Abstract
Retrograde trophic signaling of nerve growth factor (NGF) supports neuronal survival and differentiation. Dysregulated trophic signaling could lead to various neurological disorders. Charcot-Marie-Tooth type 2B (CMT2B) is one of the most common inherited peripheral neuropathies characterized by severe terminal axonal loss. Genetic analysis of human CMT2B patients has revealed four missense point mutations in Rab7, a small GTPase that regulates late endosomal/lysosomal pathways, but the exact pathological mechanism remains poorly understood. Here, we show that these Rab7 mutants dysregulated axonal transport and diminished the retrograde signaling of NGF and its TrkA receptor. We found that all CMT2B Rab7 mutants were transported significantly faster than Rab7wt in the anterograde direction, accompanied with an increased percentile of anterograde Rab7-vesicles within axons of rat E15.5 dorsal root ganglion (DRG) neurons. In PC12M cells, the CMT2B Rab7 mutants drastically reduced the level of surface TrkA and NGF binding, presumably by premature degradation of TrkA. On the other hand, siRNA knock-down of endogenous Rab7 led to the appearance of large TrkA puncta in enlarged Rab5-early endosomes within the cytoplasm, suggesting delayed TrkA degradation. We also show that CMT2B Rab7 mutants markedly impaired NGF-induced Erk1/2 activation and differentiation in PC12M cells. Further analysis revealed that CMT2B Rab7 mutants caused axonal degeneration in rat E15.5 DRG neurons. We propose that Rab7 mutants induce premature degradation of retrograde NGF-TrkA trophic signaling, which may potentially contribute to the CMT2B disease.
Introduction
Charcot-Marie-Tooth type 2B (CMT2B) peripheral neuropathy is characterized by loss of distal pain sensation, muscle weakness, recurrent foot ulcers, and a frequent need for amputation of the lower limbs (Mersiyanova et al., 2000; Kuhlenbaumer et al., 2002; Verhoeven et al., 2003; Auer-Grumbach, 2004; Houlden et al., 2004). In CMT2B, the primary pathological feature is axonal degeneration (Auer-Grumbach et al., 2000, 2006; Axelrod and Gold-von Simson, 2007; Barisic et al., 2008). The time course and pattern of clinical manifestations are highly correlated with the neuron's axonal length; neurons with the longest axons are affected first. The pathology then spreads to neurons with shorter axons. Genomic analysis of patients with CMT2B disease has discovered four missense point mutations in the ubiquitous Rab7 protein: L129F (Verhoeven et al., 2003), V162M (Verhoeven et al., 2003), N161T (Houlden et al., 2004), and K157N (Meggouh et al., 2006).
By cycling between an active form (GTP-bound) and an inactive form (GDP-bound), Rab7, a member of the small GTPase family (Asbury, 1987; Bucci et al., 2000; Stenmark and Gillooly, 2001; Stenmark and Olkkonen, 2001), regulates the trafficking and maturation of late endosomes, lysosomes, and autophagosomes (Bucci et al., 2000). The CMT2B Rab7 mutations all occur within the highly conserved domain(s) in close proximity to the GTP-binding pocket (Cogli et al., 2009, 2010). A recent study revealed that these mutations loosen the regulation of GDP to GTP exchange by Rab7 guanidine nucleotide exchange factor, thus making them more prone to bind GTP (McCray et al., 2010) and behave like Rab7Q67L, a constitutively activated Rab7 mutant (Spinosa et al., 2008). Rab7Q67L induces aggregation and fusion of late endosomes/lysosomes to produce enlarged vesicles in HeLa cells, some as large as 10 μm in diameter (Bucci et al., 2000). It has been reported that Rab7 activation induced by growth factor withdrawal contributed to cell death (Romero Rosales et al., 2009). These hyperactivated CMT2B Rab7 mutants may thus interrupt normal cellular signaling and function (Bronfman et al., 2007). CMT2B Rab7 mutants appeared to disrupt normal NGF/TrkA signaling and block NGF-induced differentiation in PC12 cells (BasuRay et al., 2010; Cogli et al., 2010) and in DRG neurons (Yamauchi et al., 2010). However, the mechanism(s) by which these CMT2B Rab7 mutants cause axonal degeneration remains unclear. Because neurons with long axons are most susceptible to dysfunction of axonal transport, hyperactive Rab7 mutants may lead to axonal degeneration in CMT2B by disrupting the axonal transport process (Cogli et al., 2009).
Using primary DRG neurons and PC12M cells, we examined the effects of human CMT2B Rab7 mutations on signaling and axonal trafficking of NGF/TrkA. In DRG neurons, CMT2B Rab7 mutants moved significantly faster than wild-type Rab7 because of accelerated anterograde transport. These mutants impaired axonal trafficking of the TrkA receptor. CMT2B Rab7 mutants also markedly suppressed NGF-induced activation of the Erk1/2 signaling pathway, possibly by promoting degradation of surface TrkA receptors. These results suggest that hyperactivated CMT2B Rab7 mutants alter trafficking and signaling of retrograde NGF/TrkA trophic signals that could contribute to axonal degeneration in CMT2B disease.
Materials and Methods
Chemicals, reagents, media, and antibodies.
