Abstract
BACKGROUNDS:
Helicobacter pylori colonize the gastric mucosa of half of the world's population. Although it is classified as a definitive type I carcinogen by World Health Organization, there is no effective vaccine against this bacterium. H. pylori evade the host immune response by avoiding toll-like detection, such as detection via toll-like receptor-5 (TLR-5). Thus, a chimeric construct consisting of selected epitopes from virulence factors that is incorporated into a TLR-5 ligand (Pseudomonas flagellin) could result in more potent innate and adaptive immune responses.
MATERIALS AND METHODS:
Based on the histocompatibility antigens of BALB/c mice, in silico techniques were used to select several fragments from H. pylori virulence factors with a high density of B- and T-cell epitopes.
RESULTS:
These segments consist of cytotoxin-associated geneA (residue 162-283), neutrophil activating protein (residue 30-135) and outer inflammatory protein A (residue 155-268). The secondary and tertiary structure of the chimeric constructs and other bioinformatics analyses such as stability, solubility, and antigenicity were performed. The chimeric construct containing antigenic segments of H. pylori proteins was fused with the D3 domain of Pseudomonas flagellin. This recombinant chimeric gene was optimized for expression in Escherichia coli. The in silico results showed that the conserved C- and N-terminal domains of flagellin and the antigenicity of selected fragments were retained.
DISCUSSION:
In silico analysis showed that Pseudomonas flagellin is a suitable platform for incorporation of an antigenic construct from H. pylori. This strategy may be an effective tool for the control of H. pylori and other persistent infections.
Keywords: Cytotoxin-associated gene A, Helicobacter pylori, multi-epitope vaccine, neutrophil activating protein, outer inflammatory protein A
Introduction
Approximately half of the world's population carry Helicobacter pylori in their upper gastrointestinal tract, with those in developing countries being the most affected.[1] Infection with this bacterium can lead to several clinical outcomes ranging from gastritis and duodenal ulcer to gastric cancer.[2,3] This infection usually affects the human stomach in childhood and persists for a lifetime, with the majority of infected persons (80-90%) remaining asymptomatic.[4] Furthermore, H. pylori was the first bacterium to be classified as a carcinogen by the World Health Organization.[5] Although the current multi-drug treatment is very effective at eradicating H. pylori, with cure rates higher than 90%, there remains a serious problem of increasing antibiotic resistance and reinfection with H. pylori after antimicrobial therapy, which occurs frequently in countries where infection rates are high.[6] Therefore, immunization against H. pylori, both to prevent and to treat infection, appears to be a better approach to control this widespread infection.[7] Great progress has been made in understanding the details of H. pylori pathogenesis and the role of its virulence factors in gastric disease and cancer. It has been shown that the main virulence factors, such as cytotoxin-associated gene A (CagA), neutrophil activating protein (Nap), and outer inflammatory protein A (OipA or HopH), in their native or recombinant forms can confer protection against H. pylori challenge in animal models.[2] CagA is the most important virulence factor associated with peptic ulcer or gastric adeno-carcinoma.[8] CagA-positive strains are associated with more severe clinical outcomes, especially in Western countries.[9] Nap is associated with severe gastro-duodenal mucosal injury and gastric cancer.[10] This protein (a toll-like receptor-2 [TLR-2] agonist) is able to elicit innate immunity and the production of interleukin IL-12 and IL-23, which promote a Th1 immune response,[11,12] and is therefore, a strong candidate as part of a multi-component recombinant vaccine against H. pylori.[13] OipA is another important protein that may be linked to gastro-duodenal diseases because of its association with other virulence factors, such as CagA[14] and associations with increased bacterial colonization.[15] Immunological studies have shown that H. pylori infection leads to a Th1 immune response but that this response is inadequate to eradicate the organism and as result, infection can persist. Insufficient adaptive immunity can be explained by incomplete activation of TLRs, suppressive effects of bacterial factors and, finally, the induction of regulatory T-cell responses.[16] Recent advances in the field of innate immunity have resulted in the identification of the TLR receptor family and their pathogen associated molecular pattern (PAMP) agonists.[17,18] Flagellin, as a PAMP (TLR-5 agonist), can polarize the immune response towards a Th1 response and lead to adjuvant properties and effects on dendritic cells.[19,20] A major role for TLR-5 in the innate immune response can be excluded by the fact that H. pylori flagellins have very low intrinsic activity to trigger TLR-5 receptors.[21,22] Because it has been proven that the Th1 immunity is necessary for protection and clearance of H. pylori,[23] vaccine research has been focused on induction of Th1 immunity. It has been demonstrated that although Th2 promoting vaccines induced strong systemic and local immune responses, only Th1 promoting vaccines are protective.[16] The intracellular location of H. pylori strengthens the argument that a Th1 immune response is necessary to eradicate infection.[24,25] Classical protocols have relied on a mixture of both antigen and adjuvant, but the disadvantage of this strategy was that the real-time loading and activation of antigen presenting cells (APCs) was not guaranteed. Due to the nature of the flagellin protein, it can be encoded into a protein fusion combination providing both constituents to the same APCs to generate a homogeneous antigen-presenting and activated cell population.[26] It was demonstrated that the immune response induced by H. pylori did not efficiently clear these bacteria.[16] Therefore, it is favorable to trigger the immune response in a way that differs from natural infection. For this purpose a newly designed antigen that differs from the naturally occurring antigen should be constructed.[27] Epitope-based vaccines represent a new strategy for generating a specific immune response and avoiding the side-effects of unfavorable epitopes in the complete antigen. Furthermore, an epitope-based immunogen could also include single antigenic molecules combined from different epitopes for increased potency.[28,29] Moreover, it has been shown that the immunogenicity of predefined epitopes is significantly increased by this strategy[30] for tuberculosis bacilli[31] and influenza virus.[32] Here, we designed a new chimeric construct containing the flagellin protein, replacing its D3 domain with a combination of selected antigenic segments of H. pylori virulence factors (CagA, Nap, and OipA) directly fused together. For efficient expression in an Escherichia coli host, this synthetic construct gene was analyzed by bioinformatics algorithms. Finally, a novel in silico approach was used to analyze the structural characteristics of this chimeric protein.
