Abstract
Personality traits are complex phenotypes related to psychosomatic health. Individually, various gene finding methods have not achieved much success in finding genetic variants associated with personality traits. We performed a meta-analysis of four genome-wide linkage scans (N=6149 subjects) of five basic personality traits assessed with the NEO Five-Factor Inventory. We compared the significant regions from the meta-analysis of linkage scans with the results of a meta-analysis of genome-wide association studies (GWAS) (N∼17 000). We found significant evidence of linkage of neuroticism to chromosome 3p14 (rs1490265, LOD=4.67) and to chromosome 19q13 (rs628604, LOD=3.55); of extraversion to 14q32 (ATGG002, LOD=3.3); and of agreeableness to 3p25 (rs709160, LOD=3.67) and to two adjacent regions on chromosome 15, including 15q13 (rs970408, LOD=4.07) and 15q14 (rs1055356, LOD=3.52) in the individual scans. In the meta-analysis, we found strong evidence of linkage of extraversion to 4q34, 9q34, 10q24 and 11q22, openness to 2p25, 3q26, 9p21, 11q24, 15q26 and 19q13 and agreeableness to 4q34 and 19p13. Significant evidence of association in the GWAS was detected between openness and rs677035 at 11q24 (P-value=2.6 × 10−06, KCNJ1). The findings of our linkage meta-analysis and those of the GWAS suggest that 11q24 is a susceptible locus for openness, with KCNJ1 as the possible candidate gene.
Keywords: personality, KCNJ1, NEO, linkage, GSMA
Introduction
Human personality is a compound of complex traits that are associated with several psychiatric,1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 and somatic disorders.17, 18, 19, 20, 21, 22, 23, 24, 25, 26 Despite high heritability ranging from 33 to 60%, the understanding of the genetic origins of personality trait variation is extremely limited. Where linkage analyses have identified several large regions, very few overlap.27 Candidate gene studies have their own concerns in that the findings have generally not been replicated.27 A recent genome-wide association study (GWAS), including up to 18 000 individuals, yielded only a few loci that attained genome-wide significance.28
The general success of GWAS in gene discovery and failure to replicate most of the significant linkage peaks for complex traits and diseases shifted interest towards GWAS. Association studies have benefitted from precise estimation of the locus, reproducibility of the findings and the availability of large population-based cohorts, which has led to powerful studies for gene discovery as well as replication using meta-analyses. However, the GWAS have not been very successful in finding genes of complex psychiatric/behavioral traits. The ‘case of the missing heritability29 has led to the view that common traits may be driven by relatively rare variants.30 Despite the fact that rare variants are weakly tagged by the common single-nucleotide polymorphisms (SNPs) present in the current arrays,30, 31 association signals in GWAS at loci including rare variants have been seen in particular for lipids.29 Also, two recent papers argued that common variants may tag more rare ones from a theoretical perspective,30, 31 implying that linkage and association analyses should be able to identify the same loci. For complex traits, it was suggested to use genome-wide linkage and association analyses in two steps to maximize power.32 Linkage may point towards the regions in the GWAS to look for possible associated variants, reducing the number of tests and relaxing the significance threshold, thus resulting in a higher chance of finding genetic variants.
In this study, we aimed to discover genetic variants, both rare and common, that affect personality traits, including neuroticism, extraversion, openness, agreeableness and conscientiousness. We first perform a quantitative trait linkage analysis in four independent cohorts – assuming that this would pick signals at loci containing large effects with relatively rare (population-specific) variants, and then combine these in a meta-analysis – assuming that the meta-analysis would pick linkage signals at loci harboring genes with relatively moderate to small effects. Finally, we compare the findings of our meta-analysis of linkage scans with those of the largest meta-analysis of GWAS of the same traits (which included the linkage samples examined)33 to localize the possible linked gene. This is by far the largest linkage study conducted for the NEO personality traits combined with the largest GWAS of these traits conducted to date.
Materials and methods
We performed a meta-analysis of linkage studies that used the NEO Five Factor Inventory (NEO-FFI) or the Revised NEO personality inventory (NEO-PI-R)34 to assess the five basic personality traits including: (1) neuroticism – a trait that refers to the tendency to experience negative emotions; (2) extraversion – a measure of sociability, positive emotions and action; (3) openness – a measure of intellectual curiosity and preference for variety; (4) agreeableness – a measure of altruism, cooperation and harmony; and (5) conscientiousness – a measure of an individual's tendency to plan, organize and direct his impulses.34 This five-factor model is a hierarchical model where each of these five traits is defined by six underlying facets assessed by the NEO-PI-R. These five traits, also known as the Big Five, are considered universal,35 stable in adulthood36 and orthogonal, but correlations appear possibly due to self-report.37 Women score generally higher on neuroticism and agreeableness38, 39, 40 compared with men. The NEO-FFI consists of 60 items, 12 for each trait, whereas the NEO-PI-R consists of 240 questions inclusive of the ones in the NEO-FFI. This study included linkage scans from four independent populations, including the Erasmus Rucphen Family (ERF), the Netherlands Twin Register (NTR), the Australian Adolescent sample (QIMR_adolescent) and the Australian adult sample (QIMR_adult) (Table1). ERF and NTR used the NEO-FFI for personality assessment, whereas the Australian samples were assessed with either NEO-FFI or NEO-PI-R; however, for this study the 60 items of the NEO-FFI was used for the final assessment. Within each cohort the scores were considered invalid if an individual answered fewer than nine questions, otherwise the missing data were imputed by taking the individual's average score for the valid items of that dimension. The descriptive statistics of the study samples are provided in the Supplementary Table 1. Variance component (VC) linkage analysis, adjusted for age and gender, was performed in each cohort with MERLIN (Center for Statistical Genetics, University of Michigan, Ann Arbor, MI, USA) (Table 1). A power analysis for a quantitative trait locus (QTL) explaining 1, 5 and 10% of the trait variance was also performed in each cohort with POLY (Chen and Abecasis)41 at fixed type 1 error rates of 1 and 5%.