Hanks Balanced Salt Solution, neurobasal, Opti-MEM, trypsin, B27, l-glutamine, Glutamax, penicillin-streptomycin, and Lipofectamine 2000 were purchased from Invitrogen. DMEM-high glucose (Cellgro) was purchased from Mediatech. FBS was purchased from Phoenix Research Products. HEPES, poly-L-lysine, and AraC were purchased from Sigma. Electro-transfection reagents were purchased from Lonza Walkersville. PDMS was purchased from Sylgard (Dow Corning). NGF was purified from mouse submaxillary glands following a previously published protocol (Mobley et al., 1986). The following antibodies were used: rabbit anti-LAMP3, rabbit anti-GFP, anti-Rab5B (Santa Cruz Biotechnology); mouse anti-pErk1/2 (Santa Cruz Biotechnology); mouse anti-pAkt, rabbit anti-Akt, rabbit anti-Erk1/2, and rabbit anti-pTrkA (pY490) (Cell Signaling Technology); and rabbit anti-TrkA (UBI). Mouse anti-GAPDH (GT239) was from GeneTex, rabbit monoclonal antibodies against Rab5A, Rab7A, and Rab11A were from Epitomics. A mouse anti-β-tubulin antibody and Bisbenzimide H (Hoechst33258) were from Sigma. The rabbit p62 ployclonal antibody was from Medical & Biological Laboratories. The rabbit anti-rat trkA extracellular domain antibody (RTA) was generously provided by Dr. L. Reichardt of University of California, San Francisco. Goat anti-rabbit or anti-mouse IgG-Alexa568 conjugates were purchased from Invitrogen. GTP agarose beads were purchased from Innova Biosciences.
Plasmid construction and siRNA sequences.
The dog EGFP-Rab7 fusion constructs (EGFP-Rab7Q69L, EGFP-Rab7N125I, and EGFP-Rab7T22N) were generous gifts from Prof. C. Bucci (Università del Salento, Italy). The EGFP-Rab5Awt plasmid was from Professor M. Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). We cloned the full-length wild-type human Rab7 from a human cDNA library by PCR. The four single point mutation constructs (L129F, K157N, N161T, and V162M) were introduced by site-directed mutagenesis into the human Rab7 cDNA sequence by PCR. All constructs (digested with XhoI-EcoRI) were in-frame fused to the C terminus of the EGFP plasmid (pEGFP-N1 vector, Clontech). All sequences were verified by sequencing. These human constructs are designated as follows: EGFP-Rab7wt, EGFP-Rab7L129F, EGFP-Rab7K157N, EGFP-Rab7N161T, and EGFP-Rab7V162M). To make the expression vector of pcDNA3-TrkA-mCherry, the cDNA sequence of mCherry (a generous gift from Dr. R. Tsien of University of California, San Diego) was fused to the C terminus of rat TrkA cDNA (a gift from Dr. S. Meakin of University of Western Ontario) using the Gateway system (Invitrogen). TrkA-GFP was a kind gift from Dr. B. Rudkin (Centre National de la Recherche Scientifique, Paris, France). The siRNA pools were generated using Dicer-mediated cleavage of double-stranded RNAs (Chen et al., 2012). Rab7 (NP_076440) coding sequence 1 (CDS1) was amplified using the T7 forward primer: gcgtaatacgactcactataggggtgttggaaagacatcgc and the T7 reverse primer: gcgtaatacgactcactataggttgtacagctccacttccg; coding sequence 2 (CDS2) was amplified using the T7 forward primer: gcgtaatacgactcactataggccaagaacacacacgtagg and the T7 reverse primer: gcgtaatacgactcactataggggttgcatatattaacatgtac. The PCR products were transcribed using the T7 RNA polymerase, and the resulting sense and antisense strand mRNAs were annealed, followed by digestion using recombinant Dicer enzyme. The samples were purified to generate a pool of siRNAs. The GL3 siRNAs were made against Luciferase and were used as a nonspecific control for Rab7 siRNAs (Chen et al., 2012).
DRG dissection buffer and cell culture media.
The buffers and media were prepared as follows: dissection buffer: 1× Hanks Balanced Salt Solution with 10 mm HEPES, 200 U/ml penicillin, and 200 μg/ml streptomycin; quenching medium: 1× DMEM with 15% FBS, 200 U/ml penicillin, and 200 μg/ml streptomycin; plating medium: 1× DMEM with 10% FBS, 200 U/ml penicillin, 200 μg/ml streptomycin, and 50 ng/ml NGF; maintenance medium: 1× Neurobasal with 1× B27, 0.375 mm Glutamax, 200 U/ml penicillin, 200 μg/ml streptomycin, and 50 ng/ml NGF; and antimitotic medium: 1× Neurobasal with 1× B27, 0.125 mm l-glutamine, 0.375 mm Glutamax, 200 U/ml penicillin, 200 μg/ml streptomycin, 50 ng/ml NGF, and 4 μm AraC. PC12M cells were maintained in DMEM supplemented with 4.5 g/L glucose, 10% horse serum, 5% FBS, 200 U/ml penicillin, and 200 μg/ml streptomycin.
Primary DRG neuronal culture in microfluidic chambers.
DRG neurons were harvested and cultured from rat E15.5 embryos of either sex according to a published protocol (Chan et al., 2000). Briefly, dissociated cells were transfected by electroporation (Amaxa) and plated into the channeled PDMS microfluidic chambers (Cui et al., 2007). Ten minutes after plating, maintenance medium was added to the cell culture and incubated for 24 h before being replaced with antimitotic medium to suppress the growth of non-neuronal cells. After incubating with antimitotic medium for 12 h, maintenance medium was added back to the cell culture. Half of maintenance medium was then changed every other day, and cells were cultured until imaging.
Cell culture and transient transfection.
PC12M cells were obtained from Dr. M. White (University of Texas Southwestern Medical Center, Dallas, Texas). These cells were maintained in DMEM-high glucose, 10% horse serum, 5% FBS, and 1% penicillin/streptomycin. Before transfection, PC12 cells were subcultured on glass coverslips precoated with poly-L-lysine for 24 h to achieve ∼90% confluency. Immediately before transfection, cells were changed to Opti-MEM for 30 min. A total of 0.5 μg DNA plasmid was mixed with 2 μl Lipofectamine 2000 in 100 μl Opti-MEM for 25 min at room temperature. For cotransfection with siRNA sequences, 1.0 μg TrkA-mCherry or TrkA-GFP plasmids were premixed with 1.0 μg siRNA sequences followed by incubation with 3 μl Lipofectamine 2000 for 25 min at room temperature. The mixture was added drop-wise into the cell culture. Cells were switched back to maintenance media 3–4 h after transfection. Cells were imaged 36–48 h thereafter. For NGF-induced differentiation, cells were treated with 50 ng/ml NGF for 3 d and were fixed and stained for analysis.
