Abstract
Reactive nitrogen species derived from NO have been implicated in cancer and other diseases, but their intracellular concentrations are largely unknown. To estimate them under steady-state conditions representative of inflamed tissues, a kinetic model was developed that included the effects of cellular antioxidants, amino acids, proteins, and lipids. For an NO concentration of 1 µM, total peroxynitrite (Per, the sum of ONOO− and ONOOH), NO2·, and N2O3 were calculated to have concentrations in the nM, pM, and fM ranges, respectively. The concentrations of NO2· and N2O3 were predicted to decrease markedly with increases in glutathione (GSH) levels, due to the scavenging of each by GSH. Although lipids accelerate the oxidation of NO by O2 (because of the high solubility of each in hydrophobic media), lipid-phase reactions were calculated to have little effect on NO2· or N2O3 concentrations. The major sources of intracellular NO2· were found to be the reaction of Per with metals and with CO2, whereas the major sinks were its reactions with GSH and ascorbate (AH−). The radical-scavenging ability of GSH and AH− caused 3-nitrotyrosine to be the only tyrosine derivative predicted to be formed at a significant rate. The major GSH reaction product was S-nitrosoglutathione. Analytical (algebraic) expressions are provided for the concentrations of the key reactive intermediates, allowing the calculations to be extended readily.
Keywords: nitric oxide, nitrogen dioxide, nitrous anhydride, peroxynitrite, inflammation
Introduction
Nitric oxide (NO) synthesis by activated macrophages, which is part of the nonspecific immune response [1], leads to the formation of other reactive nitrogen species. In particular, adding NO to a solution that contains O2 and (such as cytosol) will produce nitrogen dioxide (NO2·), nitrous anhydride (N2O3), and peroxynitrite (ONOO−), which exhibit a variety of reactivities with cellular components, including proteins, nucleic acids, and lipids [2]. Although NO and its oxidation products can assist in killing invading microorganisms, the concurrent chemical damage to host cells may initiate or contribute to diseases associated with chronic inflammation, such as certain forms of cancer [3–5]. In other words, high local rates of NO production, if sustained, may constitute a significant health risk. Efforts to examine that risk have been hampered by the paucity of information on the concentrations of NO and other reactive nitrogen species in inflamed tissues. In macrophage cultures in vitro, NO concentrations have been measured and shown to be as high as 1 µM [3]. However, the reactivity of NO2·, N2O3, and ONOO− leads to concentrations that are too small for direct measurements in cells or culture media. In the absence of even an order-of-magnitude estimate of its concentration, it is difficult to assess the pathophysiological role of a particular compound. Given the impracticality of direct measurements, the most promising approach for quantifying reactive nitrogen species in cells is to develop kinetic models that embody what is known about the rates of key reactions. The predictions of such models can be tested by measuring the concentrations of suitable biomarkers (i.e., stable end products of nitrogen oxide chemistry).
A reaction-diffusion model was developed previously to calculate steady state concentrations of NO and ONOO− in cells exposed to external sources of NO and/or ONOO−, based on estimated rates of production in cytosol and mitochondria [6]. For this purpose, the only important reactions were the formation of ONOO− from NO and , the decomposition of ONOOH and ONOO− (the latter catalyzed by CO2), and the scavenging of by superoxide dismutase. It was shown that rates of diffusion are fast enough to cause the intracellular concentrations of NO, O2, and CO2 to each closely approximate those in the adjacent extracellular fluid. In other words, for a given cell, the concentrations of these dissolved gases may be viewed as being imposed by the surroundings. Other reactive nitrogen species, such as NO2· and N2O3, were not considered.
More recently, Lancaster [7] proposed a much more comprehensive model of intracellular chemistry related to NO, including oxidation, nitrosation, and nitration pathways. Among the 51 reactions considered were several involving glutathione, which is a major intracellular antioxidant, and tyrosine, which has derivatives that have been used as biomarkers. Although this pioneering model contained enough chemical detail to predict intracellular concentrations of reactive nitrogen species, such concentration estimates were not reported. Also, the model was formulated to describe time-dependent responses to the sudden introduction of key reactants. That is, cells were modeled as transient batch reactors. While such a formulation may be appropriate for simulating certain in vitro experiments, the time scale for inflammatory processes is such that the various chemical species will be at steady state, with rates of formation and consumption in continuous balance.
The objective of the present work was to develop a model for intracellular nitrogen oxide chemistry that combines a reasonably comprehensive set of reactions with a steady-state formulation designed to simulate what occurs in vivo. The chemical system considered by Lancaster [7] was extended by including additional antioxidants, additional amino acids, and certain lipid-phase reactions. As will be shown, radical scavenging by ascorbate, in particular, has important effects on the concentrations of several species of interest. Concerning lipid-phase chemistry, it has been reported that oxidation of NO by O2 is greatly accelerated in hydrophobic media by the relatively high solubilities of NO and O2 [8, 9]. Also, the reaction of NO2· with polyunsaturated fatty acids in membranes [10] might be an important sink for reactive nitrogen species, a possibility which is automatically neglected if only aqueous reactions are included. Concentrations of the reactive nitrogen species and related radicals are estimated, and the biological implications of the results are discussed.
Model Development
Overview
This section begins with a description of the chemical pathways that are included in the kinetic model. The physical assumptions are then identified and the general form of the mass balance equations is presented. Following that are derivations of the concentration expressions for specific species. Those derivations are grouped according to what is needed algebraically to proceed to subsequent steps: ONOO− and carbonate and hyroxyl radicals are considered first; then, NO2· and N2O3; and finally, glutathione and tyrosine derivatives.
Reactions
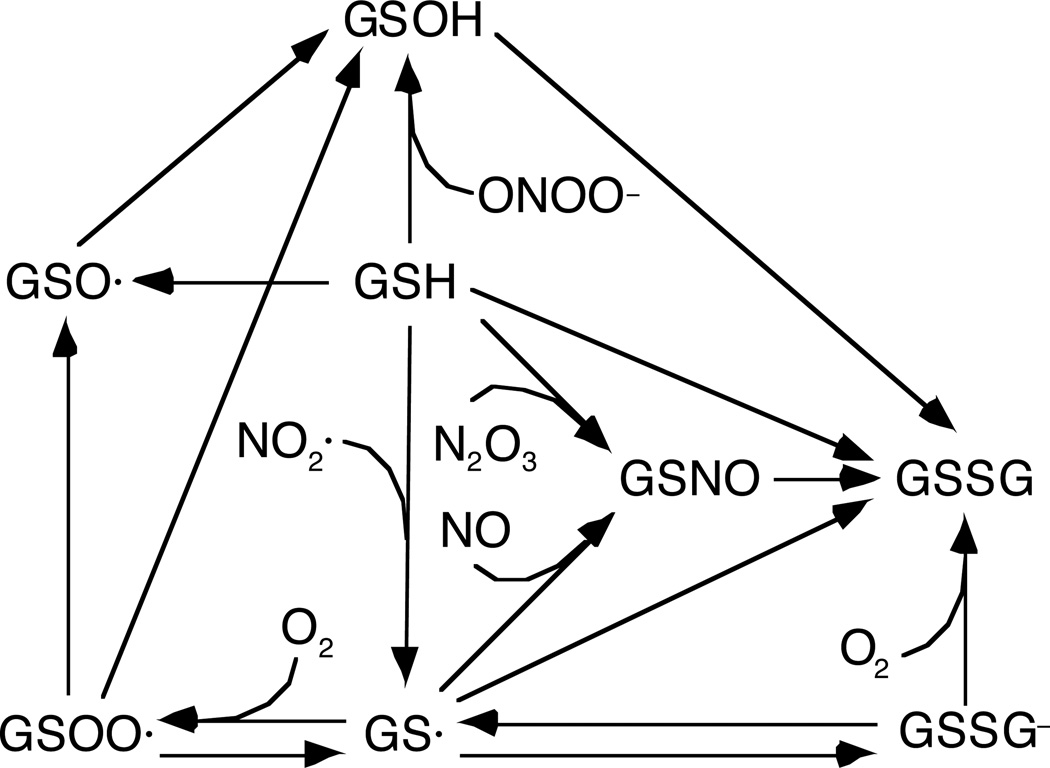
Figure 1 summarizes the nitrogen oxide reactions that were included in modeling the aqueous phase, arranged so as to emphasize the central role of NO2·. As shown, NO2· may be formed from the reaction of NO with O2, the decomposition of N2O3, the decomposition of peroxynitrous acid (ONOOH), the reaction of ONOO− with transition metal centers or selenium-containing proteins (denoted collectively as Mn+), and the decomposition of nitrosoperoxycarbonate (ONOOCO2−). The consumption of NO2· is via its reactions with NO (to form N2O3), antioxidants, and amino acids, all of which lead eventually to nitrite (NO2−). The antioxidants considered were glutathione (GSH), ascorbate (AH−), and urate (UH2−); the amino acids were cysteine (Cys), tyrosine (Tyr), and tryptophan (Trp), each of which is known to be reactive with NO2· [11]. As indicated by the bold arrows, and as shown later, cytosolic NO2· appears to be derived mainly from peroxynitrite, and consumed mainly by GSH and AH−.
Figure 1.
Nitrogen oxide chemistry, emphasizing reactions that produce or consume NO2·. The more important sources and sinks are denoted by thicker arrows. NO2· is derived mainly from peroxynitrite and consumed mainly by antioxidants.