Table1. General features of independent genome scan.
| Cohort | ERF | NTR | QIMR_adolescents | QIMR_adults |
|---|---|---|---|---|
| Origin | Dutch | Dutch | Australiana | Australiana |
| Design | Extended families | sibpairs | sibpairs | sibpairs+extended families |
| Sample size (%women) | 2244 (55.0) | 1507 (61.6) | 1096 (56.5) | 1349 (55.7) |
| Total no. of markers used | 5250 | 864 | 5479 | 5479 |
| Marker type | Single neucleotide | Microsatellites | Single-neucleotide polymorphisms | Single-neucleotide polymorphisms |
| Analysis type | Variance components | Variance components | Variance components | Variance components |
| Analysis software | Merlin | Merlin | Merlin | Merlin |
| Personality assessment | NEO-FFI | NEO-FFI | NEO-PI-R/NEO-FFI | NEO-FFI |
Majority of the sample is Causasian, predominantly Anglo-Celtic (ancestry outliers, identified using HapMap3 and GenomeEUTwin individuals as a reference panel, were excluded).
Erasmus Rucphen Family
The study sample consisted of 2657 individuals who participated in ERF study.42 The study population essentially consists of one extended family spanning over 23 generations and including more than 23 000 individuals descending from 20 related couples who lived in the region between 1850 and 1900. All descendants were ascertained and descendants of 18 years and older were invited to participate. Spouses were invited only for family members who had children of 18 years and older.
For all participants, genomic DNA was extracted from peripheral venous blood utilizing the salting out method.43 For genome-wide linkage analysis, genotyping was performed using the Illumina 6K linkage panel that includes markers distributed evenly across the human genome (median distance between markers is 301 kb). Of the 6000 single-nucleotide polymorphisms (SNPs), we used 5250, after quality control and excluding X-chromosomal SNPs. The genotyping was performed at the Center National de Génotypage in France according to the standard protocol.
Multipoint VC linkage analysis was performed using MERLIN v.1.0.1 software44 for all the five personality traits. Marker allele frequencies were estimated from the data. The pedigrees were split on non-overlapping fragments of no more than 18 bits with the help of two programs: Jenti45 and PedSTR.46 Three sets of subpedigrees were obtained with different parameters with the help of these programs, which were then analyzed separately. These three sets differed one from another by number, size and structure of pedigree fragments. However, they demonstrated similar profiles of LODs for all analyzed traits (Spearman's correlations varied from 0.6 (P<0.001) to 0.8 (P<0.001)), which allowed us to use the maximum of three values of LOD for each marker locus.47 In accordance with Bonferroni correction, suggestive and significant thresholds were estimated as 2.34 and 3.75, respectively. The analysis was based on 2244 genotyped and phenotyped persons from ERF.
Netherlands Twin Register
The NTR48 sample consists of 711 families with 3412 non-clone individuals (1438 founders, 1870 females) with an average of 4.8 subjects per family. In all, 282 of these families have both founders genotyped and 138 families had one genotyped founder. In addition, there are 290 nuclear families with no genotyped founders, and one extended pedigree with four founders without genotypes. Autosomal genomes had 757 markers spaced at an average of 4.76 cM (range 0.0–20.59 cM), with average heterozygosity of 0.76. Founders had the genetic data for 446 autosomal microsatellites. NEO measures together with the age and gender information were available for a total of 1507 subjects (998 non-founder; 509 non-founders) from 409 families with genetic data. Of 409 families, 270 had two phenotyped siblings, 113 had three, 19 had four, 1 family had five, 4 families had six and 2 families had seven phenotyped siblings, resulting in a total of 835 sibling pairs. In addition, this sample also features 83 phenotyped MZ clones.
The genetic maps were obtained through the Rutgers University Map Interpolator.49 The allele frequencies were estimated with the Mendel v.10.0 software package (Kenneth Lange, University of California, CA, USA).50 VC linkage scan of the autosomal genome was conducted with MERLIN v.1.1.2.
Queensland Institute of Medical Research, Australian Study Sample
NEO personality data (NEO-PI-R or NEO-FFI) were collected as part of two independent research streams – one focused on an adult sample (QIMR_adult) and the other with an adolescent/young adult focus (QIMR_adolescent). The QIMR_adult data were collected as part of the Nicotine Addiction Genetics (NAG) study (2001–2005), which targeted families based on heavy smoking index cases identified in earlier interviews and questionnaires51 and was itself part of the Interactive Research Project Grants (IRPG). This sample comprised 1349 genotyped individuals aged 21 to 85 years (M=45.5±13.1) from 519 families, and included 15 complete MZ pairs for whom data were averaged.
The QIMR_adolescent data were collected from two population studies under the umbrella of the Brisbane Adolescent and Young Adult Twin Studies, specifically, studies of cognition (1996–ongoing)52 and health and well-being (2002–2003),52 and from a study of borderline personality disorder (2004–2006).53 This sample comprised 1096 genotyped individuals aged 16 to 27 years (M=19.4±2.7), from 563 families, and included 127 complete MZ pairs for whom data were averaged.
Participants were typically Caucasian, predominantly Anglo-Celtic (ancestry outliers, identified using HapMap3 and GenomeEUTwin individuals as a reference panel, were excluded). Written, informed consent was obtained from all participants and from a parent or guardian for those aged under 18 years. Ethics approval was received from the institutional review boards appropriate to each study (QIMR and Washington University School of Medicine).