Materials and Methods
Sequence analysis
Related sequences of H. pylori virulence factors (CagA, Nap, and OipA) and P. aeroginosa flagellin type A (flaA) were obtained from GenBank. Using Clustal W software http://www.ebi.ac.uk/Tools/clustalw2/, multiple sequence alignment was performed to identify the common and conserved antigenic fragments in putative H. pylori strain sequences.
Segment selection in virulence factors
Based on BALB/c histocompatibility molecules, all virulence factor sequences were analyzed separately using DNASTAR (DNAstar, Inc. www.dnastar.com) to identify the best segments containing both B and T-cell epitopes. Furthermore, the presence of appropriate epitopes was confirmed with other web-based B- and T-cell epitope prediction algorithms in single or assembled forms of selected segments.
Prediction of immunogenic epitopes
B-cell epitopes
The amino acid sequences were analyzed to predict continuous and discontinuous B cell epitopes using the Bcepred program (http://www.imtech.res.in/raghava/bcepred)[33] and the Disco Tope 1.2 server (http://www.cbs.dtu.dk/services/DiscoTope/), respectively.[34]
T-cell epitopes prediction based on BALB/c mice major histocompatibility molecules (H2d)
Among different haplotypes of major histocompatibility molecules (MHC) in mice, those of the BALB/c mice harboring H2d were selected. These alleles contain I-Ad and I-Ed of class II and H2-Kd, H2-Ld, H2-Dd of class I MHC.[35]
Based on position specific scoring matrices or profiles from sets of aligned peptides to bind to given histocompatibility molecules, analyses of peptides related to H2 class II sequences were performed with the web-based program Rankpep (http://imed.med.ucm.es/Tools/rankpep.html). Similarly, the cytotoxic-T lymphocyte epitopes (CTL) related to H2 class-I was predicted with the web-based CTL-Pred program (http://www.imtech.res.in/raghava/ctlpred).[36]
CagA-Nap-OipA constructs design
The H. pylori antigenic construct (HAC) was assembled by fusing three selected segments into a single engineered molecule without any linker. The N-terminal of cagA (NP_207343.1), middle fragment of Nap (NP_207041.1) and C-terminal of OipA (AAQ57665.1) segments were linked together. The synthetic construct was deposited into GenBank (Accession No.: 1343925).
HAC design, replacement with the D3 domain and codon optimization
Upon three-dimensional (3D) study of Pseudomonas flagellin and homology comparison with Salmonella flagellin by protein visualizer, the D3 domain of this molecule was predicted and replaced with the antigenic construct (HAC) to develop complete flaA-HAC.
The designed construct was optimized with the Genscript Optimum Gene8482; algorithm (www.genescript.com, Piscataway, New Jersy, USA). The chimeric gene was prepared for cloning and expression in the E. coli prokaryotic system.
Bioinformatics analysis of chimeric protein
The messenger RNA secondary structure of the designed construct was predicted by the centroidfold program[37] and was compared before and after gene optimization. The prediction of the secondary structure of the recombinant protein was performed using the Advanced Protein Secondary Structure Prediction Server 2 (http://www.imtech.res.in/raghava/apssp2/) and GOR-IV.[38] For prediction of 3D structure of each selected segment and the complete designed construct, several ab-initio online programs such as 3Dpro http://scratch.proteomics.ics.uci.edu/ and Robetta http://robetta.bakerlab.org/submit.jsp were used. The 3D protein models calculated by comparative modeling such as Mod-Base http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi and LOOPP server (cbsuapps.tc.cornell.edu/loopp) were used. The stability of the 3D structure of the synthetic protein was further analyzed by swiss-PDB viewer for energy minimization. Prediction of solubility upon overexpression[39] and antigenicity of recombinant proteins were performed with online programs such as SOLpro and ANTIGENpro http://scratch.proteomics.ics.uci.edu/.
Results
Antigenic segment selection
The alignment of CagA, Nap and OipA amino acid sequences of different H. pylori strains resulted in the identification of three conserved segments. DNAstar software analysis primarily showed they were contained within both the B- and T-cell epitopes [Figure 1]. These segments, covering 342 residues, consist of 122 residues of the N-terminal portion of CagA (162-283), 106 residues of the middle portion of Nap (30-135) and 114 residues of the C-terminal portion of OipA (155-268). Without using any extra amino acids, these fragments were directly linked together to produce the synthetic HAC.
Figure 1.
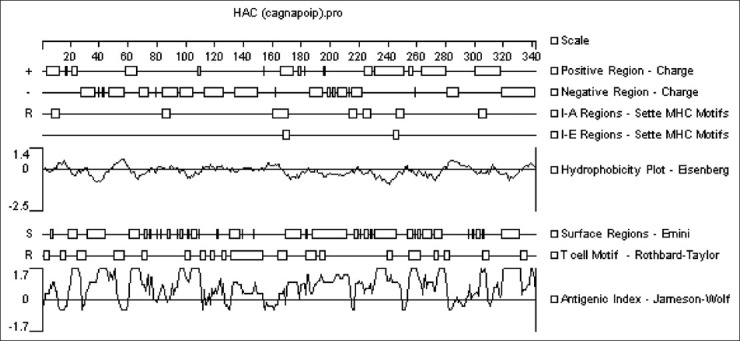
Analysis of the H. pylori antigenic construct by DNAstar software (see the text for details)
HAC replacement with D3 domain of flaA
There are four conserved domains consisting of D0, D1, D2, and D3 in the P. aeroginosa flaA structure. The conserved D0 domain consists of the C- and N-terminal of the flagellin hairpin molecule and has been reported as the agonist of TLR-5, and the D3 domain is a variable region. Upon 3D structural prediction of Pseudomonas flaA (Accession No.: Gu060499) and homology comparison with Salmonella flagellin (Accession No.: P06179), it was clear that the D3 domain in flaA molecule consists of 50 amino acid residues, which were replaced with the HAC. The schematic arrangement of the chimeric flaA-HAC molecule is shown in Figure 2.