The nitrogen oxide chemistry considered in the lipid phase was largely a subset of that shown in Figure 1. The central process there was the autoxidation sequence leading from NO and O2 to NO2 and N2O3. The one new pathway in the lipid part of the model was the consumption of NO2 by its reaction with polyunsaturated fatty acids. Ions were assumed to be completely excluded from the lipid phase, so that no reactions involving charged species were considered there.
Figure 2 provides another view of nitrogen oxide chemistry, focusing now on peroxynitrite. Peroxynitrite ion is formed from the rapid reaction of NO with and is in near-equilibrium with ONOOH. The three pathways that lead from ONOO− to NO2· have been mentioned already. Additional reactions yield NO2−, including ONOO− with GSH and ONOOH with proteins. Another feature in Figure 2 not shown in Figure 1 is that the decomposition of ONOOCO2− and ONOOH each yield nitrate (NO3−) as a stable end product, in addition to the reactive product NO2.
Figure 2.
Nitrogen oxide chemistry, emphasizing reactions that produce or consume peroxynitrite. The more important reactions are denoted by thicker arrows.
The glutathione chemistry that was considered is shown in Figure 3. Of particular importance is that GSH reacts with each of the trace nitrogen oxides, yielding glutathionyl radical (GS·) from NO2·, S-nitrosoglutathione (GSNO) from N2O3, and glutathione sulfenic acid (GSOH) from ONOO−. Other intermediates or products include glutathione sulfinyl radical (GSO·), glutathione peroxysulfenyl radical (GSOO·), glutathione disulfide (GSSG), and the disulfide anion (GSSG−).
Figure 3.
Glutathione reactions that involve nitrogen oxides or oxygen.
Tyrosine nitration chemistry is depicted in Figure 4. Tyrosyl radical (Tyr·) is produced by the one-electron oxidation of Tyr by radicals such as GS ·, , and NO2. Tyrosyl radical reacts with NO, NO2·, and itself, to form 3-nitrosotyrosine (Tyr-NO), 3-nitrotyrosine (NO2-Tyr), and 3,3-dityrosine (diTyr), respectively. It is scavenged by antioxidants, especially AH−, resulting in the recovery of Tyr.
Figure 4.
Reactions that lead to nitration or nitrosation of tyrosine. The more important reactions are denoted by thicker arrows.
For clarity, certain minor reactions included in the model were omitted from Figures 1–4. The full set of 66 reactions (excluding acid-base dissociations) is shown in Table 1. Also shown are the rate constants and literature sources. The rate constant for the hydrolysis of N2O3 (k3) includes the catalytic effects of HCO3- but neglects the effects of phosphate. Each rate constant in the lipid phase was assumed to be the same as in water. There is evidence that this is true for the reaction of NO with O2 [8,9], although for the other reactions this assumption is untested.
Table 1.
Reactions and rate constants.
| # | Reaction | Rate Constant | Reference | ||
|---|---|---|---|---|---|
| 1 | k1 = 2.4×106M−2s−1 | [12] | |||
| 2 |
|
k
2= 1.1×109
M
−1
s
−1
k − 2 = 8.4×104 s −1 |
[13] [14] |
||
| 3 |
|
k 3 = 2.1×104 s −1 | [15] | ||
| 4 |
|
k 4 = 2.0×107 M −1 s −1 | [16] | ||
| 5 |
|
k 5 =1.0×109 M −1 s −1 | [17] | ||
| 6 |
|
k 6 = 7.5×108 M −1 s −1 | [18] | ||
| 7 |
|
k
7=2×109
M
−1
s
−1
k −7=6.2×105 s −1 |
[19] [19] |
||
| 8 |
|
k
8 = 6×108
M
−1
s
−1
k − 8 = 1.6×105 s −1 |
[19] [19] |
||
| 9 |
|
k 9 = 5×109 M −1 s −1 | [19] | ||
| 10 |
|
k 10 = 1.7×109 M −1 s −1 | [20] | ||
| 11 |
|
k11 = 5.5×10−3M−1s−1 | [21] | ||
| 12 | k12 = 6×108M−2s−1 | [22] | |||
| 13 |
|
k13 = 3.8×108M−1s−1 | [20] | ||
| 14 |
|
k14 =1×109M−1s−1 k−14 = 0.75s−1 |
[23] [23] |
||
| 15 |
|
k15 =4×108M−1s−1 x = 0.56 |
[23] | ||
| 16 | k16 = 3×109M−1s−1 | [23] | |||
| 17 | k17 = 2×106M−1s−1 | [24] | |||
| 18 | k18 = 6.6×107M−1s−1 | [25] | |||
| 19 |
|
k19 = 6.6×102M−1s−1 | [26] | ||
| 20 |
|
k20 = 5.3×106M−1s−1 (pH=7.0) | [27] | ||
| 21 | k21 = 200M−1s−1 | [28] | |||
| 22 | k22 = 720M−1s−1 | [29] | |||
| 23 |
k23 = 3.5×105M−1s−1 k−23 = 3.5×105M−1s−1 |
[30] [30] |
|||
| 24 |
|
k24 =1×105M−1s−1 | [24] | ||
| 25 | k25 =1×104M−1s−1 | [31] | |||
|
|
[31] | ||||
| 26 | k26 = 4×108M−1s−1 | [32] | |||
| 27 | k27 = 3.5×109M−1s−1 | [33] | |||
| 28 |
|
k28 = 4.5×107M−1s−1 | [15] | ||
| 29 | k29 = 3.2×105M−1s−1 (pH=7.5) | [10] | |||
| 30 |
k30 =1×109M−1s−1 k−30 =1×103s−1 |
[15] [15] |
|||
| 31 | k31 = 0.5s−1 | [15] | |||
| 32 |
|
k32 =1.5×109M−1s−1 | [15] | ||
| 33 |
|
k 33 =1.4×1010 M −1 s −1 | [34] | ||
| 34 |
|
k34 = 1.8×104s−1 | [34] | ||
| 35 |
|
k 35 = 3×108 M −1 s −1 | [34] | ||
| 36 |
|
k36 = 4.0 ×107M−1s−1 | [35] | ||
| 37 |
|
k37 = 1.7×109M−1s−1 | [15] | ||
| 38 | k38 = 1.3×109M−1s−1 | [15] | |||
| 39 | k39 = 0.06s−1 (pH = 7.4) | [26] | |||
|
|
[26] | ||||
| 40 | k40 =1×1010M−1s−1 | [36] | |||
| 41 |
|
k41 =1×1010M−1s−1 | [37] | ||
| 42 |
|
k42 = 4.8×109M−1s−1 | [38] | ||
| 43 |
|
k43 = 5.3×109M−1s−1 | [36] | ||
| 44 |
|
k44 = 3.0×108M−1s−1 | [39] | ||
| 45 | k45 = 8.5×106M−1s−1 | [39] | |||
| 46 |
|
k46 =1.4×1010M−1s−1 | [14] | ||
| 47 | k47 = 3.3×109M−1s−1 | [14] | |||
| 48 | k48 = 7.2×109M−1s−1 (pH=6−7) | [14] | |||
| 49 | k49 =1.3×1010M−1s−1 | [14] | |||
| 50 |
|
k50 = 4.7×1010M−1s−1 (pH=7.0) | [14] | ||
| 51 | k51 = 7.0×108M−1s−1 (pH=7.0) | [27] | |||
| 52 | k52 = 4.6×107M−1s−1 (pH=7.0) | [27] | |||
| 53 | k53 =1.1×109M−1s−1 (pH=11.0) | [14] | |||
| 54 | k54 =1.0×106M−1s−1 (pH=6.5) | [10] | |||
| 55 | k55 = 5.0×107M−1s−1 (pH=7.4) | [16] | |||
| 56 | k56 = 3.5×107M−1s−1 (pH=6.7) | [14] | |||
| 57 | k57 = 2.0×107M−1s−1 (pH=7.4) | [16] | |||
| 58 |
|
k58 = 1.0×105M−1s−1 | [40] | ||
| 59 | k59 = 5.0×103M−1s−1 | [40] | |||
| 60 |
|
k60 = 1.0×1010M−1s−1 | [41] | ||
| 61 | k61 =2.0×105 (pH=9.5) | [10] | |||
| 62 |
|
k62 =1.0×106 (pH=9.0) | [10] | ||
| 63 |
|
k63 = 4.5×108M−1s−1 k−63 = 6.9×103s−1 |
[42,43] | ||
| 64 |
|
k64 = 1.0×103s−1 | [44] | ||
| 65 | k65 = 6.0×108M−1s−1 | [19] | |||
| 66 |
|
k66 = 4.4×108M−1s−1 | [45] |
Mass Balance Equations
Based on our previous finding that intracellular concentrations of key species vary by only small amounts from the center to the periphery of a cell [6], we did not attempt to model spatial variations. Although we estimated that concentrations of and total peroxynitrite (ONOO− plus ONOOH) are about 3 and 8 times higher, respectively, in mitochondria than in cytosol [6], for simplicity we chose here not to make such distinctions. That is, all intracellular aqueous compartments were lumped together. However, a distinction was made between aqueous and lipid concentrations.
All species were assumed to have time-independent concentrations. The species considered fell into three categories: (i) major dissolved gases (NO, O2, CO2); (ii) biomolecular targets, including antioxidants, amino acids, and ; and (iii) reactive intermediates. The concentrations for groups (i) and (ii) were specified directly, based on the literature. Those for group (iii) constituted the unknowns in the set of governing equations. Radicals and other reactive intermediates tend to be present at such low concentrations that their ability to escape a cell is very limited. That is, permeation across the plasma membrane will tend to be slow relative to intracellular reactions, because the concentration driving force for diffusion is so small. Accordingly, for group (iii) species the cell as a whole was regarded as a closed system, with rates of formation exactly balancing rates of consumption. However, internal exchanges between the aqueous and lipid phases were included.