Genotyping was carried out using the Illumina (San Diego, CA, USA) 610K or 370K SNP platform and Illumina BeadStudio software, with 269 840 SNPs common to the subsamples passing QC (28% of the SNPs selected for linkage were from this set of SNPs).54 Data were imputed using HapMap I+II (CEU, build 36 release 22) using the MACH software (Center for Statistical Genetics, Ann Arbor, MI, USA).54 SNP selection for linkage analysis was matched as far as possible to the SNP set used with the ERF sample. The final selection contained 5479 SNPs, of which 5181 had a direct match with our genotyped or imputed data. For the remaining 298 SNPs, proxies based on linkage disequilibrium (>0.8), or position, were used. Multipoint VC linkage analysis was performed using MERLIN 1.1.244 in both samples.
Meta-analysis of linkage scans
Results from individual genome-wide linkage scans were combined together in a meta-analysis using the Genome Search Meta-Analysis (GSMA).55, 56 The GSMA method divides the genome into several bins of equal width (chosen arbitrarily such that smallest chromosome should have at least two bins), and is robust to differences between studies in ascertainment schemes, marker maps and statistical methods used to detect linkage. Customarily a bin width of 30 cM, which gives 120 bins across the autosomal genome, is used. The bins are then ranked in descending order in each study, with the bin with the highest LOD score getting the highest rank. The ranks within a bin are summed across each study to get a summed rank (SR). The SR statistic is tested for significance using its distribution function or by simulation,55 which gives the probability of observing a given SR for a bin (PSR). PSR only gives the point-wise probability for the SR for a certain bin. A genome-wide interpretation of the results is obtained through the ordered statistic (OR), which determines the probability (POR) of a given SR for a bin by chance when bins are assigned ranks randomly in multiple simulations.57 Simulations show that a bin with a significant PSR and a significant POR (PSR<0.05 and POR<0.05) has a high probability of containing a true susceptibility locus.57 For an individual bin, the genome-wide significance is defined as PSR<0.05/number of bins and suggestive as PSR<1/number of bins. For an individual scan, we used 3.3 as significant and 1.9 as suggestive linkage threshold.58
In this study, except for the NTR sample genome-wide linkage results were reported against SNP markers, which helped us map all the results directly to the base pair positions. For NTR, we mapped all the results to base pair positions by interpolation and using base pair positions from Rutgers map.49 For each study, we divided the autosomal genome into 125 bins of width 25 million base pairs (mbp) each, giving a maximum of 10 bins on chromosome 1 and a minimum of 2 bins on chromosome 22. To evaluate the possibility of correlation between adjacent bins, we also performed the analysis using a bin width of 40 mbp. We performed weighted meta-analysis, where weights were calculated as the square root of the sample size in each study. A total of 10 000 permutations were performed to get the PSR and POR. A bin was considered significant if (1) either the Bonferroni-corrected significance was achieved, that is, PSR<0.0004 for a bin or (2) if both PSR and POR were nominally significant for multiple bins.57 Suggestive linkage threshold was set at PSR<0.008. Heterogeneity testing was performed using the HEGESMA59 software (School of Medicine, University of Thessaly, Thessaly, Greece).
In an attempt to discover the variant that might be causing the linkage signals, we fine mapped the interesting regions from the meta-analysis of linkage scans with the results of a meta-analysis of GWAS of the NEO personality traits.28 The meta-analysis of GWAS (n>17 000) included the samples used in this study (and many others as well), but the marker sets used were different. Brief description of the studies included in the meta-analysis of GWAS is provided in Supplementary Table 2.
Results
Individual scans
Results of the power analysis are provided in the Supplementary Table 3. The results from individual genome-wide linkage scans are provided in Supplementary Figures 1–5 and significant and suggestive findings are summarized in Supplementary Table 4. Significant evidence of linkage was observed for neuroticism in the ERF study at chromosome 3p14 (rs1490265, LOD=4.67) and at chromosome 19q13 (rs628604, LOD=3.55) in the QIMR (adolescent) sample; for extraversion at 14q32 (ATGG002, LOD=3.3) in the NTR sample; for agreeableness at 3p25 (rs709160, LOD=3.67) and two adjacent regions on chromosome 15, including 15q13 (rs970408, LOD=4.07) and 15q14 (rs1055356, LOD=3.52) in the QIMR (adolescents) study. Considering the suggestive findings, there was an overlap at chromosome 2q14 between ERF (LOD=2.1) and NTR (LOD=2.03) for neuroticism and at 12q23 between ERF (LOD=2.85) and NTR (LOD=1.96) for openness (Supplementary Table 4).
