Abstract
The Gram-negative, opportunistic pathogen Acinetobacter baumannii has recently captured headlines due to its ability to circumvent current antibiotic therapies. Herein we show that the multi-drug resistant (MDR) AYE strain of A. baumannii contains a gene locus that encodes three enzymes responsible for the biosynthesis of the highly-modified bacterial nucleotide sugar, UDP-N,N -diacetylbacillosamine (UDP-diNAcBac). Previously, this UDP-sugar has been implicated in the pgl pathway of Campylobacter jejuni. Here we report the overexpression, purification, and biochemical characterization of the A. baumannii enzymes WeeK, WeeJ, and WeeI that are responsible for the production of UDP-diNAcBac. We also demonstrate the function of the phosphoglycosyltransferase (WeeH), which transfers the diNAcBac moiety to undecaprenyl-phosphate. UDP-diNAcBac biosynthesis in A. baumannii is also directly compared to the homologous pathways in the pathogens C. jejuni and Neisseria gonorrhoeae. This work demonstrates for the first time the ability of A. baumannii to generate the highly-modified, UDP-diNAcBac nucleotide sugar found previously in other bacteria adding to the growing list of pathogens that assemble glycoconjugates including bacillosamine. Additionally, characterization of these pathway enzymes highlights the opportunity for investigating the significance of highly-modified sugars in bacterial pathogenesis.
Keywords: bacillosamine, Acinetobacter baumannii, acetyltransferase, aminotransferase, bacterial O-linked glycosylation, biosynthesis, UDP-diNAcBac, glycosylation
Introduction
An alarming trend in antibiotic resistance continues to escalate among human pathogens. A prime example is Acinetobacter baumannii, which has garnered a great deal of attention from the medical community stemming from its capacity to resist a majority of antimicrobial therapies.1A. baumannii is a Gram-negative, aerobic, non-motile opportunistic pathogen that affects immunocompromised patients in a hospital setting. While much effort has been invested in uncovering the mechanism of action of antibiotic resistance,2,3 little has been accomplished in the understanding of pathogenicity. The AYE strain of A. baumannii was originally isolated from the 2001 epidemic outbreak in France resulting in a 26% mortality rate among infected individuals.4,5 Disturbingly, this strain contains an 86-kb genomic island that encodes for 45 of its 52 resistance genes.6 This resistance island, the largest identified to date, is responsible for the inactivation of β-lactams, aminoglycosides, chloramphenicol, rifampin, and tetracycline.7
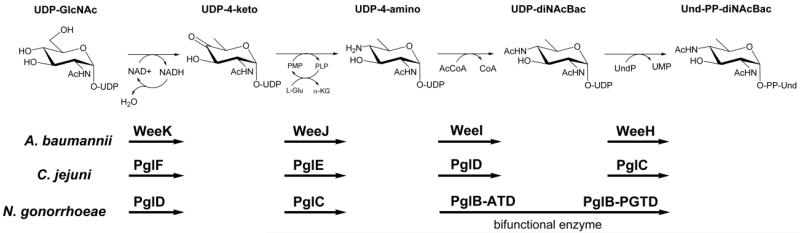
Extensive work has corroborated the link between virulence and bacterial glycosylation in the model system C. jejuni.8 An interesting characteristic of virulence in this pathogen is the biosynthesis of the unique, bacterial UDP-diNAcBac sugar and its incorporation into complex glycoconjugates. Importantly, the disruption of the enzymes responsible for its production results not only in the diminished ability of C. jejuni to adhere to and invade human epithelial cells, but also a reduction in chick and mouse colonization.9,10 Three distinct enzymes are employed in the biosynthesis of UDP-diNAcBac. First, a dehydratase catalyzes the NAD+ dependent C4 oxidation, which promotes elimination of water across the C5-C6 glycosyl bond. This is followed by re-reduction of the α,β-unsaturated system at C6 to generate the UDP-4-keto sugar .11 Subsequently, an aminotransferase catalyzes the transfer of the amino group from l-glutamic acid to the C4 position of UDP-4-keto in a pyridoxal-dependent manner to generate the UDP-4-amino sugar.11 Lastly, an acetyl-coenzyme A (AcCoA)-dependent acetyltransferase generates the UDP-diNAcBac sugar nucleotide.12 Phospho-diNAcBac is then enzymatically transferred to undecaprenyl phosphate and serves as the starting membrane-bound monosaccharide building block for the assembly of more complex oligosaccharides. In C. jejuni, the pathway that utilizes UDP-diNAcBac culminates in the transfer of a heptasaccharide onto the side-chain amide nitrogen of asparagine (N-linked), whereas the system in N. gonorrhoeae transfers a trisaccharide onto a serine or threonine residue (O-linked).13 Importantly, the first sugar in glycan biosynthesis for this O-linked system has been confirmed as UDP-diNAcBac.14
Bioinformatic analysis of the C .jejuni and N. gonorrhoeae UDP-diNAcBac systems resulted in the identification of a series of enzymes that catalyze the biosynthesis of UDP-diNAcBac in the AYE strain of A. baumannii. The ultimate protein glycosylation steps in these pathways can be classified by the distinct oligosaccharyltransferases; PglB in C. jejuni (N-linked) and PglO in N. gonorrhoeae (O-linked). Comparative assessment of their respective oligosaccharyltransferases supported the hypothesis that the AYE strain of A. baumannii was an O-linked system as it bears a close resemblance to PglO in N. gonorrhoeae. Furthermore, genetic analyses based upon sequence homology to their respective C. jejuni and N. gonorrhoeae enzymes were consistent with a series of analogous proteins (WeeK, WeeJ, and WeeI) responsible for UDP-diNAcBac biosynthesis (Figure 1). Additionally, a phosphoglycosyltransferase (WeeH) that catalyzes the transfer of phospho-diNAcBac to an undecaprenol phosphate (Und-P) polyisoprenyl carrier was also identified. Given that less virulent strains of A. baumannii exist in nature, these genomes were searched for the existence of this biosynthetic pathway to determine its prevalence. While multiple strains contained this particular pathway, the antibiotic susceptible A. baumannii strain (ATCC 17978) did not. Instead, this strain contains a distinct O-linked glycosylation system with a core GalNAc sugar anchoring a branched pentasaccharide.15 The terminal O-acetylated glucuronic acid sugar (GlcNAc3NAcAOAc) shares homology to a similar pathway in the PAO1 strain of Pseudomonas aeruginosa16 however these enzymes are absent in the AYE strain of A. baumannii.
Figure 1.
The UDP-diNAcBac biosynthetic pathway in the AYE strain of A. baumannii.
Here we report on the expression, purification, and kinetic characterization of the three enzymes (WeeK, WeeJ, and WeeI) responsible for the biosynthesis of UDP-diNAcBac in the AYE strain of A. baumannii. We also determine the substrate specificity of the phosphoglycosyltransferase (WeeH) that catalyzes the transfer of the UDP-activated bacterial sugar onto an Und-P lipid carrier. Furthermore, we discuss the active site homology between O- and N-linked UDP-diNAcBac pathway proteins in the context of binding and catalysis. This work establishes the presence of the biosynthetic machinery necessary for the production of the UDP-diNAcBac nucleotide sugar in A. baumannii. Biochemical characterization of the UDP-diNAcBac biosynthesis pathway in A. baumannii is significant in the context of its potential relationship to the more pathogenic and antibiotic resistant strains of this serious human pathogen.