With these assumptions, the steady-state mass balance equation for species i was
| (1) |
where v is the volume fraction of the lipid phase (estimated as 0.03, or 3% of cell volume [8,9]) and and are the net rates of formation of species i in the aqueous and lipid (membrane) phases, respectively. These reaction rates are per unit volume of the phase indicated, include all reactions in which species i participates, and are defined as positive for formation and negative for consumption.
The remaining key assumption was that lipid-aqueous mass transfer is rapid enough that concentration ratios between the two phases are very near their equilibrium values.
Thus, it was assumed that
| (2) |
where [i] and [i]m denote concentrations of i in cytosol and membranes, respectively, and Qi is the membrane/cytosol partition coefficient. For each species Qi was viewed as a known constant. Equations 1 and 2, combined with the rate laws for each reaction, provided the set of algebraic equations that governed the concentrations of all reactive intermediates. Equation 2 was used also, where needed, to obtain membrane concentrations from the aqueous values specified for dissolved gases and biomolecular targets.
Estimates of the remaining parameters in the model are given in Table 2. These include the cytosolic concentrations that were fixed as inputs, various lipid-aqueous partition coefficients, and certain pK values. The NO concentration of 1 µM is intended to be representative of an inflamed tissue, and the O2 and CO2 concentrations each correspond to partial pressures of 40 mmHg. The O2 value is that in venous blood or typical body tissues, and is about one-fourth of that generally used in cell cultures. The , and H+ concentrations each correspond to an intracellular pH of 7.0. The concentration was estimated by us previously [6], by equating the respiratory production of with its consumption by superoxide dismutase (SOD). The rate of production was assumed to be 20% of the rate of H2O2 production reported in liver cells, and literature values for SOD activity were employed. The antioxidant and amino acid concentrations are all directly from the literature. The amino acid values are for free amino acids only. The metal and protein concentrations are those to be used in conjunction with the rate constants k58 and k59; the basis for those estimates is discussed in detail in Alvarez and Radi (40).
Table 2.
Concentrations, partition coefficients, and pK values.
| Species | Concentration (cytosol) [Ref.] |
Partition Coefficient [Ref.] |
pK | |
|---|---|---|---|---|
| NO | 1 µM [3] | 9 [8] | ||
| O2 | 50 µM [24] | 3 [8] | ||
| CO2 | 1.2 mM [40] | 6.8 [49] | ||
| 10 mM [46] | ||||
| 4.8 µM | ||||
| H+ | 0.1 µM [46] | |||
| 20 pM [6] | ||||
| NO2· | 0.3 [50] | |||
| GSH | 5 mM [16] | 3.9 × 10−6 [50] | 9.2 | |
| AH2 | 0.5 mM [40, 16] | 4.7 | ||
| UH3 | 0.1 mM [40] | 5.4 | ||
| Tyr | 100 µM [47] | 5.5 × 10−3 [49] | ||
| Trp | 0.3 µM [48] | 8.7 × 10−2 [49] | ||
| CysSH | 10 µM [47] | 3.2 × 10−3 [49] | ||
| ONOOH | 6.8 | |||
| Proteins | 15 mM [40] | |||
| Metals (Mn+) | 0.5 mM [40] | |||
| Linoleic acid | 1.0 M* | |||
| Arachidonic acid | 0.34 M* |
Estimated concentration in membrane phase.
The lipid-aqueous partition coefficients tend to be more uncertain. For NO, we chose the higher of two reported values (QNO= 9), to obtain an upper bound on the effects of lipid autoxidation; the effects of using the lower value instead (QNO = 3) will be discussed. Given the absence of direct measurements, QNO2 and QGSH were estimated using group-contribution correlations [50]. However, all of the amino acid values are based on experimental data [49]. Missing from the table is QN2,O3, for which there is no literature value. As will be shown, the results are insensitive to the choice of QN2,O3. The partition coefficients of all ions were assumed to be zero, as already mentioned. Based on pH 7.0 and the pK values shown, most of the glutathione will be in the neutral form (99.4%), whereas ascorbic acid and uric acid will be mainly anions (99.5% and 97.5%, respectively). With a pK of 6.8 for peroxynitrous acid, 39% of total peroxynitrite will be neutral and 61% anionic. Unless indicated otherwise, all calculations were based on the parameter values in Tables 1 and 2, and a membrane volume fraction of v = 0.03.
Peroxynitrite, Hydroxyl Radical, and Carbonate Radical
It is simplest algebraically to begin with peroxynitrite. Equation 1 was applied first to total peroxynitrite, where [Per] = [ONOO−] + [ONOOH]. The principal reactions that involve either the anion or acid are Reactions 25 and 58–60, which are limited to the aqueous phase. Thus,
| (3) |
Letting f = 1 / (1 + 10pK-pH) represent the fraction of total peroxynitrite that is present as the anion and 1 − f the fraction as the acid, Equation 3 was rearrranged to
| (4) |
Other reactions that involve peroxynitrite are 19, 39, and 42. A comparison with the other sink terms in Equation 3 showed the rates of Reactions 19 and 39 to be negligible. Because Reaction 39 is negligible in cytosol, and because QONOOH is unlikely to be large, that reaction will be negligible also in the membrane phase. Using Equation 4 and the result for [·OH] obtained below, reaction 42 was also confirmed to be unimportant.
A similar approach was used to estimate the concentration of ·OH radical, which participates in reactions 33 and 39–50. Retaining only the dominant reactions, which are 33, 46 and 47 in the aqueous phase and 40 in the membrane phase, the net rates of formation in the aqueous and membrane phases are
| (5) |
| (6) |
Inserting these expressions into Equation 1, using [·OH]m= QOH[·OH] and [ONOOH]m= QONOOH[ONOOH], and rearranging, gives
| (7) |
Equation 7 can be simplified further by noting that QONOOH should not be large, in which case v (1 − f)QONOOH << (1 − v)f. Accordingly, the membrane source term in the numerator is negligible. Likewise, the membrane sink term in the denominator (that involving QOH) will also be negligible. Thus, the final expression for the OH concentration is
| (8) |
The major sink for ·OH was found to be GSH, which accounts for >90% of the denominator in Equation 8. It should be noted that this expression will underestimate [·OH], because ·OH is generated by other intracellular reactions. However, as will be discussed, our other results are unaffected even if [·OH] is several orders of magnitude larger than implied by Equation 8.
For radical, the sources are Reactions 25, 44 and 45 and the sinks are Reactions 20, 26–28, and 51–53. Because is an anion, only aqueous reactions were considered. Retaining only the dominant terms, it was found that
| (9) |
For the major sink is its reaction with ascorbate, which accounts for >90% of the denominator in Equation 9.
Nitrogen Dioxide and Nitrous Anhydride
For N2O3, Reactions 2, 3, and 18 can occur in the cytosol, and Reactions 2 and 18 can also take place in the lipid membranes. However, by reasoning similar to that used for ·OH, the membrane sink terms (Reaction 18 and reverse of Reaction 2) were each found to be negligible. The simplified expression for the aqueous N2O3 concentration is
| (10) |
The term in the numerator that contains QNO,QNO2 represents the augmentation of N2O3 production due to intramembrane autoxidation of NO. The concentration of N2O3 is depressed by GSH in two ways: via the sink term in the denominator of Equation 10 and via the influence of GSH on the NO2 concentration in the numerator (see below).
The reactions involving NO2. are much more numerous: 1, 2, 4, 14, 25, 29, 37–39, 43, 54–58, and 61–62. Simplifications were obtained by applying a steady state condition to GSOONO2 in Reaction 14, and assuming that [Tyr·] is small enough to neglect Reactions 37 and 38 for NO2· (an assumption verified using the tyrosine results discussed later). Retaining only the dominant sources and sinks in the remaining reactions in each phase, and neglecting Reaction 2 in the membrane, the aqueous NO2 concentration was found to be
| (11a) |
| (11b) |
| (11c) |
The term in the numerator of Equation 11a that involves represents the increased rate of NO2· formation due to membrane autoxidation. The NO2· concentration is reduced by the presence of antioxidants and unsaturated fatty acids (LA, linoleic acid; AA, arachidonic acid), as shown by the various terms in the denominator.
Glutathionyl Radical, S-Nitroso Glutathione, and Tyrosyl Radical
The reactions that involve GS· are 4–8, 10, 20, 21, 23, 24, 46, and 65. Of these, Reactions 8, 20, 21 and 65 will occur only in the cytosol. Simplifications were obtained by applying steady state assumptions to GSOO·, GSSG−, and GSO·. For GSOO·, it was found that only Reaction 7 was important; for GSO·, Reaction 6 could be ignored because it is a second-order reaction between radicals, each of which will have a very small concentration. Reactions 20, 21 and 46 were found to be negligible sources of GS· and reverse Reaction 23 was a negligible sink. Also, all membrane reactions were found to be negligible compared to those in the aqueous phase. After considerable manipulation, it was found that
| (12a) |
| (12b) |
| (12c) |
Note that [GS−] = 10pH-pK [GSH] = 0.0063 [GSH] for pH 7.0 and pK = 9.2. Before [GS·] can be evaluated, it is necessary to calculate [GSNO] and [Tyr·], as described next.
