Abstract
A cell-based high-throughput screen to identify small molecular weight stimulators of the innate immune system revealed substituted pyrimido[5,4-b]indoles as potent NFκB activators. The most potent hit compound selectively stimulated Toll-like receptor 4 (TLR4) in human and mouse cells. Synthetic modifications of the pyrimido[5,4-b]indole scaffold at the carboxamide, N-3, and N-5 positions revealed differential TLR4 dependent production of NFκB and type I interferon associated cytokines, IL-6 and interferon γ-induced protein 10 (IP-10) respectively. Specifically, a subset of compounds bearing phenyl and substituted phenyl carboxamides induced lower IL-6 release while maintaining higher IP-10 production, skewing toward the type I interferon pathway. Substitution at N-5 with short alkyl substituents reduced the cytotoxicity of the leading hit compound. Computational studies supported that active compounds appeared to bind primarily to MD-2 in the TLR4/MD-2 complex. These small molecules, which stimulate innate immune cells with minimal toxicity, could potentially be used as adjuvants or immune modulators.
Introduction
The innate immune response is the first line of defense against microbial pathogens such as viruses, bacteria, fungi, and protozoa. A critical component of the innate immune response is the NFκB family of transcription factors.1,2 The Toll-like receptors (TLRs) are critical components of the innate immune system that regulate NFκB activation. In general, the TLRs recognize macromolecules that are associated with pathogens and with cell stress. These pathogen-associated molecular patterns (PAMPs) and their corresponding TLRs include: lipopeptides (TLR2), double-stranded RNA (TLR3), lipopolysaccharide (LPS, TLR4),3 bacterial flagellin (TLR5), guanine and uridine-rich single-stranded RNA (TLR7, 8), and hypo-methylated CpG rich DNA (TLR9).4 Innate immune cells use pattern recognition receptors (PRRs) such as TLRs to promote a rapid response to perceived threats before pathogen-specific adaptive immunity can be established. Indeed, this rapid response, involving multiple components of the innate immune system, has been recognized to guide the type of adaptive immune response that is most effective for the specific pathogenic threat.
The discovery that certain small molecules could serve as specific innate immune receptor ligands has expanded the possibility of designing drugs that may modulate the immune response at its earliest stages. Notable examples of such small molecules are the TLR7 agonists imiquimod,5 isotorabine,6 and the more recent 8-oxo-9-benzyladenines,7 as well as the TLR7/8 agonist resiquimod.8 A useful application of this concept is the incorporation of adjuvants into vaccines. Adjuvants are added to antigens in a vaccine setting to provide enhanced response to poorly immunogenic antigens, to increase seroconversion rates in populations with reduced responsiveness due to age (infants and the elderly) or disease (diabetes, renal failure), to facilitate the use of smaller doses of antigen, and to permit immunization with fewer doses of vaccine.9 Macromolecular activators of the innate immune system, such as monophosphoryl lipid A (MPLA), a TLR4 ligand, are utilized as adjuvants,10 but development of small molecular weight nonlipid ligands might have several advantages to address different immunological requirements for vaccines or immune therapeutics.
As part of our studies on small molecules that can activate TLRs, we conducted a high through-put screening (HTS) campaign in which a commercially available library of compounds was screened in a human cell-based NFκB activation assay. Many innate signaling pathways converge on the transcription factor NFκB. Hence it was used in the primary screen as a broad indicator of small molecule agonism of innate immunity. The specific innate receptor for the lead compounds was determined using genetically modified cells and primary mouse and human blood or bone marrow mononuclear cells. Here we report our discovery and initial structure–activity relationship (SAR) studies of the pyrimido[5,4-b]indole class of ligands and their characterization as TLR4/MD-2 agonists.
Results and Discussion
High-Throughput Screen Study Design
A library of compounds was acquired from the University of California, San Francisco, Small Molecule Discovery Center consisting of about 170000 compounds from eight suppliers (https://smdc.ucsf.edu). The library was screened in three phases at a commercial HTS screening facility using the THP-1 human monocytic leukemia cell line, which contained a β-lactamase reporter gene under the control of the NFκB response element that had been stably integrated into the cells. All screens were performed in activator mode using LPS as a positive control, achieving typical Z′ values above 0.75. The three-phase screening process consisted of (1) a pilot screen of about 10000 compounds selected as representative of the entire primary library, (2) the primary screen of the entire library, and (3) a confirmation screen of about 2000 hits found in the primary screen. Compounds identified as active in two screens were considered to be confirmed hits. An analysis of the cluster enrichment methods for hit selection has been recently reported.11
Discovery of Pyrimido[5,4-b]indoles as Activators of NFκB
Following the cluster enrichment analysis, 225 compounds were selected for further in vitro biological evaluation involving cytokine induction assays in primary cells, including human peripheral blood mononuclear cells (hPBMC), mouse splenocytes, mouse bone marrow derived dendritic cells (mBMDC), and mouse bone marrow derived macrophages (mBMDM). These cells were incubated in triplicate, with each of the 225 compounds at a single concentration (1 μM for splenocytes and 5 μM for all other mouse cells and human cells), and the supernatants were tested for the presence of NFκB dependent cytokines, IL-8 or IL-6, released from the human or mouse cells, respectively. Thirty-nine of the 225 compounds stimulated the human and mouse cells to secrete IL-8 or IL-6 above the detectable limit. To further confirm activity, these compounds were repurchased and retested by stimulating hPBMC and mBMDC with titrated doses and assaying for IL-8 and IL-6.
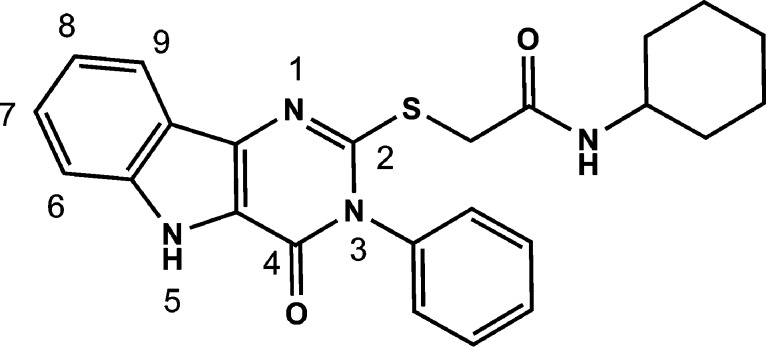
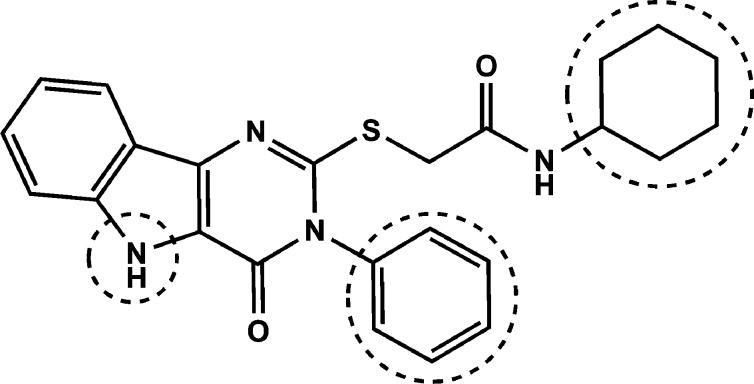
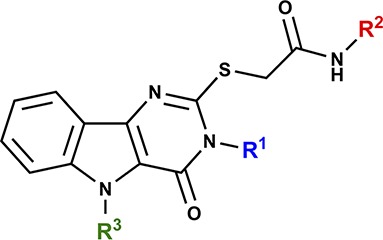
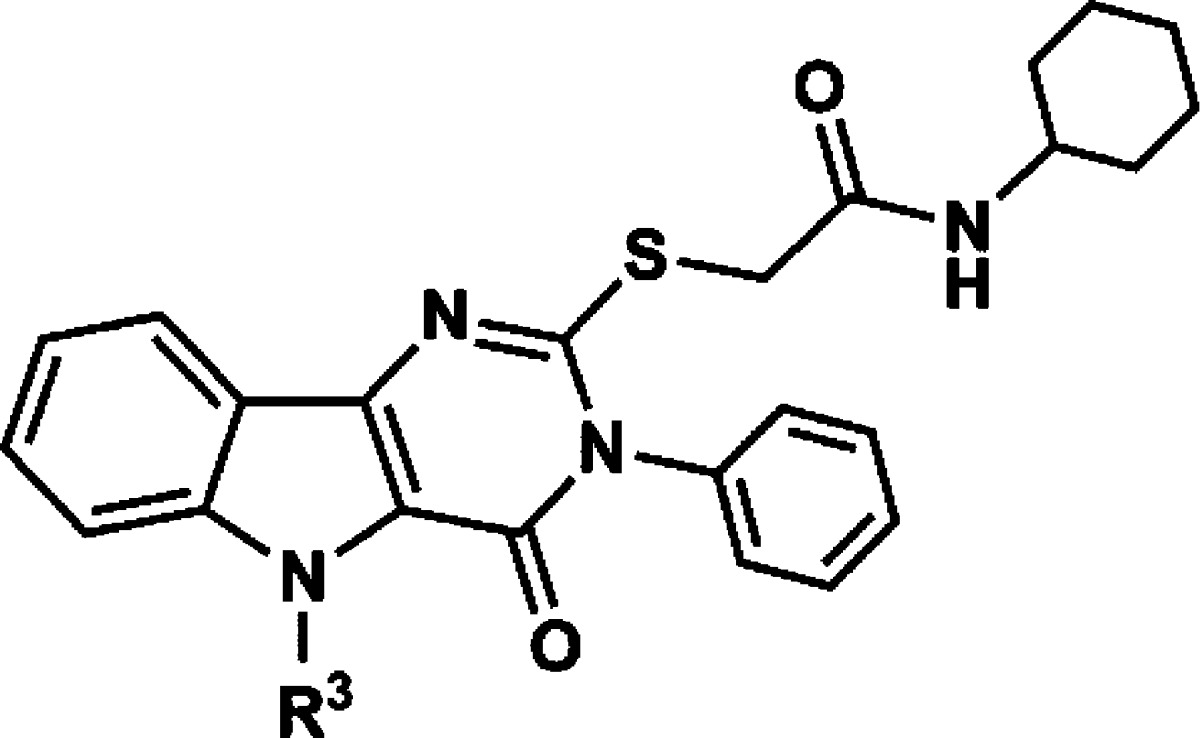
A few structurally diverse library scaffolds were identified in these cytokine assays as having reproducible responses in mouse and human stimulation assays. Among these scaffolds, the pyrimido[5,4-b]indoles emerged as the most potent and diverse class of compounds in the mouse cell assays with clear evidence of SAR. Within this scaffold cluster, the leading hit from the initial primary and secondary screens was a substituted acetamide attached to the pyrimidoindole ring system through a thioether linkage (Figure 1). Thus, compound 1 provided a starting point for structure–activity studies.
Figure 1.
Structure of hit compound 1.
Target Receptor Identification
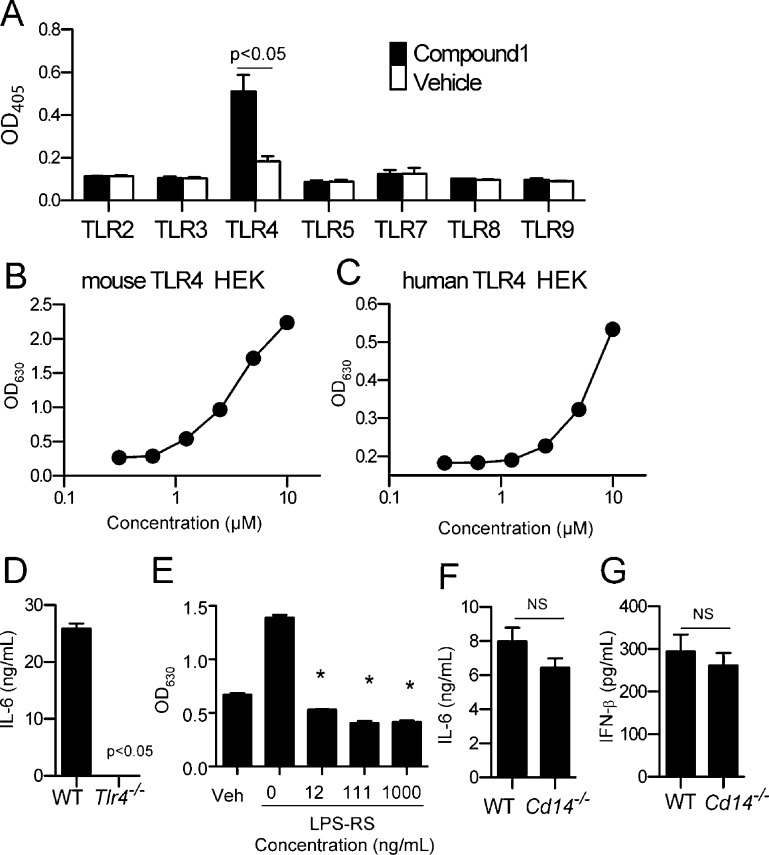
To identify the target receptor, we used HEK293 cells stably transfected with individual human (h) TLRs: TLR2, TLR3, TLR4/MD-2/CD14, TLR5, TLR7, TLR8, or TLR9 expressing HEK293 cells, along with an NFκB activation reporter producing secreted embryonic alkaline phosphatase (SEAP). Among the tested TLR transfected cells, only those expressing TLR4/MD-2/CD14 responded to pyrimidoindoles, as shown in Figure 2A for compound 1. Because TLR4/MD-2/CD14 was the receptor complex for the active compounds in this series, it was important to rule out the possibility that activity might have been caused by LPS contamination. Therefore, compound 1 (and all active derivatives) was assayed for LPS (endotoxin) levels using a commercially available detection system and found to contain less than 10 endotoxin units (EU)/μmol compound. To further exclude contamination, compound 1 was resynthesized according to Scheme 1. Both samples of compound 1 displayed indistinguishable physicochemical and biological properties, indicating that the positive biological activity was not due to LPS or another contaminant. Full titration curves of the resynthesized compound were performed using the SEAP assay with mouse and human TLR4 transfected HEK293 cell lines, confirming dose-dependent activation (Figure 2B, C).
Figure 2.
Target identification of compound 1 using human TLR HEK293 reporter cell lines and genetically deficient cells. (A) Human TLR2, TLR3, TLR4/MD-2/CD14, TLR5, TLR7, TLR8, and TLR9 HEK 293 Blue cells or NF-κB/SEAPorter cells were incubated with compound 1 (10 μM) for 20–24 h, and activation was evaluated by SEAP secretion in the culture supernatants by colorimetric assay at OD405. Data shown are mean ± SEM of triplicates and representative of two to three independent experiments showing similar results. p < 0.05 was considered significant compared to the vehicle control using Student’s t test. (B,C) Mouse (B) or human (C) TLR4 HEK transfectomas were incubated with graded concentrations of compound 1. TLR4 mediated NFκB activation was measured by SEAP secretion in the culture supernatant. (D) WT and Tlr4–/– mBMDC were incubated with compound 1 (10 μM) for18 h. IL-6 in the culture supernatants was measured by ELISA. (E) Mouse TLR4 transfectoma cells were incubated with 2.5 μM compound 1 in the presence or absence of TLR4 antagonist LPS-RS (12, 111, 1000 ng/mL). Activation of the TLR4/NFκB pathway was evaluated by SEAP secretion in the culture supernatants. * denotes p < 0.05 considered as significant compared to vehicle using one way ANOVA with Dunnett’s post hoc testing. (F,G) WT and Cd14–/– mBMDC were incubated with compound 1 (3.7 μM) overnight. IL-6 in the culture supernatants was measured by ELISA (F) and type I IFN (IFN-β) measured by luciferase release in from an ISRE reporter cell line (G). p < 0.05 was considered as significant compared to vehicle using Student’s t test. NS denotes “not significant”. Data shown are mean ± SEM of triplicates and representative of two independent experiments showing similar results.
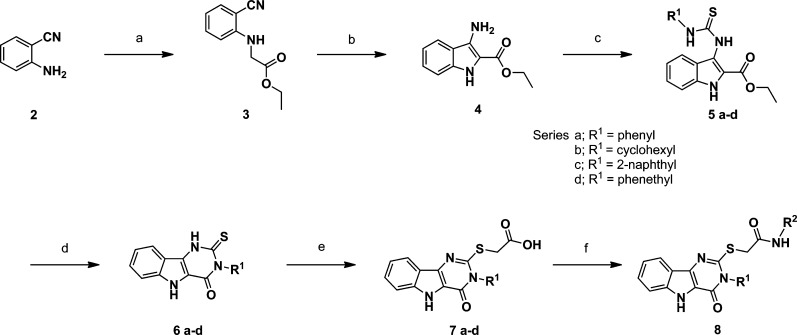
Scheme 1. General Synthetic Route.
Reagents and conditions: (a) BrCH2COOEt, NaHCO3, EtOH, reflux; (b) tert-BuOK, THF, <30 °C; (c) R1-NCS, EtOH, reflux; (d) PPA, 110 °C; (e) ClCH2COOH, KOH/EtOH, reflux; (f) R2-NH2 (for R2 substitutions refer to Table 1), HATU, DMF, room temp.
To confirm that TLR4 was indeed the receptor, compound 1 was assayed for IL-6 production in mBMDCs from wild-type and TLR4 deficient mice (Figure 2D). Genetic disruption of TLR4 completely abrogated IL-6 secretion induced by compound 1. The binding of compound 1 to the TLR4/MD-2/CD14 complex was further confirmed using a competitive antagonist for the TLR4 binding complex, LPS-RS (LPS from Rhodococcus sphaeroides).12 LPS-RS inhibited the activation by compound 1 in a dose-dependent manner, indicating that compound 1 bound to the TLR4 complex (Figure 2E).