Figure 2.

Schematic representation of the H. pylori antigenic construct consisting of flagellin type A and the H. pylori antigenic construct fragment. The lengths of the amino acid residues are listed in parentheses
Secondary and tertiary structure prediction
Secondary structure was predicted by several on line programs, and the best result was achieved by GOR-IV, as shown in Figure 3. The accuracy and quality of the model depends on two factors: First, the sequence alignment of query and template proteins and, second, the structures of the loop regions.
Figure 3.
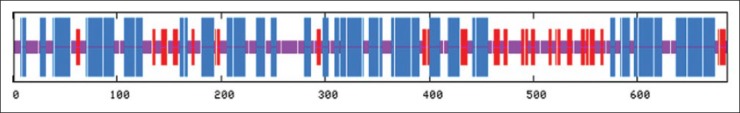
Secondary structure analysis of the flagellin type A/H. pylori antigenic construct protein helix: Blue, sheet: Red, coil: Violet
Three hundred models were predicted by LOOPP (learning, Observing and Outputting Protein Pattern) online software. Based on the analysis of the structures, two models were selected. Further analysis of selected models by Discovery Studio viewer showed two essential C- and N-terminals of the alpha helix structures (D0) that are needed for TLR-5 interaction and were retained in these models. Moreover, the HAC replacement within the D3 domain has no effect on the conformation of the C- and N-terminal domains of the flaA molecule [Figure 4].
Figure 4.
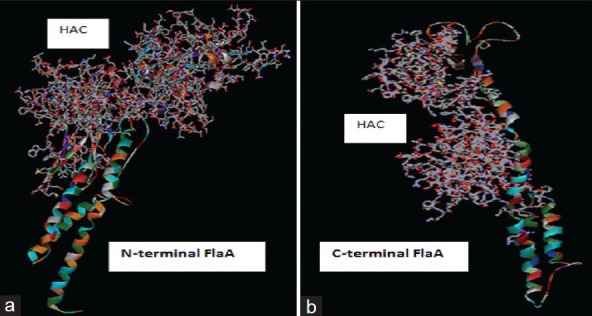
The position of the replaced H. pylori antigenic construct and flagellin segments in models predicted by LOOPP server and visualized by Discovery Studio viewer. (a) Intact N-terminus of flagellin type A (flaA) (183 residues) attached to the complete H. pylori antigenic construct construct and only a small fragment of C-terminal flaA (1-68 out of 163 residues). (b) The incomplete H. pylori antigenic construct consisting of a partial cytotoxin-associated gene A fragment (64-122) and intact C-terminus of flaA
Model stability for flaA-HAC
The stability of the model was first assessed by the computation of a Ramachandran plot based on each residue and then by energy minimization based on the total energy of the model. The majority of residues are in the stable region in the Ramachandran plot, which is shown in Figure 5.
Figure 5.
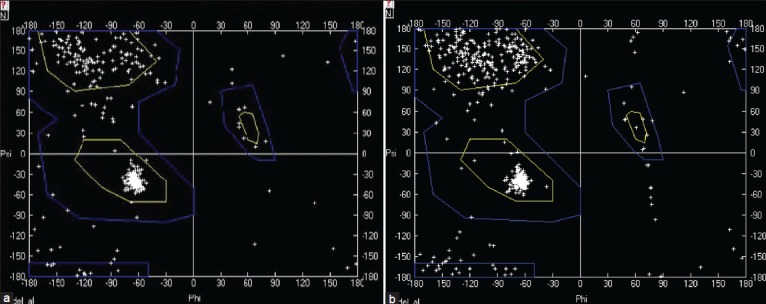
Ramachandran plot for two selected flagellin type A- H. pylori antigenic construct models. The a and b models are the same as those presented in Figure 4
Prediction of B- and T- cells epitopes for HAC
The predicted epitopes were candidates for inducing humoral or cell-mediated immunity against H. pylori in a BALB/c mouse model. For continuous B-cell epitope prediction, several physico-chemical properties of the HAC sequence, including hydrophilicity, flexibility, accessibility, exposed surface, polarity, and antigen propensity were analyzed [Table 1]. The results showed there were no continuous epitope sites in the junction of different fragments. Furthermore, with the Disco Tope server and based on model A [Figure 4], the discontinuous B-cell epitopes were predicted [Table 2]. In comparison with model B [Figure 4], the Nap segment in the model A shows more epitope pattern similarity with the standard NapA protein (PDB code: 1JI4). Based on SOLpro and ANTIGENpro software, the solvent accessibility and antigenicity were estimated with probabilities of 0.51 and 0.89, respectively.
Table 1.
HAC continuous B- cell epitopes prediction in flaA-HAC chimeric protein based on physical and chemical properties by Bcepred
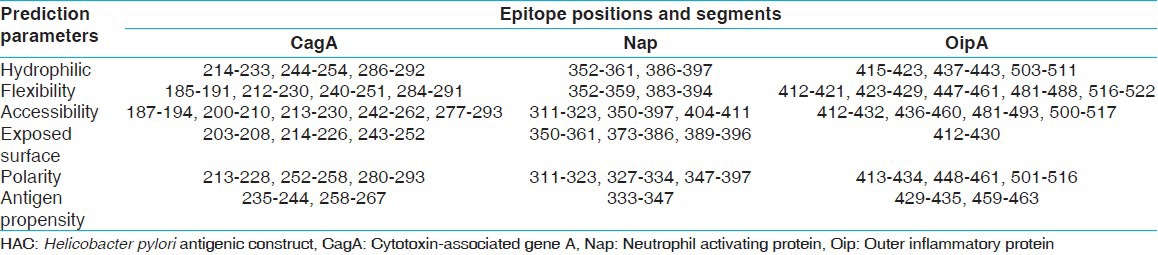
Table 2.