For GSNO, the relevant reactions were 5, 10–13, and 18. Reaction 12 could be ignored because it is a third-order reaction among trace species. All membrane reactions were negligible here because of the small Qi values. Applying Equation 1 once again, it was found that
| (13a) |
| (13b) |
The reactions that involve Tyr· radical are 23, 28–30, 32–34, 36–38, and 66. Simplifications were obtained by neglecting Reaction 36 (a second-order reaction among trace species), applying steady state conditions to Tyr-NO and Tyr(OH)·, and identifying the dominant sources and sinks among the terms that remained. All reactions in the membrane were found to be negligible in this case. The result was
| (14) |
As seen in Equations 12a and 14, the expressions for [GS·] and [Tyr·] are coupled. However, it was found that, for physiological concentrations of GSH and Tyr, the terms that cause the coupling are negligible. Also, for physiological concentrations of AH−, the ascorbate terms in the denominators of Equations 12a and 14 were easily the dominant sinks for GS· and Tyr·. In addition, because is greatly depressed at physiological levels of AH− (Equation 9), radical is ordinarily not an important source for GS.
Accordingly, for normal values of [AH-], Equations 12a and 14 could be simplified to
| (15) |
| (16) |
Likewise, it was found that Reaction 11 is a neglible sink for GSNO, allowing Equation 13a to be siimplified to
| (17) |
Reaction Rates
In addition to providing estimates of the concentrations of key intermediates, the analysis permitted us to calculate the rate of each reaction in Table 1. The rates of formation of nitrotyrosine and dityrosine are of particular interest, as these end products have been proposed as biomarkers for NO2· and peroxynitrite activity. The rates of formation of these tyrosine derivatives are given by
| (18) |
| (19) |
Results
Effects of NO on RNS Concentrations
In the kinetic model all reactive nitrogen species (RNS) are derived ultimately from NO. Figure 5 shows the predicted concentrations of total peroxynitrite (Per), NO2·, and N2O3 as functions of the NO concentration. Those concentrations are linear, weakly quadratic (almost linear) and cubic, respectively, in [NO], and differ by several orders of magnitude. Whereas [NO] in macrophage cultures (and presumably sites of inflammation) is on the order of 1 µM, the concentrations of Per, NO2·, and N2O3 were found to be in the nM, pM and fM ranges, respectively. The prediction that [Per] is in the nM range is consistent with our previous peroxynitrite model [6]. To our knowledge, these are the first estimates of [NO2·] and [N2O3] that take into account the effects of antioxidants, proteins, lipids and other cellular components.
Figure 5.
Effects of NO concentration on RNS concentrations. Values are shown for total peroxynitrite (Per), NO2·, and N2O3. The concentrations in this and subsequent plots are volume-weighted averages of cytosolic and membrane values. The values for Per, NO2·, and N2O3 are in the nM, pM and fM ranges, respectively.
Effects of GSH on RNS concentrations
Among the various antioxidants, GSH is predicted to have a particularly strong effect on RNS concentrations, because of its relatively high intracellular concentrations (mM range). The expected effect of [GSH] on [NO2·] is shown in Figure 6, for three scenarios. For the baseline conditions (QNO2 = 0.3 and lipids included), [NO2·] declined sharply with increasing [GSH], the relationship being nearly hyperbolic ([NO2·] ∝ [GSH]−1). When the membrane phase was omitted (v = 0), the results were nearly the same, indicating that lipid reactions do not strongly influence the NO2· concentration. Only when the relative solubility of NO2· in the membrane phase was assumed to be much larger than the baseline value (QNO2 = 0.3 instead of 0.3) was there a significant reduction in [NO2·], relative to the baseline curve. Varying QN2, O3 = 0.3 from 0 to 10 (not shown) did not have a noticeable effect on either [NO2·] or [N2O3], indicating that this partition coefficient is not an important parameter.
Figure 6.
Effects of glutathione on the concentration of NO2·. Results are shown for three situations: the baseline case (QNO2 = 0.3); a high relative solubility of NO2· in the lipid phase (QNO2 = 3); and a simplified model in which the lipid phase was omitted.
Figure 7 shows the effects of [GSH] on [N2O3], for the same three scenarios just discussed. As with [NO2·], there was a marked decrease in [N2O3] with increasing [GSH]. Close examination reveals that at high GSH concentrations, [N2O3] was even more sensitive to [GSH] than was [NO2·] (i.e., [N2O3] ∝[GSH]−2 ). This increased sensitivity stems from the fact that GSH influences [N2O3] both by scavenging NO2· (needed to produce N2O3) and by scavenging N2O3 itself. Except at low values of [GSH], neither increases in QNO2 nor the removal of lipids had a significant effect on [N2O3].
Figure 7.
Effects of glutathione on the concentration of N2O3, for the same conditions as in Figure 6.
As indicated by Equation 4, peroxynitrite concentrations are predicted to be unaffected by GSH levels.
RNS Sources and Sinks
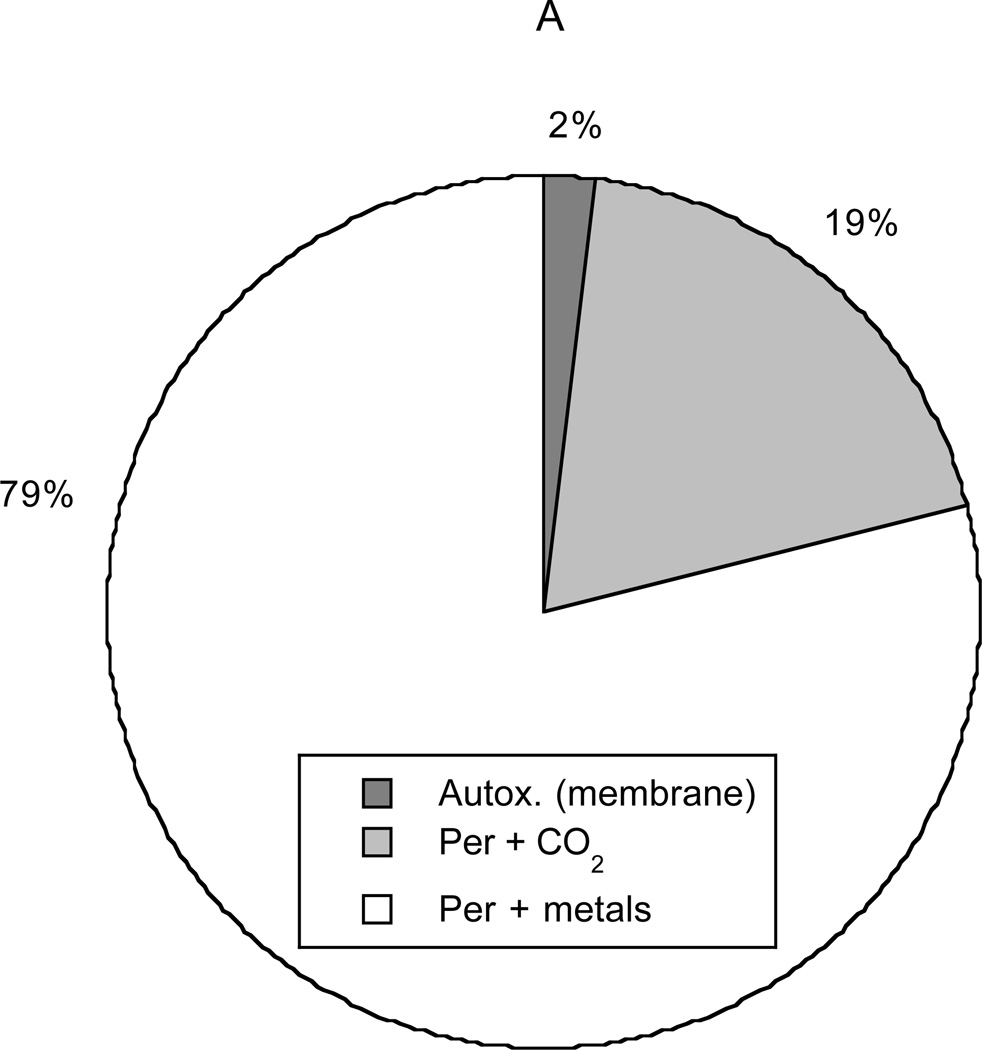
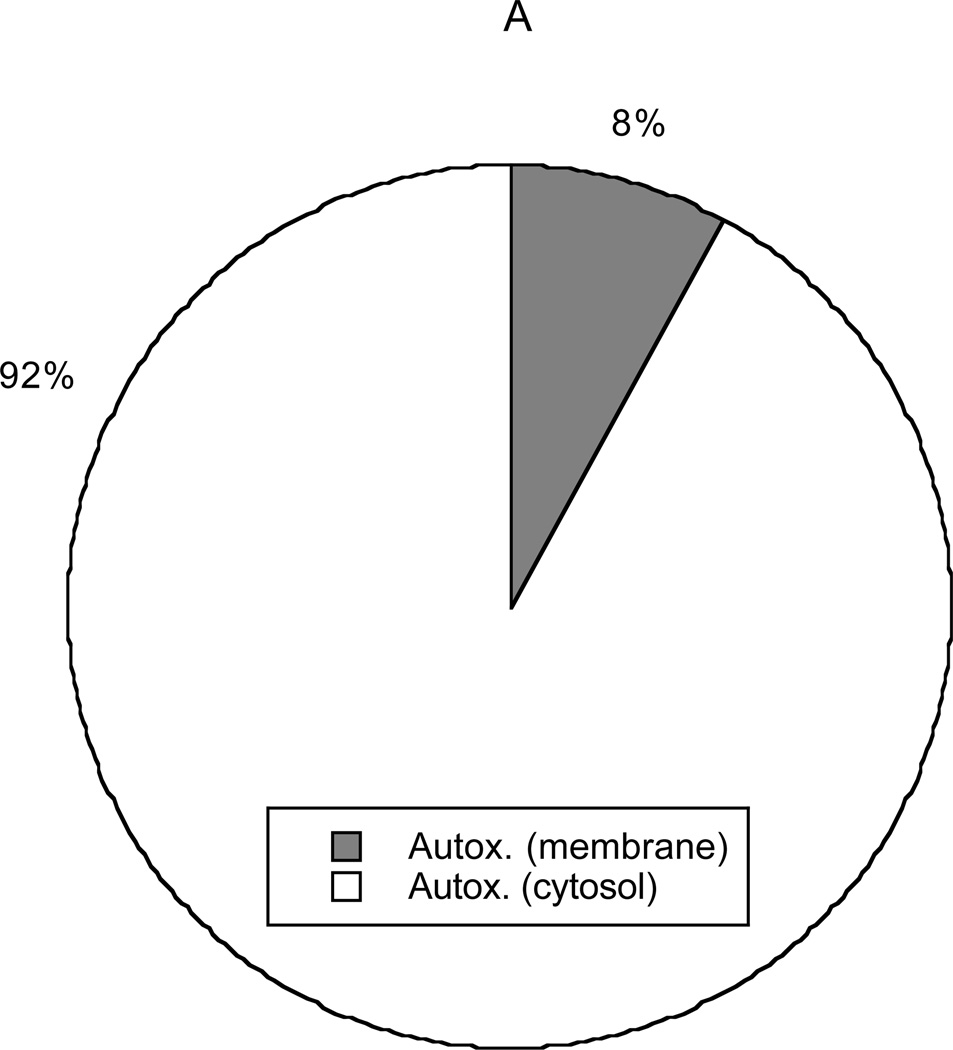
Figure 8 depicts the primary sources and sinks for NO2·. Conversion of peroxynitrite, mediated either by metals or CO2, was calculated to account for 98% of intracellular NO2· production. Autoxidation in cytosol (not shown in the graph) produced only 0.2% of the NO2· and autoxidation in membranes only 1.8%. Autoxidation in membranes was calculated to be more important than that in cytosol, despite the relatively small membrane volume (3% of cell volume), because of the favorable partitioning of NO and O2 into lipids. Scavenging of NO2· by GSH was found to be the dominant sink (79%), followed by reaction with ascorbate (14%). Reactions of NO2 with polyunsaturated fatty acids (PUFA, 4%) and other targets (3%) were found to be minor sinks. These results reemphasize the importance of GSH as a scavenger of NO2·, as indicated already by Figure 6.