TLR4 signals through two distinct pathways, leading respectively to NFκB dependent cytokines and type I interferon (IFN) production. Several naturally occurring TLR4 ligands and MPLA have been reported to require CD14 to activate the type I IFN regulatory pathway.13
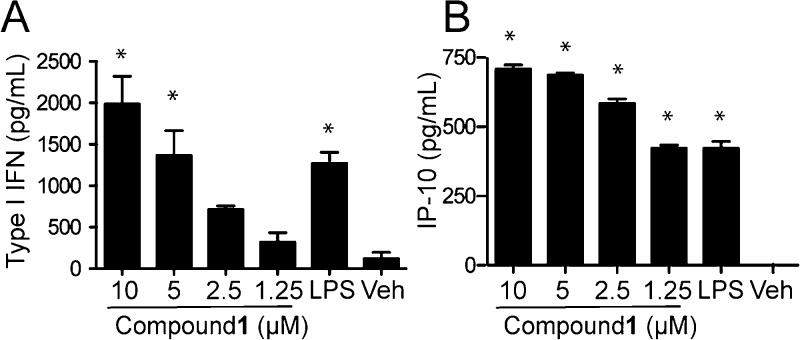
The hTLR4 transfected HEK293 cell line (Figure 2A) also overexpresses MD-2 and CD14, which are TLR4 accessory proteins.14,15 Compound 1, however, was not dependent on CD14 for either IL-6 or type I IFN production, as demonstrated using CD14 deficient cells (Figure 2F, G). The supernatants from mBMDCs stimulated with graded doses of compound 1 were also tested for IP-10 as a surrogate marker of type I IFN release. Results showed a dose-dependent response for type I IFN (Figure 3A) production, which paralleled that of IP-10 (Figure 3B).
Figure 3.
Type I IFN induction by compound 1. Wild-type mBMDCs were incubated with graded concentrations of compound 1 overnight. DMSO 0.5% served as the vehicle control. The levels of type I IFN were determined using an L929-ISRE luciferase reporter cell line and a mIFNβ standard (A). IP-10 levels were determined by ELISA (B). Data shown are mean ± SEM of triplicates and representative of two independent experiments showing similar results. * denotes p < 0.05 compared to vehicle using one way ANOVA with Dunnett’s post hoc testing.
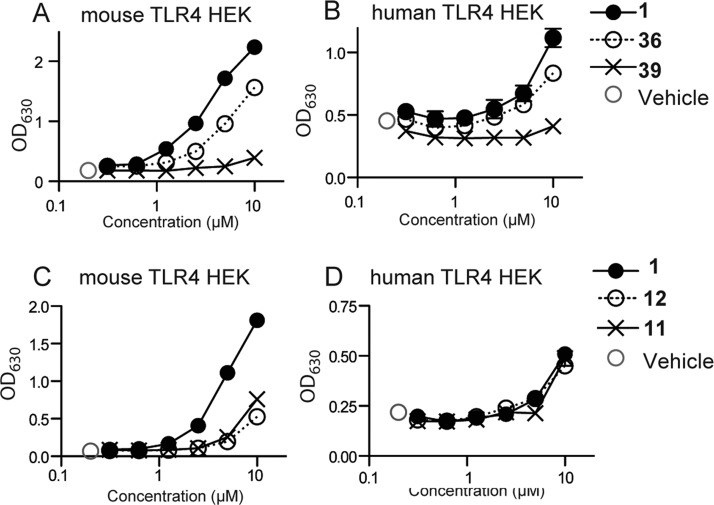
The above assays were utilized to compare the derivatives of the lead pyrimidoindole for their ability to activate mouse and human TLR4. Mouse TLR4 activation was assessed using primary mBMDC and IL-6 release (Figure 4) and confirmed using mTLR4 transfected HEK293 cells (Figure 5A, C). The values shown in the Tables 1–3 for IL-6 release, and mTLR4 activation are area under the curve (AUC) values for titrated doses of compounds from 312 nM to 10 μM. Each cytokine induction curve was first converted to a percent activity curve, and then the AUC of the percent activity curve was calculated. The process of converting to a percent activity curve allowed subtracting background and adjusting for plate-to-plate variation. Finally, the AUC values were normalized to the activity of compound 1 within each experiment, set at 100. Human TLR4 activation is shown for stimulation of PBMC (IL-8) and hTLR4 HEK293 transfectomas (Figure 5B,D) at 10 μM, as these assays were not sufficiently sensitive at lower concentrations to make AUC comparisons. The levels of hTLR4 activation by SAR derivatives are expressed relative to compound 1, set at 100.
Figure 4.
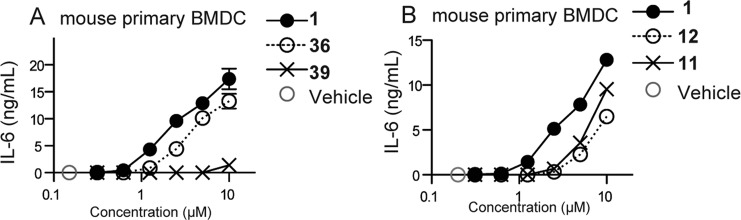
Representative data of SAR compound screening using mouse primary dendritic cells. Biological screening of SAR compounds was conducted using primary mBMDC. The cells were incubated with graded concentrations of the indicated compounds 1, 36, and 39 (A) or 11 and 12 (B) for 18 h. DMSO 0.5% served as the vehicle control. IL-6 levels in culture supernatants were measured by ELISA. Data shown are mean ± SEM of triplicate.
Figure 5.
Representative data of SAR compound screening using TLR4 transfectomas. Mouse TLR4 (A,C), and hTLR4 (B,D) HEK transfectomas were incubated with graded concentrations of the indicated compounds 1, 36, and 39 (A) or 11 and 12 (B) for 18 h. DMSO 0.5% served as the vehicle control. The specific activation of the reporter cell lines was measured by SEAP activity in the supernatant by absorption at 630 nm. Data shown are mean ± SEM of triplicate data.
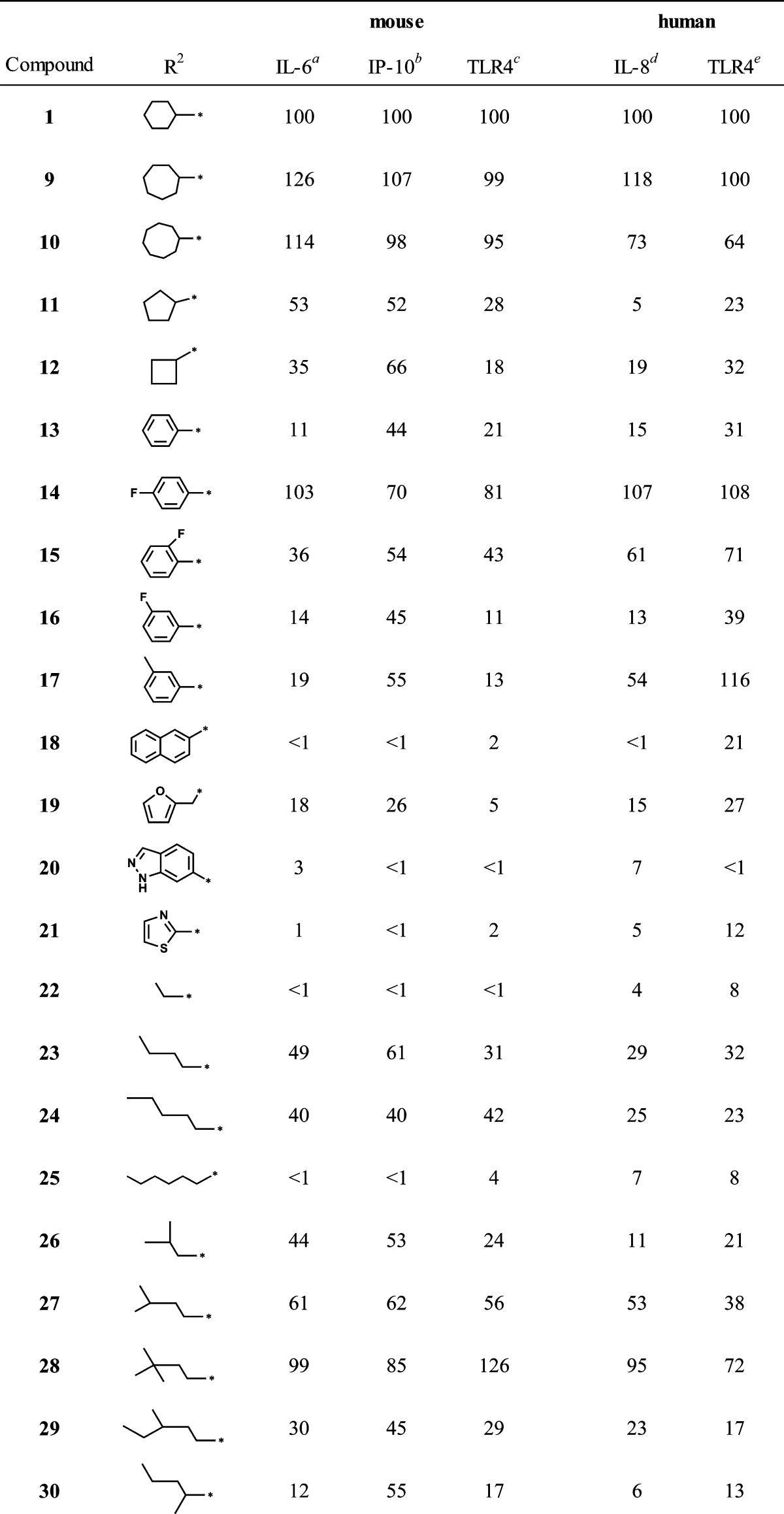
Table 1. N-Substituted Carboxamide Derivatives.
AUC values normalized to compound 1. Stimulation with LPS (10 ng/mL), and 5 μM of 1 released an average of 20.6 ± 4.8 and 10.5 ± 1.3 ng/mL of IL-6, respectively.
All compounds tested at 5 μM, values normalized to 1. Stimulation with LPS 10 ng/mL and 5 μM of 1 released averages of 234.8 ± 8.6 and 590 ± 11 pg/mL of IP-10, respectively.
AUC values normalized to compound 1. TLR4 cell activation stimulated with 10 ng/mL LPS and 5 μM compound 1 resulted in OD630 of 1.69 ± 0.08 and 1.52 ± 0.03, respectively.
All compounds tested at 10 μM, values normalized to 1. Stimulation with LPS 10 ng/mL and 10 μM of 1 released averages of 14.5 ± 0.6 and 7.8 ± 0.8 ng/mL of IL-8, respectively.
All compounds tested at 10 μM, values normalized to 1. TLR4 cell activation stimulated with 10 ng/mL LPS and 10 μM compound 1 resulted in OD630 of 1.90 ± 0.04 and 0.88 ± 0.02, respectively.
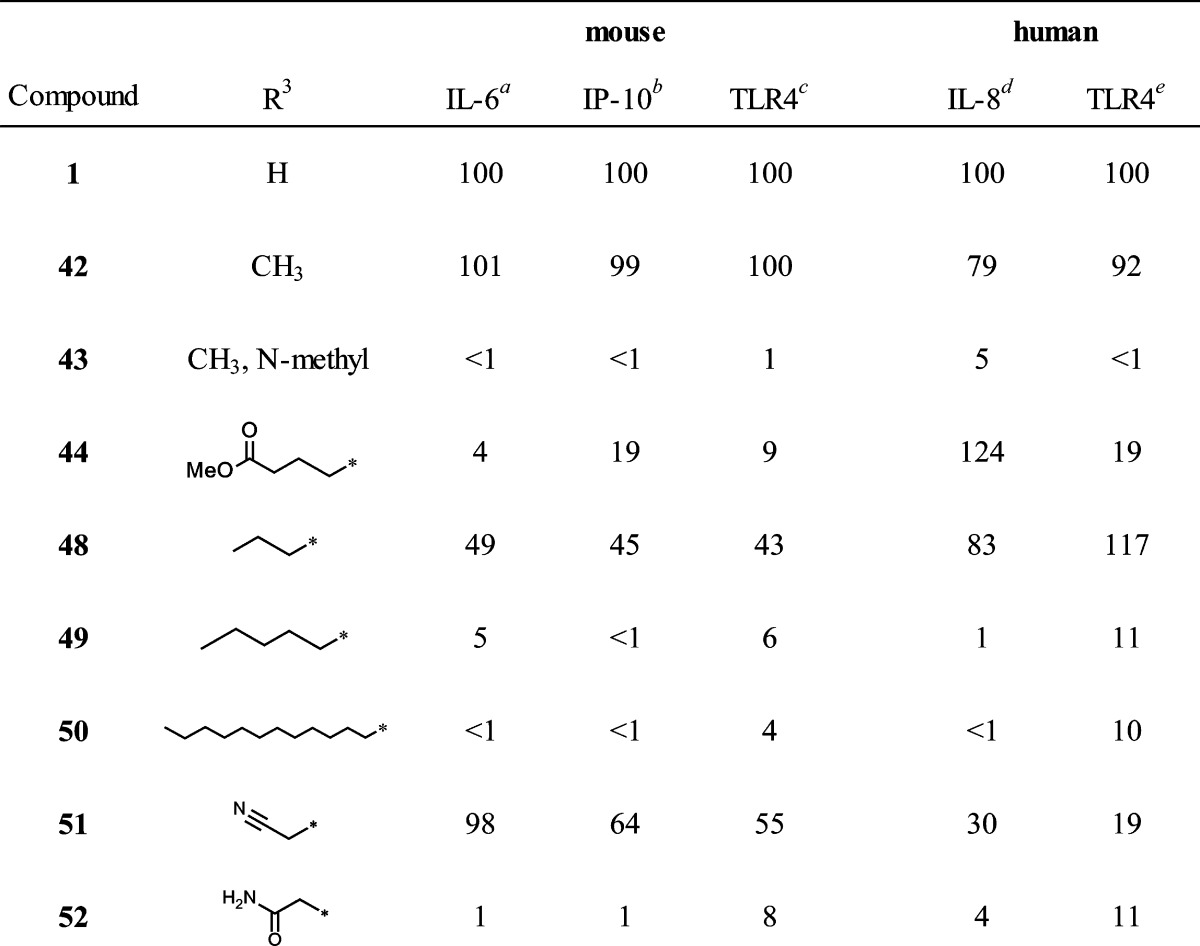
Table 3. N-5 Derivatives.
AUC values normalized to compound 1. Stimulation with LPS (10 ng/mL) and 5 μM of 1 released an average of 20.6 ± 4.8 and 10.5 ± 1.3 ng/mL of IL-6, respectively.
All compounds tested at 5 μM, values normalized to 1. Stimulation with LPS 10 ng/mL and 5 μM of 1 released averages of 234.8 ± 8.6 and 590 ± 11 pg/mL of IP-10, respectively.
AUC values normalized to compound 1. TLR4 cell activation stimulated with 10 ng/mL LPS and 5 μM compound 1 resulted in OD630 of 1.69 ± 0.08 and 1.52 ± 0.03, respectively.
All compounds tested at 10 μM, values normalized to 1. Stimulation with LPS 10 ng/mL and 10 μM of 1 released averages of 14.5 ± 0.6 and 7.8 ± 0.8 ng/mL of IL-8, respectively.
All compounds tested at 10 μM, values normalized to 1. TLR4 cell activation stimulated with 10 ng/mL LPS and 10 μM compound 1 resulted in OD630 of 1.90 ± 0.04 and 0.88 ± 0.02, respectively.
SAR Studies
The primary HTS library contained a family of 452 compounds in the pyrimido[5,4-b]indole class and therefore represented a valuable initial indication of structural features important for activation of NFκB. Upon inspection of the NFκB activation values in the initial HTS relative to the various structural features, several trends were apparent. There was a requirement for a hydrophobic moiety in the region of the cyclohexyl group of compound 1. In this same region, the carboxamide function was essential. Furthermore, a hydrophobic group at N-3, preferably a phenyl group, was also required for activity. Substitutions on the N-3 phenyl, other than fluorine atoms, resulted in loss of activity. Removal of the benzo ring of the indole portion of the scaffold also resulted in loss of activity. When the C-4 oxo was replaced by NH to form a triazine ring, loss of activity was observed. Finally, exchange of the entire acetamide moiety on the C-2 thiol with the N-3 phenyl group, such that the acetamide was attached at N-3 and the phenyl was attached at the C-2 thiol, resulted in loss of activity. With these SAR features as an initial guide, we elected to investigate modifications of hit compound 1 at three regions while maintaining the core pyrimido[5,4-b]indole ring system: the N-substitution of the S-acetamide, the N-3 substituent, and the N-5 substituent (Figure 6).
Figure 6.
SAR regions of modification of hit compound 1.