Material and Methods
Common Materials
All chemicals were purchased from Sigma-Aldrich unless otherwise stated. The UDP-4-keto, UDP-4-amino, and UDP-diNAcBac sugars were biosynthesized as described previously from the C. jejuni enzymes PglF, PglE, and PglD.12
Cloning, Expression, and Purification
The WeeK, WeeJ, WeeI and WeeH genes were amplified via the polymerase chain reaction (PCR) from the genomic DNA of the AYE strain4 of Acinetobacter baumannii (ATCC). BamHI and XhoI restriction sites were engineered to facilitate cloning of each construct into the pET-24a(+) vector (Novagen). Amplifications were accomplished with the PfuTurbo DNA Polymerase (Stratagene) as described by the manufacturer. Amplicons were purified and double-digested with BamHI and XhoI restriction enzymes (NE Biolabs). Digested inserts and linearized vectors were fractionated by agarose gel electrophoresis and purified with the Wizard SV Gel and PCR Cleanup Kit (Promega). Ligations were conducted with the T4 DNA ligase kit (Promega) using a 15 minute incubation at room temperature. Sequencing by Genewiz (Cambridge, MA) confirmed the presence of all gene products.
The pET24a(+) plasmids containing each gene were used to transform E. coli BL21(DE3)pLysS RIL competent cells (Stratagene). 1 L of LB media containing 50 μg/mL kanamycin and 30 μg/mL chloramphenicol was inoculated with 8 mL of an overnight culture of cells. The cells were then allowed to grow at 37 °C while shaking until they reached an optical density of ~0.8 (λ = 600 nm). The culture was cooled to 16 °C and induced with 0.5 mM iso-β-D-thiogalactosylpyranoside (IPTG). After incubating for 18 hours with shaking, the cells were harvested (2600 × g) and stored at −80 °C until needed.
Each protein purification step was carried out at 4 °C. The cell pellet (~3 g) was resuspended in 40 mL of 50 mM HEPES pH 7.4/100 mM NaCl/30 mM imidazole (Buffer A) and then lysed by sonication. WeeK resuspension buffer was supplemented with 200 μM NAD+ and WeeJ with 200 μM pyridoxal 5′-phosphate. For WeeJ and WeeI, the lysate was cleared by centrifugation (145000 × g, 60 min) and added to 2 mL of Ni-NTA resin (Qiagen). The slurry was allowed to tumble for 3 hours and then packed into a fritted PolyPrep column (Biorad). The resin was washed with 20 column volumes of Buffer A and then eluted with a buffer containing 50 mM HEPES pH 7.4/100 mM NaCl/300 mM imidazole (Buffer B). Fractions containing the purified protein by SDS-PAGE were pooled, dialyzed against 50 mM HEPES pH 7.4/100 mM NaCl (Buffer C) to remove the imidazole, and then supplemented with 15% glycerol. Protein concentrations were calculated based upon the predicted extinction coefficients at λ = 280 nm. Aliquots of the protein were stored at −80 °C until needed.
The membrane-associated proteins WeeK and WeeH required additional purification steps to those of the soluble proteins presented above. Following sonication, cellular debris was cleared at 8000 × g (45 min) and the supernatant was transferred to a clean centrifuge tube. Further centrifugation took place at 145000 × g (60 min) to pellet the cell envelope fraction (CEF). The CEF was homogenized in 5 mL of Buffer A supplemented with 1% Triton X-100. This solution was allowed to tumble overnight and then subjected to centrifugation 145000 × g (60 min) to remove any unsolubilized material. The supernatant was combined with 2 mL of Ni-NTA resin and tumbled for 3 hours. The slurry was added to a PolyPrep column and washed with 20 column volumes of Buffer A supplemented with 0.1% Triton X-100. The protein was eluted from the resin with Buffer B containing 0.1% Triton X-100. Imidazole was removed through dialysis with Buffer C/0.1% Triton X-100. In the case of WeeH, these dialysis conditions resulted protein precipitation. This issue was remediated by increasing the NaCl concentration in the dialysis buffer to 350 mM. Due to the addition of a UV-active detergent, protein concentrations were calculated with the DC Protein Assay Kit (Biorad). Purified protein was supplemented with 15% glycerol and stored at −80 °C.
Dehydratase (WeeK) Activity Assay
Kinetic characterization of WeeK utilized capillary electrophoresis (CE) to directly determine UDP-4-keto product formation. The assay contained 50 mM Tris-HCl pH 8.5, 0.005% Triton X-100, 1 mM NAD+, and varying amounts of UDP-GlcNAc. The reaction was initiated with 0.5 μM WeeK and time points were taken over a time span of 180 minutes at 25°C. The reaction was stopped by filtration through a 10K MWCO membrane (Millipore) to remove the enzyme and the filtrate was injected for 15 s at 30 mbar on a P/ACE MDQ system (Beckman Coulter). Separation of analytes occurred at 20 kV over a 45 minute time period on a bare silica capillary (75 μm × 80 cm) with a 25 mM sodium tetraborate (pH 9.3) running buffer and monitored at a λ = 254 nm. Substrate and product peaks were manually integrated utilizing the Beckman 32 Karat software suite. Steady-state rate parameters were calculated from equation 1 using the program GraFit 6.0.12 (Erithacus Software). The kinetic parameters are a result of duplicate measurements at each substrate concentration.
| (1) |
Aminotransferase (WeeJ) Activity Assay
Calculation of kinetic constants was carried out as described previously (3). Briefly, the generation of the UDP-4-amino product from the WeeJ reaction was coupled to an excess of the C. jejuni acetyltransferase PglD and the activity of WeeJ was determined by following the production of CoASH at 25 °C. In a flat, clear bottom 96-well plate (Falcon) was added 50 mM HEPES pH 7.4, 0.05% BSA, 0.001% Triton X-100, 1 μM PglD, 400 μM AcCoA, and 400 nM WeeJ. The substrate concentrations of l-glutamate and UDP-4-keto were varied separately to determine kinetic parameters utilizing initial velocity measurements while keeping the other substrate at saturation (10 × Km). Reactions were initiated with the l-glutamate substrate and quenched with 20% n-propanol, 2 mM DTNB (5,5′-dithio-bis-(2-nitrobenzoic acid), and 1 mM EDTA over a 30 minute time period, which provides a spectroscopic readout for the production of CoA. The absorbance at 415 nm was monitored on an Ultramark EX microplate imaging system (BioRad). Reactions were performed in duplicate and a blank reaction without WeeJ was set up as a background control and subtracted from the final observed reaction rate.