Figure 8.
Sources (panel A) and sinks (panel B) for NO2·. In decreasing order of importance, the sources are the reaction of peroxynitrite with metals or selenium, the reaction of peroxynitrite with CO2, and autoxidation of NO in membranes. Autoxidation in the cytosol (not shown) is negligible. The sinks (also in decreasing order) are the reaction with glutathione, the reaction with ascorbate, the reaction with unsaturated fatty acids, and all other reactions.
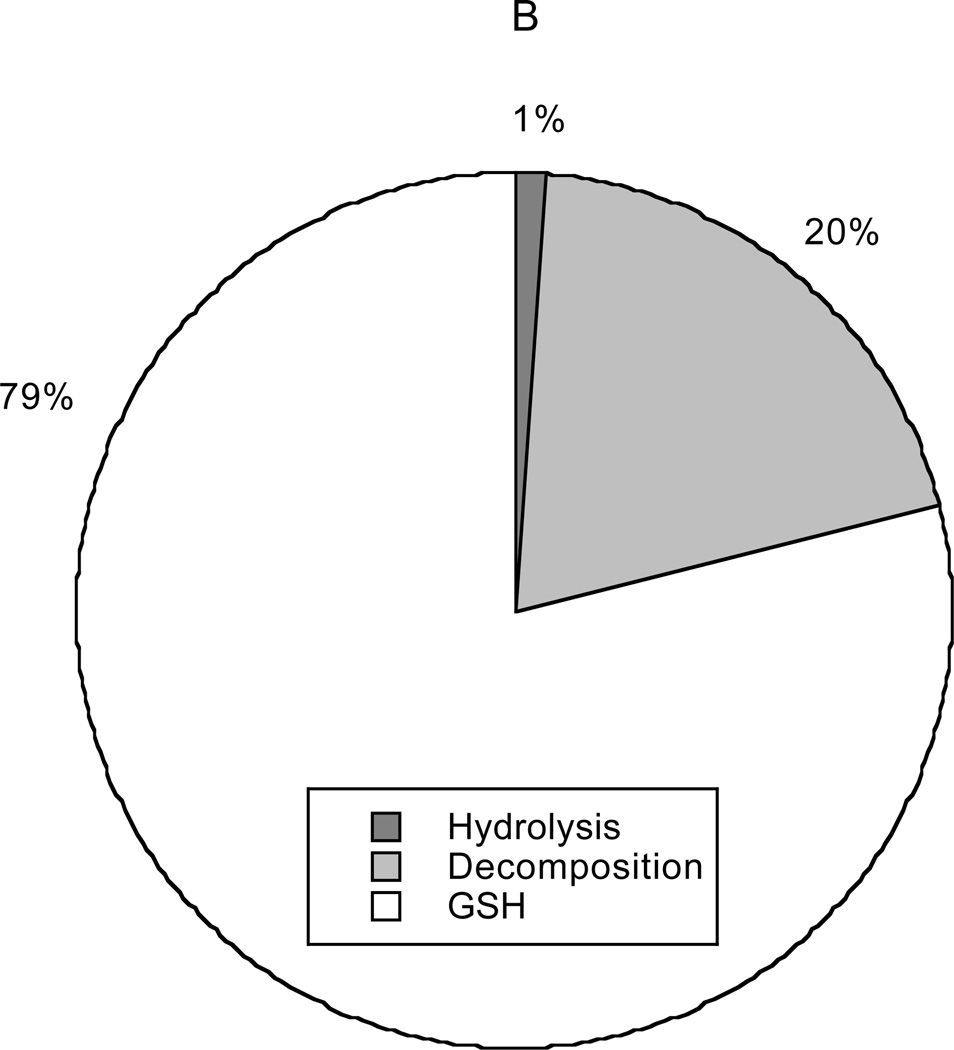
The origin and fate of N2O3 are depicted in Figure 9. In this case the only source was autoxidation. Again, the lipid contribution was disproportionate to membrane volume (8% of N2O3 produced in 3% of cell volume). However, unlike NO2· formation, the total rate of production of N2O3 via autoxidation in cytosol was much larger than that via membrane autoxidation. The reason for this is that partitioning of NO2· into the membrane is unfavorable (QNO2 = 0.3), and NO2· is required to produce N2O3. All N2O3 consumption in the model was cytosolic, with scavenging by GSH being most important (79%), followed by decomposition into NO and NO2· (20%). Only 1% of the N2O3 formed was calculated to be hydrolyzed to NO2−.
Figure 9.
Sources (panel A) and sinks (panel B) for N2O3. The dominant source is autoxidation of NO in cytosol, the contribution from autoxidation in membranes being minor. The sinks (in decreasing order of importance) are the reaction with glutathione, decomposition to NO and NO2·, and hydrolysis to .
Glutathione Products
Of the various reaction products of GSH with RNS, GSNO was found to be present at the highest concentration. For [GSH] = 1 mM, 5 mM, and 10 mM, [GSNO] = 5.4 µM, 2.2 µM, and 1.4 µM, respectively. The reason that [GSNO] varies inversely with [GSH] is that N2O3 produces GSNO, and [N2O3] decreases rapidly with increasing [GSH] (Figure 7). The concentrations of GSOH, GSO·, and GSOO· were all found to be negligible relative to that of GSNO. Depending on the GSH/GSSG redox potential, each cellular compartment (cytosol, mitochondria, nucleus) has its own GSH/GSSG concentration ratio [51]. Because GSSG levels are maintained by enzymatic processes, and because the concentration of GSSG is at most 3% of that of GSH, concentrations of GSSG were not calculated in our analysis. Concerning the lumping together of aqueous compartments in the model, it is worth noting that mitochondrial GSH concentrations are generally similar to cytosolic values [52].
Tyrosine Products
In the presence of ascorbate (our baseline conditions) it was found that 3-nitrotyrosine (NO2-Tyr) was the only significant tyrosine product; dityrosine (diTyr) was nearly absent, accounting for only 0.01% of the moles of Tyr going to either product. The reason for this is that the rate of diTyr formation is proportional to the square of the Tyr· radical concentration (Equation 18), and ascorbate is an extremely effective Tyr· scavenger (Equation 16). When ascorbate was assumed to be absent, NO2-Tyr remained the major product; however, diTyr was no longer entirely negligible, accounting then for 6% of the molar consumption of Tyr. For all combinations of antioxidant levels considered (1 mM ≤ [GSH] ≤ 10 mM and 0 ≤ [AH-] ≤ 0.5 mM), NO2-Tyr formation accounted for at least 91% of the consumption of Tyr.
