Abstract
BACKGROUND:
Over the last few decades, obesity, diabetes, and hypertension have become main health evils. The health problems of obesity are well-recognized. However, the fact that all obese individuals are not at the same risk of developing a disease is also recognized. The apolipoprotein B (APOB) plays a central role in lipid metabolism. So we compare the association of APOB XbaI gene polymorphism and lipid profile total in obese north Indian population.
MATERIALS AND METHODS:
A total of 132 obese (body mass index [BMI] >25 kg/m2) and 132 age matched non-obese (BMI ≤ 25 kg/m2) subjects were studied after taking detailed clinical profile. Lipid profile in serum/plasma was done using commercial kits. Genetic analysis of APOB XbaI was done using Polymerase Chain Reaction-Restriction Fragment Leanth polymorphism (PCR-RFLP).
STATISTICAL ANALYSIS:
Statistical analysis was performed by Statistical Package for the Social Sciences (SPSS) (version 11.5) software (IBM Corporation). All continuous variables were expressed as mean ± SD and tested by analysis of variance test. Comparisons of categorical variables were assessed using χ2 tests or Fisher's exact test. P < 0.05 was considered as significant.
RESULTS:
Analysis showed that obese subjects had significantly higher value of the waist-to-hip ratio, blood pressure (systolic and diastolic), and lipid profile. In APOB XbaI gene polymorphism, we did not find significant differences in genotype or allele frequencies. Moreover, none of the studied metabolic parameters (lipid profile) showed any association with the gene polymorphism.
CONCLUSIONS:
Study reveals no considerable association of APOB XbaI gene polymorphism with obesity and lipid profile in north Indians.
Keywords: Body mass index, lipid profile, obesity, polymorphism
Introduction
Obesity has a negative effect on health, being associated with cardiovascular disease, hypertension, and diabetes.[1] Obesity is also associated with adverse changes in plasma lipoprotein metabolism. Various lipid/lipoprotein abnormalities have been observed in obese individuals, including elevated cholesterol, triglycerides (TG), and lower high-density lipoprotein cholesterol (HDL-C) levels. Of these indicators, changes in TG and HDL-C levels are most consistent and pronounced.[2,3]
The apolipoprotein B (APOB) plays a central role in lipid metabolism as the main protein component of very low-density lipoprotein (VLDL) and low-density lipoprotein (LDL). It also serves as the ligand for removal of LDL from the circulation by receptor-mediated endocytosis via the LDL receptor.[4] Variants of the APOB gene may, therefore, be involved in the pathogenesis of obesity.
The APOB gene is located on the short arm of chromosome[2] (2p23-24)[5] and several single nucleotide polymorphisms in the APOB gene have been described, including XbaI[6] and EcoRI.[7] The XbaI polymorphism arises due to a single base variation in exon 26 (at 2488th position ACC→ACT) of the APOB gene that does not lead to change in amino acid sequence.[5] This polymorphism has been found to be associated with inter individual variability of lipid levels, but the results are conflicting.[8] Epidemiological studies have shown an association between the APOB gene polymorphisms with generalized and regional obesity and an increase in various lipoprotein subfractions (total cholesterol [TC], low density lipoprotein cholesterol [LDL-C], and TG), and atherosclerosis[9,10] though most of these studies have been carried out in Caucasian subjects. On the other hand, Liu et al.[11] reported that X(+) allele of APOB gene is a risk factor for the development of gallstone in Chinese patients, in another study no association exists between the APOB XbaI polymorphism and cholelithiasis in Mexican population.[12] Saha et al.[10] observed a significant association of APOB XbaI gene polymorphism with obesity and serum lipid levels while contradictory results also available, Misra et al.[13] reported that APOB XbaI gene polymorphism did not associate with obesity. We have, therefore, attempted to correlate APOB XbaI gene polymorphism with obesity and lipid profile in obese and non-obese subjects of northern Indian population.