Acetyltransferase (WeeI) Activity Assay
Kinetic characterization of WeeI was carried out using a previously modified procedure.14 CoASH generation resulting from the acetyltransferase reaction carried out by WeeI was monitored in the presence of Ellman’s reagent (DTNB) through the generation of the TNB2− chromophore in a continuous fashion. To a flat, clear bottom 96-well plate (Falcon) was added 50 mM HEPES pH 7.4, 5 mM MgCl2, 0.05% BSA, 0.001% Triton X-100, 1 mM DTNB, and 1 nM WeeI. Reactions were completed in duplicate and initial rates were measured in the linear portion of the reaction curve over a 5 minute time period at 25 °C. The substrate concentrations of AcCoA and UDP-4-amino were varied separately to determine kinetic parameters using initial velocity measurements while holding the other substrate at a saturating level. Due to the solubility and poor binding of UDP-4-amino to WeeI, the AcCoA Km was determined at Km of the sugar substrate. A background control in the absence of UDP-4-amino was subtracted from each reaction rate.
Phosphoglycosyltransferase (WeeH) Activity Assay
A radioactive assay17 was utilized to establish the UDP-sugar specificity of WeeH. In a 1.5 mL eppendorf tube, 2 nmol of undecaprenyl phosphate was solubilized in 3% DMSO and 1% Triton X-100 by sonication. To this solution was added 30 mM Tris-acetate pH 8.0, 50 mM MgCl2, and 20 μM UDP-sugar (20 mCi/mmol), in a final volume of 100 μL. The reaction was initiated with 200 nM WeeH and time points taken over a 60 minute duration at 25 °C. Aliquots of 15 μL were quenched in 1 mL of 2:1 chloroform:methanol mixture and extracted 3 times using 400 μL PSUP (3% chloroform, 49% methanol, 48% water). Following extraction, 5 mL Ecolite(+) scintillation fluid (MP Biomedicals) and 5 mL OptiFluor (Perkin Elmer) were added to the aqueous and organic layers respectively. A Beckman scintillation counter (LS6500) was employed to determine the radioactivity in each sample.
Results
Expression and Purification of WeeK, WeeJ, WeeI, and WeeH
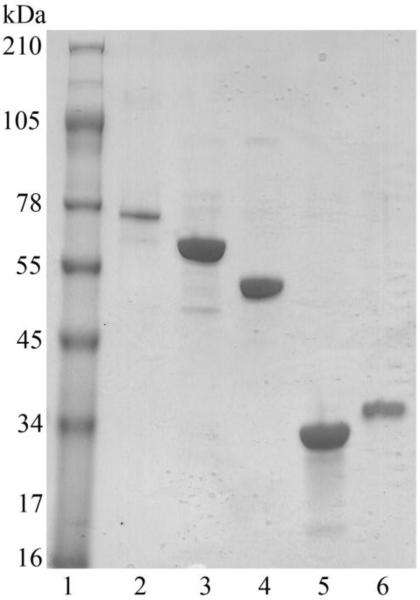
Full-length WeeK, WeeJ, WeeI, and WeeH were cloned from the AYE genomic DNA and ligated into the pET-24a(+) vector. Each protein contained an N-terminal T7 tag and a C-terminal His6 tag for purification purposes. Following overexpression in E. coli BL21 RIL cells and purification with Ni-NTA resin, multi-mg quantities were achieved for each protein from 1 L of culture: WeeK (3 mg), WeeJ (62 mg), WeeI (81 mg), WeeH (2.9 mg). WeeJ was preincubated with excess PLP throughout the entire purification procedure. The stoichiometry of bound PLP to WeeJ ratio was established as 0.9:1 based upon the extinction coefficient of the cofactor (6600 M−1 cm−1) at an absorbance of 390 nm in 0.1 M NaOH. Membrane proteins WeeK and WeeH were solubilized from the lipid membrane with Triton X-100 detergent. Purity for each protein was assessed by SDS-PAGE to be >95% (Figure 2). Full-length constructs were confirmed through Western blot analysis probing with antibody for the T7 and His6 tags. Storage of proteins for >6 months at −80 °C had no affect on enzyme activity.
Figure 2.
SDS-PAGE gradient gel (4-20%) of a MW standard (lane 1), WeeK (lane 2), WeeK140 (lane 3), WeeJ (lane 4), WeeI (lane 5), and WeeH (lane 6).
Functional and Kinetic Characterization of the Dehydratase WeeK
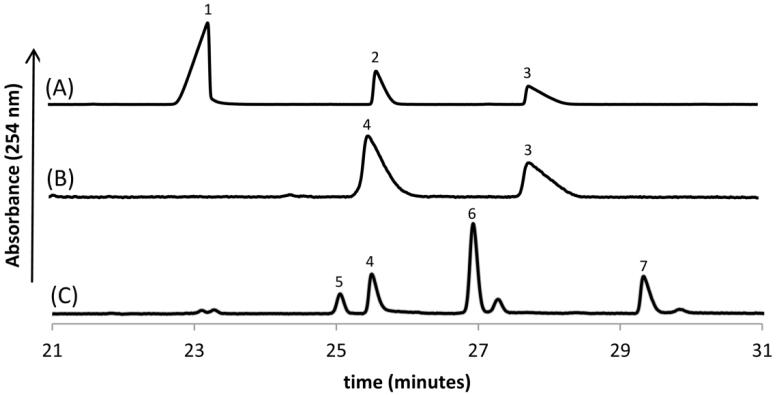
Activity of the full-length WeeK was first investigated with the previously characterized C. jejuni dehydratase substrates NAD+ and UDP-GlcNAc. Substrate turnover was followed by capillary electrophoresis utilizing the UV absorbance of uridine at 254 nm (Figure 3A). After an overnight reaction at 1 μM of WeeK, <5% of the UDP-4-keto product was observed. A control reaction containing the UDP-4-keto sugar product in the same reaction buffer was run in parallel to ensure that this component was stable for the duration of the assay. Steps were taken to understand the very poor of activity for this enzyme. The presence of divalent metals (MgCl2, MnCl2, ZnCl2, and CaCl2) yielded no improvement in product formation (data not shown). Furthermore, varying amounts of salt (NaCl and KCl) produced a similar result. WeeK was also unable to catalyze the reaction containing the substrate pairs UDP-GalNAc/NAD+ or UDP-GlcNAc/NADP+. Two additional detergents were utilized for purification in the anticipation of stabilizing a soluble, active protein. In both cases, n-dodecyl-β-D-maltopyranoside (DDM) and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) resulted in less substrate turnover than the aforementioned Triton X-100 purified material. Lastly, buffer and pH were examined for their effect on dehydratase activity in an in vitro activity assay. While alternate buffer solutions yielded no improvement in activity, pH had a substantial effect. Increasing the pH from 7.4 to 8.5 resulted in a 10-fold increase of the UDP-4-keto product formation. All further experiments utilized the Triton X-100 detergent purified protein at a buffer pH of 8.5.
Figure 3.
Electropherogram trace representing the WeeK (A), WeeJ (B), and WeeI reactions (C). Each numbered peak corresponds to a specific analyte: (1) NAD+; (2) UDP-GlcNAc; (3) UDP-4-keto; (4) UDP-4-amino; (5) UDP-diNAcBac; (6) AcCoA; (7) CoA.