Discussion
The present kinetic analysis provides what seem to be the first estimates of the intracellular concentrations of NO2· and N2O3, two key reactive species derived from NO. Incorporating the effects of the antioxidants, proteins, and lipids present in cells yielded concentrations which were much smaller than those in simple aqueous solutions containing the same levels of NO. For example, with [NO] = 1.0 µM and [O2] = 50 µM, which should be representative of inflamed tissues, we found that [NO2·] = 0.8 pM and [N2O3] = 2.1 fM when the kinetic effects of cellular constituents were included (Figure 5). The corresponding steady-state concentrations in simple aqueous solutions, where only autoxidation (Reactions 1–3) occurs, are calculated to be 40 pM and 500 fM at ambient O2 levels ([O2] = 200 µM) and 10 pM and 120 fM at tissue O2 levels ([O2] = 50 µM). The latter are still some 10 and 60 times the respective intracellular values. In a 0.01 M phosphate buffer at pH 7.0 and ambient O2, the accelerated hydrolysis of N2O3 (Reaction 3) is calculated to yield [NO2·] = 9 pM and [N2O3] = 110 fM, concentrations similar to those at tissue O2 levels in simple solutions without phosphate. Differences in [O2] aside, the main reason for the lower intracellular concentrations is the reaction of both NO2· and N2O3 with glutathione (Reactions 4 and 18). As shown in Figures 6 and 7, GSH has a particularly strong effect on [N2O3], because it scavenges NO2· (needed to form N2O3) as well as N2O3 itself. The marked ability of GSH to scavenge NO2·, and thereby prevent the formation of N2O3, has been recognized previously [53–56].
In addition to containing antioxidants such as glutathione and ascorbate, a distinctive feature of cells is the possibility of lipid-phase reactions as well as aqueous ones. It has been shown by Lancaster and co-workers that, in various hydrophobic media, the rate of autoxidation per unit volume is greatly amplified by the high solubilities of NO and O2 in such media relative to water [8, 9]. Thus, the favorable partitioning of NO and O2 into lipids may lead to large rates of autoxidation in cell membranes per unit volume, even though the rate constants in the two phases are equal or nearly so [8, 9]. The present analysis included the effects of membrane autoxidation, and allowed also for the possibility of intramembrane reactions involving other uncharged species. Because reported rate constants for lipid phases were usually lacking, we assumed each to be equal to the corresponding aqueous value. Thus, the predicted magnitude of the lipid effects depended entirely on the membrane/cytosol partition coefficients (Qi for species i) and the fraction of cell volume occupied by lipids (v, fixed at 0.03 or 3%).
Autoxidation in the membrane (lipid) phase was found not to have a significant effect on the predicted RNS concentrations. For NO2· and N2O3, the results are affected somewhat by uncertainties in QNO, with Liu et al. [8] giving a value of 9 and Miller et al. [9] a value of 3. This corresponds to an order-of-magnitude difference in the rate of NO consumption by O2 in membranes, which is proportional to . However, even adopting the higher value of QNO as a baseline parameter (Table 2), we found the effects of membrane autoxidation on NO2· and N2O3 levels to be small. As shown in Figures 8 and 9, only about 2% and 8%, respectively, of the synthesis of NO2· and N2O3 was associated with membrane autoxidation. If it was assumed that QNO = 3, the membrane contributions to synthesis were reduced to 0.2% for NO2· and 3% for N2O3. Another partition coefficient that affected the levels of NO2 and N2O3 was QNO2. However, as shown in Figures 6 and 7, it became significant only when it was assumed to greatly exceed the baseline estimate of 0.3. Uncertainties in the other relevant partition coefficient, QN2,O3, were found to have negligible effect.
In addition to accelerating autoxidation, membranes might alter the concentrations of RNS by reacting with them directly. The possibility considered in the analysis was the reaction of NO2· with polyunsaturated fatty acids (PUFA, Reactions 61 and 62). As shown in Figure 8, the reaction with PUFA was found to be only a minor sink for NO2·. In summary, although they influence the rate of NO consumption by autoxidation, intramembrane reactions appear to have little effect on the concentrations of RNS. Although having a negligible effect on RNS concentrations, membrane reactions with RNS might still be an important pathway for cellular damage.
A similarly detailed treatment of intracellular nitrogen oxide chemistry was provided by Lancaster [7]. Most of the reactions found in Table 1 were included also in that analysis and the two sets of calculations employ similar or identical values of the rate constants. However, the present model includes a number of additional reactions, namely, Reactions 11, 12, 21, 22, 24, 33–35, 42, 44–59, 61–66. Of these, it seems especially important to include the reaction of peroxynitrite with transition metal centers or selenium-containing compounds (Reaction 58) as a source for NO2·, as pointed out by Radi et al. [40, 57]. As shown in Figure 8, this accounted for nearly 79% of our calculated NO2· production. The reaction of peroxynitrite with proteins (Reaction 59) is also noteworthy, as a sink for peroxynitrite. Ascorbate should not be ignored, mainly because of its ability to scavenge tyrosyl radicals (see below), but also because it is a moderately important sink for NO2· (Figure 8). Lancaster [7] also chose to consider only aqueous chemistry, but this decision is supported by our finding that membrane reactions do not strongly influence RNS levels.
According to the present analysis, the only significant products of glutathione and tyrosine are GSNO and NO2-Tyr, respectively. This differs from the findings of Lancaster [7], where the main glutathione products predicted were GSOH, GSOO· and GSO·, and dityrosine (dityr) was important, in addition to NO2-tyr. These differences stem not only from the inclusion of somewhat different sets of reactions in the two models, but also from a large discrepancy in the assumed levels of . The synthesis rates used by Lancaster [7] were two orders of magnitude larger than what led to the concentration in Table 2 (as much as 200 µM/s, as compared with 1 µM/s). A high concentration, for example, will favor the reaction of Tyr· with over the reaction of Tyr· with NO.
With regard to glutathione products, the model predicts [GSNO] ranging from 1 to 5 µM for [GSH] of 1 to 10 mM, under conditions of steady-state exposure to 1 µM NO and 50 µM O2. These predictions for [GSNO] are reasonable, given the micromolar levels determined in various tissues from humans and animal models of human disease. For example, Kluge et al. measured [GSNO] of 6–8 µM in normal rat cerebellum [58], while Stamler and coworkers have found nitrosothiol levels in general ranging from 7 µM in blood to 15–20 µM in pulmonary fluids [59,60]. Further studies are needed to assess [GSNO] under conditions of inflammation in vivo.
The ratio of the rate of NO2-Tyr formation to that of diTyr is predicted to vary as [NO2·]/[Tyr·], as indicated by Equations 17 and 18. Carbonate radical tends to be the dominant source of Tyr· (more important than NO2· or GS·), and ascorbate its main scavenger. Accordingly, [Tyr·] is very nearly proportional to [Tyr]/[AH-] (Equation 16). The concentration of , in turn, varies as 1/[AH-] (Equation 9). Accordingly, when ascorbate is present, the rate of NO2-Tyr formation relative to that of diTyr will vary nearly as [NO2·][AH-]2/[Tyr]. This illustrates why NO2-Tyr is predicted to be favored so strongly over diTyr at typical intracellular levels of ascorbate, which range from 0.5–2 mM [61]. It should be noted, however, that may be underestimated in the present model to some extent, because is formed also from the reaction of H2O2 with and Cu-Zn SOD [62–64]; a higher would favor more diTyr formation.
The concentration of ·OH radical may be underestimated considerably by Equation 8, because there are sources of ·OH in addition to those included in the model. For example, the reaction of reduced transition metal ions such as copper (I) and iron (II) with H2O2 can give rise to ·OH [65]. However, none of the other results are affected noticeably even if [·OH] is increased by a factor of 103, and the effect on RNS concentrations is negligible even if [·OH] is increased by a factor of 109.
Another pathway for generating NO2 · in an inflamed tissue is the conversion of to NO2· by the extracellular myeloperoxidase (MPO) activity associated with neutrophils [66]. As we are unaware of any data that would allow quantification of this source of NO2·, it was not included in the present model. It is conceivable that, in some circumstances, MPO activity might lead to NO2· concentrations that exceed those predicted here. If extracellular concentrations of NO2· could be estimated, along with a cell membrane permeability to NO2·, then the steady-state mass balance in Equation 1 could be altered to include a source term that corresponds to NO2· diffusion into the cell.
The list of RNS sinks that were considered is also incomplete. For example, N2O3 is known to react with DNA, nucleotides, and amino acids [67]. Of the potential N2O3 targets that were not included in the model, perhaps the most abundant is ATP, which may be present at concentrations as high as 10 mM [68]. Based on preliminary data obtained using dATP (V. Dendroulakis and P.C. Dedon, unpublished) the rate constant for N2O3 with ATP is estimated as 5.0 × 106 M−1s−1. However, even at the high ATP concentration just mentioned, including ATP decreased the predicted value of [N2O3] only from 2.2 fM to 1.9 fM. In summary, the effect of GSH scavenging on [N2O3] is already so strong that reactions with other biomolecular targets are unlikely to depress [N2O3] below the predicted fM range.
The model predicts a roughly order-of-magnitude increase in [N2O3] as [GSH] decreases from 10 to 1 mM (Figure 7), and a corresponding 3- to 4-fold increase in [GSNO]. Such a large increase in [N2O3] would be expected to lead to increased reactions with DNA and other cellular molecules. However, we previously observed that the level of nitrosatively-induced deamination products in DNA increased by at most 40 percent in cells exposed to a steady-state level of 0.6 µM NO for 8 hr when GSH levels were decreased by more than 10-fold with buthionine sulfoximine treatment [69]. This relatively small increase in N2O3-induced DNA damage is consistent with a role for GSH in scavenging N2O3, but suggests that other factors intervene to possibly mask the effects of increased N2O3, such as a balance between DNA repair and N2O3-induced damage. Indeed, the level of nitrosative DNA damage may not serve as a useful index of [N2O3], given our observation of a 3- to 4-fold protective effect when comparing levels of nitrosative DNA damage in cells and purified DNA, both exposed to identical [NO] and [O2] [70, 71], and evidence for significantly higher levels of nitrosative damage in RNA compared to DNA (Pang et al., manuscript in preparation).