Materials and Methods
Subjects
A total of 634 subjects were enrolled initially from the out patients department of Chatrapati Shahuji Maharaj Medical University, Lucknow and volunteers from the general population of Lucknow (North India). Out of these, 88.20% were Hindus (Hindi speaking and residing in Lucknow for at least two generations) while 21.8% belonged to other religions. Individuals of South, East, and Central Indian origin were excluded. A process of disproportionate stratified and systematic sampling was used to select individuals between 19 and 60 years old, oversampling of the majority groups to ensure that prevalence estimates for the majority groups were reliable and to allow statistical comparison. Every individual has been classified as Hindu North Indian (Hindi speaking) depending on self-reported family origin from two generations. Moreover, the possibility of population admixture is slight because in this part of country, inter-religion marriage or consanguinity is rare.
Out of these subjects, only 132 obese (body mass index [BMI] >25 kg/m2) and 132 non-obese (BMI ≤ 25 kg/m2) individual were selected befitting the strict inclusion criteria. All subjects were asked for detailed clinical history, and required measurements were done for height, weight, BMI, and waist-to-hip ratio (WHR). Height was measured to the nearest centimeter using a rigid stadiometer. Weight was measured in minimum clothing to the nearest 0.1 kg using a calibrated balance scale. BMI was calculated as the weight in kilogram divided by meter square of height. Only normotensive (systolic < 130 mmHg; diastolic < 85 mmHg) and non-diabetic (fasting blood sugar < 110 mg/dl) subjects were included. Subjects were considered as normal if they fall in normal ranges of various parameters. For example, leptin = 2-11 mg/ml, serum insulin = 0-30 μU/ml. Only non-smoker, non-diabetic, normotensive subjects who did not have a history of coronary artery disease, neoplasia, congenital and mental disorders, and endocrine disorders such as myxoedema and Cushing syndrome were included. The study was approved by an institutional ethics committee, and informed consent was obtained from each subject in accordance with principles of the declaration of Helsinki.
Sample collection
After an informed consent, overnight fasting blood samples (5 ml) were taken from all subjects. Two milliliter blood was taken in Ethylenediamine tetraacetic acid (EDTA) for analysis of DNA. The genomic DNA was extracted from peripheral blood leucocytes pellet using the standard salting out method (Miller 1988). Remaining 3 ml blood was used for serum/plasma isolation.
Anthropometry
Body weight was measured to the nearest 0.1 kg, and height was measured to the nearest 0.01 m.[14] The waist circumference was measured half way between the lower rib and iliac crest, the hip circumference was measured over the widest part in the gluteal region, and the WHR was calculated.[15]
Biochemical parameters
Lipid profile, total five biochemical parameters namely, TC, TG, HDL, LDL, and VLDL were estimated in obese and non-obese subjects. Fasting blood sugar was assayed by glucose oxidase-peroxidase) method.[16]
APOB XbaI polymorphism
Genomic DNA was isolated from peripheral blood according to standard procedures. The − 866A/G polymorphism in the promoter of human APOB gene was determined by digesting PCR products with restriction enzyme XbaI (Fermantas Inc., USA) as previously described.[17] The primers used were 5’- GGAGACTATTCAGAAGCTAA-3’ as upstream primer and 5’-GAAGAGCCTGAAGACTGACT-3’ as downstream primer.
Quality control
Quality control and assessment was done at every step of the study. The amount of isolated DNA was of good quality (absorbance 260 nm/280 nm, ratio > 1.75). One sample with known genotype and a reagent blank were included after every 20 samples in the PCR. A 50 base-pair marker was included during electrophoresis. Twenty percent of samples from patients and controls including samples of each genotype were re-genotyped by other laboratory personnel and no discrepancy was found.
Statistical analysis
All the statistical calculation for the continuous data of biochemical factors were performed using Statistical Package for the Social Sciences (SPSS) version 11.5 statistical software packages (IBM Corporation). For each variable, the values were expressed as mean ± SD. Data was evaluated by t test and one-way analysis of variance. Allele and genotypic frequencies for APOB XbaI was calculated with the gene counting method. Comparison of the categorical data i.e., different APOB genotypes among controls and obese subjects was done by Fischer's exact test and χ2 test. Odd's ratios were calculated with a 95% confidence interval limit using 2 × 2 contingency table. P < 0.05 was considered significant. The observed genotype frequencies were compared with the expected frequencies to check for the Hardy–Weinberg equilibrium and P < 0.05 was used as the level of significance.