WeeK is annotated in the NCBI protein database as a UDP-glucose 4-epimerase, which would reversibly convert UDP-glucose into UDP-galactose. To test this, WeeK was incubated with 1 mM of either UDP-glucose or UDP-galactose for 18 hours at room temperature in the presence of 1 mM NAD+. Following analysis of these reactions by CE, it was concluded that WeeK does not convert UDP-glucose/UDP-galactose to their respective C4 epimers (data not shown). Based upon experimental evidence, we conclude that WeeK is not an epimerase and instead exhibits only NAD+ dependent dehydratase activity
With a reliable and robust WeeK CE assay in place, steps were taken to measure the kinetic rate constants of UDP-GlcNAc. The UDP-GlcNAc was varied over a substrate concentration of 0.8 – 200 μM at 1 mM NAD+. UDP-4-keto formation was quantified by integrating the area under the product peak from the CE electropherogram trace. The reaction velocities generated from product formation were an average from two separate experiments. These data were plotted versus substrate concentration with equation 1 (Figure S1A) to yield the kinetic parameters in Table 1. For comparison, the previously calculated C. jejuni NAD+-dependent dehydratase (PglF) values are included in the table. Inhibition of WeeK activity was observed at high concentrations of substrate (>500 μM) and therefore not included in the final analysis.
Table 1.
Steady-State Kinetic Parameters for A. baumannii and C. jejuni Dehydratase Enzymes
| dehydratase | substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| WeeK | UDP-GlcNAc | 5.8 ± 1.2 | 2.7 × 10−3 ± 1.2 × 10−4 | 466 |
| WeeK140 | UDP-GlcNAc | 23 ± 4.5 | 4.7 × 10−4 ± 1.7 × 10−5 | 20 |
| PglFa | UDP-GlcNAc | 7000 | 0.12 | 17 |
Kinetic parameters published in reference 12.
Based upon previous studies with the C. jejuni PglF,11,12 the transmembrane domain was removed through cloning to yield the soluble domain of WeeK. This soluble construct improves upon the low yield from full-length construct purification and allows for a facile way to biosynthesize large amounts of UDP-4-keto. To define the appropriate truncation, a sequence alignment with PglF130 and examination of a hydropathy plot with TMHMM18 resulted in the removal of 140 amino acids from the N-terminus (WeeK140). Expression and purification in the pET-24a(+) vector resulted in a 28 mg/L of culture yield at >95% purity by SDS-PAGE This construct catalyzed the conversion of UDP-GlcNAc to UDP-4-keto, however at a reduced rate with respect to the full-length protein. Kinetic characterization of UDP-GlcNAc for WeeK140 was carried out (Figure S1B) and compared directly to the full-length construct in Table 1.
Functional and Kinetic Characterization of the Aminotransferase WeeJ
Capillary electrophoresis was initially utilized to confirm the conversion of UDP-4-keto to UDP-4-amino by WeeJ (Figure 3B). Since this readout allows for direct comparison of substrates and products, standards of each UDP-sugar were run in parallel with this reaction. UDP-4-keto sugar biosynthesized from the C. jejuni and N. gonorrhoeae pathways resulted in the production of UDP-4-amino by WeeJ in both cases. These results confirm that the A. baumannii enzyme exhibits aminotransferase activity with same stereospecificity observed previously from the C. jejuni (PglE) and N. gonorrhoeae (PglC) aminotransferases and confirms our analysis that WeeK does not show epimerase activity. To determine the kinetic constants for the substrates l-glutamate and UDP-4-keto, an in vitro assay coupling the production of UDP-4-amino to the C. jejuni acetyltransferase PglD was developed. In the presence of Ellman’s reagent, generation of the TNB2− chromophore (ε412nm = 14,150 M−1 cm−1) indicates that acetylation of the UDP-4-amino sugar has transpired. Kinetic characterization of each substrate occurred at saturating levels (10 × Km) of the other substrate to ensure the rate of reaction was dependent only upon the concentration of varying substrate. Typical Michaelis-Menten kinetics were observed for all concentrations of l-glutamate (1.6 – 200 mM) and UDP-4-keto sugar (0.03 – 4 mM) (Figure S2). Initial velocity measurements were averaged between two separate runs and plotted to yield the final kinetic parameters in Table 2.
Table 2.
Steady-State Kinetic Parameters for A. baumannii, C. jejuni, and N. gonorrhoeae Aminotransferase Enzymes.
| aminotransferase | substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| WeeJ | UDP-4-keto | 1003 ± 110 | 0.030 ± 0.002 | 30 |
| WeeJ | L-glutamate | 25,000 ± 1900 | 0.17 ± 0.004 | 6.7 |
| PglE | UDP-4-keto | 366 ± 57 | 2.4 ± 0.1 | 6600 |
| PglE | L-glutamate | 11,000 ± 340 | 0.028 ± 0.0003 | 2.6 |
| PglCa | UDP-4-keto | 233 ± 35 | 0.038 ± 0.001 | 164 |
| PglCa | L-glutamate | 4900 ± 900 | 0.025 ± 0.01 | 5.1 |
Kinetic parameters published in reference 14.
Functional and Kinetic Characterization of the Acetyltransferase WeeI
Similar to the characterization of WeeJ, a capillary electrophoresis assay confirmed that WeeI produced UDP-diNAcBac from the UDP-4-amino substrate generated by the C. jejuni and N. gonorrhoeae pathways (Figure 3C). A continuous, in vitro assay relying on the generation of the TNB2− chromophore from Ellman’s reagent was again employed. Initial attempts to determine kinetic parameters for UDP-4-amino resulted in an atypical sigmoidal binding curve suggestive of positive cooperativity (Hill coefficient = 2) (data not shown). Further experiments were applied to establish that WeeI activity was dependent upon MgCl2. In the presence of EDTA, WeeI retained the ability for substrate turnover establishing that MgCl2 is not essential for catalytic function of this enzyme. Comparison of the reaction rates indicated a 6.4-fold increase in activity with the addition of 5 mM divalent magnesium. The presence of MgCl2 resulted in typical Michaelis-Menten kinetics over a range of AcCoA (0.02 – 2 mM) and UDP-4-amino (0.08 - 10 mM) concentrations (Figure S3). As a result of the poor binding of UDP-4-amino to WeeI, the apparent AcCoA Km was determined at a UDP-4-amino concentration of 4 mM (at Km). Kinetic parameters listed in Table 3 are the outcome of initial velocity measurements repeated in duplicate.
Table 3.