Meta-analysis
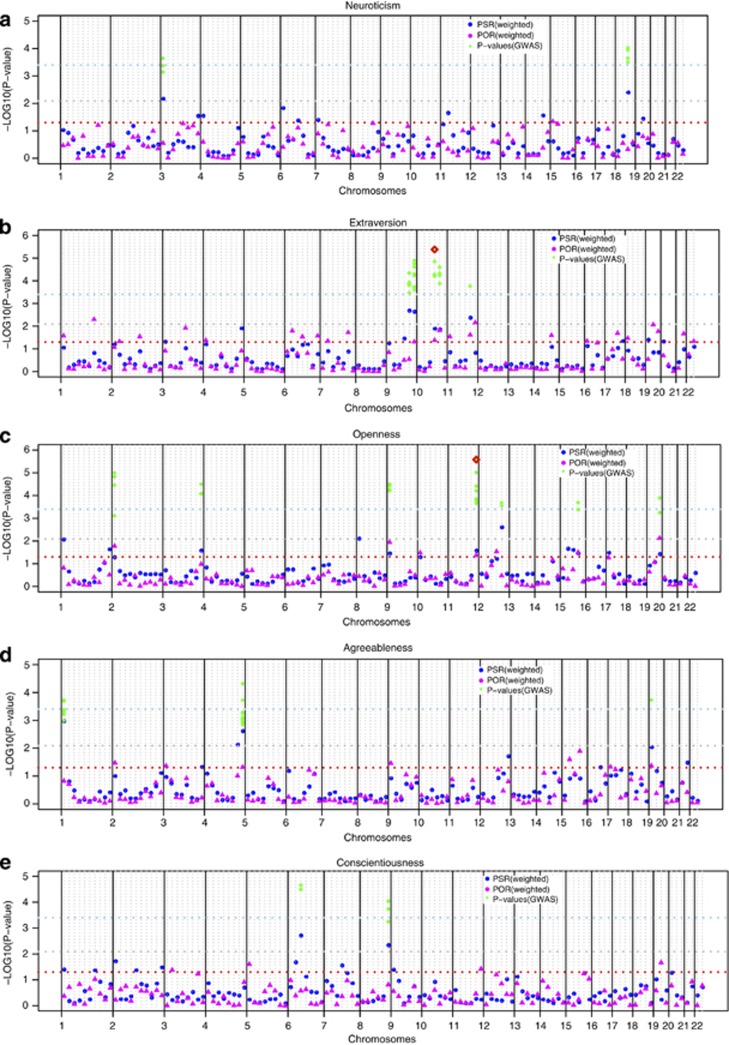
Genome-wide results of the meta-analysis are illustrated in Figure 1. Table 2 provides a summary of the bins with significant evidence of linkage. None of the bins crossed the Bonferroni-corrected genome-wide significance threshold. However, there were multiple bins for which both PSR and POR were nominally significant (Table 2 and Figure 1).
Figure 1.
Results of the meta-analysis of the linkage scans. (a) For neuroticism, (b) for extraversion, (c) for openness, (d) for agreeableness and (e) for conscientiousness. Dots represent PSR and triangles represent POR. The X axis shows the whole autosomal genome divided by solid vertical lines into chromosomes, which are further divided into bins by dotted gray lines. The Y axis shows the negative principal log of the P-values. The red horizontal dotted line represents the nominal threshold P-value=0.5, the gray dotted horizontal line represents the suggestive threshold P-value=0.008 and the light blue horizontal dotted line represents the Bonferroni-corrected threshold P-value=0.0004. A bin is considered significant if a circle for that bin surpasses the sky blue line or if both the circle and the triangle for a specific bin are above the dotted red line. The green diamonds represent the P-values of the SNPs from the results of the meta-analysis of the GWAS of the same traits falling in the bins of interest. The green diamonds with the red outline are the P-values significant/borderline significant after correcting for the number of total SNPs in the bin.
Table 2. Comparison of interesting regions of linkage meta-analysis with the personality GWAS meta-analysis.
| Trait | Bin | Cyto | SRP-value | ORP-value | PHET | Approx. region (mbp) | SNP GWAS (N>17 000) | β | Effect allele | MAF | Effective no. of SNPs in the regiona | P-value (nominal) | Bonferroni thresholdb | Candidate gene | Distance of the SNP to the candidate gene (kb)c |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Extraversion | 9.6 | 9q34 | 0.003 | 0.049 | 0.036 | 125–150 | rs11103814 | −0.35 | A | 0.33 | 2049 | 1.3 × 1−05 | 4.2 × 1−06 | OLFM1, KIAA0649 | 204.8, 153.8 |
| 10.4 | 10q24 | 0.017 | 0.042 | 0.063 | 75–100 | rs7922066 | −0.44 | A | 0.18 | 2986 | 4.2 × 1−06 | 2.1 × 1−06 | CRTAC1 | Intron(NM_001206528.1) | |
| 10.5 | 10q24 | 0.018 | 0.015 | 0.042 | 100–125 | rs7905091 | 0.47 | T | 0.13 | 3073 | 2.5 × 1−05 | 2.1 × 1−06 | LOXL4, PYROXD2 | 18.1, 97.2 | |
| 11.5 | 11q22 | 0.005 | 0.024 | 0.073 | 100–125 | rs12419614 | 1.65 | A | 0.03 | 3177 | 1.1 × 1−04 | 2.0 × 1−06 | GRIA4 | Intron(NM_000829.3) | |
| Openness | 2.1 | 2p25 | 0.067 | 0.016 | 0.186 | 0–25 | rs11685318 | −0.38 | A | 0.20 | 3056 | 1.0 × 1−05 | 1.8 × 1−06 | PDIA6, KCNF1 | 40.4, 58.7 |
| 9.1 | 9p21 | 0.045 | 0.011 | 0.053 | 0–25 | rs10966354 | −0.30 | T | 0.37 | 3865 | 3.1 × 1−05 | 1.4 × 1−06 | ELAVL2, TUSC1 | 810.7, 1039.6 | |
| 11.6 | 11q24 | 0.034 | 0.043 | 0.625 | 125–150 | rs677035 | −0.56 | T | 0.08 | 1393 | 2.6 × 1−06 | 4.4 × 1−06 | FLI1, KCNJ1 | 13.9, 10.9 | |
| 15.4 | 15q26 | 0.045 | 0.031 | 0.280 | 75–100 | rs8034376 | −0.47 | T | 0.11 | 3135 | 2.0 × 1−04 | 2.0 × 1−06 | FAM169B, IGF1R | 6.5, 128.6 | |
| 19.3 | 19q13 | 0.048 | 0.007 | 0.165 | 50–75 | rs2287826 | −0.62 | A | 0.07 | 2106 | 1.2 × 1−04 | 4.8 × 1−06 | LILRA4, LAIR1 | 12.8, 1.9 | |
| Agreeableness | 4.8 | 4q34 | 0.003 | 0.048 | 0.008 | 175–200 | rs13113475 | 0.28 | A | 0.20 | 1959 | 4.8 × 1−05 | 3.1 × 1−06 | SPCS3, VEGFC | 228.8, 122.5 |
| 19.1 | 19p13 | 0.011 | 0.042 | 0.134 | 0–25 | rs10413538 | 0.49 | T | 0.28 | 3209 | 3.7 × 1−04 | 3.6 × 1−06 | ZNF826 | Utr-5(NM_001039884.1) |
P-values from GWAS significant after Bonferroni correction in bold italics.