HAC discontinuous B-cell epitopes prediction in flaA-HAC chimeric protein by Discotope server
The HAC sequence was analyzed, and the T-cell epitopes sites related to H2 class II [Table 3] and class I [Table 4] of BALB/c mice histocompatibility molecules were predicted.
Table 3.
HAC T- cell epitopes prediction based on BALB/c histocompatibility molecules by rankpep
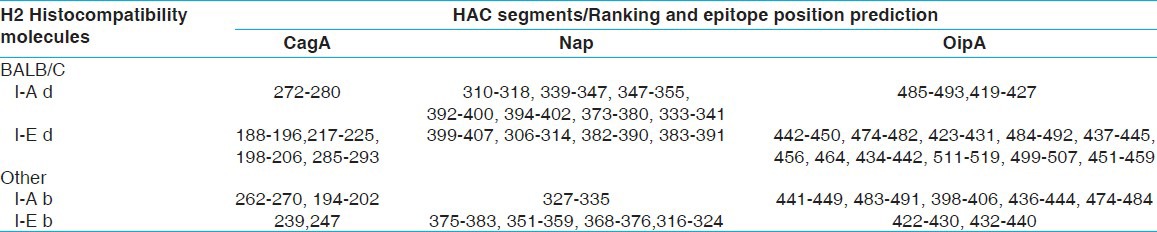
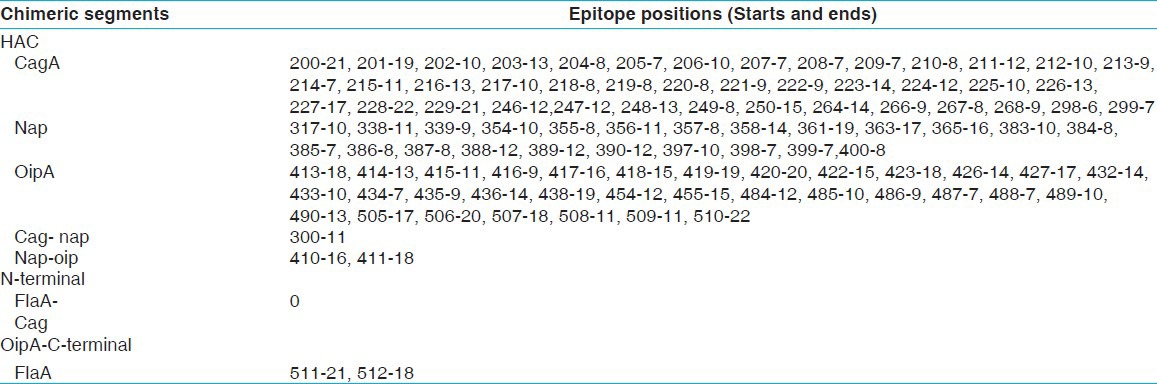
Table 4.
HAC cytotoxicity T-cell epitopes prediction based on different histocompatibility molecules by CTLpred and nHLApred
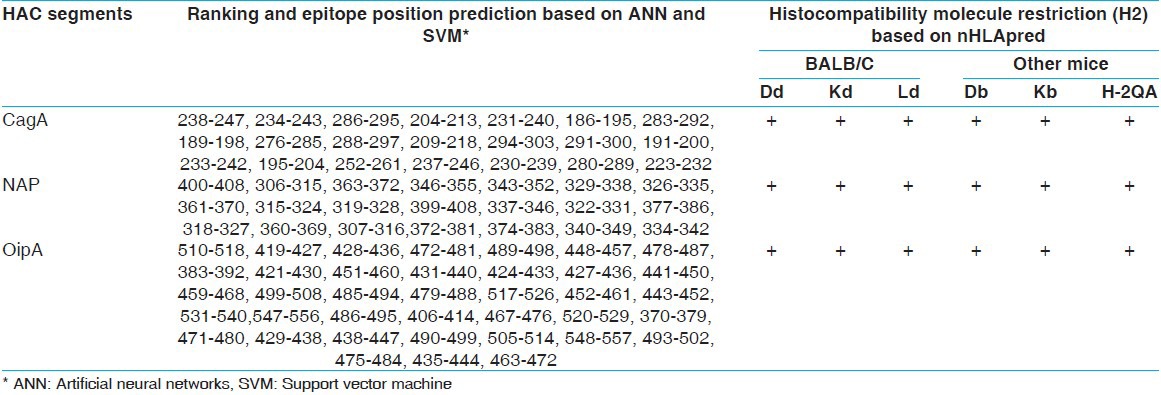
Optimization and synthesis of chimeric construct
The sequence encoding the flaA-HAC chimeric gene was optimized by changing a variety of factors that could regulate and influence gene expression.
The analysis of the wild-type gene and the optimized synthetic chimera is shown in Figure 6.
Figure 6.