Steady-State Kinetic Parameters for A. baumannii, C. jejuni, and N. gonorrhoeae Acetyltransferase Enzymes
| acetyltransferase | substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Weel | UDP-4-amino | 4300 ± 140 | 1.1 × 104 ± 1.7 × 102 | 2.6 × 106 |
| Weel | AcCoA | 110 ± 6.0 | 3.5 × 103 ± 5.5 × 101 | 3.2 × 107 |
| PglD | UDP-4-amino | 311 ± 23 | 1.2 × 104 ± 3.8 × 102 | 4.0 × 107 |
| PglD | AcCoA | 194 ± 30 | 1.1 × 104 ± 5.0 × 102 | 5.5 × 107 |
| PglB-ATD | UDP-4-amino | 192 ± 26 | 2.0 × 103 ± 8.2 × 101 | 1.0 × 107 |
| PglB-ATD | AcCoA | 338 ± 47 | 2.3 × 103 ± 1.1 × 102 | 6.9 × 106 |
Substrate Specificity of the Phosphoglycosyltransferase WeeH
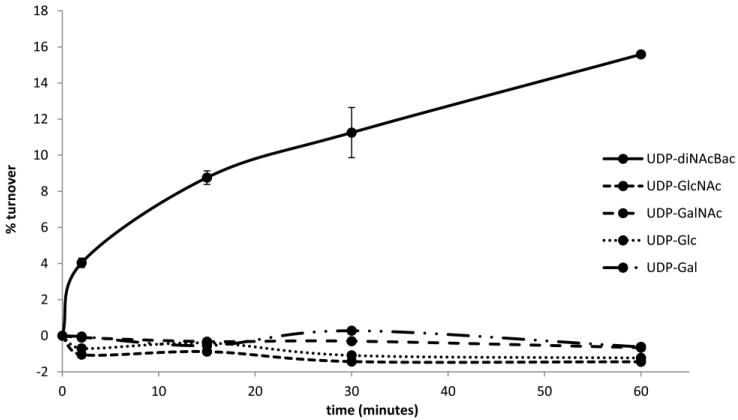
Substrate specificity of WeeH was determined using a previously established radioactivity-based assay.14,19 This method relied on the transfer of a tritium-labeled phospho-sugar (from the UDP-activated substrate) to a hydrophobic undecaprenyl phosphate. The polyprenyldiphosphate-monosaccharide product (Und-PP-diNAcBac) is extracted into the organic phase separating it from the aqueous soluble unreacted radioactive UDP-diNAcBac. In total, five UDP-sugars were analyzed for their ability to act as a substrate for WeeH. This phosphoglycosyltransferase exhibited clear selectivity for UDP-diNAcBac over all other UDP-sugars (UDP-GlcNAc, UDP-GalNAc, UDP-Glc, UDP-Gal) (Figure 4).
Figure 4.
Specificity of the A. baumannii phosphoglycosyltransferase WeeH in the presence of an assortment of UDP-sugars. Aliquots of the reaction containing 2 nmol Und-P, 1% Triton X-100, 3% DMSO, 30 mM TRIS-acetate pH 8.0, 50 mM MgCl2, 40 μM UDP-sugar, 20 μM UDP-GalNAc (20 mCi/mmol), 4.5 μM C. jejuni PglA, and 200 nM WeeH were taken over a 30 minute time course. Error bars represent standard deviation from triplicate measurements.
Active Site Comparison Between O- and N-linked UDP-diNAcBac Pathway Proteins
To better understand the relationship between UDP-diNAcBac pathways in the O- and N-linked systems, active site sequence homology was investigated with an emphasis on the aminotransferase and acetyltransferase enzymes in the C. jejuni, N. gonorrhoeae, and A. baumannii pathways. The NAD+-dependent dehydratase enzymes were excluded from this comparative analysis since structures of these enzymes have not yet been determined.
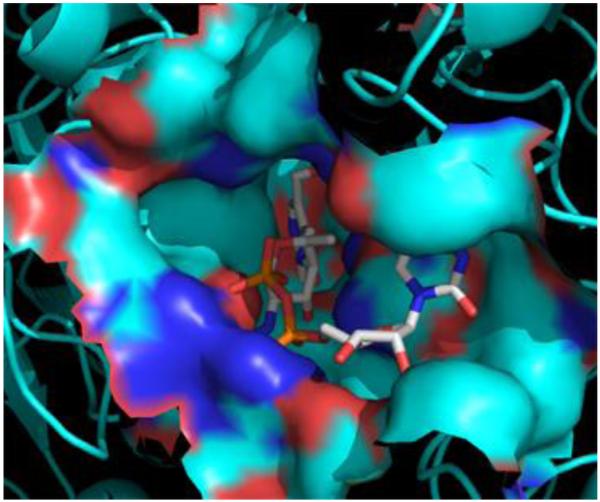
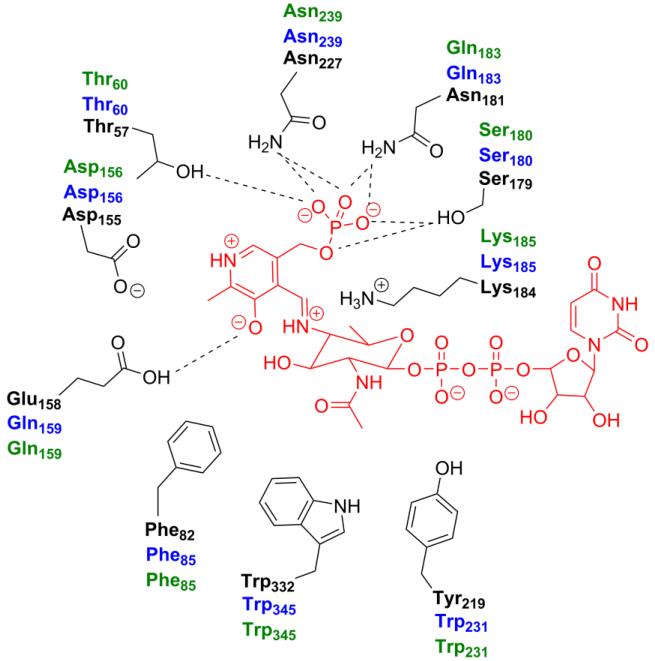
To define the binding pocket for the aminotransferase PglE from the N-linked glycosylation pathway, the structural analysis of a homologous enzyme from Helicobacter pylori (PseC) was employed.20 A crystal structure of PseC with the bound external aldimine (2FNU) was aligned with the PLP bound PglE (1O61) crystal structure to define the active site residues. PseC catalyzes the transamination reaction at the C4 position of a UDP-activated sugar similar to the PglE UDP-4-keto substrate. In this case, the only variation is the stereochemistry of the methyl group at the C5 position (β-l-arabino-hexulose as opposed to α-d-xylo-hexulose). Alignment of the PseC and PglE structures resulted in a root mean square (rms) value of 1.1 Å for the monomer and a rms value of 0.6 Å for the active site residues. A second aminotransferase structure from Pseudomonas aeruginosa (WbpE) containing the bound external aldimine was used to provide further support for the PseC findings.22 The residues defining the PglE active site were identical in both examples. The aminotransferase binding pocket was defined to a 5 Å distance from the external aldimine-bound molecule in the structural visualization program PyMOL (Figure 5).23 Sequence alignment of the N. gonorrhoeae (PglC) and A. baumannii (WeeJ) aminotransferases to PglE was accomplished using Clustal Omega. The final alignment among the three aminotransferases was visually represented by the program Jalview (Figure S4).24 While the overall sequence identity between O-linked and N-linked aminotransferases is relatively low, the enzymes from the O-linked glycosylation pathway (PglC and WeeJ) exhibit exceptionally high homology (67% sequence identity) (Table 4). This observation is even more apparent when comparing the residues within the active site. Not surprisingly, the catalytic lysine residue responsible for product formation is completely conserved among the C. jejuni (K184), N. gonorrhoeae (K185), and A. baumannii (K185) aminotransferases (Figure 6). Six of the ten PseC-binding residues interact with PLP and homologous amino acids can be accounted for in the PglE structure. Of note, the PglE residues D155, S179, N227 and T57 are completely conserved among the three aminotransferases and associate directly with PLP. While E158 and N181 are not identical in the N. gonorrhoeae and A. baumannii model, a similar role can be hypothesized by glutamine at both positions. Only Y316 in the PseC structure has direct interaction with the sugar moiety. No obvious counterpart can be identified in the sequence alignment with PglE, PglC, and WeeJ.