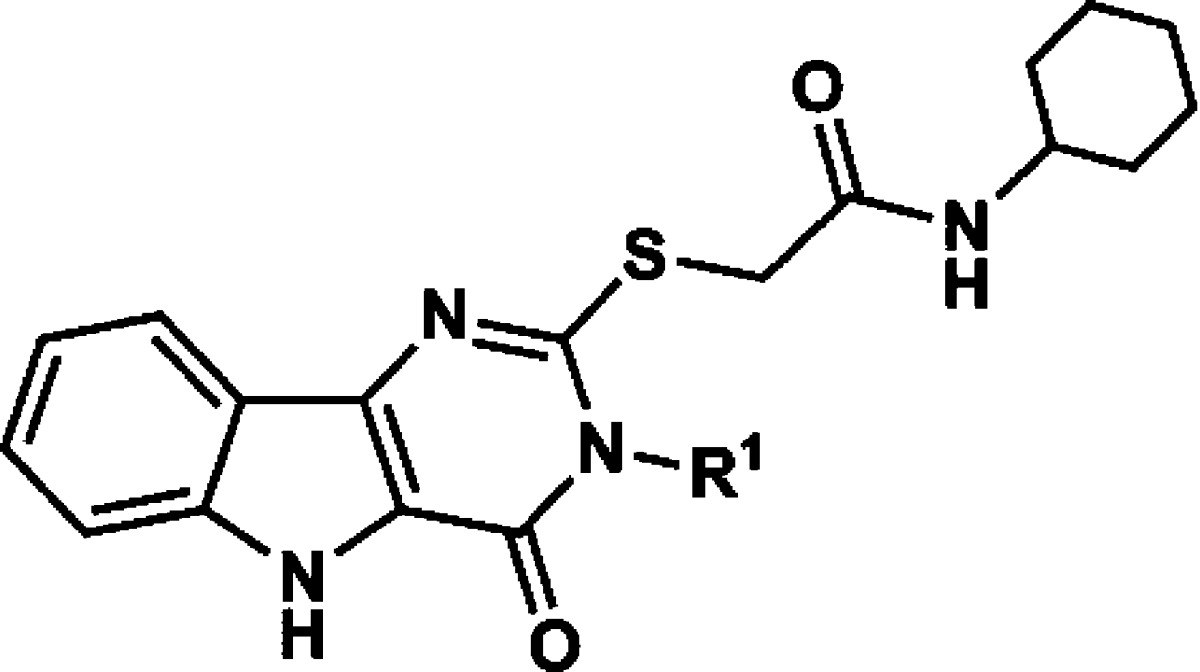
N-Substitutions at the carboxamide moiety were first undertaken to probe the limitations of the hydrophobic group requirement at this position with respect to optimization of cytokine induction (Table 1). While keeping all other structural features of hit compound 1 constant, we prepared a series of carboxamides substituted with various alkyl, cycloalkyl, aromatic, and heteroaromatic groups. The synthesis began with construction of the appropriately substituted pyrimido[5,4-b]indole ring system as shown in Scheme 1. 2-Aminobenzonitrile (2) was reacted with ethyl bromoacetate to yield ethyl 2-((2-cyanophenyl)amino)acetate (3) followed by base catalyzed ring closure to the aminoindole16,17 (4). At this point in the synthesis, a variety of substituents, represented by R1 in the scheme, may be introduced that will determine the respective N-3 substituent following annulation of the pyrimidine ring. Thus, reaction of compound 4 with an isothiocyanate, such as phenylisothiocyanate, provided the substituted thioureidoindole16,18 (5), where R1was phenyl in this example. Ring closure of 5 using polyphosphoric acid yielded the pyrimido[5,4-b]indole (6) bearing the N-3 substituent R1. Alkylation of the 2-thioxo function with chloroacetic acid provided the versatile intermediate 7, which was then used to prepare the final test compounds (8) bearing a variety of N-substitutions at the acetamide moiety, designated as R2 in Scheme 1. Compounds 1, 13, 14, 15, 16, 17, as well as the N-3 derivatives 37 and 38, discussed later, were registered with Chemical Abstracts Service, but no literature references were found for any of these compounds. The hit compound 1 was resynthesized according to Scheme 1, as discussed above. Thus, by this synthetic method, compounds shown in Table 1, with R1 held constant as a phenyl group, were prepared and evaluated in a cytokine induction assay for IL-6 in mBMDC and compared to compound 1. Examples of the data obtained from typical cytokine induction assays and NFκB activation assays are shown in Figures 4 and 5, wherein compounds 1, 11, 12, 36, and 39 are compared for IL-6 production by mBMDC and hPBMC.
Inspection of the IL-6 AUC values relative to the hydrophobic group R2 revealed that in general, compounds bearing the larger cycloalkyl groups, such as cyclooctyl (9), cycloheptyl (10), and cyclohexyl (1), were the most active, followed by branched alkyls and then straight chain alkyls and aromatic and heteroaromatic groups. A notable exception would be the p-fluorophenyl (14) and o-fluorophenyl (15) compounds. For the R2 group, there appeared to be a strict “hydrophobic volume” requirement for activity. Hence, the 3,3-dimethylbutyl compound (28) was among the most active of the alkyls, with isopentyl (27), butyl (23), and isobutyl (26) being somewhat less active. Interestingly, when the alkyl chain length of R2 was extended to more than five carbons or reduced to three or fewer carbons, whether branched or not, significant loss of activity was observed (Table 1).
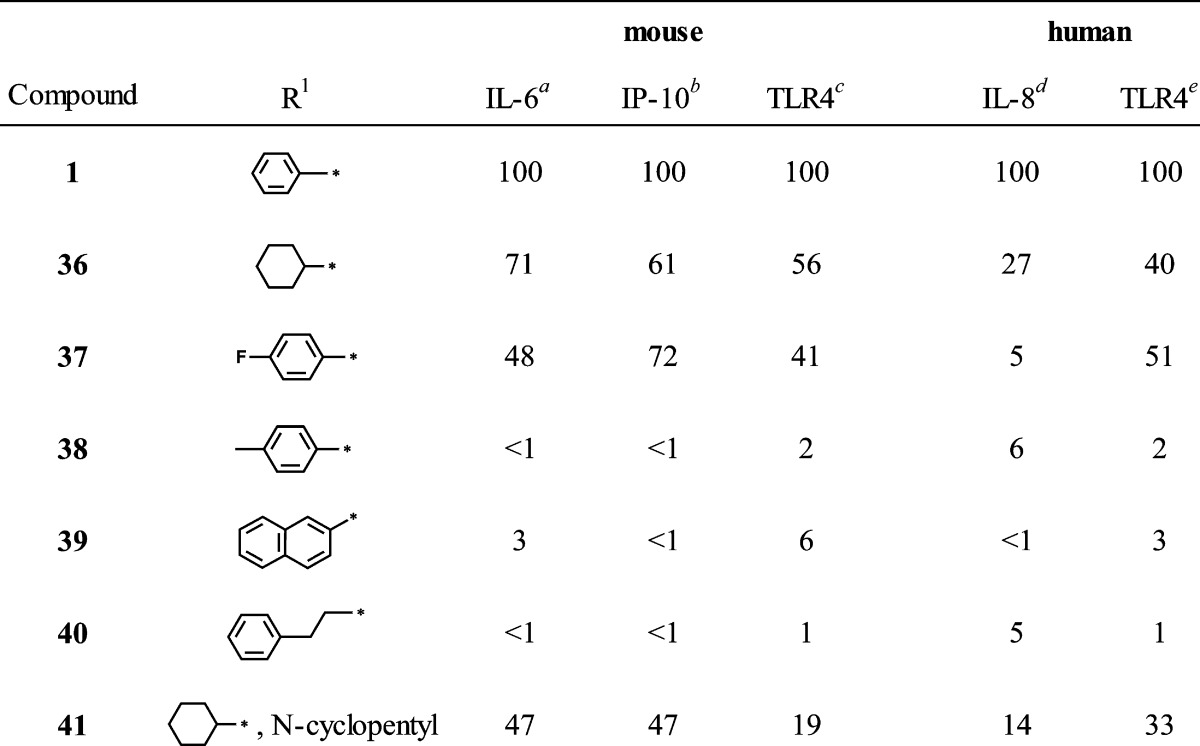
Encouraged by the SAR trends at the carboxamide moiety, we then addressed the N-3 position (Table 2). As mentioned above, data from the NFκB primary screen indicated that a hydrophobic group at N-3, preferably an unsubstituted phenyl, was required for activity. However, to draw that conclusion with confidence, more examples of compounds bearing similar hydrophobic groups than were represented in the primary compound collection were needed. We therefore elected to prepare and evaluate a few additional derivatives of hit compound 1 with variation only at N-3. Thus, the R1 group in compound 5 was varied by reaction of 4 with the appropriate isothiocyanate, and the remaining three steps for each derivative were completed as outlined in Scheme 1 while maintaining the R2 group as cyclohexyl in each case. Table 2 compares the IL-6 inducing activity of compound 1 with the N-3 derivatives. As the IL-6 data indicate, replacing the N-3 phenyl in compound 1 with a cyclohexyl group (36) caused some loss of activity, while substitution on the N-3 phenyl (37 and 38) resulted in greater loss of activity. Replacing N-3 phenyl with a larger aromatic, such as naphthyl (39), or extending the phenyl with an alkyl chain, as in phenethyl (40), yielded total loss of activity. The combined substitution of R1 with cyclohexyl and R2 with cyclopentyl (41) resulted in partial loss of activity.
Table 2. N-3 Derivatives.
AUC values normalized to compound 1. Stimulation with LPS (10 ng/mL) and 5 μM of 1 released an average of 20.6 ± 4.8 and 10.5 ± 1.3 ng/mL of IL-6, respectively.
All compounds tested at 5 μM, values normalized to 1. Stimulation with LPS 10 ng/mL and 5 μM of 1 released averages of 234.8 ± 8.6 and 590 ± 11 pg/mL of IP-10, respectively.
AUC values normalized to compound 1. TLR4 cell activation stimulated with 10 ng/mL LPS and 5 μM compound 1 resulted in OD630 of 1.69 ± 0.08 and 1.52 ± 0.03, respectively.
All compounds tested at 10 μM, values normalized to 1. Stimulation with LPS 10 ng/mL and 10 μM of 1 released averages of 14.5 ± 0.6 and 7.8 ± 0.8 ng/mL of IL-8, respectively.
All compounds tested at 10 μM, values normalized to 1. TLR4 cell activation stimulated with 10 ng/mL LPS and 10 μM compound 1 resulted in OD630 of 1.90 ± 0.04 and 0.88 ± 0.02, respectively.
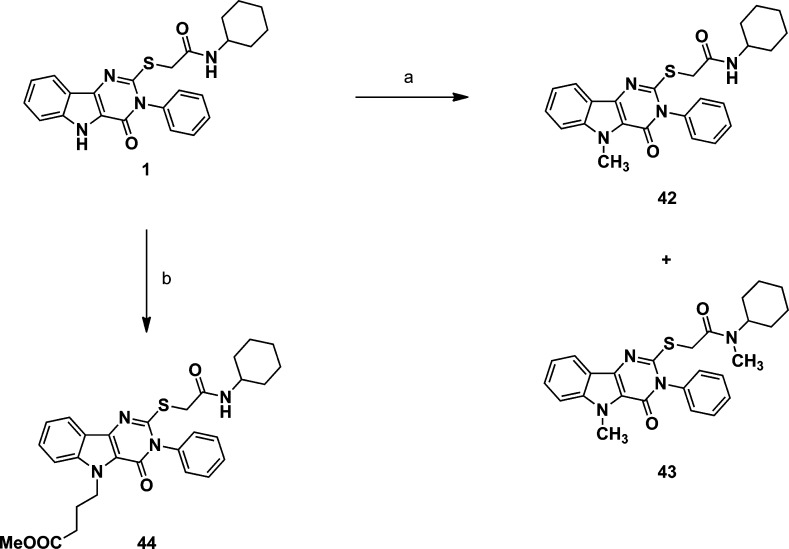
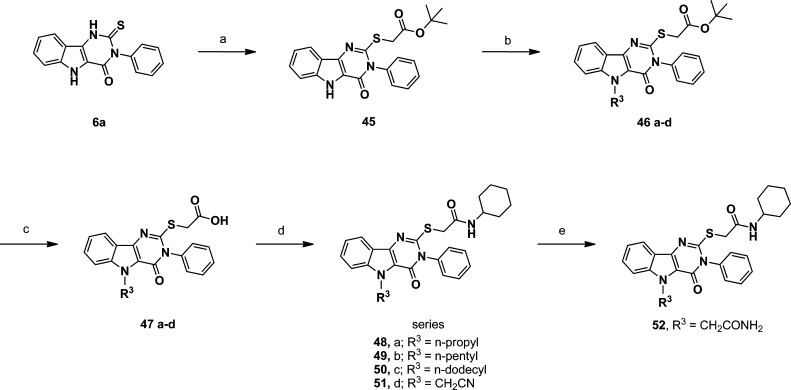
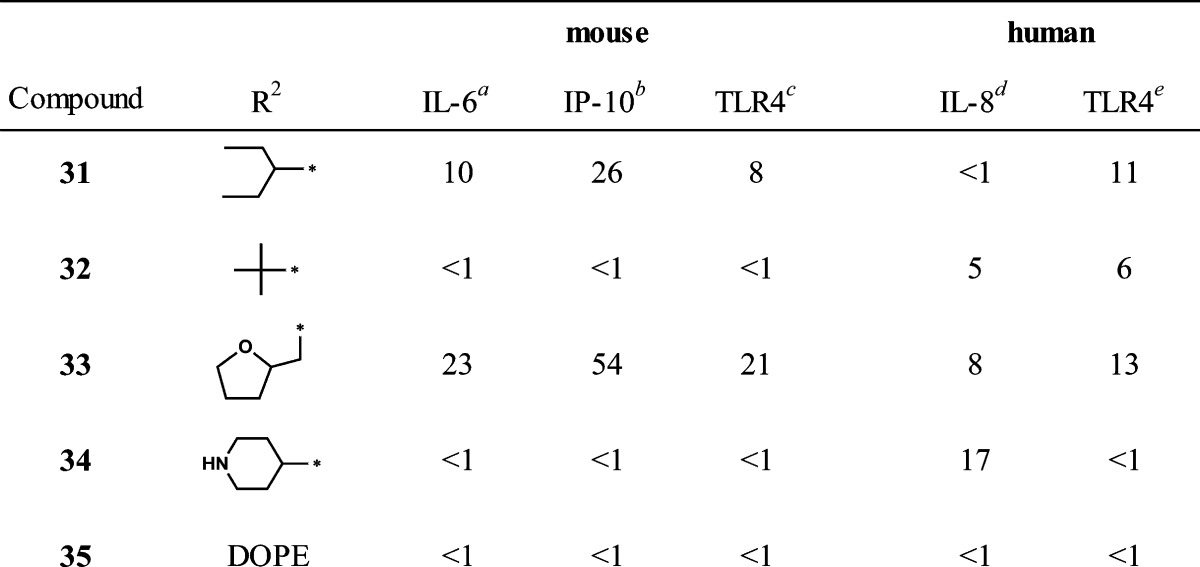
The third area of interest for modification was the N-5 position. None of the compounds in the original HTS library had modifications at this position to guide the SAR. We elected to prepare a few simple N-5 alkyl derivatives of compound 1. The N-5 indole-like nitrogen can be alkylated if the proton is first removed by a strong base, such as sodium hydride. Starting from compound 1, Scheme 2 shows the preparation of the N-5 methyl derivative (42) that also produced a dimethyl side product (43) wherein the second methylation occurred at the carboxamide function, yielding the N,N-disubstituted derivative. The N-5 methyl butyrate ester (44) of compound 1 was also prepared by this method. Table 3 compares the IL-6 inducing activity of the N-5 derivatives relative to compound 1. Interestingly, simple methylation at N-5 did not abrogate the immune stimulatory activity of compound 1. Moreover, methylation at N-5 decreased the toxicity of compound 1 (Figure 7) as measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) viability assay. Dimethylation, on the other hand, resulted in total loss of IL-6 activity (compound 43). To further probe the N-5 position, we elected to study a few N-5 alkylated derivatives of compound 1 while avoiding dialkylation in the process. Accordingly, we prepared the t-butyl ester of 7a (compound 45) and then alkylated the N-5 position with several primary alkyl halides, as depicted in Scheme 3. The resulting t-butyl esters (46a–d) were easily hydrolyzed to the corresponding free carboxylic acids (47a–d) and then converted to the N-cyclohexylcarboxamides (48–51) by the same method as described for compound 1. Thus, using this strategy, the N-5 n-propyl, n-pentyl, n-dodecyl, and cyanomethyl derivatives were prepared. Finally, the cyanomethyl derivative (51) was converted to the N-5 acetamide derivative (52). All N-5 alkyl derivatives were found to be less active than the N-5 methyl (42), with the n-propyl (48) being the next most active of the series. The trend in the toxicity profile for these N-5 alkyl derivatives was confirmed, at least for the shorter chain alkyls, in that the N-5 methyl (42), cyanomethyl (51), and n-propyl (48) derivatives did not reduce cell viability at concentrations up to 10 μM (Figure 7, shown for 42, data not shown for 48 and 51).
Scheme 2. N-5 Derivatives.
(a) NaH, DMF, room temp, then CH3I; (b) NaH, DMF, then Br(CH2)3COOCH3
Figure 7.
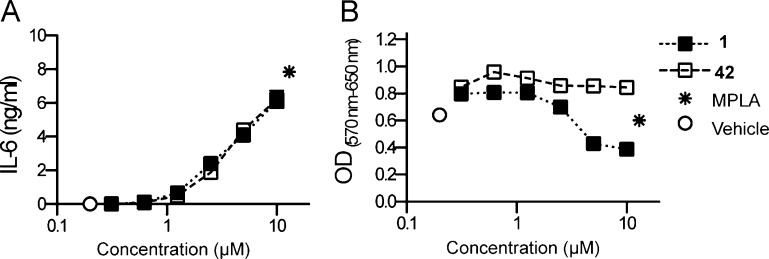
Assessment of cytotoxicity of compounds 1 and 42. Cytotoxicity of the compounds was evaluated by MTT assay as a measure of viability. mBMDC (105/well) were incubated with graded concentrations of the compounds for 18 h and compared with vehicle (0.5% DMSO) and MPLA (1 μg/mL). (A) IL-6 levels in the culture supernatants were determined by ELISA. (B) The cells were lysed after the overnight incubation with MTT reagents, and absorbance at 570 nm was measured, subtracting the reference absorbance at 650 nm. Data shown are mean ± SEM of triplicates and are representative of two independent experiments showing similar results.
Scheme 3. N-5 Alkyl Derivatives.
(a) ClCH2COO t-Bu, KOH, DMA,H2O; (b) NaH, DMF, then R3X (iodopropane or iodopentane or bromododecane or bromoacetonitrile); (c) TFA, DCM or CH3CN; (d) cyclohexyl-NH2, HATU, DMF, room temp; (e) compound 51, H2SO4, H2O.
The TLR4 transfectoma lines transmit their reporter signal after activation of the proinflammatory transcription factor, NFκB. As one might expect, the IL-6 secretion induced by the compounds in primary mBMDCs correlated well with their respective activities in mouse TLR4 transfectoma cell lines. With few exceptions, the most active compounds in the IL-6 assays were also those that showed the highest activity in the mTLR4 transfectoma line (Tables 1–3). In addition to stimulating the production of inflammatory cytokines (e.g., IL-6) by NFκB activation, TLR4 signaling can induce the production of type I IFN via the TRIF pathway.19,20 This pathway results in the activation of interferon regulatory factors (IRF), which are nuclear transcription factors that promote the transcription of IFNs. Type I IFN is important for the activation of antigen presenting dendritic cells, leading to good adjuvant activity, and also promotes cellular defenses against a variety of pathogens, particularly RNA viruses. This host defense is aided by circulating interferon inducible factors such as IP-10. The TLR4 transfectoma reporter lines in this study did not utilize an interferon reporter system; hence, the IP-10 data does not correlate as closely as the IL-6 data.