An Amaxa nucleoporation device (Lonza) was used for transfection of DRG neurons. Rat E15 DRG neurons were prepared as described above. For each transfection, ∼0.3 million cells were resuspended in 20 μl of the transfection buffer (Amaxa rat neuron transfection kit, catalog #VSPI-1003). A total amount of ∼0.6 μg of DNA plasmid was added to the cell suspension and thoroughly mixed in a test tube. The cell-plasmid solution was then transferred to a cuvette for transfection by electroporation using Amaxa's built-in SCN Program 4. The reaction was immediately quenched by the addition of 0.5 ml plating medium to the cell-plasmid solution for recovery. Cells were centrifuged, resuspended in 20 μl of plating media, and loaded into the PDMS microfluidic devices.
For measuring NGF-induced neurite outgrowth, PC12M cells were transfected as described above and were treated with 50 ng/ml NGF for 72 h. Images of transfected cells were captured with a Leica Microscope (Leica, DMI6000B) equipped with a 20× objective lens and a CCD camera (Photometrics, Coolsnap HQ2). Neurites were measured and analyzed using ImageJ.
To assay for axonal degeneration in DRG neurons, E15–E16 rat DRG neurons were cultured on glass coverslips for 3 d, at which point the neuron's axons were well extended. We then transfected the neurons with the various Rab7 constructs using Magnefect-NanoII (Nanotherics), a technique that allowed us to transfect adhered neurons preserving the existing axons/neurites. After 72 h, the length of the axons was measured and analyzed using ImageJ.
Immunostaining.
Cells were grown on glass coverslips and transfected as described earlier. At 24–36 h after transfection, cells were fixed in 4% PFA for 15 min at room temperature. Samples were then permeabilized and blocked with 0.15% Triton X-100, 1% BSA in PBS for 10 min. The cells were incubated with diluted primary antibodies (1/100 for LAMP-3, 1/200 for p62, and 1/200 for RTA, respectively) for 1 h at room temperature, rinsed, and incubated with goat anti-rabbit IgG-Alexa568 conjugates (1/800) for 45 min at room temperature. Nuclei were stained with Hoechst 33258 (0.1 μg/ml) in PBS at room temperature for 10 min. To stain for surface TrkA with the RTA IgGs, the same protocol was followed, except that cells were treated in blocking buffer, from which 0.15% Triton X-100 was omitted. All images were collected with a Leica microscope (Leica, DMI6000B) equipped with an 100× oil objective lens and a CCD camera (Photometrics, Coolsnap HQ2).
GTP-agarose pulldown assays, SDS-PAGE/immunoblotting.
We adopted a novel assay system for measuring the amount of GTP-bound forms of Rab7 using GTP agarose beads (Innova Biosciences) following a published protocol (Baloh et al., 2007) with some modifications. Briefly, 48 h after transfection, cells were collected and lysed in 50 mm Tris-HCl pH 7.5, 250 mm NaCl, 5 mm Mg acetate, 0.5% Triton X-100, and protease inhibitors. The samples were centrifuged, and clear supernatants were incubated with GTP-agarose beads for 2 h at 4°C with rotation. The GTP-agarose beads were washed and boiled in SDS-loading buffer before SDS-PAGE. The amount of EGFP-Rab7 that bound to GTP was measured by blotting with a specific antibody to GFP for EGFP-tagged Rab7 constructs.
Established protocols were followed for SDS-PAGE/immunoblotting (Wu et al., 2001, 2007). Equal amounts of proteins were separated on 4–12% NuPAGE Bis-Tris precast gel (Invitrogen) and then electro-transferred to PVDF membranes. The membranes were blocked with 5% nonfat milk, and probed with specific antibodies as indicated. All blots were captured using ChemiDoc XRS+ (Bio-Rad), and blots within linear ranges were quantitated using the Image Lab version 3.0.1 software (Bio-Rad).
Live cell imaging and measuring the sizes of TrkA-mCherry endosomes and data analysis.
PC12M cells and DRG neurons were observed using an inverse microscope (Leica, DMI6000B), and images were captured using a CCD camera (Photometrics, Coolsnap HQ2). A 100× oil-immersion objective (HCX PL Fluotar, NA 1.3) was used for DIC and fluorescence imaging. Immediately before imaging, maintenance medium was replaced by warm CO2-independent medium to maintain the pH during data acquisition. Fluorescently labeled endosomes were first located, followed by 60 consecutive snapshots at a capturing speed of 2 s/frame. Kymographs were generated using ImageJ (National Institutes of Health), and speeds from individual traces were calculated using the Matlab program. ImageJ was used to measure the size distribution of TrkA-mCherry endosomes.
Statistical analysis.
Statistical significance of quantitative data was determined by applying a two-tailed Student's t test to the mean values.