Figure 5.
Surface representation of C. jejuni PglE binding pocket. The crystal structure of the homologous aminotransferase PseC bound to the PMP-UDP-l-AltNAc external aldimine (2FNU) was utilized to define the active site. Following alignment with the PLP-bound PglE crystal structure (1O61), amino acids within 5 Å of the external aldimine were identified as binding-pocket residues.
Table 4.
Sequence Identity for A. baumannii (Ab), C. jejuni (Cj), and N. gonorrhoeae (Ng) Aminotransferase Enzymes.
| aminotransferase pair | total protein (%) | external aldimine active site (%) |
|---|---|---|
| PglE(Cj)/PglC(Ng) | 22 | 38 |
| PglE(Cj)/WeeJ(Ab) | 18 | 38 |
| WeeJ(Ab)/PglC(Ng) | 67 | 90 |
Figure 6.
Illustration of the relevant amino acids responsible for the aminotransferase binding pocket in PglE(Cj). Residues labeled in blue and green represent analogous positions in PglC(Ng) and WeeJ(Ab) respectively.
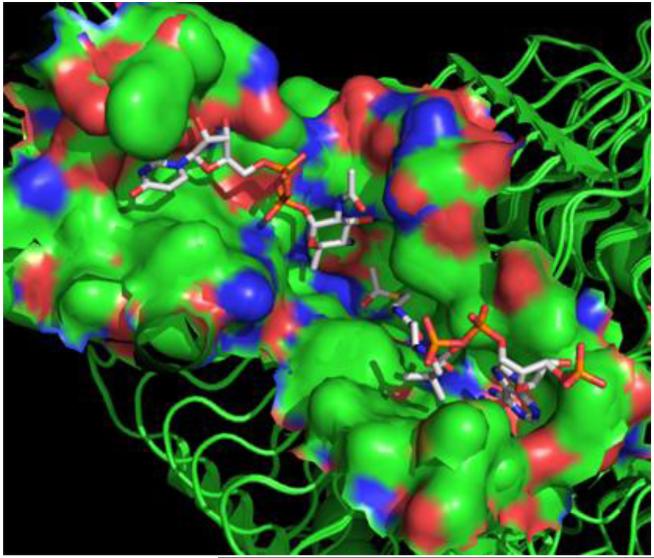
In order to establish the acetyltransferase binding pockets of WeeI and PglB-ATD, the C. jejuni PglD crystal structures containing bound UDP-4-amino (3BSS) and AcCoA (3BSY) were utilized.25 Each binding site was defined as for the aminotransferases, with a 5 Å distance surrounding the respective substrate (Figure 7). Sequence alignment and visualization were again accomplished utilizing Clustal Omega and Jalview (Figure S5). Interestingly, the sequence identity of both active sites varies highly depending upon the specific substrate-binding pocket and acetyltransferase pair being examined (Table 5). The AcCoA binding site exhibits more homology between the O-linked (WeeI/PglB-ATD) pathway enzymes. The majority of interactions between AcCoA and protein side chains occur at the carbonyl oxygen of the thioester (Figure 8A). The nucleotide and pantetheine moieties of AcCoA are held in the substrate-binding site by a network of water molecules and backbone interactions from I155 and G173, which are represented by threonine and glycine in the O-linked acetyltransferase enzyme sequences. Unexpectedly, the UDP-4-amino binding pocket shares more similarity between N-linked PglD and O-linked PglB-ATD. Interactions between the pyranose C4 amine (H125), the ribosyl 3′ hydroxyl group (D35), and the uridine imide (D36) are exclusively conserved (Figure 8B). The uracil of the NDP sugar is stabilized by a similar hydrophobic pocket (Y10, I55, I60, and I64) in PglB-ATD and WeeI model structures. There are two major differences in the proposed WeeI UDP-4-amino binding pocket with respect to PglD. The asparagine at position 162 that interacts with the carbonyl oxygen of the pyranose C2 acetyl group is replaced with a glycine (G175). Importantly, the H15 residue that interacts with the sugar substrate β-phosphate moiety and inserts into the pocket to accommodate UDP-4-amino is replaced with a phenylalanine (F13).
Figure 7.
Surface representation of the C. jejuni PglD binding pocket. Amino acid residues within 5 Å of the UDP-4-amino (top left) and AcCoA (bottom right) substrates were classified as contributing to the acetyltransferase active site. Binding pocket identification relied on the UDP-4-amino (3BSS) and AcCoA (3BSY) bound PglD crystal structures.
Table 5.
Sequence Identity for A. baumannii (Ab), C. jejuni (Cj), and N. gonorrhoeae (Ng) Acetyltransferase Enzymes
| acetyltransferase pair | total protein (%) | AcCoA active site (%) | UDP-4-amino active site (%) |
|---|---|---|---|
| PglD(Cj)/PglB-ATD(Ng) | 34 | 37 | 64 |
| PglD(Cj)/WeeI(Ab) | 26 | 34 | 48 |
| WeeI(Ab)/PglB-ATD(Ng) | 26 | 56 | 42 |
Figure 8.
Illustration of the relevant amino acids responsible for the AcCoA (A) and UDP-4-amino (B) binding pockets in PglD(Cj). Residues labeled in blue and green represent analogous positions in PglB(Ng)ATD and WeeI(Ab) respectively.
Discussion
The A. baumannii enzymes WeeK, J, and I produce UDP-diNAcBac
Previous studies have focused on characterizing the UDP-diNAcBac glycosylation pathway enzymes in N-linked (C. jejuni)12 and O-linked (N. gonorrhoeae)14 systems. The finding that the AYE strain of A. baumannii contains an O-linked bacillosamine biosynthesis pathway further confirms the connection between the sugar and glycoconjugates that may be involved in pathogenicity while adding to the growing number of bacteria with this system. This strain of A. baumannii is of particular interest due to its extreme antibiotic resistance. Understanding the virulence factors associated with A. baumannii is of great importance to the medical community particularly as a potential new target in efforts to address the ever-growing resistance to current antibiotics. The A. baumannii dehydratase (WeeK), aminotransferase (WeeJ), and acetyltransferase (WeeI) were individually investigated for their ability to catalyze their anticipated substrates and characterized kinetically. Activity assays with UDP-sugar substrates generated from the C. jejuni and N. gonorrhoeae glycosylation pathways confirmed that the A. baumannii enzymes utilize the same general mechanism to produce UDP-diNAcBac. From a kinetic viewpoint, it appears that this pathway uses a similar overall strategy employed by homologous enzymes in C. jejuni and N. gonorrhoeae. Specifically, the committed step in UDP-diNAcBac biosynthesis is controlled by the first and rate-limiting enzyme on this pathway (WeeK) while the final acetylation of UDP-4-amino by WeeI provides the most catalytically efficient reaction. Collectively, these enzymes are responsible for the biosynthesis of the highly-modified bacterial NDP sugar, UDP-diNAcBac. While the main focus of this paper has been on the characterization of the early pathway enzymes responsible for UDP-diNAcBac biosynthesis, composition of the final protein-linked oligosaccharide is still unknown.