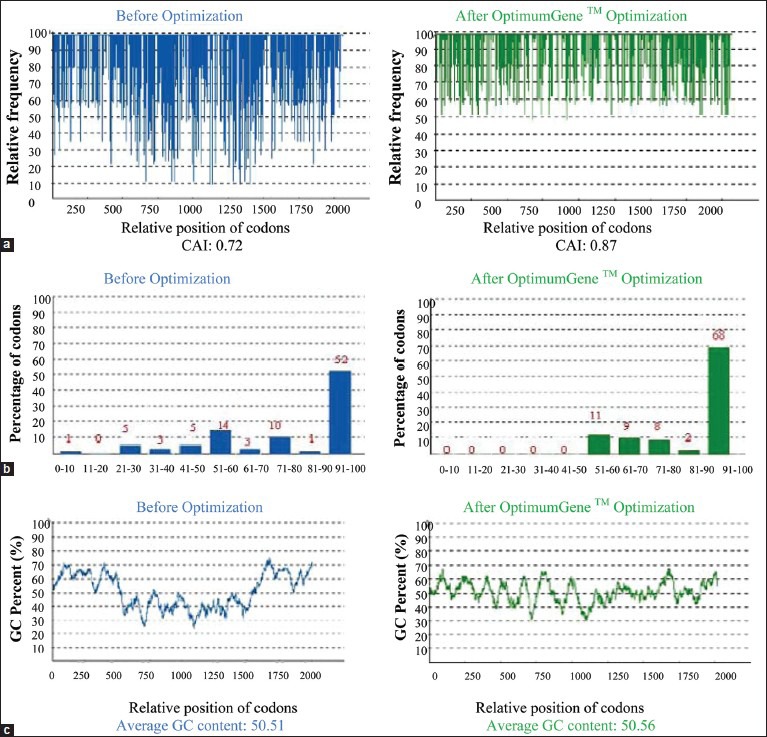
The sequence of the flagellin type A - H. pylori antigenic construct gene was optimized by changing some factors to increase gene expression. (a) Codon adaptation index, (b) frequency of optimal codons, and (c) G+C content adjustment
The codon usage bias in E. coli was increased by upgrading the codon adaptation index (CAI) to 0.87. The GC (guanine plus cytosine) content and unfavorable peaks have been optimized to prolong the half-life of the mRNA. The Stem-Loop structures, which impact ribosomal binding and stability of mRNA, were changed. In addition, the optimization process has screened and successfully modified any negative cis-acting sites.
In brief, within the synthetic construct, the splice sites, instability elements and all the cis-acting sites that may have a negative influence on the expression rate were removed. Furthermore, the necessary restriction enzyme sites (BamHI and HindIII) were introduced at the ends of the sequence for subsequent cloning [Figure 6].
Discussion
Most investigations in vaccine candidate development against H. pylori involve whole bacteria lysates,[40,41] recombinant purified subunit antigens,[41] and DNA vaccines, but no major breakthroughs have been achieved. Recently, the focus of vaccine development has shifted to multi-epitope vaccines as a new strategy to control certain severe and chronic infectious diseases, such as AIDS,[42] meningitis,[43] malaria,[44] tuberculosis,[31] and hemorrhagic diarrhea.[45] Potential advantages of epitope-based vaccines could result in this type of vaccine being an effective strategy to control H. pylori. Earlier studies revealed that many bacterial flagellins had adjuvant behavior for inducing humoral and cellular immunity.[46] Biochemical analysis of flagellin showed that conserved domains, D1 and D2,[47] in the amino and carboxy, terminus were sufficient for TLR-5 receptor activation and concomitant nuclear factor kappa B signaling, which resulted in expression of cytokines, such as tumor necrosis factor-α, chemokine including IL-8 and free radicals such as nitric-oxide. Moreover, the hyper variable domain (D3) of flagellin was not involved in activation of this receptor.[48] It was shown that the leucine-rich region of TLR-5 containing amino acid residues 386-407 was a likely binding site for flagellin.[49]
In this study, special emphasis was placed on designing a new single construct that consisted of Pseudomonas flaA as an adjuvant and H. pylori antigens (HAC) to elicit an innate and adaptive response. Based on mechanisms of antigen selection in dendritic cells,[50,51] the fusion of adjuvant-antigen in this construct is a crucial advantage for simultaneous loading and activation of professional APCs, which could potentially create the optimal immune response. Moreover, many studies have shown that bacterial flagellin was a good platform[52] for B- and T-cell epitopes to elicit cross presentation of antigen.[26,53,54,55,56]
In this design, the HAC construct was replaced with the D3 domain of the Pseudomonas flaA. The 3D structure predictions confirmed that the HAC created separate domains in the flagellin structure. Moreover, the conserved N- and C-terminal segments of the molecule, which are necessary for TLR-5 activation, was retained [Figure 2]. Recent evidence in support of this design demonstrated that the D3 domain of flagellin was structurally independent and that its replacement with other antigen such as the hemagglutinin of swine influenza[57] virus did not interfere with TLR-5 activation and was more efficient than other domain fusions. In addition to deficient intrinsic TLR-5 activity of flagellin in H. pylori,[22] the secretory protein VacA (vacuolating cytotoxin A) suppresses host adaptive immunity, so this virulence factor was not incorporated into the HAC antigen.[23,58] This construct contains the main virulence factors that are involved in the pathogenesis of H. pylori. CagA, as an intracellular bacterial cytotoxin, is injected into host epithelial cells by a bacterial type IV secretory system. This antigen would be expressed by MHC class I molecules to induce a cell-mediated immune response. In this design, although we used recombinant cagA as an external antigen, which is mainly presented by the MHC class II molecule, integration of this protein into flagellin would increase cross-presentation and result in increase in MHC class I presentation. The highly conserved immunogenic amino terminal of CagA was the subject of segment selection in this study. Nap, as a ferritin like protein, has important roles in attachment to the carbohydrates of the host cell membrane matrix. The induction of an immune response against the immunogenic segment of this protein may result in effective protection by preventing bacterial host cell attachment[59] and induction of a Th1 immune response.