Results
Expression of CMT2B Rab7 mutants induces enlarged vesicles in PC12M cells but not in DRG neurons
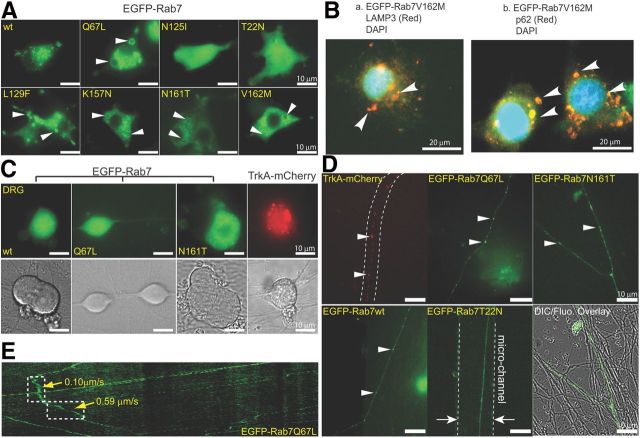
Expression of all Rab7 mutants was first performed in PC12M cells using human Rab7 constructs that were all N-terminally tagged with EGFP. As shown in Figure 1A, wild-type Rab7 (EGFP-Rab7wt) was mainly located in small vesicular structures, presumably late endosomes and lysosomes (Bucci et al., 2000). However, all four CMT2B Rab7 mutants (EGFP-Rab7L129F, EGFP-Rab7K157N, EGFP-Rab7N161T, and EGFP-Rab7V162M) and the dog constitutively active mutant (EGFP-Rab7Q67L) exhibited large donut-ring-like vacuolar structures in conjunction with small vesicles dispersed throughout the cytoplasm. On the other hand, the dominant-negative dog Rab7 mutants (EGFP-Rab7N125I and EGFP-Rab7T22N) showed diffused green fluorescence signals with few discernible vesicular structures (Fig. 1A). The enlarged Rab7 vesicles in PC12M cells transfected with the CMT2B mutants and the constitutively active mutant confirmed previous reports that those CMT2B Rab7 mutants were overly active (Bucci et al., 2000; Spinosa et al., 2008) (see Fig. 6B). To investigate whether these abnormally enlarged vesicles were lysosomes/autophagosomes, we fixed cells that expressed EGFP-Rab7V162M, and the samples were stained with specific antibodies for LAMP3 and p62, markers for lysosomes and autophagosomes, respectively. As shown in the representative images, signals for EGFP-Rab7V162M showed marked colocalization with LAMP3 (Fig. 1Ba, arrowheads) and with p62 (Fig. 1Bb, arrowheads). These results are evidence that the enlarged vesicles seen in CMT2B Rab7 mutant-expressing cells are lysosomes/autophagosomes.
Figure 1.
Expression profiles of CMT2B Rab7 mutants in PC12M cells and in rat DRG neurons. A, Fluorescent images of EGFP labeled wild-type (wt), constitutively active (Q67L), dominant-negative (N125I and T22N), and the four CMT2B Rab7 mutants (L129F, K157N, N161T, and V162M) expressed in PC12M cell lines. Rab7wt accumulates as small puncta structures, whereas the constitutively active and CMT2B mutants generate ring-shape vesicular structures. The dominant-negative Rab7 mutants are diffused within the cytoplasm. B, Cells that expressed EGFP-Rab7V162M were fixed and stained for lysosomes with rabbit anti-LAMP3 IgGs or for autophagosomes with rabbit anti-p62 IgGs. Nuclear staining was also performed. Representative images are shown: Ba, LAMP3. Bb, p62. Arrowheads indicate colocalization. C, Expression of the wild-type, the constitutively active (Q67L), the CMT2B mutant N161T, and TrkA-mCherry in DRG neurons visualized by fluorescent microscopy with corresponding DIC images shown. D, Snapshots of endosomes containing TrkA-mCherry or EGFP-Rab7 in DRG axons. TrkA-mCherry signals show a puncta pattern in DRG axons (top left), and a similar pattern is seen for EGFP-Rab7Q67L and EGFP-Rab7N161T (top middle and right). The puncta pattern for EGFP-Rab7wt is less pronounced than EGFP-Rab7Q67L and EGFP-Rab7N161T (bottom left). The dominant-negative form, EGFP-Rab7T22N, shows a diffused pattern (bottom middle). A micro-channel in a microfluidic device is indicated to determine the direction of axonal transport (bottom middle). A merged image between the EGFP-Rab7N161T channel and its DIC channel is also presented to indicate typical transfection efficiency (bottom right). E, Representative kymographs of axonal transport of endosomes containing EGFP-Rab7Q67L are shown with the transport speeds of two segments computed.
Figure 6.
CMT2B Rab7 mutants show enhanced activation (i.e., GTP-bound) and markedly suppress the NGF-induced Erk1/2 activation. As in Figure 5, PC12M cells were cultured and transfected with the indicated constructs. Control and transfected cells were treated with 50 ng/ml NGF for 30 min and cell lysates were harvested for immunoblotting. A, The lysates were analyzed using an antibody against GFP to show that all the EGFP-Rab7 constructs were expressed at about the same level. B, Cell lysates were incubated with GTP-agarose beads to pull down activated Rab7 proteins. The amounts of EGFP-Rab7 proteins were analyzed by SDS-PAGE and blotted with an antibody against GFP. A, The blot was then reprobed with an antibody against Rab7A to indicate the endogenous levels of Rab7A under all conditions (C). D, The lysates are probed with antibodies against pAkt, total Akt, and (E) with antibodies against pErk1/2 and total Erk1/2. A nontreated sample was also included as a control. D, E, Representative blots within linear exposure were quantitated using ImageLab (Bio-Rad), and the results are shown. The level of activated pAkt (D) and pErk1/2 (E) in response to NGF treatment in untransfected cells was set at 100%.
Expression of these same Rab7 constructs in DRG neurons, however, did not show the same effect as in PC12M cells. Although the dominant-negative mutants (EGFP-Rab7N125I and EGFP-Rab7T22N) still showed diffused patterns in DRG neurons as in PC12M cells, we did not detect large ring-shaped vesicles for either CMT2B Rab7 mutants (i.e., EGFP-Rab7N161T) or for the EGFP-Rab7Q67L constitutively active mutant in DRG neurons (Fig. 1C). Instead, those mutants showed small vesicular structures, similar to those from EGFP-Rab7wt (Fig. 1C). As a comparison, TrkA-mCherry, a fluorescently labeled NGF receptor, also exhibited similar puncta structures in the cell bodies of DRG neurons (Fig. 1C).