Results
Two hundred sixty four subjects comprising obese (60 male and 72 female) and non-obese (79 male and 53 female) adults were evaluated. The mean BMI of the obese and non-obese subjects was 29.40 ± 4.13 and 21.73 ± 2.11, respectively. In obese subjects, the WHR (P < 0.001), Systolic (P < 0.001) and diastolic (P < 0.002) blood pressures, though in normal range, were significantly higher in obese subjects as compared to non-obese subjects. Fasting blood glucose was significantly higher in obese subjects as compared to non-obese subjects. TC, TG, LDL, and VLDL was also higher (P ≤ 0.001) in obese subjects as compared to non-obese subjects. On the contrary, HDL was significantly higher (P < 0.001) in non-obese as compared to obese subjects [Table 1].
Table 1.
Basic characteristic of study subjects
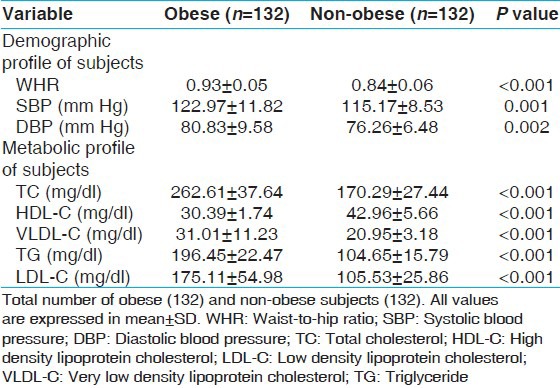
Association of APOB XbaI gene polymorphism with obesity
This polymorphism was in Hardy–Weinberg equilibrium in non-obese population. The allele frequencies were X−, 75.4% versus 71.2%; and X+, 24.6% versus 28.8% in obese and non-obese subjects. The frequencies of X+X+ genotype did not differ significantly in obese and non-obese. Similarly, frequency of X+ allele was not significantly different in the two groups [Table 2].
Table 2.
Association of APOB XbaI gene polymorphism with obesity
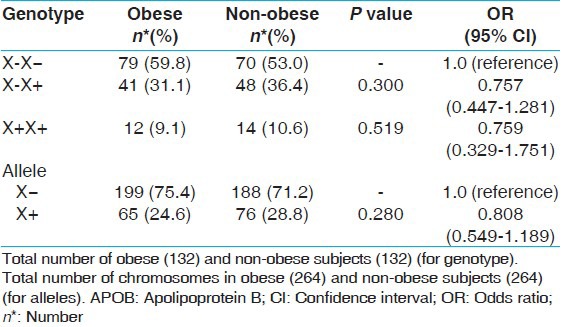
Association of APOB XbaI gene polymorphism with obesity related parameters
In obese and in non-obese subjects, none of the obesity related parameters like lipid levels specially TC, TG, LDL, HDL, and VLDL were associated with genotypes of APOB [Table 3]. The APOB X+ allele also did not show any statistically significant difference with clinical profile of obese or non-obese subjects [Table 4].
Table 3.
Association of APOB XbaI gene polymorphism with obesity related parameters (lipid levels)
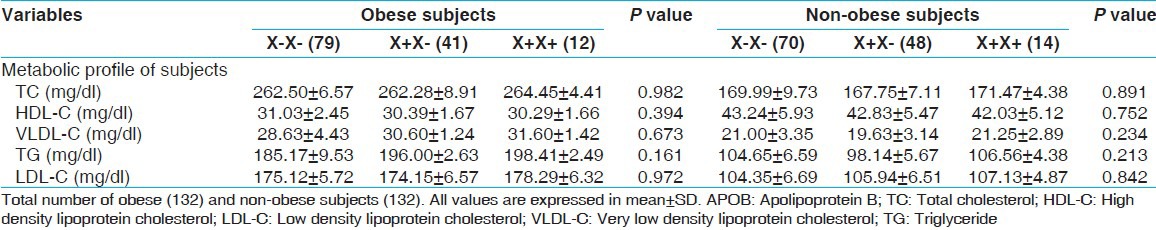
Table 4.