Effective number of SNPs from the GWAS present in the bin.
Bonferroni threshold based on the total number of SNPs (both genotyped and imputed) from the GWAS present in the bin.
Positions are taken from dbSNP (http://www.ncbi.nlm.nih.gov/snp) and SCAN (http://www.scandb.org/newinterface/about.html) databases.
For extraversion, five bins showed nominally significant PSR and POR. These included bins 9.6, 11.5 and two adjacent bins on chromosome 10 (10.4 and 10.5). The linkage signals on the adjacent bins on chromosome 10 were being caused by the same peaks that extended over 40 cM (Supplementary Figure S2). When the bin width was increased to 40 mbp, the finding for the new bin on chromosome 10 covering the previous 10.4 bin and partially covering the 10.5 bin remained significant and the adjacent bin did not show linkage signals. When comparing the significant bins with the meta-analysis of GWAS, we identified clusters of SNPs with low P-values for the bin in chromosome 10 (rs7088779, P-value=4.2 × 10−06) (Table 2 and Figure 1b). Rs7088779 was marginally significant after being corrected for the number of SNPs in the bin. This SNP is located between CRTAC1 (cartilage acidic protein 1) and C10orf28, a region previously implicated in Alzheimer's disease.
For openness, five bins (9.1, 11.6, 15.4 and 19.3) were significant in that they showed both PSR and POR<0.05 (Table 2 and Figure 1c). A cluster of eight SNPs with very low P-values was identified from the GWAS for the bin 11.6 (Figure 1c), which maps to 11q24 region. The most significant SNP (rs677035) showed a P-value of 2.6 × 10−06, which passed the Bonferroni threshold. Rs677035 is an intergenic SNP located between FLI1 and KCNJ1.
Two bins (4.8 and 19.1) showed nominally significant PSR and POR for agreeableness, but were not supported by association (Table 2 and Figure 1d). Heterogeneity between studies was detected for bin 4.8. None of the bins for neuroticism or conscientiousness showed significant linkage (Figure 1e).
Discussion
We found significant evidence of linkage of neuroticism with chromosomes 3p14 (rs1490265, LOD=4.67) and 19q13 (rs628604, LOD=3.55); of extraversion with 14q32 (ATGG002, LOD=3.3); and of agreeableness with 3p25 (rs709160, LOD=3.67) and with two adjacent regions on chromosome 15, including15q13 (rs970408, LOD=4.07) and 15q14 (rs1055356, LOD=3.52) in the individual scans. In the meta-analysis, we discovered one region on chromosome 11q24 significantly linked to openness, which was also strongly supported by the results of the largest GWAS of personality traits performed to date.
Our meta-analysis included 6149 individuals for multiple extended families and families with sibships. There are a number of methodological issues relevant for the interpretation of the findings. We used a physical map (base pair positions) and bin width of 25 mbp (which would roughly translate to 25 cM) as opposed to the traditional 30 cM bin width used in all previous studies. The choice of 25 mbp was made after taking into account the genetic maps of all four studies and specially the position of last reported markers on chromosome 22 (49 mbp), giving two bins of about equal size on the smallest chromosome as was required, thus avoiding manipulation of data. However, this selection could lead to a correlation between two adjacent bins. Interestingly, this correlation was observed only for extraversion in two pairs of adjacent bins on chromosomes 9 and 10. The size of the linkage peak on chromosome 9 extends to about 50 cM (Supplememtary Figure S2), implying that choice of a bin width of more than 30 cM could not have removed this correlation. For chromosome 10, the significance remained even after increasing the bin width to 40 mbp, which suggests that our result on 10q24 for extraversion is consistent. The GSMA gave broad linkage regions spread over 25 mbp, but we made an effort to localize the susceptibility genes by using additional information from the meta-analysis of GWAS.
It is interesting to note that none of the significant regions from the individual scans showed any evidence of linkage in the meta-analysis. There may be several explanations: first, our significant findings of individual studies may just be false positives. This may be a possibility, but it is difficult to believe that all significant findings are false, as VC linkage analysis is usually robust in detecting linkage.60 Second, this may due to the differences in the power of the various studies (see Supplementary Table 3). For example, the reason why the linkage of neuroticism to 3p14 was detected in the ERF study only may be the size of the study, which was twice as large as any other included study. Third and more likely, is possible locus heterogeneity across populations. High locus heterogeneity, which results in inconsistent linkage peaks, is one characteristic of complex traits like personality. However, the heterogeneity analysis did not provide evidence of heterogeneity at this locus. Also, the rank-based test used for the meta-analysis of linkage scans is insensitive to the significance of a linkage peak in an individual study and is more adept at finding subtle linkage peaks present in all studies included in the meta-analysis. For instance, the meta-analysis ignores the overlap at chromosome 2q14 between ERF (LOD=2.01) and NTR (LOD=2.03) for neuroticism despite falling in the same bin. Similarly, two adjacent regions for openness at chromosome 11q25 for ERF (LOD=2.05) and 11q24 for QIMR_adolescent (LOD=2.55) and chromosome 12q23 for ERF (LOD=2.85) and 12q24 for NTR (LOD=1.96) that showed suggestive evidence of linkage were not picked up in the meta-analysis. Such results were also observed in studies that used parametric methods (Fisher's method) to meta-analyze genome-wide linkage scans.61
The most interesting finding of the meta-analysis is 11q24 that showed nominally significant PSR and POR and also significant association in the GWAS after adjusting for multiple testing. The 11q24 region is implicated in mental retardation62 and migraine.63 The significant SNP in the GWAS rs677035 is located between FLI1 and KCNJ1. Previously, we have found linkage to other potassium channel genes including KCNJ2, KCNJ6 and KCNJ16,27 making KCNJ1 the most interesting gene in the region. The other regions of interest include 10q24 for extraversion. The region showed both PSR and POR<0.05, and additionally showed borderline significant evidence of association in the meta-analysis of GWAS after adjusting for multiple testing. The region has been implicated in spinocerebellar ataxia,64 AD and cognitive function.65, 66
We cannot discard other regions that showed a high probability of containing true susceptibility loci in the linkage analyses as being false positives based on insignificant association results of the GWAS. Our multiple testing corrections for association within a linkage region were based on total number of SNPs in the region (rather than independent SNPs), which made the Bonferroni threshold very conservative. Secondly, the power of the linkage analysis and association analysis to detect a QTL depends on the effect size of the QTL and the linkage disequilibrium between the candidate loci,32 as the former is more powerful to detect relatively rare large effect loci, while the latter is more powerful for finding frequent loci with subtle effects. The power of association analysis to detect a QTL declines rapidly with the decreasing degree of linkage disequilibrium between the QTL and the candidate locus.32 It is also possible that because of arbitrary binning of the linkage peaks for the meta-analysis, which is not very precise, we might have completely missed the association signal.