Along the axon, TrkA-mCherry, EGFP-Rab7Q67L, and EGFP-Rab7N161T showed punctuated patterns (Fig. 1D). The punctuated pattern for EGFP-Rab7wt was less pronounced (Fig. 1D). As expected, signals for the dominant-negative mutant, EGFP-Rab7T22N, were diffused throughout the axon (Fig. 1D). We observed no gross morphological differences of axons between DRG neurons transfected with either TrkA or Rab7 mutants and nontransfected neurons in the same culture (Fig. 1D). To quantitate axonal transport, we generated kymographs based on time-lapsed imaging series. The transport speed of each segment in the kymograph was calculated as depicted for EGFP-Rab7Q67L (Fig. 1E), and the mean velocity was computed as the time-weighted average over all endosomes.
CMT2B Rab7 mutants move within axons of DRG neurons at a faster speed than their wild-type counterpart
We next examined the overall mobility of CMT2B Rab7 mutant-containing vesicles within axons of DRG neurons by calculating the mean speed of their retrograde and anterograde axonal transport. Judging from the enlarged vesicles induced by CMT2B Rab7 mutants in PC12M cells, we initially speculated that mutants may slow down their axonal transport because of steric hindrance. Because axonal transport is very sensitive to the healthy status of cells and other environmental factors, such as temperature, we needed to have an intrinsic calibration. We chose to use TrkA for this calibration because axonal transport of TrkA has been well established and extensively characterized in sympathetic and DRG neurons (Ehlers et al., 1995; Riccio et al., 1997; Tsui-Pierchala and Ginty, 1999; Delcroix et al., 2003; Huang and Reichardt, 2003; Jullien et al., 2003; Sorkin, 2005; Deinhardt et al., 2006; Moises et al., 2007). To achieve this, DRG neurons were singly transfected with either TrkA-mCherry plasmid or with individual EGFP-Rab7 plasmids using electroporation. Cells transfected with each EGFP-Rab7 construct were then separately mixed together with cells transfected with TrkA-mCherry before being plated onto poly-L-lysine-coated coverslips. As such, individually transfected DRG neurons in the same culture expressed either TrkA-mCherry or the particular Rab7 construct alone. Axonal transport of both TrkA-mCherry and EGFP-Rab7 signals was then recorded. The transport speeds of Rab7 variants were validated and collected only for those cultures with consistent TrkA-mCherry transport speeds. At room temperature, TrkA-mCherry showed a consistent average transporting speed of ∼0.7–0.8 μm/s from all the healthy cell cultures (Fig. 2A). All Rab7-containing endosomes traveled with an average speed of ∼0.3–0.5 μm/s, significantly slower than the TrkA-containing endosomes (Fig. 2A). As expected, signals for both EGFP-Rab7N125I and EGFP-Rab7T22N were diffused; thus, the transport speeds were not recorded.
Figure 2.
CMT2B Rab7 mutants move faster than Rab7wt within DRG axons but slower than TrkA-mCherry. A, Average transport speeds of singly transfected TrkA-mCherry and various EGFP-Rab7 constructs (wt, Q67L, L129F, K157N, N161T, V162M, N125I, and T22N). The different Rab7 constructs (Q67L, L129F, K157N, N161T, and V162M) moved significantly faster than Rab7wt, but much slower than TrkA-mCherry at room temperature. Axonal transport of the N125I and the T22N mutants were not registered because neither showed punctuated patterns. Similarly, at 37°C, the Q67L and N161T mutants moved significantly faster than the wt (inset). ***p < 0.001. B, A detailed comparison of retrograde and anterograde axonal transport speed between EGFP-Rab7wt, EGFP-Rab7Q67L, and EGFP-Rab7N161T for the inset of A. EGFP-Rab7wt, EGFP-Rab7Q67L, and EGFP-Rab7N161T moved at similar speeds in the retrograde direction. However, both EGFP-Rab7Q67L and EGFP-Rab7N161T traveled much faster than EGFP-Rab7wt in the anterograde direction. C, Percentile of endosomes transported in both directions. Percentage of anterograde endosomes increases for EGFP-Rab7N161T compared with EGFP-Rab7wt and EGFP-Rab7Q67L. D, Histograms of retrograde versus anterograde speed for EGFP-Rab7wt, Rab7Q67L, and EGFP-Rab7N161T. A fast anterograde transporting population (2.0–4.0 μm/s) appears for both EGFP-Rab7Q67L and EGFP-Rab7N161T compared with EGFP-Rab7wt.
As mentioned above, we expected slower axonal transport of CMT2B Rab7 mutants compared with the wild-type. Surprisingly, we observed the opposite: all CMT2B Rab7 mutants and the constitutively active EGFP-Rab7Q67L moved significantly faster than EGFP-Rab7wt. At room temperature, the average transport speed of EGFP-Rab7wt was 0.25 μm/s, significantly slower (p < 0.001) than the speed of EGFP-Rab7Q67L (0.4–0.5 μm/s) and all CMT2B Rab7 mutants (Fig. 2A). For each data point, we averaged the traveling speeds of 80–400 endosomes to ensure statistical significance of the measured differences (Table 1).
Table 1.
Summary of the mean speed and number of endosomes for TrkA and Rab7 endosomes at 25 ± 1°Ca
| Mean speed (μm/s) |
Endosome counts |
|||
|---|---|---|---|---|
| TrkA | Rab7 | TrkA | Rab7 | |
| Wild-type | 0.78 ± 0.02 | 0.25 ± 0.02 | 354 | 359 |
| Q67L | 0.78 ± 0.03 | 0.42 ± 0.02 | 465 | 390 |
| Ll29F | 0.76 ± 0.04 | 0.53 ± 0.03 | 229 | 171 |
| K157N | 0.80 ± 0.04 | 0.48 ± 0.03 | 226 | 153 |
| N161T | 0.77 ± 0.04 | 0.42 ± 0.03 | 254 | 81 |
| V162M | 0.84 ± 0.03 | 0.49 ± 0.07 | 250 | 86 |
| N125I | 0.77 ± 0.03 | NA | 221 | NA |
| T22N | 0.79 ± 0.08 | NA | 46 | NA |
aAverage axonal transport speed (mean ± SEM) of TrkA-mCherry and EGFP-Rab7 in DRG neurons at 25 ± 1°C.