Association of APOB XbaI polymorphisms with obesity related parameters (lipid levels) (X+carrier and non-carrier)
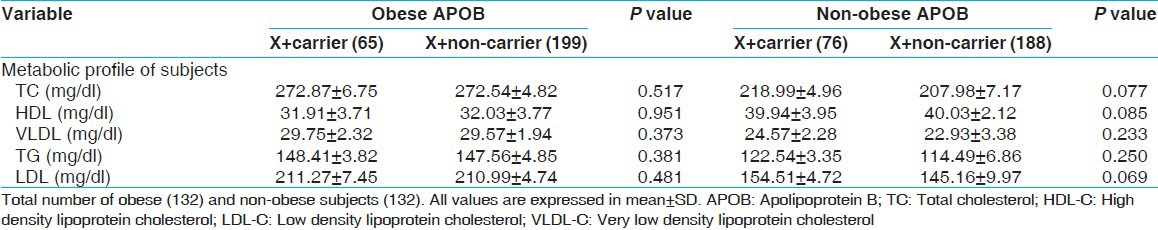
Lipid levels like TC, TG, LDL, HDL, and VLDL were found significantly different between obese and non-obese subjects. When we analyzed the association between genotypes of APOB XbaI gene polymorphism with lipid profile in obese and non-obese subjects, none of the variables associated with APOB XbaI polymorphism [Table 4].
Discussion
The present study examined the genotypic and allelic frequencies and the effect of APOB XbaI polymorphism on obesity and lipid profile in the northern Indian obese and non-obese population. The APOB XbaI polymorphism was selected for the present study by virtue of its documented association with obesity and dyslipidemia in North Indian population.[10] Various studies have shown an association between the APOB gene polymorphisms with lipoprotein subfractions (TC, LDL-C, and TG).[18,19] Asian Indians are prone to develop dyslipidemia and accelerated atherosclerosis.[20] Genetic investigations of the Asian Indian populations established in other part of the world show a correlation of APOB gene polymorphisms with hyperlipidemia.[10]
Present study shows that there was no significant difference in the genotype and allele frequencies of APOB XbaI polymorphism between obese and non-obese subjects. Saha et al.,[19] also reported no significant difference in allelic frequencies of XbaI gene polymorphism of the APOB gene.
The association of the XbaI polymorphism with serum lipid levels has been found in several studies.[21,22,23] On the other hand, we did not find any association of APOB XbaI gene polymorphism with serum lipid levels in obese and non-obese subjects. In the present study, a comparison of clinical variables was also done in relation to the genotypes of the APOB XbaI gene in obese and non-obese subjects but no association was found with serum lipid levels. Our results were in agreement with a previous study,[24] while Saha et al.,[19] reported no association between the APOB XbaI gene and serum lipids levels.
On the other hand it appears that APOB XbaI polymorphism exhibits population specific variation, which may be due to gene and environment interactions. APOB XbaI polymorphism does not lead to changes in the amino acid sequence and cannot be implicated at structure level. It is possible that some other polymorphism in its vicinity might be present, which is in linkage disequilibrium with APOB XbaI polymorphism and accountable for the observed association with obesity and lipid levels in other studies. The effect of APOB XbaI polymorphism on the lipid levels is due to linkage disequilibrium with ins/del polymorphism, which causes an amino acid change in the signal peptide of the APOB gene. Saha et al.,[10] showed a strong disequilibrium between ins/del polymorphism and the XbaI polymorphism of APOB gene. Moreover, gene–environment interaction may also responsible for the inconsistency of data due to differences in the diet and lifestyle of populations of various part of the world.
In summary, there was no evidence of association of APOB polymorphisms (XbaI) with obesity and serum lipid levels. There may be several reasons for the differences observed in the various studies. The populations studied by in different studies were genetically different.[9,10,19] In view of the strong associations reported in some studies, studies are needed with a larger sample size to study association of XbaI polymorphism with obesity.