UDP-diNAcBac enzyme diversity in N- and O-linked glycosylation
Previous kinetic characterization of the C. jejuni dehydratase PglF resulted in a Km of 7 mM and a kcat of 0.12 s−1.12 Surprisingly, the A. baumannii WeeK binds to UDP-GlcNAc with a significantly higher affinity (1000-fold) however catalyzes this reaction at an appreciably reduced rate (44-fold). As a result, WeeK is a catalytically more efficient enzyme (kcat/Km = 466 M−1 s−1) relative to PglF (kcat/Km = 17 M−1 s−1). The two NAD+-dependent dehydratases have a sequence identity (31%) that is similar to other homologous proteins on this pathway yet exhibit contrasting kinetic parameters. It is interesting that WeeK binds UDP-GlcNAc with such high affinity since this substrate is utilized for many other pathways within the cell including biofilm formation,26 lipooligosaccharide,27 and various cell envelope components.
To define the WeeJ aminotransferase UDP-sugar binding pocket, a homologous structure from Helicobacter pylori (PseC) was employed (Figure 5). When comparing the sequence identities of the three aminotransferases (overall and active site), a trend emerges (Table 4). While the N-linked C. jejuni aminotransferase catalyzes the same reaction, it shares little in identity to its O-linked relatives. This observation however is not reflected in the catalytic efficiency for the substrate l-glutamate (Table 2) as all three aminotransferases share comparable kinetic parameters. When evaluating catalytic efficiency for the UDP-4-keto substrate in relation to its sequence, a different story unfolds. PglE has an elevated rate of turnover in comparison to the O-linked pathway proteins that result in a 39-fold (PglC) and 217-fold (WeeJ) increase in catalytic efficiency. In contrast, PglC and WeeJ share a similar albeit lower efficiency for UDP-4-keto catalysis.
The final step in the biosynthesis of UDP-diNAcBac is catalyzed by the acetyltransferase WeeI. The AcCoA and UDP-4-amino binding pockets have been well established through C. jejuni PglD crystallographic analysis (Figure 7).25 While a clear trend was established when comparing O-linked versus N-linked aminotransferases, a different picture emerges with the acetyltransferase enzymes. With respect to sequence identities, PglD and PglB-ATD from N. gonorrhoeae share greater homology in all aspects apart from the AcCoA binding pocket (Table 5). This is a surprising observation when relating these results with homology between both the dehydratases and aminotransferases. It is apparent that the gene products of UDP-diNAcBac biosynthesis in A. baumannii were acquired collectively as they are located consecutively in the same operon. It is not currently understood why changes to the acetyltransferase binding pocket may have occurred with respect to the substrate affinity and fitness of this particular strain of the bacterium. The similarity in AcCoA kinetic parameters (Table 3) is directly reflected in the conserved residues for cofactor binding. Unexpectedly, PglD and PglB-ATD share a higher homology in their UDP-4-amino binding sites relative to WeeI (Table 5). Many of the residues that interact with the UDP-sugar are strictly conserved across all three acetyltransferases. Surprisingly, WeeI exhibits poor affinity for UDP-4-amino with respect to the other acetyltransferases (> 10-fold). Only two major differences are observed in the sugar-binding pocket (Figure 8B). First, an asparagine side chain that interacts with the carbonyl oxygen of the pyranose C2 acetyl group in the PglD structure is replaced with glycine. However, an adjacent glutamine in WeeI may serve as a hydrogen-bond donor depending upon its location within the tertiary structure. Second and seemingly more noteworthy is the replacement of histidine (H15) with phenylalanine in WeeI (F13). This residue interacts with the sugar substrate β-phosphate and repositions in the pocket to accommodate UDP-4-amino. Therefore, this amino acid is positioned to act as a gatekeeper for sugar substrate binding. The hydrophobicity and size of this residue with respect to histidine may partially explain the poor affinity of this substrate towards WeeI (Table 3).
Enzymatic flux through the UDP-diNAcBac pathway
To eliminate adverse byproducts and off-pathway reactions, nature often exploits substrate channeling, wherein intermediates are shuttled to successive enzymes to increase the efficiency of a particular pathway. In the case of the UDP-diNAcBac biosynthesis, the initial NAD+-dependent dehydratase is catalytically inefficient with respect to the subsequent enzymes in the pathway.12,28 Nevertheless, interpretation of these assay results must be viewed with some caution since the in vitro analysis conditions may not reflect the true kinetic potential of the membrane-bound dehydratase in its natural cellular environment. WeeK appears to function as a gatekeeper to UDP-diNAcBac production as the formation of the UDP-4-keto sugar is the rate-limiting step. In order to drive this pathway forward, downstream enzymes appear to be tuned to increase their catalytic efficiency. In all three acetyltransferases examined in this paper, the catalysis of UDP-4-amino to UDP-diNAcBac is significantly elevated with respect to the earlier pathway enzymes. The high catalytic efficiency of the acetyltransferase drives the production of UDP-diNAcBac by rapidly consuming UDP-4-amino and in turn promotes the conversion of more UDP-GlcNAc to UDP-4-keto. A similar effect can be observed in the biosynthesis of UDP-ManNAc(3NAc)A in P. aeruginosa;16 additionally, this type of flux is prevalent in metabolic pathways. For example, glycolytic flux in bacteria utilizes a feed-forward loop where high levels of fructose-1,6-bisphosphate signal for increased activity of glyceraldehyde 3-phosphate dehydrogenase (GAPDH).29,30 Pyruvate kinase (PK) is also part of this loop and its activity is responsible for flux into the lower half of glycolysis. Metabolic flux control is also elicited through the pyruvate node during anaerobic growth in Escherichia coli to maintain redox balance in the cell.31 UDP-diNAcBac biosynthesis is yet another example of flux created by a highly active enzyme at the terminal end of the pathway that can overcome the deficient catalytic efficiency of preceding enzymes.
These studies establish details of the characterization of the early UDP-diNAcBac pathway proteins WeeK, WeeJ, WeeI, and WeeH. Comparison to the analogous C. jejuni and N. gonorrhoeae systems has resulted in an understanding of the similarities and differences between N- and O-linked glycosylation pathway enzymes. Although a direct correlation between pathogenicity and O-linked glycosylation in the AYE strain of A. baumannii remains to be elucidated, this work highlights an analogous pathway previously shown to diminish infectivity when disrupted. Future work focusing on inhibiting the A. baumannii enzymes responsible for UDP-diNAcBac biosynthesis will strengthen the correlation between pathogenicity and bacterial glycosylation. The rise of this multi-drug resistance strain in the hospital environment is cause for alarm and makes the search for novel virulence targets all that more important. The enzymes responsible for UDP-diNAcBac biosynthesis may well represent novel targets in the struggle against A. baumannii resistance.