The last virulence factor was OipA. The evidence suggests that OipA participates in bacterial colonization[15] and adherence. This coupled with its association with other virulence factors suggested including the OipA segment as a component of the HAC.[15] For some limitations in gene expression, such as the size of synthetic gene and the capacity of a heterologous host to express a recombinant protein, only small segments of CagA, Nap, and OipA with the highest density of B- and T-cell epitopes were selected. In the flaA-HAC, the Ab-initio (data not shown) and homology structural prediction confirmed the presence of a random coil strand in the N- and C-terminus of each selected segment, which caused the effective separation of different domains. Because of the presenting N- and C-terminal coil strand in each segment, three selected fragments were linked directly in the synthetic HAC. The analysis also showed that three distinct parts were developed completely in this chimeric protein. The CagA-Nap-OipA combination presented in Figure 2 was more stable than any other possible arrangement of the three segments (Data not shown). Both the Nap segment in the HAC and the D2 domain of flaA revealed an α-helix structure. To avoid the linking between these two α-helices and to maintain the flagellin structure, the Nap segment was located between CagA and OipA as the central fragment of the HAC.
The α-helical structure of the Nap segment suggested it as the central fragment in the HAC. Using this strategy, the linking of the two α helix structures related to this segment and the D2 domain of flagellin was avoided, and the flagellin structure was maintained. The accuracy of the construct was also supported by the epitope prediction results. There were no undesired continuous B- and T-cell epitopes in the different segment junctions and only a few undesired conformational epitopes, including one epitope in the CagA-Nap junction and two other epitopes in each of the Nap-OipA and OipA-amino terminal flaA junctions. It is possible that these undesired epitopes could raise a few nonspecific immune responses, but they have no effect on protection against H. pylori infection. The presence segments with differing G+C percentages and codon usage variations could reduce the overall expression level of the HAC in an E. coli host. Therefore, the chimeric flaA-HAC was codon optimized. To that end, the CAI, G+C percent and half-life of the mRNA were also analyzed. Additionally, the stem-loop structures of the mRNA that impact ribosomal binding and mRNA stability were modified, and negative cis-acting sites were avoided [Figure 6]. The prediction of the minimum free energy of the chimeric protein compared to its original structure showed that the protein had adequate stability. This prediction study also revealed that the chimeric construct had the potential to induce humoral and cellular (CD4+ and CD8+) immune responses against this pathogen in the BALB/c model.[60] These responses were demonstrated to be the crucial factors for protective immunity against this pathogen.[16]. In addition, some of the predicted HAC epitopes [Table 1] could be presented to other mouse and human immune systems. This potential may be valuable for a heterogeneity study of this construct.[60]
In conclusion, to overcome the persistent infection, induction of targeted innate and adaptive immunity using a novel strategy is essential. The synthetic chimeric polytope containing several B- and T-cell epitopes appears to be a convincing method for this purpose. Here, we have described multi-epitopes of several H. pylori virulence factors fused to Pseudomonas flagellin as a TLR-5 agonist for effective induction of innate and adaptive responses against this pathogen in an animal model. This strategy may be a useful tool to control persistent infection by this bacterium.
Footnotes
Source of Support: Tarbiat modares University
Conflict of Interest: None declared.
References
- 1.Domínguez-Bello MG, Pérez ME, Bortolini MC, Salzano FM, Pericchi LR, O, et al. Amerindian Helicobacter pylori strains go extinct, as European strains expand their host range. PLoS One. 2008;3:e3307. doi: 10.1371/journal.pone.0003307. Zambrano-Guzmán. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Giudice G, Malfertheiner P, Rappuoli R. Development of vaccines against Helicobacter pylori. Expert Rev Vaccines. 2009;8:1037–49. doi: 10.1586/erv.09.62. [DOI] [PubMed] [Google Scholar]
- 3.Pellicano R, Franceschi F, Saracco G, Fagoonee S, Roccarina D, Gasbarrini A. Helicobacters and extragastric diseases. Helicobacter. 2009;14:58–68. doi: 10.1111/j.1523-5378.2009.00699.x. [DOI] [PubMed] [Google Scholar]
- 4.Ramírez Ramos A, Sánchez Sánchez R. Helicobacter pylori 25 years after (1983 -2008): Epidemiology, microbiology, pathogenics, diagnostics and treatment. Rev Gastroenterol Peru. 2009;29:158–70. [PubMed] [Google Scholar]
- 5.Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–23. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Lee YC, Lin JT, Chen TH, Wu MS. Is eradication of Helicobacter pylori the feasible way to prevent gastric cancer? New evidence and progress, but still a long way to go. J Formos Med Assoc. 2008;107:591–9. doi: 10.1016/S0929-6646(08)60176-X. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal K, Agarwal S. Helicobacter pylori vaccine: From past to future. Mayo Clin Proc. 2008;83:169–75. doi: 10.4065/83.2.169. [DOI] [PubMed] [Google Scholar]