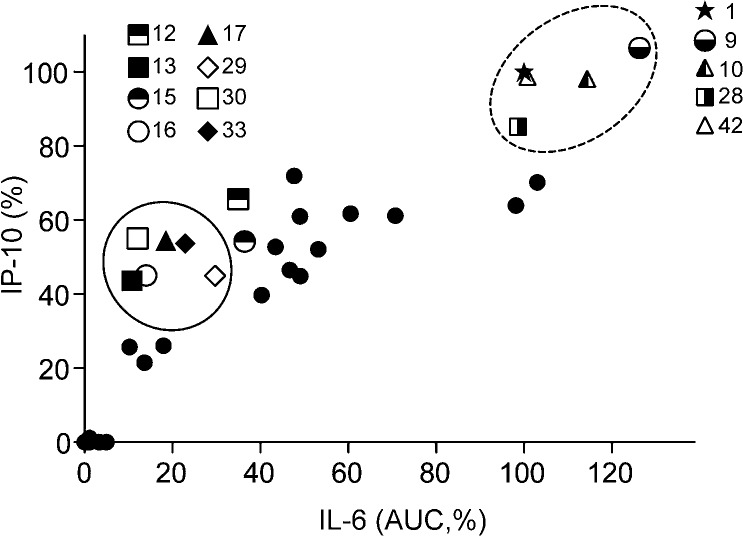
Interestingly, several of the compounds, namely 12, 13, 15, 16, 17, 29, 30, and 33, induced diminished IL-6 release compared to compound 1 but had nearly equivalent induction of IP-10 (Tables 1 and 2). These results suggest that there may be a possibility of separating the two activities on the basis of structural features. The correlation of IL-6 and IP-10 production of the selected compounds is shown graphically in Figure 8. Two distinct groups of compounds are observed, shown as circled regions of the plot. Thus, the group at the upper right area (dotted circle) comprises compounds that induce high IL-6 and high IP-10 production, including compounds 1, 9, 10, 28, and 42. The other group is composed of compounds that induce low IL-6 but relatively higher IP-10 production. These two clusters were selected based on mean ± SD of the normalized IL-6 and IP-10 values of all SAR compounds. For IL-6, the mean was 32 ± 37. For IP-10, the mean was 37 ± 33. The cluster in the dotted circle includes those compounds whose values were at least one SD above the mean for both cytokines. The other cluster includes those compounds whose values were below the mean for IL-6 and were above the mean for IP-10. It was notable that when the carboxamide substituent was phenyl or substituted phenyl (13, 15, 16, 17), with the exception of p-fluorophenyl (14), IL-6 release was reduced while IP-10 production was maintained at a relatively higher level. A few compounds bearing branched aliphatic carboxamide substituents showed a similar trend, as noted above for 29, 30, and 33.
Figure 8.
Correlation plot of IL-6 versus IP-10 induced by SAR derivatives. IL-6 (AUC) and IP-10 values in Tables 1–3 were plotted. Two distinct groups, compounds in the dotted circled (upper right) and in solid (left) circled areas, were selected based on the induction of IL-6 and IP-10. Numbers in the legends indicate compound ID. These clusters were selected based on mean ± SD of the normalized IL-6 and IP-10 values of all SAR compounds. For IL-6. the mean ± SD was 32 ± 37. For IP-10, the mean ± SD was 37 ± 33. The group at the upper right area comprises compounds that induce high IL-6 and high IP-10 production, including compounds 1, 9, 10, 28, and 42, whose values were at least one SD above the mean for both cytokines. The other cluster is composed of compounds (13, 16, 17, 29, 30, and 33) that induce low IL-6 but relatively higher IP-10 production, whose values were below the mean for IL-6 and were above the mean for IP-10.
In general, the active compounds exhibited greater activity in the murine cell systems than in human cells. Although the innate immune system is highly conserved among species, there are differences between human and mouse TLR4.21−23 Similar to the active compounds reported here nonsynthetic ligands have been noted to have species specific behavior. Notably, tetraacylated lipid IVa, a synthetic lipid A precursor, has been reported to act as a weak agonist to mouse TLR4/MD-2 but as an antagonist to human TLR4/MD-2.24,25 Recently mouse and human TLR4/MD2 crystal structures with this ligand have suggested that the different charge distributions of mouse and human TLR4/MD-2 affect the positions of the phosphate groups of lipid IVa, orienting them in a manner that would limit receptor dimerization by human TLR4.22,26
Computational Studies
TLR4, in association with MD-2, is responsible for the physiological recognition of LPS.27,28 The structural basis of receptor specificity and of the mechanism of activation by LPS have recently been elucidated by determining the crystal structure of the TLR4/MD-2-LPS complex at 3.1 A resolution.29 Binding of agonistic ligands such as LPS causes dimerization of the extracellular domains to form a TLR4/MD-2-LPS macromolecular complex. Like the extracellular domains of other TLRs, TLR4 contains leucine-rich repeats and adopts a characteristic horseshoe-like shape. MD-2 is noncovalently bound to the side of the horseshoe ring and also directly interfaces with the ligand. MD-2 has a β-cup fold structure composed of two antiparallel β-sheets forming a large hydrophobic pocket for ligand binding. LPS binds to this pocket and directly mediates dimerization of the two TLR4/MD-2 complexes. TLR4 can be activated by structurally diverse LPS molecules, which have been predicted to occupy this pocket in MD-2.
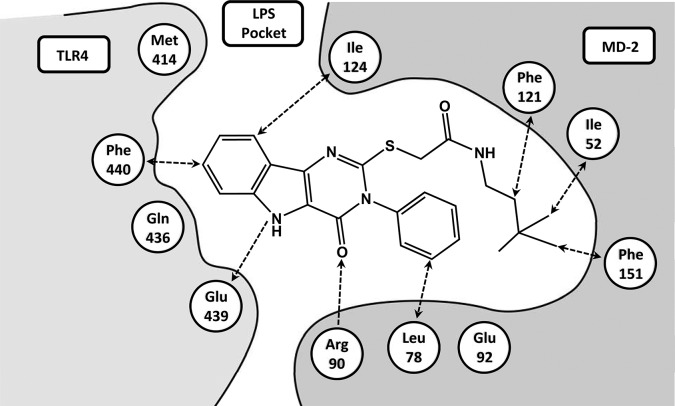
Small molecule hit compounds discovered in the present study were also thought to bind to the TLR4/MD-2 complex in such a way as to facilitate dimerization. Accordingly, we examined the predicted binding mode(s) of one of the most active compounds to the murine TLR4/MD-2 complex by conducting molecular docking of compound 28 to the crystal structure of the mouse complex (PDB 2Z64) using the programs HEX30 and AMPAC.31 We selected the best configurations of 28 bound to this complex based on molecular surface shape complementarity and the most favorable intermolecular energy of interactions. It is noteworthy that the best docking position for 28 was within the LPS pocket.29 Figure 9 shows the predicted binding mode of compound 28 in the TLR4/MD-2 model, while the set of binding interactions that may keep the compound in the MD-2 pocket bound to both TLR4 and MD-2 is depicted in Figure 10. There is a set of favorable electrostatic interactions resulting in possible hydrogen bonds formed by the residues Glu439 of TLR4 and Arg90 of MD-2 with compound 28, although the former would approach a hydrogen bond interaction due to the flexibility of the Glu439 side chain. There are also multiple potential hydrophobic interactions with MD-2 and TLR4. Such interactions of the compound with two proteins can improve the free energy of complex formation by approximately 8–10 kcal/mol.
Figure 9.
Predicted binding mode of compound 28 to mouse TLR4/MD-2 complex.
Figure 10.
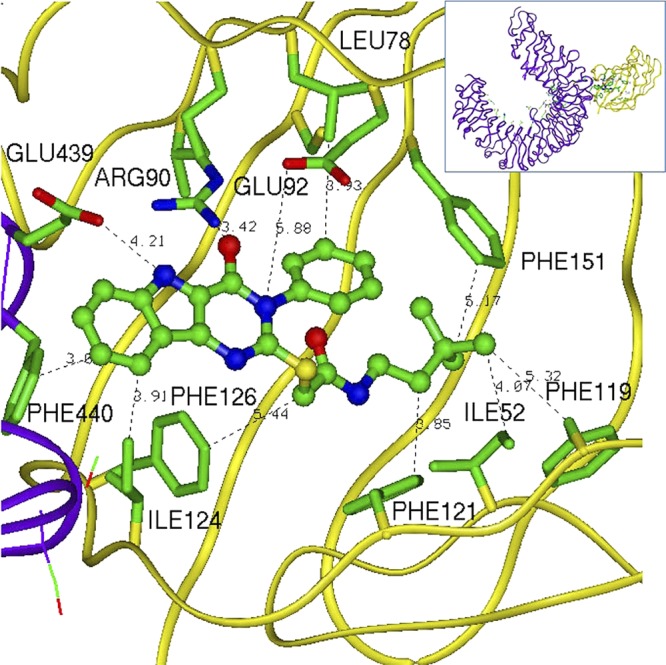
Predicted binding interactions of compound 28 with mouse TLR4/MD-2 complex (PDB 2Z64).
Interestingly, the predicted binding model for the pyrimidoindoles is similar to that proposed for tricyclic antidepressant, amitriptyline.32 This molecule also has some TLR4/MD-2 binding and has a similar three-ring scaffold. Other small molecules such as paclitaxel,33 opioids,34 and a peptide35 have also been reported to bind to the MD-2 binding pocket.
Conclusion
In the course of an HTS designed to identify activators of innate immunity, a series of substituted pyrimido[5,4-b]indoles were discovered as selective TLR4 ligands. Small molecules of this class are unique among TLR4 activators in that they are “non-lipid-like”. Structure–activity evaluation in both mouse and human cells revealed that, to maintain activity, the carboxamide region of this scaffold must contain a hydrophobic moiety of significant “volume”. Interestingly, a subset of the compounds bearing phenyl and substituted phenyl carboxamides induced lower NFκB-dependent inflammatory cytokine release while maintaining interferon-dependent IP-10 production. Varying the substituents at N-3 indicated an even greater restriction for a hydrophobic moiety at this position, with a phenyl group being preferred for activity. Finally, N-5 substitution revealed that short alkyl substituents at this position attenuate cell toxicity relative to the corresponding nonsubstituted derivative while maintaining TLR4 activity. Both the inflammatory cytokine and type I IFN inducing activities of these compounds were CD14-independent. Computational studies with one of the active compounds predicted binding primarily to MD-2 in the murine TLR4/MD-2 complex. Lead optimization studies to improve the activity of the compounds using computational methods are currently underway in our laboratories. To date, no single adjuvant can elicit the optimal immune response to all pathogens in vaccines. Here we have described a panel of small molecules that stimulate immune cells to produce distinct profiles of NFκB and interferon associated cytokines. This panel of molecules may allow for differential analysis of the relative pathway induction needed for adequate immunoprotection and immunotherapy for diverse pathogens.
Experimental Section
Chemistry
Materials
Reagents were purchased as at least reagent grade from Sigma-Aldrich (St. Louis, MO) unless otherwise specified and used without further purification. Solvents were purchased from Fischer Scientific (Pittsburgh, PA) and were either used as purchased or redistilled with an appropriate drying agent. HTS compound library was obtained from UCSF Small Molecule Discovery Center (San Francisco, CA). Commercial HTS service was provided by Invitrogen (Grand Island, NY). Compounds 13, 14, 15, 16, 17, 37, and 38 were purchased from Life Chemicals (Burlington, ON, Canada). All synthesized compounds, intermediates, and purchased compounds from Life Chemicals were determined to be >95% pure by HPLC utilizing an Agilent 1100 LC/MSD. Endotoxin levels of active compounds were measured with Endosafe-PTS (Charles River, Wilmington, MA) and found to have less than 10 EU/μmol. Reaction room temperature was maintained between 22 and 24 °C.
Instrumentation
Analytical TLC was performed using precoated TLC silica gel 60 F254 aluminum sheets purchased from EMD (Gibbstown, NJ) and visualized using UV light. Flash chromatography was carried out on EMD silica gel 60 (40–63 μm) system or with a Biotage Isolera One (Charlotte, NC) using the specified solvent. Reactions were monitored using an Agilent 1100 LC/MSD (Santa Clara, CA) with either a Supelco Discovery HS C18 column (Sigma-Aldrich) or an Onyx Monolithic C18 (Phenomenex, Torrance, CA) with purity above 98% by percent area. All synthesized compounds and intermediates were analyzed by high-resolution MS using an Agilent 6230 ESI-TOFMS (Santa Clara, CA). Selected compounds were analyzed by IR (KBr window method) to confirm presence of functional groups having useful diagnostic frequencies using a Perkin-Elmer 1600 series. 1H NMR spectra were obtained on a Varian Mercury 300 or 400 (Varian, Inc., Palo Alto, CA). 13C spectra were obtained on Varian 500 with Xsens probe. The chemical shifts are expressed in parts per million (ppm) using suitable deuterated NMR solvents in reference to TMS at 0 ppm. The 3D structures were prepared and optimized using the AMPAC semiempirical quantum chemistry program (Accelrys, San Diego, CA).
General Procedure A for the Synthesis of Compound 5
To a solution of compound 4 (1 equiv) in warm EtOH was added the appropriate isothiocyanate (1.1 equiv) dropwise with stirring. The reaction was refluxed for 6 h and cooled overnight. Solids were filtered, washed with EtOH, and dried overnight in vacuo to give compound 5.
General Procedure B for the Synthesis of Compound 6b–d
Compound 5 was dissolved in polyphosphoric acid and stirred at 110 °C for 3.5 h. Solution was added to ice-cold water and extracted with EtOAc and dried over MgSO4. The solid was then dried in vacuo overnight to give compound 6.
General Procedure C for the Synthesis of Compound 7
In a flame-dried flask, compound 6a (1 equiv) and KOH (2 equiv) were dissolved in anhydrous EtOH with heat. In a separate flame-dried flask, chloroacetic acid (1 equiv) was added to anhydrous EtOH. Chloroacetic acid solution was then added to the reaction mixture and refluxed for 6 h. The reaction was concentrated by half and acidified with 3 M HCl to pH 4. The solids were collected, washed with water, and dried in vacuo to give compound 7.
General Procedure D for the Synthesis of Compound 8
Compound 7 (1 equiv), triethylamine (2 equiv), and the appropriate amine (1.1 equiv) were dissolved in anhydrous DMF. To this solution, HATU (1.1 equiv) dissolved in DMF was added and stirred until complete and concentrated in vacuo. The crude material was then recrystallized in MeOH to give compound 8.
General Procedure E for the Synthesis of Compound 46
NaH (1.1 equiv) was added to a solution of compound 45 (1 equiv) in DMF. The reaction mixture was stirred for 5 min, and then the appropriate alkyl halide (1.1 equiv) was added and stirred until complete. The crude product was extracted with EtOAc and dried over MgSO4. Crude material was finally recrystallized with MeOH to give compound 46.
General Procedure F for the Synthesis of Compound 47
Compound 46 was dissolved in 1:1 acetonitrile/triflouroacetic acid and stirred at room temperature overnight. Depending on the N-alkyl substitution, crude material either precipitated as pure product or was purified by chromatography to give compound 47.
N-Cyclohexyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (1)
Compound 7a (50 mg, 0.14 mmol), triethylamine (40 μL, 0.28 mmol), and cyclohexylamine (18 μL, 0.16 mmol) were dissolved in anhydrous DMF (1 mL). HATU (59.5 mg, 0.16 mmol) dissolved in 0.2 mL of DMF was added to the reaction mixture and stirred for 20 min and then concentrated in vacuo. The crude material was recrystallized with MeOH to give 61 mg in near quantitative yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 0.97–1.33 (m, 4 H), 1.51 (d, J = 8.53 Hz, 1 H), 1.56–1.80 (m, 4 H), 3.48 (m, 1 H), 3.87 (s, 2 H), 7.24 (t, J = 7.29 Hz, 1 H), 7.38–7.67 (m, 6 H), 8.05 (d, J = 7.98 Hz, 1 H), 8.15 (d, J = 7.70 Hz, 1 H), 12.09 (s, 1 H). HRMS calcd for C24H24N4O2SNa (M + Na)+, 455.1512; found, 455.1511.
Ethyl 2-((2-Cyanophenyl)amino)acetate (3)
Anthranilonitrile (30.29 g, 256 mmol), ethyl bromoacetate (29.46 mL, 267 mmol), and sodium bicarbonate (25.6 g, 300 mmol) were combined in anhydrous EtOH (90 mL) and refluxed for 42 h. After cooling slightly, solution was decanted from precipitate into a prewarmed flask. Further cooling of the decantate yielded crystals, which were then filtered and washed with cold water to give 23.6 g of white crystalline product. 1H NMR (500 MHz, DMSO-d6) δ ppm 1.13–1.27 (m, 3 H), 4.04 (d, J = 6.41 Hz, 2 H), 4.12 (q, J = 7.22 Hz, 2 H), 6.36 (t, J = 6.25 Hz, 1 H), 6.64 (d, J = 8.54 Hz, 1 H), 6.69 (t, J = 7.47 Hz, 1 H), 7.41 (t, J = 7.78 Hz, 1 H), 7.49 (dd, J = 7.93, 1.53 Hz, 1 H). HRMS calcd for C11H12N2O2Na (M + Na)+, 227.0791; found, 227.0794.