The present model can be modified to develop more accurate predictions of intracellular RNS concentrations as the chemistry becomes more completely understood. The algebraic expressions that are presented for the concentrations of RNS and other reactive intermediates permit one to easily update the calculations as new data concerning any of the rate constants or partition coefficients become available. Those expressions also provide a way to assess the importance of including additional intracellular reactions. In general, for an additional source to be important, it must be at least comparable to terms already present in the numerator of a given concentration expression; new sinks may be compared with the terms in the denominator. Because of the analytical form of the results, no special software is needed to assess the effects either of new parameter values or additional reactions.
Acknowledgments
This work was supported by a grant from the National Cancer Institute (PO1 CA26731).
References
- 1.Moncada S, Palmer RMJ, Higgs EA. Nitric oxide: Physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 2.Tamir S, Burney S, Tannenbaum SR. DNA damage by nitric oxide. Chem. Res. Toxicol. 1996;9:821–827. doi: 10.1021/tx9600311. [DOI] [PubMed] [Google Scholar]
- 3.Lewis RS, Tamir S, Deen WM. Kinetic analysis of the fate of nitric oxide synthesized by macrophages in vitro. J. Biol. Chem. 1995;270:29350–29355. doi: 10.1074/jbc.270.49.29350. [DOI] [PubMed] [Google Scholar]
- 4.deRojas-Walker T, Tamir S, Ji H, Wishnok JS, Tannenbaum SR. Nitric oxide induces oxidative damage in addition to deamination in macrophage DNA. Chem. Res. Toxicol. 1995;8:473–477. doi: 10.1021/tx00045a020. [DOI] [PubMed] [Google Scholar]
- 5.Tamir S, Tannenbaum SR. The role of nitric oxide in the carcinogenic process. Biochim. Biophys. Acta. 1996;1288:F31–F36. doi: 10.1016/0304-419x(96)00021-2. [DOI] [PubMed] [Google Scholar]
- 6.Nalwaya N, Deen WM. Analysis of cellular exposure to peroxynitrite in suspension cultures. Chem. Res. Toxicol. 2003;16:920–32. doi: 10.1021/tx025664w. [DOI] [PubMed] [Google Scholar]
- 7.Lancaster JR., Jr. Nitroxidative, nitrosative, and nitrative stress: kinetic predictions of reactive nitrogen species chemistry under biological conditions. Chem. Res. Toxicol. 2006;19:1160–1174. doi: 10.1021/tx060061w. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Joshi MJS, Thomas DD. Accelerated reaction of nitric oxide with O2 within the hydrophobic interior of biological membranes. Proc. Natl. Acad. Sci. USA. 1998;95:2175–2179. doi: 10.1073/pnas.95.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moller MN, Li Q, Vitturi DA, Robinson JM, Lancaster JR, Denicola A. Membrane “lens effect”: focusing the formation of reactive nitrogen oxides from the NO/O2 reaction. Chem. Res. Toxicol. 2007;20:709–714. doi: 10.1021/tx700010h. [DOI] [PubMed] [Google Scholar]
- 10.Prutz WA, Monig H, Butler J, Land EJ. Reactions of nitrogen dioxide in aqueous model systems: oxidation of tyrosine units in peptides and proteins. Arch. Biochem. Biophys. 1985;243:125–134. doi: 10.1016/0003-9861(85)90780-5. [DOI] [PubMed] [Google Scholar]
- 11.Kikugawa K, Kato T, Okamoto Y. Damage of amino acids and proteins induced by nitrogen dioxide, a free radical toxin, in air. Free Radic. Biol. Med. 1994;16:373–382. doi: 10.1016/0891-5849(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 12.Lewis RS, Deen WM. Kinetics of the reaction of nitric oxide with oxygen in aqueous solutions. Chem. Res. Toxicol. 1994;7:568–574. doi: 10.1021/tx00040a013. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz SE. Advances in Environmental Science and Technology. N.Y.: Wiley; Trace atmospheric constituents: properties, transformations, and fates; pp. 1–115. [Google Scholar]
- 14.Ross AB, Mallard WG, Helman WP, Buxton GV, Hurie RE, Neta P. NDRL/NIST Solution Kinetics Database ver 3. Notre Dame Radiation Laboratory. Gaithersburg, MD: Notre Dame; 1998. http://www.rcdc.nd.edu/browse_compil.html. [Google Scholar]
- 15.Goldstein S, Czapski G, Lind J, Merenyi G. Tyrosine nitration by simultaneous generation of NO and O2 − under physiological conditions. How the radicals do the job. J. Biol. Chem. 2000;275:3031–3036. doi: 10.1074/jbc.275.5.3031. [DOI] [PubMed] [Google Scholar]
- 16.Ford E, Hughes MN, Wardman P. Kinetics of the reactions of nitrogen dioxide with glutathione, cysteine, and uric acid at physiological pH. Free Radic. Biol. Med. 2002;32:1314–1323. doi: 10.1016/s0891-5849(02)00850-x. [DOI] [PubMed] [Google Scholar]
- 17.Kirsch M, Lehnig M, Korth H-G, Sustmann R, de Groot H. Inhibition of peroxynitrite-induced nitration of tyrosine by glutathione in the presence of carbon dioxide through both radical repair and peroxynitrite formation. Chem. Eur. J. 2001;15:3313–3320. doi: 10.1002/1521-3765(20010803)7:15<3313::aid-chem3313>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman M, Hayon E. Pulse radiolysis study of sulfhydryl compounds in aqueous solution. J. Phys. Chem. 1973;77:990–996. [Google Scholar]
- 19.Wardman P, von Sonntag C. Kinetic factors that control the fate of thiyl radicals in cells. Methods Enzymol. 1995;251:31–45. doi: 10.1016/0076-6879(95)51108-3. [DOI] [PubMed] [Google Scholar]
- 20.Wood PD, Mutus B, Redmond RW. The mechanism of photochemical release of nitric oxide from S-nitrosoglutathione. Photochem. Photobiol. 1996;64:518–524. [Google Scholar]
- 21.Shin HW, George C. Steven. Microscopic modeling of NO and Snitrosoglutathione kinetics and transport in human airways. J. Appl. Physiol. 2001;90:777–788. doi: 10.1152/jappl.2001.90.3.777. [DOI] [PubMed] [Google Scholar]
- 22.Jourd’heuil D, Mai CT, Laroux FS, Wink DA, Grisham MB. The reaction of S-nitrosoglutathione with superoxide. Biochem. Biophys. Res. Commun. 1998;246:525–530. [PubMed] [Google Scholar]
- 23.Goldstein S, Lind J, Merenyi G. Reaction of organic peroxyl radicals with NO2 and NO in aqueous solution: Intermediacy of organicperoxynitrate and peroxynitrite species. J. Phys. Chem. 2004;108:1719–1725. [Google Scholar]
- 24.Wardman P. Evaluation of the ‘radical sink’ hypothesis from a chemical-kinetic viewpoint. J. Radioanal. Nucl. Chem. 1998;232:23–27. [Google Scholar]
- 25.Keshive M, Singh S, Wishnok JS, Tannenbaum SR, Deen WM. Kinetics of S-nitrosation of thiols in nitric oxide solutions. Chem. Res. Toxicol. 1996;9:988–993. doi: 10.1021/tx960036y. [DOI] [PubMed] [Google Scholar]
- 26.Bonini MG, Augusto O. Carbon dioxide stimulates the production of thiyl, sulfinyl and disulfide radical anion from thiol oxidation by peroxynitrite. J. Biol. Chem. 2001;276:9749–9754. doi: 10.1074/jbc.M008456200. [DOI] [PubMed] [Google Scholar]
- 27.Chen SN, Hoffman MZ. Rate constants for the reaction of the carbonate radical with compounds of biochemical interest in neutral aqueous solution. Radiat. Res. 1973;56:40–47. [PubMed] [Google Scholar]
- 28.Jones CM, Lawrence A, Wardman P, Burkitt MJ. SocKinetics of superoxide scavenging by glutathione: an evaluation of its role in the removal of mitochondrial superoxide. Biochem. Soc. Trans. 2003;31:1337–1339. doi: 10.1042/bst0311337. [DOI] [PubMed] [Google Scholar]
- 29.Luo D, Smith SW, Anderson BD. Kinetics and mechanism of the reaction of cysteine and hydrogen peroxide in aqueous solution. J. Pharm. Sci. 2005;94:304–316. doi: 10.1002/jps.20253. [DOI] [PubMed] [Google Scholar]
- 30.Quijano C, Romero N, Radi R. Tyrosine nitration by superoxide and nitric oxide fluxes in biological systems: modeling the impact of superoxide dismutase and nitric oxide diffusion. Free Radic. Biol. Med. 2005;39:728–741. doi: 10.1016/j.freeradbiomed.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Lymar SV, Hurst JK. Rapid reaction between peroxonitrite ion and carbon dioxide: Implications for biological activity. J. Am. Chem. Soc. 1995;117:8867–8868. [Google Scholar]
- 32.Behar D, Czapski G, Duchovny I. Carbonate radical in flash photolysis and pulse radiolysis of aqueous carbonate solutions. J. Phys. Chem. 1970;74:2206–2210. [Google Scholar]
- 33.Czapski G, Holcman J, Bielski BH. Reactivity of nitric oxide with simple shortlived radicals in aqueous solutions. J. Am. Chem. Soc. 1994;116:11465–11469. [Google Scholar]