The regions we have discovered with linkage are good candidates for sequencing exomes and regulatory regions and may unveil variants with moderate to large effects contributing to the make-up of human personality. The criteria for significant evidence for linkage in the meta-analyses were not reached in this study. The highest level of significance for linkage reached, translated into a high probability (of unknown size) to include a susceptibility locus. It is obvious that false negatives may occur because of locus heterogeneity using the current methods. To overcome these problems, we combined the data from the linkage analyses of the five basic NEO personality traits in four independent populations with those of the genome wide association analyses. Here we relied on classical association analysis and genetic imputations based on HapMap. This approach will be strengthened by imputing populations that underwent GWAS using the 1000 genomes, providing a greater resolution and a better coverage of rare variants.
Acknowledgments
ERF: The genotyping for the ERF study was supported by EUROSPAN (European Special Populations Research Network) and the European Commission FP6 STRP Grant (018947; LSHG-CT-2006-01947). The ERF study was further supported by grants from the Netherlands Organization for Scientific Research, Erasmus MC, the Center for Medical Systems Biology (CMSB) and the Netherlands Brain Foundation (HersenStichting Nederland). We are grateful to all patients and their relatives, general practitioners and neurologists for their contributions, and to P Veraart for her help in genealogy, Jeannette Vergeer for the supervision of the laboratory work and P Snijders for his help in data collection.
NTR: We acknowledge financial support from the Netherlands Organization for Scientific Research (NWO): Grants 575-25-006, 480-04-004, 904-61-090; 904-61-193, 400-05-717 and Spinozapremie SPI 56-464-14192. MHMdeM is financially supported by ZonMW (Addiction) Grant number 311-60008. We further acknowledge financial support from the Center for Medical Systems Biology (NWO Genomics), Neuroscience Campus Amsterdam; EU/QLRT-2001-01254; NIMH R01 MH059160; Geestkracht program of ZonMW (10-000-1002); and matching funds from universities and mental health care institutes involved in NESDA. Genotyping was funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, and analysis was supported by grants from GAIN and the NIMH (MH081802). Genotype data were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/dbgap, accession number phs000020.v1.p1).
QIMR: We thank Marlene Grace and Ann Eldridge for sample collection; Anjali Henders, Megan Campbell, Lisa Bowdler, Steven Crooks and staff of the Molecular Epidemiology Laboratory for sample processing and preparation; Harry Beeby, David Smyth and Daniel Park for IT support. We acknowledge support from the Australian Research Council (A7960034, A79906588, A79801419, DP0212016 and DP0343921), Beyond Blue and the Borderline Personality Disorder Research Foundation. Genotyping was funded by the National Health and Medical Research Council (Medical Bioinformatics Genomics Proteomics Program, 389891). Further, we gratefully acknowledge Drs Dale R Nyholt and especially Scott Gordon for their substantial efforts involving the QC and preparation of the QIMR and NAG/IRPG GWA data sets from which SNPs were selected. Dr Nyholt also contributed 8% of the NAG/IRPG GWA cohort (NHMRC IDs 339462, 442981, 389938 and 496739). In addition, we thank him for helpful discussions regarding methodology.
GWAS cohorts: We also acknowledge the individual contributions of the cohorts included in the meta-analysis of genome-wide association scans. Paul T Costa Jr receives royalties from the NEO-FFI. Laura J Bierut is an inventor on the patent ‘Markers for Addiction' (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. Dr Bierut served as a consultant for Pfizer Inc. in 2008.