Acknowledgments
Authors acknowledge Indian Council of Medical Research, DBT, New Delhi and intramural grant from KGMU UP, Lucknow – 226 003, India (Formerly Chatrapati Shahuji Maharaj Medical University, Lucknow, Uttar Pradesh, India) for the financial support to carry out this research work.
Footnotes
Source of Support: Indian Council of Medical Research, DBT, New Delhi and intramural grant from KGMU UP, Lucknow – 226 003, India (Formerly Chatrapati Shahuji Maharaj Medical University, Lucknow, Uttar Pradesh, India)
Conflict of Interest: None declared.
References
- 1.Larsson B. Obesity, fat distribution and cardiovascular disease. Int J Obes. 1991;2:53–7. [PubMed] [Google Scholar]
- 2.Freedman DS, Serdula MK, Percy CA, Ballew C, White L. Obesity, levels of lipids and glucose, and smoking among Navajo adolescents. J Nutr. 1997;127:2120S–7. doi: 10.1093/jn/127.10.2120S. [DOI] [PubMed] [Google Scholar]
- 3.Depres JP. Obesity and lipid metabolism: Relevance of body fat distribution. Curr Opin Lipidol. 1991;2:7–15. [Google Scholar]
- 4.Goldstein JL, Brown MS. Familial hypercholesterolemia. In: Stanbury JB, Wyngaarden JB, Fredrickson DS, Goldstein JL, Brown MS, editors. The Metabolic Basis of Inherited Disease. 5th ed. New York, NY: McGraw-Hill; 1983. pp. 672–712. [Google Scholar]
- 5.Carlsson P, Darnfors C, Olofsson SO, Bjursell G. Analysis of the human apolipoprotein B gene; complete structure of the B-74 region. Gene. 1986;49:29–51. doi: 10.1016/0378-1119(86)90383-5. [DOI] [PubMed] [Google Scholar]
- 6.Priestley L, Knott T, Wallis S, Powell L, Pease R, Brunt H, et al. RFLP for the human apolipoprotein B gene: V; XbaI. Nucleic Acids Res. 1985;13:6793. doi: 10.1093/nar/13.18.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang LS, Miller DA, Bruns GA, Breslow JL. Mapping of the human APOB gene to chromosome 2p and demonstration of a two-allele restriction fragment length polymorphism. Proc Natl Acad Sci USA. 1986;83:644–8. doi: 10.1073/pnas.83.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Régis-Bailly A, Visvikis S, Steinmetz J, Feldmann L, Briançon S, Danchin N, et al. Frequencies of five genetic polymorphisms in coronarographed patients and effects on lipid levels in a supposedly healthy population. Clin Genet. 1996;50:339–47. doi: 10.1111/j.1399-0004.1996.tb02385.x. [DOI] [PubMed] [Google Scholar]
- 9.Renges HH, Wile DB, McKeigue PM, Marmot MG, Humphries SE. Apolipoprotein B gene polymorphisms are associated with lipid levels in men of South Asian descent. Atherosclerosis. 1991;91:267–75. doi: 10.1016/0021-9150(91)90174-2. [DOI] [PubMed] [Google Scholar]
- 10.Saha N, Tay JS, Heng CK, Humphries SE. DNA polymorphisms of the apolipoprotein B gene are associated with obesity and serum lipids in healthy Indians in Singapore. Clin Genet. 1993;44:113–20. doi: 10.1111/j.1399-0004.1993.tb03861.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu FL, Lu WB, Niu WX. XbaI polymorphisms of apolipoprotein B gene: Another risk factor of gallstone formation after radical gastrectomy. World J Gastroenterol. 2010;16:2549–53. doi: 10.3748/wjg.v16.i20.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez-Cuén J, Aguilar-Medina M, Arámbula-Meraz E, Romero-Navarro J, Granados J, Sicairos-Medina L, et al. ApoB-100, ApoE and CYP7A1 gene polymorphisms in Mexican patients with cholesterol gallstone disease. World J Gastroenterol. 2010;16:4685–90. doi: 10.3748/wjg.v16.i37.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misra A, Nishanth S, Pasha ST, Pandey RM, Sethi P, Rawat DS. Relationship of XbaI and EcoRI polymorphisms of apolipoprotein-B gene to dyslipidemia and obesity in Asian Indians in North India. Indian Heart J. 2001;53:177–83. [PubMed] [Google Scholar]