Supplementary Material
Highlights.
The A. baumannii enzymes WeeK, J, and I produce UDP-diNAcBac.
A. baumannii UDP-diNAcBac pathway enzymes are kinetically characterized.
UDP-diNAcBac enzymes exhibit surprising diversity in N- and O-linked glycosylation.
The terminal acetyltransferase may promote flux through the UDP-diNAcBac pathway.
Acknowledgment
We wish to thank Dr. Angelyn Larkin, Dr. Meredith Hartley, and Austin Travis for critical reading of the manuscript and useful discussions regarding experimental data.
Abbreviations
- Ab
Acinetobacter baumannii
- AcCoA
acetyl-coenzyme A
- BSA
bovine serum albumin
- CE
capillary electrophoresis
- CEF
cell envelope fraction
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- Cj
Campylobacter jejuni
- DDM
n-dodecyl-β-d-maltopyranoside
- DTNB
5,5′-dithio-bis-(2-nitrobenzoic acid) or Ellman’s reagent
- EDTA
ethylenediaminetetraacetic acid
- Gal
galactose
- GalNAc
N-acetylgalactosamine
- Glc
glucose
- GlcNAc
N-acetylglucosamine
- IPTG
iso-β-d-thiogalactosylpyranoside
- N-linked
asparagine-linked
- NAD+
nicotinamide adenine dinucleotide
- NDP
nucleotide diphosphate
- Ni-NTA
Nickel-nitrilotriacetic acid
- O-linked
serine- or threonine-linked
- Pgl
protein glycosylation
- PglB-ATD
acetyltransferase domain of PglB
- PglB-PGTD
phosphoglycosyltransferase domain of PglB
- PSUP
pure solvent upper phase
- Ng
Neisseria gonorrhoeae
- TMHMM
Tied Mixture Hidden Markov Model
- UDP
uridine diphosphate
- UDP-4-amino
UDP-2-acetamido-4-amino-2,4,6-trideoxy-α-d-glucose
- UDP-4-keto
UDP-2-acetamido-4-keto-2,4,6-trideoxy-α-d-glucose
- UDP-diNAcBac
UDP-N,N - diacetylbacillosamine or UDP-2,4-diacetamido-2,4,6-trideoxy-α-d-glucose
- Und-P
undecaprenyl phosphate
- Und-PP
undecaprenyl diphosphate
Appendix A.
Supplementary data associated with this article can be found in the online version.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by National Institutes of Health Grant GM097241 (B.I.) and the Biotechnology Training Program T32-GM08334 (M.J.M.).
References
- 1.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 2.Perez F, Huher AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Sigurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PloS One. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Menuteau O, Agoli N, Cattoen C, Nordmann P. Outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 2003;41:3542–3547. doi: 10.1128/JCM.41.8.3542-3547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genetics. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. Comparative genome sequence analysis of multi-drug resistant Acinetobacter baumannii. J. Bacteriol. 2008;190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nothaft J, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 9.Kelly J, Jarrell H, Millar L, Tessier L, Fiori LM, Lau PC, Allan B, Szymanski CM. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 2006;188:2427–2434. doi: 10.1128/JB.188.7.2427-2434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlyshev AV, Everest P, Linton D, Cawthraw S, Newell DG, Wren BW. The Campylobacter jejuni general glycosylation system is important for the attachment to human epithelial cells and in the colonization of chicks. Microbiology. 2004;150:1957–1964. doi: 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- 11.Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuck WW, Brisson JR, Logan SM. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J. Biol. Chem. 2006;281:723–732. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- 12.Olivier NB, Chen MM, Behr JR, Imperiali B. In vitro biosynthesis of UDP-N,N′-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry. 2006;45:13659–13669. doi: 10.1021/bi061456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vik A, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartley MD, Morrison MJ, Aas FE, Borud B, Koomey M, Imperiali B. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for the biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry. 2011;50:4936–4948. doi: 10.1021/bi2003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S, Feldman MF. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathogens. 2012;8:e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin A, Imperiali B. Biosynthesis of UDP-GlcNAc(3NAc)A by WbpB, WbpE, and WbpD: enzymes in the Wbp pathway responsible for O-antigen assembly in Pseudomonas aeruginosa PAO1. Biochemistry. 2009;48:5446–5455. doi: 10.1021/bi900186u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen MM, Weerapana E, Ciepichal E, Stupak J, Reid CW, Swiezewska E, Imperiali B. Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2007;46:14342–14348. doi: 10.1021/bi701956x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 19.Glover KJ, Weerapana E, Chen MM, Imperiali B. Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2006;45:5343–5350. doi: 10.1021/bi0602056. [DOI] [PubMed] [Google Scholar]
- 20.Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM, Bhatia S, Wakarchuk WW, Young NM. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J. Biol. Chem. 2006;281:8907–8916. doi: 10.1074/jbc.M512987200. [DOI] [PubMed] [Google Scholar]
- 21.Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJ. Structural analysis of a set of proteins resulting from a bacterial genome project. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 22.Larkin A, Olivier NB, Imperiali B. Structural analysis of WbpE from Pseudomonas aeruginosa PAO1: a nucleotide sugar aminotransferase involved in O-antigen assembly. Biochemistry. 2010;49:7227–7237. doi: 10.1021/bi100805b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger, LLC; [Google Scholar]
- 24.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivier NB, Imperiali B. Crystal structure and catalytic mechanism of PglD from Campylobacter jejuni. J. Biol. Chem. 2008;283:27937–27946. doi: 10.1074/jbc.M801207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litran T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St Michael F, Szymanski CM, Li J, Chan KH, Khieu NH, Larocque S, Wakarchuk WW, Brisson JR, Monteiro MA. The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur. J. Biochem. 2002;269:5119–5136. doi: 10.1046/j.1432-1033.2002.03201.x. [DOI] [PubMed] [Google Scholar]
- 28.Creuzenet C. Characterization of Cj1293, a new UDP-GlcNAc C6 dehydratase from Campylobacter jejuni. FEBS Lett. 2004;559:136–140. doi: 10.1016/S0014-5793(04)00057-2. [DOI] [PubMed] [Google Scholar]
- 29.Hynne F, Dano S, Sorensen PG. Full-scale model of glycolysis in Saccharomyces cerevisiae. Biophys. Chem. 2001;94:121–163. doi: 10.1016/s0301-4622(01)00229-0. [DOI] [PubMed] [Google Scholar]
- 30.Teusink B, Bachmann H, Molenaar D. Systems biology of lactic acid bacteria: a critical review. Microbial Cell Factories. 2011;10(Suppl 1):S11. doi: 10.1186/1475-2859-10-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Ou MS, Kim Y, Ingram LO, Shanmugam KT. Metabolic flux control at the pyruvate node in an anaerobic Escherichia coli strain with an active pyruvate dehydrogenase. Appl. Environ. Microbiol. 2010;76:2107–2114. doi: 10.1128/AEM.02545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.