Ethyl 3-Amino-1H-indole-2-carboxylate (4)
In a flame-dried flask, a suspension of potassium t-butoxide (7.124 g, 63.5 mmol) in anhydrous THF was stirred and maintained below 30 °C under argon. To this solution was added a solution of compound 3 (16 g, 63.5 mmol) in anhydrous THF over 45 min and stirred for an additional 2 h. The reaction was then poured into ice water, extracted with EtOAc, and dried over MgSO4. The solid was then dissolved in minimal EtOH, and water was added dropwise until just cloudy and allowed to precipitate at room temperature. Precipitate was filtered and washed with cold EtOH to give 6.6 g of compound 4 in 51% yield. 1H NMR (500 MHz, DMSO-d6) δ ppm 1.33 (t, J = 7.02 Hz, 3 H), 4.29 (q, J = 7.12 Hz, 2 H), 5.68 (s, 2 H), 6.88 (ddd, J = 8.08, 5.80, 1.98 Hz, 1 H), 7.15–7.25 (m, 2 H), 7.74 (d, J = 7.93 Hz, 1 H), 10.34 (s, 1 H). HRMS calcd for C11H12N2O2 (M + H)+, 205.0972; found, 205.0973.
Ethyl 3-(3-Phenylthioureido)-1H-indole-2-carboxylate (5a)
Compound 4 (5.21 g, 25.5 mmol) was reacted with phenyl isothiocyanate (3.36 mL, 28.1 mmol) in warm EtOH (75 mL) according to general procedure A to give 6.732 g compound 5a in 75% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.32 (t, J = 7.07 Hz, 3 H), 4.32 (q, J = 7.31 Hz, 2 H), 7.09 (dt, J = 14.75, 7.50 Hz, 2 H), 7.22–7.38 (m, 3 H), 7.43 (d, J = 8.29 Hz, 1 H), 7.51 (d, J = 7.80 Hz, 2 H), 7.56 (d, J = 8.29 Hz, 1 H), 9.40 (s, 1 H), 9.69 (br s, 1 H), 11.79 (s, 1 H). HRMS calcd for C18H17N3O2SNa (M + Na)+, 362.0934; found, 362.0935.
Ethyl 3-(3-Cyclohexylthioureido)-1H-indole-2-carboxylate (5b)
Compound 4 (100 mg, 0.49 mmol) was reacted with cyclohexyl isothiocyanate (169.15 mg, 0.54 mmol) in warm EtOH (750 μL) according to general procedure A to give 104 mg of compound 5b in 61.8% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.01–1.42 (m, 9 H), 1.49–1.62 (m, 1 H), 1.62–1.74 (m, 2 H), 1.77–1.96 (m, 2 H), 4.13 (br s, 1 H), 4.29 (q, J = 7.09 Hz, 2 H), 7.05 (t, J = 7.33 Hz, 1 H), 7.25 (t, J = 8.07 Hz, 1 H), 7.41 (d, J = 8.07 Hz, 1 H), 7.49 (d, J = 8.07 Hz, 1 H), 8.91 (s, 1 H), 11.71 (br s, 1 H). HRMS calcd for C18H23N3O2S (M + H)+, 345.1511; found, 345.15142.
Ethyl 3-(3-(Naphthalen-1-yl)thioureido)-1H-indole-2-carboxylate (5c)
Compound 4 (50 mg, 0.28 mmol) was reacted with 1-naphthyl isothiocyanate (49.9 mg, 0.27 mmol) in warm EtOH (750 μL) according to general procedure A to give 58.6 mg of compound 5c in 61.5% yield. HRMS calcd for C22H19N3O2SNa (M + Na)+, 412.1090; found, 412.1091.
Ethyl 3-(3-Benzylthioureido)-1H-indole-2-carboxylate (5d)
Compound 4 (100 mg, 0.49 mmol) was reacted with phenethyl isothiocyanate (80.4 μL, 0.54 mmol) in warm EtOH (1.5 mL) according to general procedure A to give 147 mg of compound 5d in 81.8% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.31 (t, J = 6.97 Hz, 3 H), 4.27 (q, J = 7.21 Hz, 2 H), 7.06 (t, J = 7.70 Hz, 1 H), 7.09–7.33 (m, 7 H), 7.43 (dd, J = 8.07, 4.03 Hz, 2 H), 7.56 (br s, 1 H), 9.12 (br s, 1 H), 11.79 (br s, 1 H). HRMS calcd for C20H22N3O2S (M + H)+, 368.1427; found, 368.1431
3-Phenyl-2-thioxo-2,3-dihydro-1H-pyrimido[5,4-b]indol-4(5H)-one (6a)
To a flame-dried flask with cold anhydrous EtOH (75 mL) was added cold acetyl chloride (7 mL, 98.5 mmol) under argon with stirring. In a separate flame-dried flask charged with argon, compound 5a (6.5 g, 19 mmol) was dissolved in anhydrous EtOH (25 mL) and added to the acetyl chloride solution. The reaction was refluxed for 12 h and cooled upon completion. Precipitate was filtered and recrystallized with EtOH to give 3.77 g of compound 6a in 75% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 7.09–7.31 (m, 3 H), 7.31–7.61 (m, 6 H), 8.20 (d, J = 8.07 Hz, 1 H), 12.17 (br s, 1 H). HRMS calcd for C16H12N3OS (M + H)+, 294.0696; found, 294.0698.
3-Cyclohexyl-2-thioxo-2,3-dihydro-1H-pyrimido[5,4-b]indol-4(5H)-one (6b)
Compound 5b (70 mg, 0.2 mmol) and 1 mL of polyphosphoric acid was reacted according to general procedure B to give 60.6 mg of compound 6b in quantitative yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.07–1.49 (m, 5 H), 1.53–1.70 (m, 1 H), 1.70–1.84 (m, 2 H), 2.01 (s, 2 H), 3.86–4.09 (m, 1 H), 7.11 (t, J = 8.07 Hz, 1 H), 7.29–7.54 (m, 2 H), 7.87 (d, J = 8.07 Hz, 1 H), 8.06 (d, J = 6.97 Hz, 1 H), 11.60 (s, 1 H). HRMS calcd for C16H18N3OS (M + H)+, 300.1165; found, 300.1169.
3-(Naphthalen-1-yl)-2-thioxo-2,3-dihydro-1H-pyrimido[5,4-b]indol-4(5H)-one (6c)
Compound 5c (150 mg, 0.38 mmol) and 1 mL of polyphosphoric acid was reacted according to general procedure B to give 132 mg of compound 6b in quantitative yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 7.02–7.18 (m, 1 H), 7.33–7.48 (m, 2 H), 7.48–7.67 (m, 3 H), 7.74 (d, J = 8.07 Hz, 1 H), 7.87 (d, J = 8.07 Hz, 1 H), 7.84 (d, J = 7.70 Hz, 1 H), 7.94–8.03 (m, 1 H), 8.03–8.18 (m, 1 H), 11.83 (s, 1 H). HRMS calcd for C20H14N3OS (M + H)+, 344.0852; found, 344.0855.
3-Phenethyl-2-thioxo-2,3-dihydro-1H-pyrimido[5,4-b]indol-4(5H)-one (6d)
Compound 5d (50.7 mg, 0.138 mmol) and 1 mL of polyphosphoric acid was reacted according to general procedure B. Crude product was purified by automated flash chromatography (Biotage) (DCM/MeOH = 99:1) to give 27.1 mg of compound 6d in 61.1% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 2.95 (t, J = 7.33 Hz, 2 H), 3.67 (t, J = 6.20 Hz, 2 H), 6.96–7.63 (m, 8 H), 7.91 (d, J = 7.70 Hz, 1 H), 8.31 (d, J = 4.77 Hz, 1 H), 11.50–11.89 (m, 1 H). HRMS calcd for C18H15N3OS (M + H)+, 321.0936; found, 321.0939.
2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (7a)
Compound 6a (3.77 g, 12.9 mmol), KOH (1.44 g, 25.8 mmol), chloroacetic acid (1.25 g, 12.9 mmol, 6.5 mL of anhydrous EtOH), and 115 mL of anhydrous EtOH were reacted according to general procedure C to give 3.66 g of compound 7a in 81% yield. IR: 3251 (OH), 1702 (CO carboxyl), 1660 (CO amide) cm–1. 1H NMR (400 MHz, DMSO-d6) δ ppm 3.94 (s, 2 H), 7.14–7.36 (m, 1 H), 7.38–7.84 (m, 7 H), 7.93 (d, J = 7.70 Hz, 1 H), 12.09 (br s, 1 H). HRMS calcd for C18H13N3O3SNa (M + Na)+, 374.0570; found, 374.0572.
2-((3-Cyclohexyl-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (7b)
Compound 6b (165.8 mg, 0.55 mmol), KOH (62 mg, 1.1 mmol), chloroacetic acid (52.3 mg, 0.55 mmol, 3 mL of anhydrous EtOH), and 62 mL of anhydrous EtOH were reacted according to general procedure C to give 35.6 mg of compound 7b in 17.9% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.05–1.52 (m, 4 H), 1.59–2.05 (m, 6 H), 3.94 (s, 2 H), 4.20–4.46 (m, 1 H), 7.15 (t, J = 7.33 Hz, 1 H), 7.30–7.59 (m, 2 H), 7.88 (d, J = 8.07 Hz, 1 H), 11.79 (br s, 1 H). HRMS calcd for C18H19N3O3SNa (M + Na)+, 380.1039; found, 380.1042.
2-((3-(Naphthalen-2-yl)-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (7c)
Compound 6c (120 mg, 0.35 mmol), KOH (39.1 mg, 70 mmol), chloroacetic acid (33 mg, 0.35 mmol, 3 mL of anhydrous EtOH), and 62 mL of anhydrous EtOH were reacted according to general procedure C to give 67.1 mg of compound 7c in 47.8% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 3.90 (s, 2 H), 7.26 (t, J = 7.40 Hz, 1 H), 7.33–7.66 (m, 5 H), 7.67–7.80 (m, 2 H), 7.98 (d, J = 7.70 Hz, 1 H), 8.09 (d, J = 8.43 Hz, 1 H), 8.17 (t, J = 4.77 Hz, 1 H), 12.14 (br s, 1 H). HRMS calcd for C22H15N3O3SNa (M + Na)+, 424.0726; found, 424.0730.
2-((4-Oxo-3-phenethyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (7d)
Compound 6d (271 mg, 0.84 mmol), KOH (94.5 mg, 1.69 mmol), chloroacetic acid (79.8 mg, 0.84 mmol, 3 mL of anhydrous EtOH), and 62 mL of anhydrous EtOH were reacted according to general procedure C to give 160 mg of compound 7d in 50% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 3.02 (t, J = 7.80 Hz, 2 H), 4.02 (s, 2 H), 4.35 (t, J = 7.31 Hz, 2 H), 6.87–7.83 (m, 8 H), 7.95 (d, J = 7.31 Hz, 1 H), 11.92 (br s, 1 H). HRMS calcd for C20H17N3O3SNa (M + Na)+, 402.0883; found, 402.0885.
N-Cycloheptyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (9)
Compound 7a (50 mg, 0.14 mmol), cycloheptylamine (39.86 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 12.46 mg in 18.5% yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 1.18–1.65 (m, 10 H), 1.66–1.88 (m, 2 H), 3.59–3.77 (m, 1 H), 3.87 (s, 2 H), 7.25 (t, J = 7.01 Hz, 1 H), 7.40–7.55 (m, 4 H), 7.56–7.68 (m, 3 H), 8.06 (d, J = 7.98 Hz, 1 H), 8.20 (d, J = 7.70 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.12, 28.28, 34.69, 37.17, 50.51, 113.31, 119.7, 120.48, 120.78, 120.81, 127.77, 130.00, 130.01, 130.32, 136.51, 137.68, 139.35, 153.01, 155.39, 165.92. HRMS calcd for C25H26N4O2SNa (M + Na)+, 469.1669; found, 469.1670.
N-Cyclooctyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (10)
Compound 7a (25 mg, 0.07 mmol), cyclooctylamine (24.15 μL, 0.08 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) were reacted according to general procedure D to give 30.77 mg in quantitative yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.23–1.89 (m, 14 H), 3.68–3.82 (m, 1 H), 3.87 (s, 2 H), 7.24 (t, J = 6.97 Hz, 1 H), 7.37–7.75 (m, 7 H), 8.06 (d, J = 8.07 Hz, 1 H), 8.18 (d, J = 7.70 Hz, 1 H), 12.10 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 23.75, 25.47, 27.25, 31.87, 37.16, 49.49, 113.32, 119.7, 120.48, 120.78, 120.81, 127.78, 130.00, 130.01, 130.32, 136.51, 137.68, 139.36, 153.02, 155.39, 165.90. HRMS calcd for C26H29N4O2S (M + H)+, 461.2006; found, 461.2010.
N-Cyclopentyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (11)
Compound 7a (50 mg, 0.14 mmol), cyclopentylamine (31 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 48.9 mg in 76% yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 1.32–1.55 (m, 4 H), 1.55–1.70 (m, 2 H), 1.71–1.86 (m, 2 H), 3.87 (s, 2 H), 3.97 (dd, J = 13.07, 6.46 Hz, 1 H), 7.25 (t, J = 7.29 Hz, 1 H), 7.35–7.72 (m, 7 H), 8.05 (d, J = 7.98 Hz, 1 H), 8.26 (d, J = 6.88 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 23.95, 32.74, 37.13, 51.12, 113.32, 119.69, 120.5, 120.76, 120.79, 127.77, 129.99, 130.01, 130.32, 136.50, 137.67, 139.35, 153.02, 155.39, 166.64. HRMS calcd for C25H26N4O2SNa (M + Na)+, 441.1356; found, 441.1367.
N-Cyclobutyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (12)
Compound 7a (50 mg, 0.14 mmol), cyclobutylamine (26.73 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 41.6 mg in 73.5% yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 1.48–1.72 (m, 2 H), 1.93 (q, J = 9.70 Hz, 2 H), 2.05–2.26 (m, 2 H), 3.85 (s, 2 H), 4.08–4.26 (m, 1 H), 7.26 (t, J = 7.29 Hz, 1 H), 7.37–7.70 (m, 6 H), 8.07 (d, J = 7.98 Hz, 1 H), 8.56 (d, J = 7.70 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 15.11, 30.72, 37.03, 44.72, 113.32, 119.70, 120.52, 120.81, 127.78, 130.00, 130.01, 130.33, 136.49, 137.69, 139.35, 152.95, 155.4, 166.21. HRMS calcd for C22H20N4O2SNa (M + Na)+, 427.1199; found, 427.1200.
N-(Naphthalen-2-yl)-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (18)
Compound 7a (50 mg, 0.14 mmol), 2-aminonaphthalene (44.82 mg, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 40.8 mg in 61% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 4.16 (br s, 2 H), 7.11 (t, J = 7.70 Hz, 1 H), 7.26–8.07 (m, 14 H), 8.31 (br s, 1 H), 10.65 (br s, 1 H), 12.10 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 37.99, 113.27, 115.48, 119.71, 120.21, 120.49, 120.68, 120.72, 125.07, 126.92, 127.71, 127.74, 127.90, 128.89, 130.01, 130.06, 130.18, 130.41, 133.84, 136.49, 137.16, 137.63, 139.30, 152.89, 155.37, 166.96. HRMS calcd for C28H20N4O2SNa (M + Na)+, 499.1199; found, 499.1201.
N-(Furan-2-ylmethyl)-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (19)
Compound 7a (50 mg, 0.14 mmol), furfurylamine (27.66 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 50.37 mg in 83.6% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 3.95 (s, 2 H), 4.29 (d, J = 5.50 Hz, 2 H), 6.25 (d, J = 3.30 Hz, 1 H), 6.32 (t, J = 2.57 Hz, 1 H), 7.24 (t, J = 6.97 Hz, 1 H), 7.35–7.72 (m, 7 H), 7.97 (d, J = 7.70 Hz, 1 H), 8.73 (t, J = 5.32 Hz, 1 H), 12.10 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 36.46, 36.76, 107.46, 107.48, 107.49, 110.86, 110.87, 110.89, 113.23, 119.71, 120.60, 120.80, 120.88, 127.76, 130.01, 130.02, 130.34, 136.50, 137.69, 139.34, 142.64, 152.32, 152.81, 155.41, 167.44. HRMS calcd for C23H18N4O3SNa (M + Na)+, 453.0992; found, 453.0995.
N-(1H-Indazol-6-yl)-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (20)
Compound 7a (50 mg, 0.14 mmol), 2-aminoindazole (41.68 mg, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 32.7 mg in 50% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 4.11 (s, 2 H), 7.09 (t, J = 7.33 Hz, 1 H), 7.17 (d, J = 8.43 Hz, 1 H), 7.32–7.77 (m, 9 H), 7.87–8.01 (m, 2 H), 8.11 (s, 1 H), 10.55 (br s, 1 H), 12.07 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 38.00, 99.22, 113.29, 114.43, 119.60, 119.65, 120.55, 120.69, 121.03, 121.24, 127.81, 129.95, 130.10, 130.45, 133.80, 136.42, 137.66, 137.73, 139.31, 140.74, 148.09, 152.88, 155.37, 162.89, 167.00. HRMS calcd for C25H18N6O2SNa (M + Na)+, 489.1104; found, 489.1106.
2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-(thiazol-2-yl)acetamide (21)
Compound 7a (50 mg, 0.14 mmol), 2-aminothiazole (31.35 mg, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 12.82 mg in 21.1% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 4.17 (s, 2 H), 6.90–7.27 (m, 2 H), 7.30–7.78 (m, 8 H), 7.83 (d, J = 7.70 Hz, 1 H), 12.03–12.16 (m, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 20.55, 28.58, 36.97, 46.95, 113.30, 119.71, 120.54, 120.77, 120.80, 127.76, 130.01, 130.32, 136.49, 137.68, 139.35, 152.91, 155.40, 167.27. HRMS calcd for C21H15N5O2S2Na (M + Na)+, 456.0559; found, 456.0562.