- 14.Bray GA. An approach to the classification and evaluation of obesity. In: Björntorp P, Brodoff BN, editors. Obesity. Philadelphia: J.B. Lippincott; 1992. pp. 294–308. [Google Scholar]
- 15.Pouliot MC, Després JP, Lemieux S, Moorjani S, Bouchard C, Tremblay A, et al. Waist circumference and abdominal sagittal diameter: Best simple anthropometric indexes of abdominal visceral adipose tissue accumulation and related cardiovascular risk in men and women. Am J Cardiol. 1994;73:460–8. doi: 10.1016/0002-9149(94)90676-9. [DOI] [PubMed] [Google Scholar]
- 16.Young DS, Pestaner LC, Gibberman V. Effects of drugs on clinical laboratory tests. Clin Chem. 1975;21:1D–432. [PubMed] [Google Scholar]
- 17.Han T, Jiang Z, Suo G, Zhang S. Apolipoprotein B-100 gene XbaI polymorphism and cholesterol gallstone disease. Clin Genet. 2000;57:304–8. doi: 10.1034/j.1399-0004.2000.570410.x. [DOI] [PubMed] [Google Scholar]
- 18.Friedlander Y, Kaufmann NA, Cedar H, Weinberg N, Kark JD. The role of XbaI polymorphism of the apolipoprotein B gene in determining levels and covariability of lipid and lipoprotein variables in a sample of Israeli offspring with family history of myocardial infarction. Atherosclerosis. 1993;98:165–77. doi: 10.1016/0021-9150(93)90126-f. [DOI] [PubMed] [Google Scholar]
- 19.Saha N, Tay JS, Humphries SE. Apolipoprotein B-gene DNA polymorphisms (XbaI and EcoRI), serum lipids, and apolipoproteins in healthy Chinese. Genet Epidemiol. 1992;9:1–10. doi: 10.1002/gepi.1370090103. [DOI] [PubMed] [Google Scholar]
- 20.Misra A, Reddy RB, Reddy KS, Mohan A, Bajaj JS. Clustering of impaired glucose tolerance, hyperinsulinemia and dyslipidemia in young north Indian patients with coronary heart disease: A preliminary case-control study. Indian Heart J. 1999;51:275–80. [PubMed] [Google Scholar]
- 21.Law A, Wallis SC, Powell LM, Pease RJ, Brunt H, Priestley LM, et al. Common DNA polymorphism within coding sequence of apolipoprotein B gene associated with altered lipid levels. Lancet. 1986;1:1301–3. doi: 10.1016/s0140-6736(86)91222-5. [DOI] [PubMed] [Google Scholar]
- 22.Talmud PJ, Barni N, Kessling AM, Carlsson P, Darnfors C, Bjursell G, et al. Apolipoprotein B gene variants are involved in the determination of serum cholesterol levels: A study in normo- and hyperlipidaemic individuals. Atherosclerosis. 1987;67:81–9. doi: 10.1016/0021-9150(87)90267-x. [DOI] [PubMed] [Google Scholar]
- 23.Berg K. DNA polymorphism at the apolipoprotein B locus is associated with lipoprotein level. Clin Genet. 1986;30:515–20. doi: 10.1111/j.1399-0004.1986.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 24.Delghandi M, Thangarajah R, Nilsen M, Grimsgaard S, Bønaa KH, Tonstad S, et al. DNA polymorphisms of the apolipoprotein B gene (XbaI, EcoRI, and MspI RFLPs) in Norwegians at risk of atherosclerosis and healthy controls. Acta Cardiol. 1999;54:215–25. [PubMed] [Google Scholar]