- 8.Wen S, Moss SF. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1–8. doi: 10.1016/j.canlet.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baghaei K, Shokrzadeh L, Jafari F, Dabiri H, Yamaoka Y, Bolfion M, et al. Determination of Helicobacter pylori virulence by analysis of the cag pathogenicity island isolated from Iranian patients. Dig Liver Dis. 2009;41:634–8. doi: 10.1016/j.dld.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long M, Luo J, Li Y, Zeng FY, Li M. Detection and evaluation of antibodies against neutrophil-activating protein of Helicobacter pylori in patients with gastric cancer. World J Gastroenterol. 2009;15:2381–8. doi: 10.3748/wjg.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amedei A, Cappon A, Codolo G, Cabrelle A, Polenghi A, Benagiano M, et al. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J Clin Invest. 2006;116:1092–101. doi: 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Elios MM, Amedei A, Cappon A, Del Prete G, de Bernard M. The neutrophil-activating protein of Helicobacter pylori (HP-NAP) as an immune modulating agent. FEMS Immunol Med Microbiol. 2007;50:157–64. doi: 10.1111/j.1574-695X.2007.00258.x. [DOI] [PubMed] [Google Scholar]
- 13.Chibing G. B-cell epitope mapping of Helicobacter pylori neutrophil-activating protein. Proceedings of the 2007 Frontiers in the Convergence of Bioscience and Information Technologies: IEEE Computer Society. 2007 [Google Scholar]
- 14.Odenbreit S, Swoboda K, Barwig I, Ruhl S, Borén T, Koletzko S, et al. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect Immun. 2009;77:3782–90. doi: 10.1128/IAI.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dossumbekova A, Prinz C, Mages J, Lang R, Kusters JG, Van Vliet AH, et al. Helicobacter pylori HopH (OipA) and bacterial pathogenicity: Genetic and functional genomic analysis of hopH gene polymorphisms. J Infect Dis. 2006;194:1346–55. doi: 10.1086/508426. [DOI] [PubMed] [Google Scholar]
- 16.Taylor JM, Ziman ME, Canfield DR, Vajdy M, Solnick JV. Effects of a Th1- versus a Th2-biased immune response in protection against Helicobacter pylori challenge in mice. Microb Pathog. 2008;44:20–7. doi: 10.1016/j.micpath.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huleatt JW, Jacobs AR, Tang J, Desai P, Kopp EB, Huang Y, et al. Vaccination with recombinant fusion proteins incorporating Toll-like receptor ligands induces rapid cellular and humoral immunity. Vaccine. 2007;25:763–75. doi: 10.1016/j.vaccine.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Rumbo M, Nempont C, Kraehenbuhl JP, Sirard JC. Mucosal interplay among commensal and pathogenic bacteria: Lessons from flagellin and Toll-like receptor 5. FEBS Lett. 2006;580:2976–84. doi: 10.1016/j.febslet.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Yedidia T, Arnon R. Effect of pre-existing carrier immunity on the efficacy of synthetic influenza vaccine. Immunol Lett. 1998;64:9–15. doi: 10.1016/s0165-2478(98)00073-x. [DOI] [PubMed] [Google Scholar]
- 20.Honko AN, Sriranganathan N, Lees CJ, Mizel SB. Flagellin is an effective adjuvant for immunization against lethal respiratory challenge with Yersinia pestis. Infect Immun. 2006;74:1113–20. doi: 10.1128/IAI.74.2.1113-1120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gewirtz AT, Yu Y, Krishna US, Israel DA, Lyons SL, Peek RM., Jr Helicobacter pylori flagellin evades toll-like receptor 5-mediated innate immunity. J Infect Dis. 2004;189:1914–20. doi: 10.1086/386289. [DOI] [PubMed] [Google Scholar]
- 22.Lee SK, Stack A, Katzowitsch E, Aizawa SI, Suerbaum S, Josenhans C. Helicobacter pylori flagellins have very low intrinsic activity to stimulate human gastric epithelial cells via TLR5. Microbes Infect. 2003;5:1345–56. doi: 10.1016/j.micinf.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Vorobjova T, Watanabe T, Chiba T. Helicobacter pylori immunology and vaccines. Helicobacter. 2008;13:18–22. doi: 10.1111/j.1523-5378.2008.00636.x. [DOI] [PubMed] [Google Scholar]
- 24.Eswarappa SM. Location of pathogenic bacteria during persistent infections: Insights from an analysis using game theory. PLoS One. 2009;4:e5383. doi: 10.1371/journal.pone.0005383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh JD, Karam SM, Gordon JI. Intracellular Helicobacter pylori in gastric epithelial progenitors. Proc Natl Acad Sci U S A. 2005;102:5186–91. doi: 10.1073/pnas.0407657102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heit A, Busch DH, Wagner H, Schmitz F. Vaccine protocols for enhanced immunogenicity of exogenous antigens. Int J Med Microbiol. 2008;298:27–32. doi: 10.1016/j.ijmm.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Zhou WY, Shi Y, Wu C, Zhang WJ, Mao XH, Guo G, et al. Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine. 2009;27:5013–9. doi: 10.1016/j.vaccine.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Sbai H, Mehta A, DeGroot AS. Use of T cell epitopes for vaccine development. Curr Drug Targets Infect Disord. 2001;1:303–13. doi: 10.2174/1568005014605955. [DOI] [PubMed] [Google Scholar]
- 29.Sette A, Fikes J. Epitope-based vaccines: An update on epitope identification, vaccine design and delivery. Curr Opin Immunol. 2003;15:461–70. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Chen YH. Design and construction of a recombinant epitope-peptide gene as a universal epitope-vaccine strategy. J Immunol Methods. 2004;285:93–7. doi: 10.1016/j.jim.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 31.De Groot AS, McMurry J, Marcon L, Franco J, Rivera D, Kutzler M, et al. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine. 2005;23:2121–31. doi: 10.1016/j.vaccine.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y, Ding J, Liu W, Chen YH. A candidate vaccine against influenza virus intensively improved the immunogenicity of a neutralizing epitope. Int Arch Allergy Immunol. 2002;127:245–50. doi: 10.1159/000053869. [DOI] [PubMed] [Google Scholar]
- 33.Nicosia G, Cutello V, Bentley PJ, Timmis J, Saha S, Raghava GP. Berlin/Heidelberg: Springer/Artificial Immune Systems; 2004. BcePred: Prediction of Continuous B-Cell Epitopes in Antigenic Sequences using Physico-chemical Properties; pp. 197–204. [Google Scholar]
- 34.Haste Andersen P, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–67. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eliopoulos N, Stagg J, Lejeune L, Galipeau J. 911 MHC class I and II mismatched marrow stromal cells from C57Bl/6 mice are immune rejected by recipient Balb/c mice. Mol Ther. 2004;9:S348. [Google Scholar]