The other authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Bienvenu OJ, Hettema JM, Neale MC, Prescott CA, Kendler KS. Low extraversion and high neuroticism as indices of genetic and environmental risk for social phobia, agoraphobia, and animal phobia. Am J Psychiatry. 2007;164:1714–1721. doi: 10.1176/appi.ajp.2007.06101667. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Svrakic DM, Przybeck TR. A psychobiological model of temperament and character. Arch Gen Psychiatry. 1993;50:975–990. doi: 10.1001/archpsyc.1993.01820240059008. [DOI] [PubMed] [Google Scholar]
- Cramer D.Type A behaviour pattern, extraversion, neuroticism and psychological distress Br J Med Psychol 199164(Part 1):73–83. [DOI] [PubMed] [Google Scholar]
- Crowe M, Andel R, Pedersen NL, Fratiglioni L, Gatz M. Personality and risk of cognitive impairment 25 years later. Psychol Aging. 2006;21:573–580. doi: 10.1037/0882-7974.21.3.573. [DOI] [PubMed] [Google Scholar]
- Fanous AH, Neale MC, Aggen SH, Kendler KS. A longitudinal study of personality and major depression in a population-based sample of male twins. Psychol Med. 2007;37:1163–1172. doi: 10.1017/S0033291707000244. [DOI] [PubMed] [Google Scholar]
- Goodwin RD, Castro M, Kovacs M. Major depression and allergy: does neuroticism explain the relationship. Psychosom Med. 2006;68:94–98. doi: 10.1097/01.psy.0000195797.78162.f4. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006;163:857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- Kaasinen V, Nurmi E, Bergman J, et al. Personality traits and brain dopaminergic function in Parkinson's disease. Proc Natl Acad Sci USA. 2001;98:13272–13277. doi: 10.1073/pnas.231313198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrae RR, Yang J, Costa PT, et al. Personality profiles and the prediction of categorical personality disorders. J Pers. 2001;69:155–174. doi: 10.1111/1467-6494.00140. [DOI] [PubMed] [Google Scholar]
- Menza M. The personality associated with Parkinson's disease. Curr Psychiatry Rep. 2000;2:421–426. doi: 10.1007/s11920-000-0027-1. [DOI] [PubMed] [Google Scholar]
- Sandahl C, Lindberg S, Bergman H. The relation between drinking habits and neuroticism and weak ego among male and female alcoholic patients. Acta Psychiatr Scand. 1987;75:500–508. doi: 10.1111/j.1600-0447.1987.tb02825.x. [DOI] [PubMed] [Google Scholar]
- Van Os J, Jones PB. Neuroticism as a risk factor for schizophrenia. Psychol Med. 2001;31:1129–1134. doi: 10.1017/s0033291701004044. [DOI] [PubMed] [Google Scholar]
- Wilson RS, Schneider JA, Arnold SE, Bienias JL, Bennett DA. Conscientiousness and the incidence of Alzheimer disease and mild cognitive impairment. Arch Gen Psychiatry. 2007;64:1204–1212. doi: 10.1001/archpsyc.64.10.1204. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Eysenck SB. Psychoticism and response to treatment in neurotic patients. Behav Res Ther. 1978;16:183–189. doi: 10.1016/0005-7967(78)90065-7. [DOI] [PubMed] [Google Scholar]
- Meins W, Dammast J. Do personality traits predict the occurrence of Alzheimer's disease. Int J Geriatr Psychiatry. 2000;15:120–124. doi: 10.1002/(sici)1099-1166(200002)15:2<120::aid-gps84>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Distel MA, Trull TJ, Willemsen G, et al. The five-factor model of personality and borderline personality disorder: a genetic analysis of comorbidity. Biol Psychiatry. 2009;66:1131–1138. doi: 10.1016/j.biopsych.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Goodwin RD, Stein MB. Peptic ulcer disease and neuroticism in the United States adult population. Psychother Psychosom. 2003;72:10–15. doi: 10.1159/000067184. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Pedersen NL, Plomin R, Faire UD, McClearn GE. Type A behavior pattern, related personality traits and self-reported coronary heart disease. Person Individ Diff. 1989;10:419–426. [Google Scholar]
- Robertson DA, Ray J, Diamond I, Edwards JG. Personality profile and affective state of patients with inflammatory bowel disease. Gut. 1989;30:623–626. doi: 10.1136/gut.30.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley BA, Weiss A, Der G, Taylor MD, Deary IJ. Neuroticism, extraversion, and mortality in the UK Health and Lifestyle Survey: a 21-year prospective cohort study. Psychosom Med. 2007;69:923–931. doi: 10.1097/PSY.0b013e31815abf83. [DOI] [PubMed] [Google Scholar]
- Winstow DJ, Wakefield JA, Goldsmith WM. The relationship between personality, health symptoms and disease. Person individ Diff. 1990;11:717–723. [Google Scholar]
- Sutin AR, Costa PT, Uda M, Ferrucci L, Schlessinger D, Terracciano A. Personality and metabolic syndrome. Age (Dordr) 2010;32:513–519. doi: 10.1007/s11357-010-9153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Sutin AR, McCrae RR, et al. Facets of personality linked to underweight and overweight. Psychosom Med. 2009;71:682–689. doi: 10.1097/PSY.0b013e3181a2925b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutin AR, Scuteri A, Lakatta EG, et al. Trait antagonism and the progression of arterial thickening: women with antagonistic traits have similar carotid arterial thickness as men. Hypertension. 2010;56:617–622. doi: 10.1161/HYPERTENSIONAHA.110.155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutin AR, Terracciano A, Deiana B, et al. Cholesterol, triglycerides, and the Five-Factor Model of personality. Biol Psychol. 2010;84:186–191. doi: 10.1016/j.biopsycho.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutin AR, Terracciano A, Deiana B, et al. High neuroticism and low conscientiousness are associated with interleukin-6. Psychol Med. 2010;40:1485–1493. doi: 10.1017/S0033291709992029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N, Schuur M, Gusareva ES, et al. A genome-wide linkage study of individuals with high scores on NEO personality traits. Mol Psychiatry. 2012;17:1031–1041. doi: 10.1038/mp.2011.97. [DOI] [PubMed] [Google Scholar]