N-Ethyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (22)
Compound 7a (50 mg, 0.14 mmol), ethylamine (17.73 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 22.9 mg in 43.3% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.02 (t, J = 7.15 Hz, 3 H), 3.04–3.16 (m, 2 H), 3.88 (s, 2 H), 7.26 (t, J = 7.33 Hz, 1 H), 7.39–7.54 (m, 3 H), 7.56–7.69 (m, 3 H), 8.05 (d, J = 8.07 Hz, 1 H), 8.26 (m, 1 H), 12.11 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 15.12, 34.34, 37.01, 113.30, 119.70, 120.59, 120.74, 120.81, 127.76, 130.01, 130.32, 136.49, 137.68, 139.35, 152.88, 155.41, 167.02. HRMS calcd for C20H18N4O2SNa (M + Na)+, 401.1043; found, 401.1046.
N-Butyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (23)
Compound 7a (50 mg, 0.14 mmol), butylamine (30.94 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 18.6 mg in 44.7% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.78 (t, J = 7.31 Hz, 3 H), 1.17–1.30 (m, 2 H), 1.32–1.46 (m, 2 H), 2.98–3.13 (m, 2 H), 3.89 (s, 2 H), 7.25 (t, J = 6.83 Hz, 1 H), 7.39–7.55 (m, 4 H), 7.60 (m, J = 5.40, 5.40 Hz, 3 H), 8.04 (d, J = 7.80 Hz, 1 H), 8.17–8.25 (m, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 13.61, 19.53, 31.2, 36.52, 38.7, 112.84, 119.26, 120.08, 120.32, 120.38, 127.3, 129.54, 129.55, 129.86, 136.05, 137.25, 138.93, 152.43, 154.96, 166.7. HRMS calcd for C22H22N4O2SNa (M + Na)+, 429.1356; found, 429.1359.
2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-pentylacetamide (24)
Compound 7a (25 mg, 0.07 mmol), n-amylamine (20 μL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) were reacted according to general procedure D to give 19.5 mg in 66% yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 0.74 (t, J = 7.15 Hz, 3 H), 1.07–1.25 (m, 4 H), 1.31–1.45 (m, 2 H), 2.98–3.15 (m, 2 H), 3.88 (s, 2 H), 7.24 (t, J = 6.88 Hz, 1 H), 7.40–7.54 (m, 3 H), 7.56–7.68 (m, 3 H), 8.04 (d, J = 7.98 Hz, 1 H), 8.20 (t, J = 5.36 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 14.26, 22.29, 29.03, 29.25, 36.92, 113.27, 119.69, 120.52, 120.78, 120.81, 127.76, 130.00, 130.32, 136.48, 137.69, 139.35, 152.85, 155.40, 167.16. HRMS calcd for C23H25N4O2S (M + H)+, 421.1693; found, 421.1689.
N-Hexyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (25)
Compound 7a (25 mg, 0.07 mmol), hexylamine (21 μL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) were reacted according to general procedure D to give 20 mg in 65% yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 0.76 (t, J = 6.46 Hz, 3 H), 1.01–1.49 (m, 8 H), 3.06 (q, J = 6.60 Hz, 2 H), 3.87 (s, 2 H), 7.24 (t, J = 6.80 Hz, 1 H), 7.41–7.55 (m, 3 H), 7.55–7.66 (m, 3 H), 8.05 (d, J = 7.70 Hz, 1 H), 8.19 (t, J = 5.23 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 14.29, 22.40, 26.49, 29.51, 31.40, 36.82, 113.28, 119.63, 120.55, 120.79, 120.81, 127.82, 129.94, 130.04, 130.36, 136.41, 137.72, 139.36, 152.80, 155.41, 167.33. HRMS calcd for C24H27N4O2S (M + H)+, 435.1849; found, 435.1846.
N-Isobutyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (26)
Compound 7a (50 mg, 0.14 mmol), isobutylamine (31.11 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 17.71 mg in 31% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.72–0.92 (m, 6 H), 1.68 (dt, J = 13.47, 6.64 Hz, 1 H), 2.91 (t, J = 6.60 Hz, 2 H), 3.93 (s, 2 H), 7.25 (t, J = 6.97 Hz, 1 H), 7.40–7.56 (m, 3 H), 7.61 (d, J = 6.23 Hz, 3 H), 8.04 (d, J = 7.70 Hz, 1 H), 8.23 (t, J = 6.23 Hz, 1 H), 12.11 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 20.55, 28.58, 36.97, 46.95, 113.30, 119.71, 120.54, 120.77, 120.80, 127.76, 130.01, 130.32, 136.49, 137.68, 139.35, 152.91, 155.40, 167.27. HRMS calcd for C22H22N4O2SNa (M + Na)+, 429.1356; found, 429.1359.
N-Isopentyl-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (27)
Compound 7a (25 mg, 0.07 mmol), isopentylamine (20 μL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) were reacted according to general procedure D to give 20 mg in 67% yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 0.78 (d, J = 6.60 Hz, 6 H), 1.29 (q, J = 6.88 Hz, 2 H), 1.54 (tt, J = 13.38, 6.84 Hz, 1 H), 3.08 (s, 2 H), 3.88 (s, 2 H), 7.24 (t, J = 7.01 Hz, 1 H), 7.36–7.68 (m, 6 H), 8.04 (d, J = 7.98 Hz, 1 H), 8.17 (t, J = 5.50 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 22.71, 25.45, 36.86, 37.68, 38.43, 113.29, 119.64, 120.55, 120.77, 120.80, 127.83, 129.95, 130.03, 130.35, 136.42, 137.71, 139.34, 152.82, 155.40, 167.23. HRMS calcd for C23H25N4O2S (M + H)+, 421.1693; found, 421.1689.
N-(3,3-Dimethylbutyl)-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (28)
Compound 7a (25 mg, 0.07 mmol)), 3,3-dimethylbutylamine (20 μL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) were reacted according to general procedure D and purified by reverse-phase chromatography (C18, 10:90 to 90:10 water:methanol gradient) to give 18 mg in 60% yield. 1H NMR (300 MHz, DMSO-d6) δ ppm 0.83 (s, 9 H), 1.26–1.47 (m, 2 H), 3.00–3.16 (m, 2 H), 3.86 (s, 2 H), 7.24 (t, J = 7.29 Hz, 1 H), 7.39–7.55 (m, 3 H), 7.56–7.70 (m, 3 H), 8.04 (d, J = 7.98 Hz, 1 H), 8.17 (t, J = 5.50 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.12, 28.28, 34.68, 37.17, 50.51, 113.31, 119.70, 120.48, 120.78, 120.81, 127.77, 130.00, 130.01, 130.32, 136.51, 137.68, 139.35, 153.01, 155.39, 165.92. HRMS calcd for C24H27N4O2S (M + H)+, 435.1849; found, 435.1846.
N-(3-Methylpentyl)-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (29)
Compound 7a (25 mg, 0.07 mmol), 3-methylpentylamine (20 μL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol) and HATU (29.76 mg, 0.08 mmol) according togeneral procedure D and purified by reverse-phase chromatography (C18, 10:90 to 90:10 water:methanol gradient) to give 17 mg in 55% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.67–0.85 (m, 6 H), 0.96–1.11 (m, 1 H), 1.13–1.26 (m, 2 H), 1.31 (dd, J = 12.65, 6.42 Hz, 1 H), 1.37–1.51 (m, 1 H), 2.99–3.17 (m, 2 H), 3.87 (s, 2 H), 7.24 (t, J = 7.52 Hz, 1 H), 7.41–7.55 (m, 4 H), 7.56–7.69 (m, 3 H), 8.04 (d, J = 8.07 Hz, 1 H), 8.17 (t, J = 5.50 Hz, 1 H), 12.10 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 11.42, 19.22, 29.22, 31.70, 36.16, 37.49, 113.27, 119.69, 120.50, 120.81, 127.77, 130.00, 130.01, 130.33, 136.48, 137.69, 139.36, 152.84, 155.40, 167.11. HRMS calcd for C24H26N4O2SNa (M + Na)+, 457.1669; found, 457.1670.
2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-(pentan-2-yl)acetamide (30)
Compound 7a (25 mg, 0.07 mmol), 2-aminopentane (18.54 μL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) according to general procedure D to give 19.1 mg in 65% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.74 (t, J = 6.97 Hz, 3 H), 1.01 (d, J = 6.60 Hz, 3 H), 1.13–1.62 (m, 4 H), 3.64–3.81 (m, 1 H), 3.89 (s, 2 H), 7.24 (t, J = 7.33 Hz, 1 H), 7.41–7.72 (m, 6 H), 8.05 (t, J = 6.60 Hz, 2 H), 12.09 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 14.25, 19.32, 21.23, 37.11, 38.84, 44.94, 113.28, 119.68, 120.47, 120.77, 120.84, 127.75, 130.01, 130.32, 136.51, 137.68, 139.35, 152.97, 155.39, 166.36. HRMS calcd for C23H24N4O2SNa (M + Na)+, 443.1512; found, 443.1514.
2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-(pentan-3-yl)acetamide (31)
Compound 7a (25 mg, 0.07 mmol), 3-aminopentane (18.24 μL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) were reacted according to general procedure D to give 19.44 mg in 65% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.77 (d, J = 6.83 Hz, 6 H), 1.21–1.62 (m, 4 H), 3.47–3.58 (m, 1 H), 3.93 (s, 1 H), 7.24 (t, J = 6.83 Hz, 1 H), 7.37–7.76 (m, 6 H), 7.93 (d, J = 7.31 Hz, 1 H), 8.05 (d, J = 7.80 Hz, 1 H), 12.10 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 10.87, 27.40, 37.00, 52.36, 113.29, 119.64, 120.50, 120.73, 120.86, 127.82, 129.95, 130.05, 130.36, 136.43, 137.71, 139.34, 152.98, 155.40, 167.06. HRMS calcd for C23H25N4O2S (M + H)+, 421.1693; found, 421.1695.
N-(tert-Butyl)-2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (32)
Compound 7a (50 mg, 0.14 mmol), t-butylamine (32.9 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 22.6 mg in 39.7% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.15–1.37 (m, 9 H), 3.85 (s, 2 H), 7.26 (t, J = 6.83 Hz, 1 H), 7.41–7.56 (m, 4 H), 7.57–7.71 (m, 3 H), 7.93 (s, 1 H), 8.07 (d, J = 7.80 Hz, 1 H), 12.11 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 28.86, 37.86, 50.89, 113.34, 119.68, 120.54, 120.74, 120.78, 127.76, 129.99, 130.00, 130.31, 136.54, 137.68, 139.37, 153.13, 155.40, 166.60. HRMS calcd for C22H22N4O2SNa (M + Na)+, 429.1356; found, 429.1359.
2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-((tetrahydrofuran-2-yl)methyl)acetamide (33)
Compound 7a (50 mg, 0.14 mmol), tetrahydrofurfurylamine (32.31 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol) and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 35 mg in 57.5% yield. IR: 3372, 3193 (NH), 1651 (CO) cm–1. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.32–1.61 (m, 1 H), 1.63–2.06 (m, 3 H), 2.96–3.26 (m, 2 H), 3.52–3.64 (m, 1 H), 3.64–3.75 (m, 1 H), 3.76–3.86 (m, 1 H), 3.92 (s, 2 H), 7.25 (t, J = 6.97 Hz, 1 H), 7.35–7.71 (m, 6 H), 8.07 (d, J = 8.06 Hz, 1 H), 8.30 (t, J = 5.50 Hz, 1 H), 12.10 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 25.12, 28.4, 36.38, 43.03, 67.15, 77.11, 112.81, 119.25, 120.11, 120.39, 127.32, 129.55, 129.57, 129.88, 136.05, 137.25, 138.92, 152.4, 154.95, 167.14. HRMS calcd for C23H22N4O3SNa (M + Na)+, 457.1305; found, 457.1308.
2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-(piperidin-4-yl)acetamide (34)
Compound 7a (25 mg, 0.07 mmol), 4-aminopiperidine (16.6 uL, 0.16 mmol), triethylamine (19.82 μL, 0.14 mmol), and HATU (29.76 mg, 0.08 mmol) were reacted according to general procedure D to give 17 mg in 65% yield. IR: 3372, 3193 (NH), 1651 (CO) cm–1. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.27 (d, J = 11.36 Hz, 1 H), 1.46 (d, J = 9.90 Hz, 1 H), 1.77–2.05 (m, 2 H), 2.64–2.85 (m, 1 H), 4.00–4.43 (m, 3 H), 7.14–7.38 (m, 2 H), 7.41–7.80 (m, 6 H), 8.04 (d, J = 7.70 Hz, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 33.41, 35.16, 36.00, 36.46, 40.91, 44.69, 45.37, 48.25, 113.30, 119.70, 120.67, 120.75, 120.79, 127.76, 130.00, 130.03, 130.33, 136.51, 137.63, 137.68, 139.36, 152.93, 152.97, 155.40, 165.44, 166.23. HRMS calcd for C23H24N5O2S (M + H)+, 434.1645; found, 434.1648.
(Z)-3-((Hydroxy(2-(2-((4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamido)ethoxy)phosphoryl)oxy)propane-1,2-diyl Dioleate (35)
Compound 7a (50 mg, 0.14 mmol), DOPE (32.9 μL, 0.16 mmol), triethylamine (39.63 μL, 0.28 mmol), and HATU (59.52 mg, 0.16 mmol) were reacted according to general procedure D to give 16.4 mg in 10.9% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.84 (t, J = 6.83 Hz, 6 H), 1.05–1.75 (m, 50 H), 1.96 (br s, 8 H), 2.23 (t, J = 7.31 Hz, 4 H), 3.02 (s, 3 H), 3.14–3.26 (m, 2 H), 3.59–3.83 (m, 3 H), 3.90 (s, 2 H), 4.02–4.17 (m, 1 H), 4.26 (d, J = 9.26 Hz, 1 H), 5.06 (br s, 1 H), 5.30 (br s, 3 H), 7.24 (t, J = 7.31 Hz, 1 H), 7.38–7.75 (m, 6 H), 8.09 (d, J = 7.80 Hz, 1 H), 8.87 (br s, 1 H), 10.12 (br s, 1 H), 12.08 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 8.84, 14.38, 14.39, 22.58, 24.85, 24.91, 27.02, 27.05, 28.88, 28.91, 28.98, 29.00, 29.07, 29.12, 29.19, 29.33, 29.43, 29.45, 29.57, 31.77, 33.82, 34.00, 37.11, 45.62, 62.78, 63.05, 70.84, 70.89, 113.19, 119.66, 120.61, 120.86, 120.89, 127.62, 129.96, 130.00, 130.27, 136.50, 137.72, 139.36, 152.72, 155.40, 167.22, 172.67, 172.91. HRMS calcd for C59H89N4O10PSNa (M + Na)+, 1099.5929; found, 1099.5930.
N-Cyclohexyl-2-((3-cyclohexyl-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (36)
Compound 7b (17.7 mg, 0.05 mmol), triethylamine (13.8 μL, 0.1 mmol), and cyclohexylamine (6.2 μL, 0.06 mmol) were dissolved in anhydrous DMF (0.5 mL). HATU (20.74 mg, 0.06 mmol) dissolved in 0.2 mL of DMF was added to the reaction mixture and stirred for 20 min and concentrated in vacuo. The crude material was recrystallized in MeOH to give 9.1 mg in 41% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.99–2.17 (m, 16 H), 2.60–2.88 (m, 4 H), 3.99 (br s, 2 H), 4.17–4.45 (m, 1 H), 7.20 (t, J = 7.33 Hz, 1 H), 7.34–7.65 (m, 2 H), 8.01 (d, J = 8.07 Hz, 1 H), 8.22 (d, J = 6.97 Hz, 1 H), 11.86 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 19.02, 24.92, 25.28, 25.63, 26.36, 28.79, 32.87, 37.21, 48.49, 56.47, 61.95, 113.22, 120.27, 120.56, 120.72, 120.82, 127.59, 136.47, 139.26, 151.90, 155.96, 166.22. HRMS calcd for C24H30N4O2SNa (M + Na)+, 461.1982; found, 461.1983.
N-Cyclohexyl-2-((3-(naphthalen-2-yl)-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (39)
Compound 7c (26.1 mg, 0.065 mmol), triethylamine (18.1 μL, 0.13 mmol), and cyclohexylamine (8.2 μL, 0.071 mmol) were dissolved in anhydrous DMF (0.1 mL). HATU (27.2 mg, 0.071 mmol) dissolved in 0.2 mL of DMF was added to the reaction mixture and stirred for 20 min and concentrated in vacuo. The crude material was recrystallized in MeOH to give 27 mg in 86% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.76–1.38 (m, 5 H), 1.42–1.95 (m, 5 H), 3.43–3.55 (m, 1 H), 3.74–4.00 (m, 2 H), 7.29 (t, J = 6.97 Hz, 1 H), 7.41 (d, J = 8.43 Hz, 1 H), 7.47–7.66 (m, 4 H), 7.68–7.81 (m, 2 H), 8.01–8.36 (m, 4 H), 12.17 (s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.82, 25.57, 31.23, 32.72, 32.75, 36.27, 36.97, 48.36, 113.39, 119.55, 120.69, 120.81, 120.84, 122.13, 126.31, 127.24, 127.99, 128.25, 128.88, 129.03, 130.07, 130.92, 132.83, 134.46, 138.05, 139.48, 153.40, 155.47, 162.87, 166.19. HRMS calcd for C28H26N4O2SNa (M + Na)+, 505.1669; found, 505.1671.
N-Cyclohexyl-2-((4-oxo-3-phenethyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (40)
Compound 7d (25 mg, 0.066 mmol), triethylamine (19.5 μL, 0.13 mmol), and cyclohexylamine (8.2 μL, 0.073 mmol) were dissolved in anhydrous DMF (0.1 mL). HATU (27.2 mg, 0.073 mmol) dissolved in 0.2 mL of DMF was added to the reaction mixture and stirred for 20 min and concentrated in vacuo. The crude material was recrystallized in MeOH to give 18.28 mg in 60% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.83–1.42 (m, 5 H), 1.44–2.08 (m, 5 H), 2.87–3.17 (m, 2 H), 3.95–4.24 (m, 2 H), 4.25–4.62 (m, 2 H), 7.33 (br s, 6 H), 7.87–8.48 (m, 3 H), 11.99 (s, 2 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.91, 25.63, 32.83, 34.09, 36.63, 45.66, 48.51, 113.26, 119.40, 120.37, 120.67, 120.84, 127.16, 127.76, 129.10, 137.30, 138.26, 139.36, 151.68, 154.92, 166.19. HRMS calcd for C26H28N4O2SNa (M + Na)+, 483.1825; found, 483.1827.