- 36.Bhasin M, Raghava GP. Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine. 2004;22:3195–204. doi: 10.1016/j.vaccine.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Sato K, Hamada M, Asai K, Mituyama T. CENTROIDFOLD: A web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–80. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–53. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 39.Pollastri G, Baldi P, Fariselli P, Casadio R. Prediction of coordination number and relative solvent accessibility in proteins. Proteins. 2002;47:142–53. doi: 10.1002/prot.10069. [DOI] [PubMed] [Google Scholar]
- 40.Eisenberg JC, Czinn SJ, Garhart CA, Redline RW, Bartholomae WC, Gottwein JM, et al. Protective efficacy of anti-Helicobacter pylori immunity following systemic immunization of neonatal mice. Infect Immun. 2003;71:1820–7. doi: 10.1128/IAI.71.4.1820-1827.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleanthous H, Tibbitts TJ, Gray HL, Myers GA, Lee CK, Ermak TH, et al. Sterilizing immunity against experimental Helicobacter pylori infection is challenge-strain dependent. Vaccine. 2001;19:4883–95. doi: 10.1016/s0264-410x(01)00248-1. [DOI] [PubMed] [Google Scholar]
- 42.Paul S, Piontkivska H. Discovery of novel targets for multi-epitope vaccines: Screening of HIV-1 genomes using association rule mining. Retrovirology. 2009;6:62. doi: 10.1186/1742-4690-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu D, Williams JN, Rice J, Stevenson FK, Heckels JE, Christodoulides M. A DNA fusion vaccine induces bactericidal antibodies to a peptide epitope from the PorA porin of Neisseria meningitidis. Infect Immun. 2008;76:334–8. doi: 10.1128/IAI.00943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai Q, Peng G, Bu L, Lin Y, Zhang L, Lustigmen S, et al. Immunogenicity and in vitro protective efficacy of a polyepitope Plasmodium falciparum candidate vaccine constructed by epitope shuffling. Vaccine. 2007;25:5155–65. doi: 10.1016/j.vaccine.2007.04.085. [DOI] [PubMed] [Google Scholar]
- 45.Amani J, Mousavi SL, Rafati S, Salmanian AH. In silico analysis of chimeric espA, eae and tir fragments of Escherichia coli O157:H7 for oral immunogenic applications. Theor Biol Med Model. 2009;6:28. doi: 10.1186/1742-4682-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braga CJ, Massis LM, Alencar BC, Rodrigues MM, Sbrogio-Almeida ME, Ferreira LC. Cytotoxic T cell adjuvant effects of three Salmonella enterica flagellins. Braz J Microbiol. 2008;39:44–9. doi: 10.1590/S1517-838220080001000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eaves-Pyles TD, Wong HR, Odoms K, Pyles RB. Salmonella flagellin-dependent proinflammatory responses are localized to the conserved amino and carboxyl regions of the protein. J Immunol. 2001;167:7009–16. doi: 10.4049/jimmunol.167.12.7009. [DOI] [PubMed] [Google Scholar]
- 48.Murthy KG, Deb A, Goonesekera S, Szabó C, Salzman AL. Identification of conserved domains in Salmonella muenchen flagellin that are essential for its ability to activate TLR5 and to induce an inflammatory response in vitro. J Biol Chem. 2004;279:5667–75. doi: 10.1074/jbc.M307759200. [DOI] [PubMed] [Google Scholar]
- 49.Mizel SB, West AP, Hantgan RR. Identification of a sequence in human toll-like receptor 5 required for the binding of Gram-negative flagellin. J Biol Chem. 2003;278:23624–9. doi: 10.1074/jbc.M303481200. [DOI] [PubMed] [Google Scholar]
- 50.Blander JM. Signalling and phagocytosis in the orchestration of host defence. Cell Microbiol. 2007;9:290–9. doi: 10.1111/j.1462-5822.2006.00864.x. [DOI] [PubMed] [Google Scholar]
- 51.Blander JM, Medzhitov R. On regulation of phagosome maturation and antigen presentation. Nat Immunol. 2006;7:1029–35. doi: 10.1038/ni1006-1029. [DOI] [PubMed] [Google Scholar]
- 52.Le Moigne V, Robreau G, Mahana W. Flagellin as a good carrier and potent adjuvant for Th1 response: Study of mice immune response to the p27 (Rv2108) Mycobacterium tuberculosis antigen. Mol Immunol. 2008;45:2499–507. doi: 10.1016/j.molimm.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Ferrero RL. Innate immune recognition of the extracellular mucosal pathogen, Helicobacter pylori. Mol Immunol. 2005;42:879–85. doi: 10.1016/j.molimm.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Neville LF, Barnea Y, Hammer-Munz O, Gur E, Kuzmenko B, Kahel-Raifer H, et al. Antibodies raised against N’- terminal Pseudomonas aeruginosa flagellin prevent mortality in lethal murine models of infection. Int J Mol Med. 2005;16:165–71. [PubMed] [Google Scholar]
- 55.Salazar-Gonzalez RM, McSorley SJ. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol Lett. 2005;101:117–22. doi: 10.1016/j.imlet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z, Louboutin JP, Weiner DJ, Goldberg JB, Wilson JM. Human airway epithelial cells sense Pseudomonas aeruginosa infection via recognition of flagellin by Toll-like receptor 5. Infect Immun. 2005;73:7151–60. doi: 10.1128/IAI.73.11.7151-7160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song L, Zhang Y, Yun NE, Poussard AL, Smith JN, Smith JK, et al. Superior efficacy of a recombinant flagellin: H5N1 HA globular head vaccine is determined by the placement of the globular head within flagellin. Vaccine. 2009;27:5875–84. doi: 10.1016/j.vaccine.2009.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, et al. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci U S A. 2007;104:16293–8. [Google Scholar]
- 59.Satin B, Del Giudice G, Della Bianca V, Dusi S, Laudanna C, Tonello F, et al. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J Exp Med. 2000;191:1467–76. doi: 10.1084/jem.191.9.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sutton P, Doidge C, Pinczower G, Wilson J, Harbour S, Swierczak A, et al. Effectiveness of vaccination with recombinant HpaA from Helicobacter pylori is influenced by host genetic background. FEMS Immunol Med Microbiol. 2007;50:213–9. doi: 10.1111/j.1574-695X.2006.00206.x. [DOI] [PubMed] [Google Scholar]