NA, Not applicable.
Next we asked whether this trend was temperature-dependent. We performed the same experiments at 37°C using an on-stage temperature controller. We chose EGFP-Rab7N161T as the representative for the CMT2B Rab7 mutants. The speeds of axonal transport for TrkA-mCherry, EGFP-Rab7wt, EGFP-Rab7Q67L, and EGFP-Rab7N161T were compared. We found that the absolute speeds for all examined constructs at 37°C increased compared with those at room temperature (Fig. 2A, inset; Table 2). Yet, the mean speed for both EGFP-Rab7Q67L (0.59 ± 0.06 μm/s) and EGFP-Rab7N161T (0.69 ± 0.03 μm/s) was significantly greater than that of EGFP-Rab7wt (0.45 ± 0.03 μm/s) (Fig. 2A, inset; Table 2).
Table 2.
Summary of the mean speed of retrograde and anterograde transport of TrkA and Rab7 endosomes at 37 ± 1°Ca
| Retrograde speed (μm/s) |
Anterograde speed (μm/s) |
Mean speed (μm/s) |
||||
|---|---|---|---|---|---|---|
| TrkA | Rab7 | TrkA | Rab7 | TrkA | Rab7 | |
| Wild-type | 0.95 ± 0.04 | 0.53 ± 0.03 | 1.32 ± 0.06 | 0.31 ± 0.06 | 1.09 ± 0.03 | 0.45 ± 0.03 |
| Q67L | 0.84 ± 0.04 | 0.57 ± 0.04 | 1.47 ± 0.11 | 0.63 ± 0.15 | 0.99 ± 0.06 | 0.59 ± 0.06 |
| N161T | 0.90 ± 0.08 | 0.63 ± 0.03 | 1.12 ± 0.15 | 0.76 ± 0.06 | 1.02 ± 0.08 | 0.69 ± 0.03 |
| TrkA | 0.91 ± 0.03 | NA | 1.32 ± 0.06 | NA | 1.06 ± 0.03 | NA |
aAverage speed (mean ± SEM) of retrograde and anterograde axonal transport of TrkA-mCherry, EGFP-Rab7wt, EGFP-Rab7Q67L, and EGFP-Rab7N161T in DRG neurons at 37 ± 1°C.
NA, Not applicable.
CMT2B Rab7 mutants show accelerated axonal transport in the anterograde direction but not in the retrograde direction
To address whether the acceleration occurred in the retrograde, anterograde, or both directions, we plated the transfected DRG cells in microfluidic chambers and allowed the axons to grow across the channels. This setup allowed us to distinguish retrograde from anterograde transport (Cui et al., 2007). Average speeds for retrograde transport between EGFP-Rab7wt (0.53 ± 0.03 μm/s), EGFP-Rab7Q67L (0.57 ± 0.04 μm/s), and EGFP-Rab7N161T (0.63 ± 0.03 μm/s) did not vary significantly (Fig. 2B,D; Table 2). However, the anterograde transport of EGFP-Rab7Q67L (0.63 ± 0.15 μm/s) and EGFP-Rab7N161T (0.76 ± 0.06 μm/s) was significantly accelerated compared with EGFP-Rab7wt (0.31 ± 0.06 μm/s) (Fig. 2B,D; Table 2). Compared with EGFP-Rab7wt, both EGFP-Rab7Q67L and EGFP-Rab7N161T showed a significant increase in the faster transport component (∼2.0–4.0 μm/s) in the anterograde direction (Fig. 2D). Furthermore, the ratio between the number of anterogradely transported endosomes and the number of retrogradely transported endosomes was 0.49 for EGFP-Rab7wt, 0.49 for EGFP-Rab7Q67L, and 0.96 for EGFP-Rab7N161T (Fig. 2C; Table 3). Therefore, a significant increase in the anterograde transport speed is accompanied by a marked increase in the ratio of anterogradely transported endosomes versus retrogradely transported endosomes for EGFP-Rab7N161T. These results suggest that EGFP-Rab7N161T exhibits marked increases of transport flux in the anterograde direction compared with EGFP-Rab7wt.
Table 3.
Count of Rab7 endosomes undergoing retrograde and anterograde transport at 37 ± 1°Ca
| Anterograde endosomes | Retrograde endosomes | Anterograde/Retrograde ratio | |
|---|---|---|---|
| Wild-type | 103 | 211 | 0.49 |
| Q67L | 51 | 105 | 0.49 |
| N161T | 211 | 219 | 0.96 |
aCount of endosomes containing EGFP-Rab7wt, EGFP-Rab7Q67L, or EGFP-Rab7N161T in DRG neurons at 37 ± 1°C.