- de Moor MHM, Costa PT, Terracciano A, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. 2012;17:337–349. doi: 10.1038/mp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Cherny SS, Purcell S, Hewitt JK. Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am J Hum Genet. 2000;66:1616–1630. doi: 10.1086/302891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MH, Costa PT, Terracciano A, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. 2012;17:337–349. doi: 10.1038/mp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PT, McCrae RR. Revised NEO Personality Inventory (NEO-PI-R) and NEO Five Factor Inventory (NEO-FFI) Professional Manual. Psychological Assessment Resources; 1992. [Google Scholar]
- McCrae RR, Terracciano A, Personality Profiles of Cultures P Universal features of personality traits from the observer's perspective: data from 50 cultures. J Pers Soc Psychol. 2005;88:547–561. doi: 10.1037/0022-3514.88.3.547. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Costa PT, McCrae RR. Personality plasticity after age 30. Pers Soc Psychol Bull. 2006;32:999–1009. doi: 10.1177/0146167206288599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesanz JC, West SG. Towards understanding assessments of the big five: multitrait-multimethod analyses of convergent and discriminant validity across measurement occasion and type of observer. J Pers. 2004;72:845–876. doi: 10.1111/j.0022-3506.2004.00282.x. [DOI] [PubMed] [Google Scholar]
- Costa PT, Terracciano A, McCrae RR. Gender differences in personality traits across cultures: robust and surprising findings. J Pers Soc Psychol. 2001;81:322–331. doi: 10.1037/0022-3514.81.2.322. [DOI] [PubMed] [Google Scholar]
- Budaev SV. Personality and Individual Differences. Vol. 26. Amsterdam: Elsevier; 1999. pp. 801–813. [Google Scholar]
- Goodwin RD, Gotlib IH. Gender differences in depression: the role of personality factors. Psychiatry Res. 2004;126:135–142. doi: 10.1016/j.psychres.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Chen WM, Abecasis GR. Estimating the power of variance component linkage analysis in large pedigrees. Genet Epidemiol. 2006;30:471–484. doi: 10.1002/gepi.20160. [DOI] [PubMed] [Google Scholar]
- Pardo LM, MacKay I, Oostra B, van Duijn CM, Aulchenko YS. The effect of genetic drift in a young genetically isolated population. Ann Hum Genet. 2005;69:288–295. doi: 10.1046/j.1529-8817.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Falchi M, Fuchsberger C. Jenti: an efficient tool for mining complex inbred genealogies. Bioinformatics. 2008;24:724–726. doi: 10.1093/bioinformatics/btm617. [DOI] [PubMed] [Google Scholar]
- Axenovich TI, Zorkoltseva IV, Liu F, Kirichenko AV, Aulchenko YS. Breaking loops in large complex pedigrees. Hum Hered. 2008;65:57–65. doi: 10.1159/000108937. [DOI] [PubMed] [Google Scholar]
- Bellenguez C, Ober C, Bourgain C. A multiple splitting approach to linkage analysis in large pedigrees identifies a linkage to asthma on chromosome 12. Genet Epidemiol. 2009;33:207–216. doi: 10.1002/gepi.20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, de Geus EJ, Vink JM, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- Matise TC, Chen F, Chen W, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, et al. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets Amer J Hum Genetics 20016950411462172 [Google Scholar]
- Pergadia ML, Agrawal A, Loukola A, et al. Genetic linkage findings for DSM-IV nicotine withdrawal in two populations. Am J Med Genet B. 2009;150B:950–959. doi: 10.1002/ajmg.b.30924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MJ, Martin NG. The Brisbane Adolescent Twin Study: outline of study methods and research projects. Aust J Psychol. 2004;56:65–78. [Google Scholar]
- Distel MA, Trull TJ, Derom CA, et al. Heritability of borderline personality disorder features is similar across three countries. Psychol Med. 2007;38:1219–1229. doi: 10.1017/S0033291707002024. [DOI] [PubMed] [Google Scholar]
- Medland SE, Nyholt DR, Painter JN, et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi F, Levinson DF, Lewis CM. GSMA: software implementation of the genome search meta-analysis method. Bioinformatics. 2005;21:4430–4431. doi: 10.1093/bioinformatics/bti725. [DOI] [PubMed] [Google Scholar]
- Wise LH, Lanchbury JS, Lewis CM. Meta-analysis of genome searches. Ann Hum Genet. 1999;63:263–272. doi: 10.1046/j.1469-1809.1999.6330263.x. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Levinson MD, Segurado R, Lewis CM. Genome scan meta-analysis of schizophrenia and bipolar disorder, part I: methods and power analysis. Am J Hum Genet. 2003;73:17–33. doi: 10.1086/376548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- Shugart YY, O'Connell JR, Wilson AF. An evaluation of the variance components approach: type I error, power and size of the estimated effect. Eur J Hum Genet. 2002;10:133–136. doi: 10.1038/sj.ejhg.5200772. [DOI] [PubMed] [Google Scholar]
- Province MA, Kardia SL, Ranade K, et al. A meta-analysis of genome-wide linkage scans for hypertension: the National Heart, Lung and Blood Institute Family Blood Pressure Program. Am J Hypertens. 2003;16:144–147. doi: 10.1016/s0895-7061(02)03248-x. [DOI] [PubMed] [Google Scholar]
- Bhalla K, Luo Y, Buchan T, et al. Alterations in CDH15 and KIRREL3 in patients with mild to severe intellectual disability. Am J Hum Genet. 2008;83:703–713. doi: 10.1016/j.ajhg.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cader ZM, Noble-Topham S, Dyment DA, et al. Significant linkage to migraine with aura on chromosome 11q24. Hum Mol Genet. 2003;12:2511–2517. doi: 10.1093/hmg/ddg252. [DOI] [PubMed] [Google Scholar]
- Nikali K, Suomalainen A, Terwilliger J, Koskinen T, Weissenbach J, Peltonen L. Random search for shared chromosomal regions in four affected individuals: the assignment of a new hereditary ataxia locus. Am J Hum Genet. 1995;56:1088–1095. [PMC free article] [PubMed] [Google Scholar]
- Liu F, Arias-Vasquez A, Sleegers K, et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81:17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Blacker D, Mullin K, et al. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.