2-((3-Cyclohexyl-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-cyclopentylacetamide (41)
Compound 7b (25 mg, 0.07 mmol), triethylamine (19.5 μL, 0.14 mmol), and cyclopentylamine (8.82 μL, 0.08 mmol) were dissolved in anhydrous DMF (0.5 mL). HATU (29.3 mg, 0.08 mmol) dissolved in 0.2 mL of DMF was added to the reaction mixture and stirred for 20 min and concentrated in vacuo. The crude material was recrystallized in MeOH to give 9.3 mg in 30.3% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.97–2.09 (m, 18 H), 2.61–2.83 (m, 4 H), 4.02 (br s, 1 H), 4.17–4.37 (m, 1 H), 7.20 (t, J = 7.30 Hz, 1 H), 7.36–7.56 (m, 2 H), 8.00 (d, J = 8.06 Hz, 1 H), 8.32 (d, J = 7.33 Hz, 1 H), 11.86 (br s, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 23.97, 25.25, 26.34, 28.78, 32.73, 37.02, 51.21, 61.99, 113.23, 120.31, 120.54, 120.67, 120.79, 127.66, 136.48, 139.24, 151.93, 155.94, 166.85. HRMS calcd for C23H28N4O2SNa (M + Na)+, 447.1825; found, 447.1827.
N-Cyclohexyl-2-((5-methyl-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (42) and N-Cyclohexyl-N-methyl-2-((5-methyl-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (43)
To a solution of compound 1 (50 mg, 0.12 mmol) in DMF (1 mL) in a flame-dried flask was added NaH 60% dispersion in mineral oil (9.24 mg, 0.24 mmol) and stirred at room temperature for 10 min. Methyl iodide (14.3 μL, 0.24 mmol) was then added to the solution and stirred overnight. Reaction mixture was concentrated and purified by preparative thin-layer chromatography (40:60 ethyl acetate:hexane) to give 18.18 mg in 33.9% yield of compound 42 and 5.3 mg in 9.6% yield of compound 43. 42: 1H NMR (400 MHz, DMSO-d6) δ ppm 0.93–1.37 (m, 5 H), 1.40–1.90 (m, 5 H), 3.44–3.58 (m, 1 H), 3.88 (s, 2 H), 4.15 (s, 3 H), 7.31 (t, J = 7.33 Hz, 1 H), 7.39–7.53 (m, 2 H), 7.53–7.78 (m, 5 H), 8.05–8.27 (m, 2 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.90, 25.62, 31.47, 32.82, 37.14, 48.41, 111.31, 119.12, 120.09, 120.74, 120.92, 128.03, 129.99, 130.34, 136.35, 137.47, 140.40, 153.27, 155.75, 166.14. HRMS calcd for C25H27N4O2S (M + H)+, 447.1849; found, 447.1852. 43: 1H NMR (400 MHz, DMSO-d6) δ ppm 0.76–1.98 (m, 10 H), 2.10 (s, 3 H), 3.97 (s, 2 H), 4.23 (s, 3 H), 7.16 (t, J = 7.52 Hz, 1 H), 7.26–7.85 (m, 8 H), 8.07 (d, J = 7.70 Hz, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.85, 25.60, 31.43, 32.28, 38.02, 50.24, 62.03, 111.44, 119.63, 120.34, 120.85, 121.08, 128.20, 129.74, 130.25, 135.43, 136.73, 139.34, 155.99, 156.50, 169.99. HRMS calcd for C26H29N4O2S (M + H)+, 461.2006; found, 461.2007.
Methyl 4-(2-((2-(Cyclohexylamino)-2-oxoethyl)thio)-4-oxo-3-phenyl-3H-pyrimido[5,4-b]indol-5(4H)-yl)butanoate (44)
To a flame-dried flask, bromobutyric acid (50 μL, 0.43 mmol) was dissolved in anhydrous MeOH (3 mL) followed by the addition of TMSCl (272.45 μL, 2.15 mmol). The solution was stirred overnight at room temperature and concentrated in vacuo to remove all trace of MeOH and HCl. In a separate flame-dried flask, compound 1 (25 mg, 0.06 mmol) was dissolved in DMF (1 mL). NaH 60% dispersion (2.31 mg, 0.06 mmol) was added to the solution. After stirring at room temperature for 10 min, methyl iodide (3.6 μL, 0.06 mmol) was added and stirred for an additional 12 h. The reaction mixture was concentrated in vacuo and purified by preparative thin-layer chromatography to give 1.3 mg in 4% yield. IR: 3293 (NH), 1732 (CO ester), 1683 (CO amide) cm–1. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.99–1.32 (m, 5 H), 1.52 (d, J = 12.19 Hz, 1 H), 1.57–1.80 (m, 4 H), 1.91–2.10 (m, 2 H), 2.29 (t, J = 7.31 Hz, 2 H), 3.47 (s, 3 H), 3.47 (br s, 1 H), 3.89 (s, 2 H), 4.62 (t, J = 6.83 Hz, 2 H), 7.30 (t, J = 7.56 Hz, 1 H), 7.39–7.51 (m, 2 H), 7.52–7.68 (m, 4 H), 7.74 (s, 1 H), 8.11 (d, J = 7.80 Hz, 1 H), 8.17 (d, J = 7.80 Hz, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.88, 25.59, 26.00, 30.71, 32.79, 37.02, 51.71, 111.34, 118.74, 120.19, 120.80, 121.09, 128.18, 129.93, 130.02, 130.37, 136.29, 137.81, 139.77, 153.45, 155.47, 166.28, 173.19. HRMS calcd for C29H32N4O4SNa (M + Na)+, 555.2036; found, 555.2036.
tert-Butyl 2-((4-Oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetate (45)
Compound 6a (100 mg, 0.34 mmol) and KOH (38.2 g, 0.68 mmol) were suspended in 2 mL of dimethylacetamide in a flame-dried flask with stirring. H2O was added dropwise until KOH was completely dissolved. t-Butyl chloroacetate (49 μL, 0.34 mmol) was immediately added to the reaction mixture and stirred at room temperature monitoring with thin-layer chromatography (1:99 MeOH:DCM). Upon completion, reaction mixture was extracted with ethyl acetate (20 mL) and water (40 mL), dried over MgSO4, and concentrated in vacuo. EtOH (5 mL) was added to the resulting viscous liquid, and pure product was filtered to give 55 mg in 40% yield. IR: 3192 (NH), 1732 (CO ester), 1674 (CO amide) cm–1. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.32–1.48 (m, 9 H), 3.91 (s, 2 H), 7.26 (t, J = 7.31 Hz, 1 H), 7.42–7.55 (m, 4 H), 7.56–7.69 (m, 3 H), 7.96 (d, J = 8.29 Hz, 1 H), 12.13 (s, 1 H). HRMS calcd for C22H21N3O3SNa (M + Na)+, 430.1196; found, 430.1197.
tert-Butyl 2-((4-Oxo-3-phenyl-5-propyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetate (46a)
Compound 45 (50 mg, 0.12 mmol), NaH 60% dispersion in mineral oil (3.3 mg, 0.14 mmol), iodopropane (13.1 μL, 0.14 mmol), and DMF (1 mL) were reacted according to general procedure E and purified by silica gel chromatography (10:90 ethyl acetate:hexane) to give 32.8 mg in 61% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.82 (t, J = 7.31 Hz, 3 H), 1.40 (s, 9 H), 1.66–1.95 (m, 2 H), 3.91 (s, 2 H), 4.56 (t, J = 6.58 Hz, 2 H), 7.30 (br s, 1 H), 7.39–7.68 (m, 6 H), 7.75 (d, J = 8.29 Hz, 1 H), 7.99 (d, J = 7.80 Hz, 1 H). HRMS calcd for C25H27N3O3SNa (M + Na)+, 472.1665; found, 472.1668.
tert-Butyl 2-((4-Oxo-5-pentyl-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetate (46b)
Compound 45 (50 mg, 0.12 mmol), NaH 60% dispersion in mineral oil (3.3 mg, 0.14 mmol), iodopentane (17.64 μL, 0.14 mmol), and DMF (1 mL) were reacted according to general procedure E and purified by silica gel chromatography (10:90 ethyl acetate:hexane) to give 33.8 mg in 59% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.80 (t, J = 6.83 Hz, 3 H), 1.25 (d, J = 6.83 Hz, 4 H), 1.32–1.55 (m, 9 H), 1.64–1.83 (m, 2 H), 3.91 (s, 2 H), 4.59 (t, J = 7.31 Hz, 2 H), 7.30 (t, J = 7.56 Hz, 1 H), 7.46 (dd, J = 7.56, 1.71 Hz, 2 H), 7.54 (t, J = 7.31 Hz, 1 H), 7.57–7.67 (m, 3 H), 7.72 (d, J = 8.78 Hz, 1 H), 7.99 (d, J = 7.80 Hz, 1 H). HRMS calcd for C27H31N3O3SNa (M + Na)+, 500.1978; found, 500.1975.
tert-Butyl 2-((5-Dodecyl-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetate (46c)
Compound 45 (50 mg, 0.12 mmol), NaH 60% dispersion in mineral oil (3.3 mg, 0.14 mmol), bromododecane (32.4 μL, 0.14 mmol), and DMF (1 mL) were reacted according to general procedure E and purified by silica gel chromatography (10:90 ethyl acetate:hexane) to give 37.4 mg in 54% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.84 (t, J = 6.83 Hz, 3 H), 1.21 (br s, 18 H), 1.40 (s, 9 H), 1.64–1.86 (m, 3 H), 3.91 (s, 2 H), 4.59 (t, J = 7.07 Hz, 2 H), 7.29 (t, J = 7.80 Hz, 1 H), 7.45 (dd, J = 7.31, 1.95 Hz, 2 H), 7.54 (t, J = 7.80 Hz, 1 H), 7.58–7.67 (m, 3 H), 7.72 (d, J = 8.29 Hz, 1 H), 7.98 (d, J = 7.80 Hz, 1 H). HRMS calcd for C34H45N3O3SNa (M + Na)+, 598.3074; found, 598.3070.
tert-Butyl 2-((5-(Cyanomethyl)-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetate (46d)
Compound 45 (318 mg, 0.78 mmol), NaH 60% dispersion (38.16 mg, 0.86 mmol), bromoacetonitrile (65.5 μL, 0.86 mmol), and DMF (20 mL) were reacted according to general procedure E to give 247.8 mg in 71% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.41 (s, 9 H), 3.94 (s, 2 H), 5.87 (s, 2 H), 7.43 (t, J = 7.31 Hz, 1 H), 7.47–7.55 (m, 2 H), 7.58–7.72 (m, 4 H), 7.88 (d, J = 8.78 Hz, 1 H), 8.03 (d, J = 8.29 Hz, 1 H). HRMS calcd for C24H22N4O3SNa (M + Na)+, 469.1305; found, 469.1303.
2-((4-Oxo-3-phenyl-5-propyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (47a)
Compound 46a (29.74 mg, 0.07 mmol), acetonitrile (1 mL), and trifluoroacetic acid (1 mL) were reacted according to general procedure F to give 26 mg in quantitative yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.81 (t, J = 6.58 Hz, 3 H), 1.75 (m, 2 H), 3.96 (s, 2 H), 4.56 (t, J = 7.07 Hz, 2 H), 7.29 (t, J = 7.31 Hz, 1 H), 7.38–7.88 (m, 7 H), 7.99 (d, J = 8.29 Hz, 1 H). HRMS calcd for C21H19N3O3SNa (M + Na)+, 416.1039; found, 416.1036.
2-((4-Oxo-5-pentyl-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (47b)
Compound 46b (30.98 mg, 0.065 mmol), acetonitrile (1 mL), and trifluoroacetic acid (1 mL) were reacted according to general procedure F to give 27 mg in quantitative yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.81 (t, J = 6.58 Hz, 3 H), 0.99–1.43 (m, 4 H), 1.62–1.84 (m, 2 H), 3.96 (s, 2 H), 4.59 (t, J = 7.07 Hz, 2 H), 7.29 (t, J = 7.31 Hz, 1 H), 7.45 (br s, 2 H), 7.51–7.68 (m, 4 H), 7.72 (d, J = 8.29 Hz, 1 H), 7.98 (d, J = 8.29 Hz, 1 H). HRMS calcd for C23H23N3O3SNa (M + Na)+, 444.1352; found, 444.1351.
2-((5-Dodecyl-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (47c)
Compound 46c (34.81 mg, 0.06 mmol), acetonitrile (1 mL), and trifluoroacetic acid (1 mL) were reacted according to general procedure F to give 28.6 mg in 92% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.82 (t, J = 6.83 Hz, 3 H), 1.19 (d, J = 7.80 Hz, 18 H), 1.71 (d, J = 6.34 Hz, 2 H), 3.94 (s, 2 H), 4.56 (t, J = 7.07 Hz, 2 H), 7.27 (t, J = 7.80 Hz, 1 H), 7.36–7.47 (m, 2 H), 7.59 (s, 4 H), 7.70 (d, J = 8.29 Hz, 1 H), 7.96 (d, J = 7.80 Hz, 1 H), 12.74 (br s, 1 H). HRMS calcd for C30H37N3O3SNa (M + Na)+, 542.2448; found, 542.2444.
2-((5-(Cyanomethyl)-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetic Acid (47d)
Compound 46a (29.74 mg, 0.07 mmol), acetonitrile (1 mL), and trifluoroacetic acid (1 mL) were reacted according to general procedure F to give 26 mg in quantitative yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 3.84–4.15 (m, 2 H), 5.87 (s, 2 H), 7.42 (t, J = 7.56 Hz, 1 H), 7.47–7.56 (m, 2 H), 7.57–7.74 (m, 4 H), 7.88 (d, J = 8.29 Hz, 1 H), 8.03 (d, J = 7.80 Hz, 1 H), 12.89 (br s, 1 H). HRMS calcd for C20H14N4O3SNa (M + Na)+, 413.0679; found, 413.0676.
N-Cyclohexyl-2-((4-oxo-3-phenyl-5-propyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (48)
Compound 47a (29.4 mg, 0.075 mmol), HATU (31.3 mg, 0.082 mmol), triethylamine (20.8 μL, 0.15 mmol), cyclohexylamine (9.43 μL, 0.082 mmol), and DMF (1 mL) were reacted according to general procedure D to give 4 mg in 11% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.82 (t, J = 7.33 Hz, 3 H), 1.00–1.37 (m, 5 H), 1.42–1.99 (m, 7 H), 3.88 (s, 2 H), 4.56 (t, J = 6.97 Hz, 2 H), 7.29 (t, J = 7.70 Hz, 1 H), 7.41–7.50 (m, 1 H), 7.50–7.68 (m, 4 H), 7.74 (d, J = 8.43 Hz, 1 H), 8.10 (d, J = 8.07 Hz, 1 H), 8.17 (d, J = 8.07 Hz, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 11.35, 24.15, 24.89, 25.61, 32.83, 37.12, 45.80, 48.40, 111.49, 118.75, 120.09, 120.61, 120.99, 127.98, 129.96, 129.98, 130.29, 136.38, 137.68, 139.83, 153.35, 155.43, 166.14. HRMS calcd for C27H31N4O2S (M + H)+, 475.2162; found, 475.2164.
N-Cyclohexyl-2-((4-oxo-5-pentyl-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (49)
Compound 47b (34 mg, 0.08 mmol), HATU (34 mg, 0.088 mmol), triethylamine (22.5 μL, 0.16 mmol), cyclohexylamine (10.2 μL, 0.088 mmol), and DMF (1 mL) were reacted according to general procedure D to give 21 mg in 52% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.80 (t, J = 6.97 Hz, 3 H), 1.00–1.40 (m, 9 H), 1.43–1.92 (m, 7 H), 3.44–3.58 (m, 1 H), 3.88 (s, 2 H), 4.58 (t, J = 7.15 Hz, 2 H), 7.29 (t, J = 7.33 Hz, 1 H), 7.40–7.68 (m, 6 H), 7.72 (d, J = 8.43 Hz, 1 H), 8.03–8.32 (m, 2 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 14.35, 22.29, 24.91, 25.62, 28.70, 30.56, 32.84, 37.11, 44.38, 48.42, 111.45, 118.70, 120.13, 120.64, 121.02, 128.02, 129.99, 130.31, 136.39, 137.70, 139.74, 153.36, 155.44, 166.16. HRMS calcd for C29H34N4O2S (M + H)+, 502.24025; found, 502.24018.
N-Cyclohexyl-2-((5-dodecyl-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)acetamide (50)
Compound 47c (28.6 mg, 0.055 mmol), HATU (23 mg, 0.061 mmol), triethylamine (15.33 μL, 0.110 mmol), cyclohexylamine (7 μL, 0.061 mmol), and DMF (1 mL) were reacted according to general procedure D and purified by silica gel chromatography (20:80 ethyl acetate:hexane) to give 22 mg in 67% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.73–0.92 (m, 4 H), 1.20 (d, J = 6.97 Hz, 22 H), 1.41–1.89 (m, 7 H), 3.45–3.61 (m, 1 H), 3.88 (s, 2 H), 4.58 (t, J = 6.60 Hz, 2 H), 7.28 (t, J = 7.52 Hz, 1 H), 7.36–7.80 (m, 7 H), 8.05–8.26 (m, 2 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 11.85, 14.50, 22.63, 24.97, 25.70, 26.59, 29.19, 29.44, 30.88, 31.82, 32.90, 37.20, 44.48, 48.42, 111.54, 118.70, 120.22, 120.69, 121.09, 128.06, 130.05, 130.39, 136.46, 137.79, 139.83, 153.41, 155.50, 166.17. HRMS calcd for C36H49N4O2S (M + H)+, 601.3571; found, 601.3570.