- 34.Solar S, Solar W, Getoff N. Reactivity of OH with tyrosine in aqueous solution studied by pulse radiolysis. J. Phys. Chem. 1984;88:2091–2095. [Google Scholar]
- 35.Hudges GR, Marwaha J, Paul T, Ingold KU. A novel procedure for generating both nitric oxide and superoxide in situ from chemical sources at any chosen mole ratio. First application: tyrosine oxidation and a comparison with performed peroxynitrite. Chem. Res. Toxicol. 2000;13:1287–1293. doi: 10.1021/tx0001272. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein S, Czapski G. Reactivity of peroxynitrite versus simultaneous generation of NO and O2 − toward NADH. Chem. Res. Toxicol. 2000;13:736–741. doi: 10.1021/tx000099n. [DOI] [PubMed] [Google Scholar]
- 37.Jourd’heuil D, Jourd’heuil FL, Kutchukian PS, Musah RA, Wink DA, Grisham MB. Reaction of superoxide and nitric oxide with peroxynitrite. Implications for peroxynitrite-mediated oxidation reactions in vivo. J. Biol. Chem. 2001;276:28799–28805. doi: 10.1074/jbc.M102341200. [DOI] [PubMed] [Google Scholar]
- 38.Santos C, Bonini MG, Augusto O. Role of the carbonate anion in tyrosine nitration and hydroxylation by peroxynitrite. Arch. Biochem. Biophys. 2000;377:146–152. doi: 10.1006/abbi.2000.1751. [DOI] [PubMed] [Google Scholar]
- 39.Buxton GV, Elliot AJ. Rate-constant for the reaction of hydroxyl radical with bicarbonate ions. Radiat. Phys. Chem. 1986;27:241–243. [Google Scholar]
- 40.Alvarez B, Radi R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 41.Goldstein S, Czapski G. The reaction of NO with O2 − and HO2: a pulse radiolysis study. Free Radic. Biol. Med. 1995;19:505–510. doi: 10.1016/0891-5849(95)00034-u. [DOI] [PubMed] [Google Scholar]
- 42.Gratzel M, Henglein A, Lilie J, Beck G. Pulsradiolytische untersuchung einigerelementarprozesse der oxidation und reduction des nitritions. Ber. Bunsenges. Phys. Chem. 1969;1969;73:646–653. [Google Scholar]
- 43.Broszkiewicz RK. The pulse radiolysis study of NaNO 2 and NaNO 3 solutions. Bull. Acad. Pol. Sci. Sedr. Sci. Chim. 1976;24:221–229. [Google Scholar]
- 44.Schwartz SE, White WH. Kinetics of reactive dissolution of nitrogen oxide in aqueous solution. Adv. Environ. Sci. Technol. 1983;12:1–116. [Google Scholar]
- 45.Hunter EPL, Desrosiers MF, Simic MG. The effect of oxygen, antioxidants, and superoxide radical on tyrosine phenoxyl radical dimerization. Free Radic. Biol. Med. 1989;6:581–585. doi: 10.1016/0891-5849(89)90064-6. [DOI] [PubMed] [Google Scholar]
- 46.Roos A, Boron WF. Intracellular pH. Physiol. Rev. 1981;61:296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- 47.Bergstrom J, Furst P, Noree LO, Vinnars E. Intracellular free amino acid concentration in human muscle tissue. J. Appl. Physiol. 1974;36:693–697. doi: 10.1152/jappl.1974.36.6.693. [DOI] [PubMed] [Google Scholar]
- 48.Fritsche E, Schäfer C, Calles C, Bernsmann T, Bernshausen T, Wurm M, Hübenthal U, Cline JE, Hajimiragha H, Schroeder P, Klotz LO, Rannug A, Fürst P, Hanenberg H, Abel J, Krutmann J. Lightening up the UV response by identification of the arylhydrocarbon receptor as a cytoplasmatic target for ultraviolet B radiation. Proc. Natl. Acad. Sci. USA. 2007;104:8851–8856. doi: 10.1073/pnas.0701764104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leo Albert, Hoekman DH, Hansch Corwin. Exploring QSAR. Washington, DC: American Chemical Society; 1995. [Google Scholar]
- 50.Meylan WM, Howard PH. Atom/fragment contribution method for estimating octanol-water partition coefficients. J. Pharm. Sci. 1995;84:83–92. doi: 10.1002/jps.2600840120. [DOI] [PubMed] [Google Scholar]
- 51.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu. Rev. Pharmacol. Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Ruiz C, Morales A, Ballesta A, Rodes J, Kaplowitz N, Fernandez-Checa JC. Effect of chronic ethanol feeding on glutathione and functional integrity of mitochondria in periportal and perivenous rat hepatocytes. J. Clin. Invest. 1994;94:193–201. doi: 10.1172/JCI117306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldstein S, Czapski G. Mechanism of the nitrosation of thiols and amines by oxygenated NO solutions: the nature of the nitrosating intermediates. J. Am. Chem. Soc. 1996;118:3419–3425. [Google Scholar]
- 54.Schrammel A, Gorren A, Schmidt K, Pfeiffer S, Mayer M. S-nitrosation of glutathione by nitric oxide, peroxynitrite, and NO/O2 − . Free Radic. Biol. Med. 2003;34:1078–1088. doi: 10.1016/s0891-5849(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 55.Sovitj P, Rose GM. Generation of thiyl radical by nitric oxide: a spin trapping study. J. Chem. Soc., Perkins Trans. 1998;2:1507–1512. [Google Scholar]
- 56.Jourd’heuil D, Jourd’heuil FL, Feelisch M. Oxidation and nitrosation of thiols at low micromolar exposure to nitric oxide. J. Biol. Chem. 2003;278:15720–15726. doi: 10.1074/jbc.M300203200. [DOI] [PubMed] [Google Scholar]
- 57.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. USA. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kluge I, Gutteck-Amsier U, Zollinger M, Quang Do K. S-nitrosoglutathione in rat cerebellum: identification and quantification by liquid chromatography-mass spectrometry. J. Neurochem. 1997;69:2599–2607. doi: 10.1046/j.1471-4159.1997.69062599.x. [DOI] [PubMed] [Google Scholar]
- 59.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singeli D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc. Natl. Acad. Sci. USA. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaston B, Reilly J, Drazen JM, Fackler J, Ramdev P, Arnelle D, Mullins ME, Sugarbaker DJ, Chee C, Singel DJ, Loscalzo J. Endogenous nitrogen oxides and bronchiolar S-nitrosothiols in human airways. Proc. Natl. Acad. Sci. USA. 1993;90:10957–10961. doi: 10.1073/pnas.90.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Washko P, Rotrosen D, Levine M. Ascorbic acid transport and accumulation in human neutrophils. J. Biol. Chem. 1989;264:18996–19002. [PubMed] [Google Scholar]
- 62.Zhang H, Joseph J, Felix C, Kalyanaraman B. Bicarbonate enhances the hydroxylation, nitration and peroxidation reactions catalyzed by copper, zinc superoxide dismutase. J. Biol. Chem. 2000;275:14038–14045. doi: 10.1074/jbc.275.19.14038. [DOI] [PubMed] [Google Scholar]
- 63.Ramirez DC, Mejiba SEC, Mason RP. Mechanism of hydrogen peroxide-induced Cu, Zn-superoxide dismutase-centered radical formation as explored by immuno-spin trapping: the role of copper and carbonate radical anion-mediated oxidations. Free Radic. Biol. Med. 2005;38:201–214. doi: 10.1016/j.freeradbiomed.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 64.Karunakaran C, Zhang H, Joseph J, Antholine WE, Kalyanaraman B. Thiol oxidase activity of copper, zinc superoxide dismutase stimulates bicarbonate-dependent peroxidase activity via formation of a carbonate radical. Chem. Res. Toxicol. 2005;18:494–500. doi: 10.1021/tx049747j. [DOI] [PubMed] [Google Scholar]
- 65.Halliwell B, Gutteridge JMC, editors. Free Radicals in Biology and Medicine. Oxford: Oxford University Press; 1998. [Google Scholar]
- 66.Burner U, Furtmüller PG, Kettle AJ, Koppenol WH, Obinger C. Mechanism of reaction of myeloperoxidase with nitrite. J. Biol. Chem. 2000;275:20597–20601. doi: 10.1074/jbc.M000181200. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen T, Brunson D, Crespi CL, Penman B, Wishnok JS, Tannenbaum SR. DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc. Natl. Acad. Sci. USA. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 69.Li CQ, Wright TL, Dong M, Dommels YE, Trudel LJ, Dedon PC, Tannenbaum SR, Wogan GN. Biological role of glutathione in nitric oxide-induced toxicity in cell culture and animal models. Free Radic. Biol. Med. 2005;39:1489–1498. doi: 10.1016/j.freeradbiomed.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 70.Dong M, Wang C, Deen WM, Dedon PC. Absence of 2-deoxyoxanosine and presence of abasic sites in DNA exposed to nitric oxide at controlled physiological concentrations. Chem. Res. Toxicol. 2003;16:1044–1055. doi: 10.1021/tx034046s. [DOI] [PubMed] [Google Scholar]
- 71.Dong M, Dedon PC. Relatively small increases in the steady-state levels of nucleobase deamination products in DNA from human TK6 cells exposed to toxic levels of nitric oxide. Chem. Res. Toxicol. 2006;19:50–57. doi: 10.1021/tx050252j. [DOI] [PMC free article] [PubMed] [Google Scholar]