 10.1093/nar/gkr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. QDMR: a quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Res. 2011;39:e58. doi: 10.1093/nar/gkr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.