2-((5-(Cyanomethyl)-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-cyclohexylacetamide (51)
Compound 47d (150 mg, 0.39 mmol), HATU (161 mg, 0.42 mmol), triethylamine (59 μL, 0.77 mmol), cyclohexylamine (49 μL, 0.42 mmol), and DMF (5 mL) were reacted according to general procedure D to give 157.3 mg in 86.7% yield. IR: 3240 (NH), 1697 (CO) cm–1. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.97–1.33 (m, 5 H), 1.45–1.87 (m, 5 H), 3.45–3.58 (m, 1 H), 3.90 (s, 2 H), 5.87 (s, 2 H), 7.42 (t, J = 7.80 Hz, 1 H), 7.47–7.56 (m, 2 H), 7.57–7.74 (m, 4 H), 7.88 (d, J = 8.29 Hz, 1 H), 8.17 (dd, J = 13.16, 7.80 Hz, 2 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.92, 25.62, 32.84, 33.04, 37.21, 48.45, 111.45, 116.75, 118.20, 121.01, 121.35, 122.12, 129.02, 129.90, 130.08, 130.56, 135.95, 138.98, 139.65, 155.44, 155.50, 166.02. HRMS calcd for C26H26N5O2S (M + H)+, 472.1802; found, 472.1803.
2-((5-(2-Amino-2-oxoethyl)-4-oxo-3-phenyl-4,5-dihydro-3H-pyrimido[5,4-b]indol-2-yl)thio)-N-cyclohexylacetamide (52)
A KOH (6 mg, 0.11 mmol) solution in water (50 μL) was added to a solution of compound 51 in dimethylacetamide (200 μL) and stirred at rt overnight. Reaction mixture was acidified with 3 M HCl, extracted with ethyl acetate and water, dried over MgSO4, and concentrated to dryness in vacuo. Further recrystallization in MeOH gives 20 mg in 37% yield. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.94–1.36 (m, 5 H), 1.43–1.89 (m, 5 H), 3.43–3.62 (m, 1 H), 3.89 (s, 2 H), 5.24 (s, 2 H), 7.12–7.22 (m, 1 H), 7.30 (t, J = 7.33 Hz, 1 H), 7.42 (m, J = 7.30, 2.20 Hz, 2 H), 7.48–7.67 (m, 5 H), 8.10 (d, J = 7.70 Hz, 1 H), 8.18 (d, J = 7.70 Hz, 1 H). 13C NMR (126 MHz, DMSO-d6) δ ppm 24.90, 25.59, 32.82, 37.05, 46.85, 48.48, 111.36, 119.30, 120.36, 120.91, 120.95, 128.13, 129.87, 130.05, 130.41, 136.14, 137.82, 140.64, 153.47, 155.65, 166.26, 169.77. HRMS calcd for C26H27N5O3SNa (M + Na)+, 512.1727; found, 512.1726.
Biological Studies
Animals
Seven–nine-week-old C57BL/6 (wild-type, WT) and Cd14–/– (C57BL/6 background) were purchased from the Jackson Laboratories (Bar Harbor, MA). Tlr4–/–mice were a gift from Dr. Shizuo Akira (Osaka University, Japan) and backcrossed for 10 generations onto the C57BL/6 background at University of California, San Diego (UCSD). All animal experiments were approved by the UCSD Institutional Animal Care and Use Committee.
In Vitro Cytokine Induction in Bone Marrow Derived Dendritic Cells (BMDC)
BMDC were prepared from C57BL/6 mice as described.36 BMDC (105cells per well) were plated in 96-well plates in 200 μL of complete RPMI1640 supplemented with 10% fetal calf serum (FCS; Sigma Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). The cells were incubated with graded concentrations of the compounds for 18 h at 37 °C, 5% CO2. After 18 h incubation, the cell culture supernatants were collected. LPS (purified LPS, Invivogen, San Diego, CA) or MPLA (1 μg/mL synthetic MPLA, Invivogen, San Diego, CA) were used as positive controls. The levels of IL-6 in the culture supernatants were determined by ELISA (BD Biosciences, La Jolla, CA).37 The AUC was calculated from BMDC dose–response curves using Prism 5 (GraphPad, San Diego). Each cytokine induction curve was first converted to a percent activity curve, and then the AUC of the percent activity curve was calculated. The process of converting to a percent activity curve allowed subtracting background and adjusting for plate-to-plate variation. Finally, the AUC values were normalized to the activity of compound 1 within each experiment, set at 100. The cultures stimulated with LPS 10 ng/mL, and 5 μM compound 1 released an average of 20.6 ng/mL ± 4.7 SD and 10.5 ng/mL ± 1.3 SD of IL-6, respectively.
In Vitro Assays Using TLR Reporter Cell Lines
Murine or humanTLR4 HEK Blue cells (Invivogen, 2.5 × 104 cells per well of a 96-well plate), or NFκB/SEAPorter HEK 293 cells (Imgenex, San Diego, CA) for human TLR2, TLR3, TLR5, TLR7, TLR8, or TLR9 (5 × 104 cells per well of 96 well plate) were incubated with graded doses of compound 1. The culture supernatants were harvested after a 20–24 h incubation period. SEAP activity in the supernatants was determined by a colorimetric assay, using either the SEAPorter Assay Kit (Imgenex), with absorbance read at 405 nm, or QuantiBlue (Invivogen), with absorbance read at 630 nm. Stimulation of the human TLR4 cells with 10 ng/mL LPS resulted in OD630 of 1.90 relative to 0.88 of cells stimulated with 10 μM compound 1. The relative reporter activation for the murine TLR4 cells for cells stimulated with 10 ng/mL LPS and 5 μM compound 1 was 1.70 ± 0.08 and 1.52 ± 0.03, respectively.
In Vitro Activities in hPBMC
Human PBMC were isolated from buffy coats obtained from the San Diego Blood Bank (San Diego, CA) as described previously.37,38 PBMC (1 × 106/mL) were incubated with various compounds in complete RPMI for 18 h at 37 °C, 5% CO2, and culture supernatants were collected. The levels of IL-8 in the supernatants were determined by ELISA (BD Biosciences, La Jolla, CA). Human cell cultures were treated with 10 ng/mL LPS as the positive control. Cultures treated with LPS 10 ng/mL and compound 1 10 μM released averages of 14.5 ± 0.6 ng/mL SD and 7.8 ± 0.8 ng/mL SD of IL-8, respectively.
Type I IFN Assay
L929 cells stably expressing an interferon sensitive response element (ISRE) luciferase reporter construct were kindly provided by Dr. B. Beutler (UT Southwestern, Texas).39 The bioactivity of type I IFN in mBMDC supernatants was measured by luciferase assay using L929-ISRE cells as described previously.39 L929-ISRE cells were plated at 5× 104 cells per well in Dulbecco’s Modified Eagle Medium (Invitrogen) supplemented with 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin (DMEM-10) in a 96-well white-walled clear-bottom plate. Thus, 50 μL of mBMDC supernatant was incubated with L929-ISRE cells in 50 μL of DMEM for 6 h. Mu-IFN Beta Standard (PBL Interferon Source, Piscataway, NJ) was used as a standard. The luciferase activities were measured by Steady-Glo luciferase assay buffer (Promega, Madison, WI).
In addition, the levels of IP-10, a surrogate marker of type I IFN, in the supernatants were determined by ELISA (R&D Systems, Minneapolis, MN). The cultures stimulated with LPS 10 ng/mL, and 5 μM compound 1 released an average of 234.8 ± 8.6 and 590.0 ± 10.5 pg/mL of IP-10 respectively.
Statistical Analysis
The data are represented as mean ± standard error of the mean (SEM). Areas-under-curve (AUCs) were calculated using the trapezoid method. Prism 5 (GraphPad Software) statistical software was used to obtain p-values for comparison between groups (p < 0.05 was considered significant). For the in vitro studies, two-tailed Student’s t test was used to compare two groups, and one-way ANOVA Dunnett’s test was used to compare multiple groups.
Acknowledgments
We are grateful for the assistance provided by Dr. Xu of the Department of Chemistry at University of California, San Diego for high resolution mass spectrometry. We acknowledge the services provided by the University of California, San Francisco Small Molecule Discovery Center. This project was supported by a contract from the National Institute of Allergy and Infectious Diseases (HHSN272200900034C) from the National Institutes of Health (to DAC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary
Abbreviations Used
- AUC
area under the curve
- HTS
high throughput screen
- TLR
Toll-like receptor
- IL
interleukin
- TNF
tumor necrosis factor
- LPS
lipopolysaccharide
- MyD88
myeloid differentiation primary response gene 88
- MD-2
myeloid differentiation protein-2
- MPLA
monophosphoryl lipid A
- PBMC
peripheral blood mononuclear cells
- EtOH
ethanol
- THF
tetrahydrofuran
- TMSCl
trimethylsilyl chloride or chlorotrimethylsilane
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DMF
N,N-dimethylformamide
- HATU
O-(7-azabenzotriazol-1-yl)-N,N,N0,N0-tetramethyluroniumhexafluorophosphate
- DCM
dichloromethane
- MeOH
methanol
- TEA
triethylamine
- BMDC
bone marrow derived dendritic cells
- BMDM
bone marrow derived macrophages
- NFκB
nuclear factor κ B
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ISRE
interferon response element
- IFN
interferon
- SEAP
secreted embryonic alkaline phosphatase
- ELISA
enzyme-linked immunosorbent assay
- IP-10
interferon γ-induced protein 10
- PRR
pattern-recognition receptors
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Dev A.; Iyer S.; Razani B.; Cheng G. NF-κB and innate immunity. Curr. Top. Microbiol. Immunol. 2011, 349, 115–143. [DOI] [PubMed] [Google Scholar]
- Li Q.; Verma I. M. NF-κB regulation in the immune system. Nature Rev. Immunol. 2002, 2, 725–734. [DOI] [PubMed] [Google Scholar]
- Fang H.; Wu Y.; Huang X.; Wang W.; Bing A.; et al. Toll-like Receptor 4 (TLR4) Is Essential for Hsp70-like Protein 1 (HSP70L1) to Activate Dendritic Cells and Induce Th1 Response. J. Biol. Chem. 2011, 286, 30393–30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H.; Takeuchi O.; Kawai T.; Kaisho T.; Sato S.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [DOI] [PubMed] [Google Scholar]
- Prins R. M.; Craft N.; Bruhn K. W.; Khan-Farooqi H.; Koya R. C.; et al. The TLR-7 Agonist, Imiquimod, Enhances Dendritic Cell Survival and Promotes Tumor Antigen-Specific T Cell Priming: Relation to Central Nervous System Antitumor Immunity. J. Immunol. 2005, 176, 157–164. [DOI] [PubMed] [Google Scholar]
- Lee J.; Chuang T.-H.; Redecke V.; She L.; Pitha P. M.; et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 6646–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Wu C. C. N.; Lee K. J.; Chuang T.-H.; Katakura K.; et al. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 1828–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits E. L. J. M.; Cools N.; Lion E.; Camp K.; Ponsaerts P.; et al. The Toll-like receptor 7/8 agonist resiquimod greatly increases the immunostimulatory capacity of human acute myeloid leukemia cells. Cancer Immunol. Immunother. 2010, 59, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman R. L.; Sher A.; Seder R. A. Vaccine Adjuvants: Putting Innate Immunity to Work. Immunity 2010, 33, 492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Haro V.; Cekic C.; Martin M.; Chilton P. M.; Casella C. R.; et al. The Vaccine Adjuvant Monophosphoryl Lipid A as a TRIF-Biased Agonist of TLR4. Science 2007, 316, 1628–1632. [DOI] [PubMed] [Google Scholar]
- Pu M.; Hayashi T.; Cottam H.; Mulvaney J.; Arkin M.; et al. Analysis of high-throughput screening assays using cluster enrichment. Stat. Med. 2012, 31, 4175–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats S. R.; Pham T.-T. T.; Bainbridge B. W.; Reife R. A.; Darveau R. P. MD-2 Mediates the Ability of Tetra-Acylated and Penta-Acylated Lipopolysaccharides to Antagonize Escherichia coli Lipopolysaccharide at the TLR4 Signaling Complex. J. Immunol. 2005, 175, 4490–4498. [DOI] [PubMed] [Google Scholar]
- Jiang Z.; Georgel P.; Du X.; Shamel L.; Sovath S.; et al. CD14 is required for MyD88-independent LPS signaling. Nature Immunol. 2005, 6, 565–570. [DOI] [PubMed] [Google Scholar]
- Wright S. D.; Ramos R. A.; Tobias P. S.; Ulevitch R. J.; Mathison J. C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 1990, 249, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Nagai Y.; Akashi S.; Nagafuku M.; Ogata M.; Iwakura Y.; et al. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nature Immunol. 2002, 3, 667–672. [DOI] [PubMed] [Google Scholar]
- Devani M. B.; Shishoo C. J.; Pathak U. S.; Parikh S. H.; Shah G. F.; et al. Synthesis of 3-substituted thieno[2,3-d]pyrimidin-4(3H)-one-2-mercaptoacetic acids and their ethyl esters for pharmacological screening. J. Pharm. Sci. 1976, 65, 660–664. [DOI] [PubMed] [Google Scholar]
- Thurkauf A.; Hutchison A.. Preparation of aryl heterocyclyl pyrimidines as GABA brain receptor ligands. (Neurogen Corp.) Cont. of U.S. Ser. No. 865,129, abandoned, 1994; 17 pp.
- Monge A.; Palop J. A.; Recalde I.; Martinez-Crespo F.; Fernandez-Alvarez E. Synthesis of 5H-pyrimido[5,4-b]indole derivatives and related compounds. An. Quim., Ser. C: Quim. Org. Bioquim. 1985, 81, 267–270. [Google Scholar]
- Kawai T.; Takeuchi O.; Fujita T.; Inoue J.-I.; Muhlradt P. F.; et al. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 2001, 167, 5887–5894. [DOI] [PubMed] [Google Scholar]
- Yamamoto M.; Sato S.; Mori K.; Hoshino K.; Takeuchi O.; et al. A novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-I2 pomoter in the Toll-like receptor signaling. J. Immunol. 2002, 169, 6668–6672. [DOI] [PubMed] [Google Scholar]
- Ohto U.; Fukase K.; Miyake K.; Shimizu T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, S7421–S7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J.; Drolet J. R.; Monks B. G.; Golenbock D. T. MD-2 Residues Tyrosine 42, Arginine 69, Aspartic Acid 122, and Leucine 125 Provide Species Specificity for Lipid IVA. J. Biol. Chem. 2010, 285, 27935–27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasl J.; Oblak A.; Gioannini T. L.; Weiss J. P.; Jerala R. Novel Roles of Lysines 122, 125, and 58 in Functional Differences between Human and Murine MD-2. J. Immunol. 2009, 183, 5138–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi M.; Tanamoto K.-i. Structural Regions of MD-2 That Determine the Agonist-Antagonist Activity of Lipid IVa. J. Biol. Chem. 2006, 281, 5484–5491. [DOI] [PubMed] [Google Scholar]
- Saitoh S.-i.; Akashi S.; Yamada T.; Tanimura N.; Kobayashi M.; et al. Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with Toll-like receptor 4 (TLR4)-MD-2 and ligand-induced TLR4 oligomerization. Int. Immunol. 2004, 16, 961–969. [DOI] [PubMed] [Google Scholar]
- Walsh C.; Gangloff M.; Monie T.; Smyth T.; Wei B.; et al. Elucidation of the MD-2/TLR4 interface required for signaling by lipid IVa. J. Immunol. 2008, 181, 1245–1254. [DOI] [PubMed] [Google Scholar]
- Shimazu R.; Akashi S.; Ogata H.; Nagai Y.; Fukudome K.; et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999, 189, 1777–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R.; Preston-Hurlburt P.; Janeway C. A. Jr. A human homolog of the Drosophila Toll protein signals activation of adaptive immunity. Natur 1997, 388, 394–396. [DOI] [PubMed] [Google Scholar]
- Park B. S.; Song D. H.; Kim H. M.; Choi B.-S.; Lee H.; et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [DOI] [PubMed] [Google Scholar]
- Macindoe G.; Mavridis L.; Venkatraman V.; Devignes M.-D.; Ritchie D. W. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010, 38, W445–W449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar M. J. S. AMPAC: a general program for chemical calculations using procedures developed by the Dewar group. Mod. Tech. Comput. Chem.: MOTECC-91 1991, 455–467. [Google Scholar]
- Hutchinson M. R.; Loram L. C.; Zhang Y.; Shridhar M.; Rezvani N.; et al. Evidence that tricyclic small molecules may possess toll-like receptor and myeloid differentiation protein 2 activity. Neuroscience 2010, 168, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer S. M.; Liu J.; Clayton J. L.; Stephens D. S.; Snyder J. P. Paclitaxel Binding to Human and Murine MD-2. J. Biol. Chem. 2008, 283, 27916–27926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson M. R.; Zhang Y.; Shridhar M.; Evans J. H.; Buchanan M. M.; et al. Evidence that opioids may have toll-like receptor 4 and MD-2 effects. Brain, Behav., Immun. 2009, 24, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Ghosh N.; Slivka P. F.; Fiorini Z.; Hutchinson M. R.; et al. An MD2 Hot-Spot-Mimicking Peptide that Suppresses TLR4-Mediated Inflammatory Response in Vitro and in Vivo. ChemBioChem 2011, 12, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C.; Hayashi T.; Takabayashi K.; Sabet M.; Smee D. F.; et al. Immunotherapeutic activity of a conjugate of a Toll-like receptor 7 ligand. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 3990–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M.; Hayashi T.; Kuy C. S.; Gray C. S.; Wu C. C.; et al. Synthesis and immunological characterization of toll-like receptor 7 agonistic conjugates. Bioconjugate Chem 2009, 20, 1194–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T.; Rao S. P.; Takabayashi K.; Van Uden J. H.; Kornbluth R. S.; et al. Enhancement of innate immunity against Mycobacterium avium infection by immunostimulatory DNA is mediated by indoleamine 2,3-dioxygenase. Infect. Immun. 2001, 69, 6156–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K.; Georgel P.; Rutschmann S.; Mann N.; Du X.; et al. Analysis of the MCMV resistome by ENU mutagenesis. Mamm. Genome 2006, 17, 398–406. [DOI] [PubMed] [Google Scholar]