Expression of CMT2B Rab7 mutants suppresses axonal transport of TrkA-mCherry in DRG neurons
To examine whether CMT2B Rab7 mutants affect axonal transport of TrkA, the receptor that is essential for retrograde transport of NGF survival signals, we cotransfected EGFP-Rab7 constructs and TrkA-mCherry in the same DRG neurons and analyzed their transport behaviors. The transport speed of vesicles containing both TrkA-mCherry and EGFP-Rab7N161T was markedly reduced (∼0.36 μm/s) compared with that of vesicles containing either TrkA-mCherry alone (0.95 μm/s) or EGFP-Rab7N161T alone (0.67 μm/s) (Fig. 3A,B). We further analyzed anterograde and retrograde transport, and our results show that both anterograde and retrograde movement of the TrkA that colocalized with Rab7 were equally reduced (Fig. 3B). The average speed for axonally transported lysosomes as labeled by YFP-LAMP has been reported to be ∼0.3 μm/s (Lee et al., 2011), which is consistent with the speed of axonally transported vesicles containing both EGFP-Rab7N161T and TrkA-mCherry. These results suggest that TrkA-mCherry vesicles overlapped with EGFP-Rab7N161T are lysosomes within axons. To determine whether the reduction of TrkA-mCherry's transport speed resulted from activation of Rab7, we examined the effect of dominant-negative Rab7 mutant (EGFP-Rab7N125I) on axonal transport of TrkA-mCherry. In neurons cotransfected with EGFP-Rab7N125I and TrkA-mCherry, axonal transport of TrkA-mCherry was similar to that of singly transfected TrkA-mCherry in both the retrograde and anterograde directions (Fig. 3C). Our results suggest that overexpression of CMT2B Rab7 mutants may suppress axonal transport of TrkA in axons of DRG neurons.
Figure 3.

Overexpression of Rab7N161T inhibits axonal transport of TrkA-mCherry, whereas a dominant-negative Rab7 mutant (Rab7N125I) has no apparent effect on axonal transport of TrkA-mCherry in DRG neurons. A, Kymographs of EGFP-Rab7N161T and TrkA-mCherry signals in cotransfected axons showing that signals that contained TrkA-mCherry only moved faster than endosomes containing both EGFP-Rab7N161T and TrkA-mCherry. B, The average transport speeds of EGFP-Rab7N161T only, TrkA-mCherry only, or cotransfected with EGFP-Rab7N161T/TrkA-mCherry in DRG neurons were quantitated and shown. R, Retrograde; A, anterograde. C, A representative axon cotransfected with the dominant-negative EGFP-Rab7N125I mutant and TrkA-mCherry as analyzed by kymograph. Transport of TrkA-mCherry was not affected by the expression of the dominant-negative EGFP-Rab7N125I.
Overexpression of CMT2B Rab7 mutants reduces surface TrkA and inhibits NGF binding and internalization
To investigate the role(s) of Rab7 in NGF-TrkA signaling, we chose to examine the binding and internalization of NGF in Rab7-transfected PC12M cells that express endogenous TrkA receptors. Cells were incubated with 0.2 nm NGF-labeled QD605 (Qdot-NGF) on ice for 45 min, washed, and further incubated at 37°C for 30 min, a condition that favors TrkA binding by NGF. If TrkA receptors were present on the plasma membrane, Qdot-NGF would bind to TrkA and some Qdot-NGF/TrkA complex would be internalized into the cytoplasm upon warming up to 37°C. This is exactly what we observed for the nontransfected cells in all samples (Fig. 4A–D, yellow-dash profiles). However, in cells expressing EGFP-Rab7L129F or EGFP-Rab7N161T, a marked reduction of Qdot-NGF signals was seen on the cell surface and in the cytoplasm (Fig. 4B,C). We observed similar reduction on the binding and internalization of Qdot-NGF in cells expressing EGFP-Rab7wt or EGFP-Rab7Q67L (Fig. 4A,D). As reported earlier, overexpression of Rab7wt would lead to increased Rab7 activity (Cogli et al., 2010), similar to the function of EGFP-Rab7Q67L and CMT2B Rab7 mutants (Bucci et al., 2000; Cogli et al., 2009; Cogli et al., 2010; McCray et al., 2010). In contrast, overexpression of dominant-negative mutant EGFP-Rab7T22N did not appear to affect the binding or internalization of Qdot-NGF (Fig. 4E). This drastic contrast of NGF-binding ability on surfaces of Rab7-overexpressing cells and nontransfected cells clearly indicates that CMT2B Rab7 mutants cause downregulation of the NGF/TrkA signaling pathway, presumably by premature degradation of TrkA receptors.
Figure 4.

Overexpression of activated Rab7 constructs, but not the dominant-negative Rab7T22N mutant, inhibits NGF binding and internalization and reduces surface TrkA. PC12M cells were transfected with EGFP-Rab7wt (A), EGFP-Rab7L129F (B), EGFP-Rab7N161T (C), and EGFP-Rab7Q67L (D) using Lipofectamine 2000 as before. Cells were pretreated with 0.2 nm Qdot-NGF for 30 min on ice, washed, and warmed up to 37°C for 20 min. Live cell imaging was performed to detect EGFP-Rab7 constructs (A–C, middle) and Qdot-NGF signals (A–C, bottom). Expression of EGFP-Rab7wt, EGFP-Rab7L129F, and EGFP-Rab7N161T resulted in a marked reduction of Qdot-NGF signals from either the membrane or the cytoplasm (white-dashed profile). In nontransfected cells (yellow-dashed profile), Qdot-NGF was readily observed. A–C, The DIC images are presented in the top panels. D, Similar results were seen in EGFP-Rab7Q67L-expressing cells. E, In contrast, expression of EGFP-Rab7T22N did not affect Qdot-NGF binding and internalization. Cells that expressed EGFP-Rab7L129F (F) or EGFP-Rab7V162M (G) were fixed and stained for surface TrkA with the rabbit RTA IgGs, followed by nuclear staining. Compared with untransfected cells, cells that expressed either EGFP-Rab7L129F (F) or EGFP-Rab7V162M (G), as indicated with arrowheads, showed a marked reduction in RTA signals. Representative images are shown.
To confirm that CMT2B Rab7 mutants resulted in reduction of surface TrkA receptor, we used an antibody that recognized the extracellular domains of TrkA (RTA) to detect surface TrkA in cells that expressed CMT2B Rab7 mutants. After transfection, PC12M cells were fixed and blocked without permeabilization. Cells were then incubated with the RTA antibody that recognizes the extracellular domains of TrkA (Grimes et al., 1996). Thus, only surface TrkA was stained under these conditions. The RTA antibody was then visualized using a goat anti-rabbit IgG Alexa-568 conjugate. As shown in Figure 4F, two cells that expressed EGFP-Rab7L129F (Fig. 4Fa, arrowheads) were almost devoid of signals for RTA (Fig. 4Fc,Fd, arrowheads). Meanwhile, untransfected cells in the same image exhibited normal levels of TrkA as detected by the RTA antibody (Fig. 4Fc,Fd). Similarly, a marked reduction in RTA signals was obtained in cells that expressed EGFP-Rab7V162M (Fig. 4Ga–Gd). In a parallel set of experiments, cells were permeabilized using Triton X-100 before incubation with the RTA antibody to detect both surface and intracellular TrkA. Again, we observed a significant reduction in RTA signals (data not shown). We thus conclude that expression of CMT2B Rab7 mutants causes a reduction in the level of surface TrkA that may well account for the reduced binding of NGF in these cells.
Knockdown of Rab7 by siRNA causes enlarged TrkA vesicles that colocalize with Rab5-early endosomes in PC12M cells
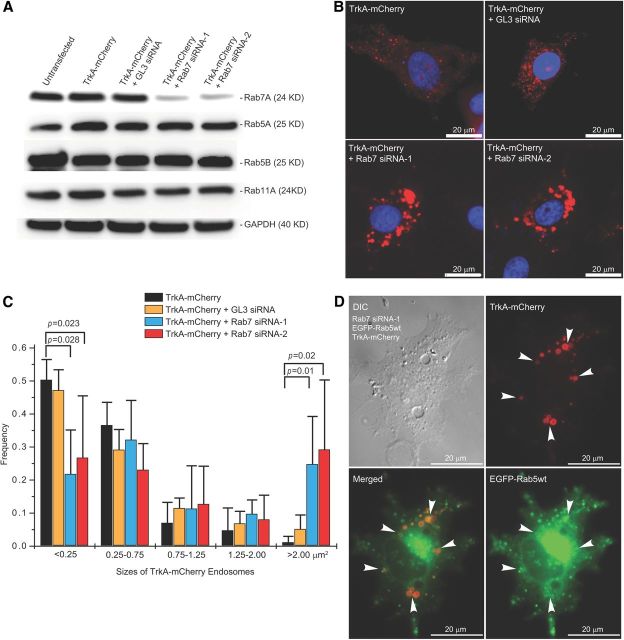
Based on the results described above, we hypothesized that knockdown of endogenous Rab7 would cause defective endocytic trafficking and protein degradation of TrkA. We designed two siRNA sequence pools against Rab7 as well as a negative control sequence (GL3). The efficiency of siRNA-mediated knockdown against Rab7 was analyzed by SDS-PAGE/immunoblotting. Transfection with Rab7 siRNA-1 and siRNA-2 reduced the Rab7 levels to ∼10–15% of those of nontransfected cells or cells transfected with the control siRNA (Fig. 5A). The knockdown was specific to Rab7 and did not disturb levels of other Rab proteins, such as Rab5A, Rab5B, and Rab11 (Fig. 5A). GAPDH was also blotted as a loading control (Fig. 5A). To investigate the effect of Rab7 knockdown on endocytic trafficking of the TrkA receptor, Rab7 siRNA-1, -2, and the negative control siRNA were separately cotransfected with TrkA-mCherry into PC12M cells. Transfected cells were examined by fluorescence microscopy 36–48 h after transfection. Suppression of Rab7 by siRNA-1 and -2 resulted in marked enlargement of TrkA-mCherry puncta compared with control cells that were singly transfected with TrkA-mCherry, an effect that was not detected in cells cotransfected with the GL3-negative control siRNA and TrkA-mCherry (Fig. 5B).
Figure 5.
RNAi-mediated knockdown of Rab7 causes abnormal accumulation of TrkA-mCherry signals within large Rab5-endocytic vesicles. PC12M cells were cultured and transfected with TrkA-mCherry alone, or together with the GL3 control siRNA, Rab7 siRNA-1, or Rab7 siRNA-2. A, Nontransfected cells as well as transfected cells were harvested for Western blotting analysis using specific antibodies as indicated. Both Rab7 SiRNA-1 and -2 specifically reduced Rab7 expression efficiently. The levels of Rab5A, Rab5B, and Rab11A were not affected. GAPDH was blotted as well and serves as a loading control. B, Representative images are shown for cells expressing TrkA-mCherry alone (top left), TrkA-mCherry + GL3 siRNA (top right), TrkA-mCherry + Rab7 siRNA-1 (bottom left), and TrkA-mCherry + Rab7 siRNA-2 (bottom right). The size distribution of TrkA-mCherry endosomes in each condition (n = 5) was quantified using ImageJ. C, Both Rab7 SiRNA-1 and -2 induced enlarged TrkA vesicles. D, Cells were cultured on glass coverslips and cotranfected with Rab7 siRNA-1, EGFP-Rab5wt, and TrkA-mCherry. Live cell imaging was performed to determine colocalization of EGFP-Rab5wt and TrkA-mCherry. A representative cell is shown. Arrowheads indicate colocalization of TrkA-mCherry with EGFP-Rab5wt.
To quantitate the size differences, we constructed the size distribution histogram of TrkA-mCherry by binning the size of TrkA-mCherry puncta into five groups (<0.25, 0.25–0.75, 0.75–1.25, 1.25–2.0, and >2.00 μm2). For the large endosomes (>2.0 μm2), a significant population of TrkA-mCherry-containing endosomes emerged: ∼25% for Rab7 siRNA-1 and ∼30% for Rab7 siRNA-2 compared with 5% for the negative control GL3 siRNA. In contrast, only ∼2% of TrkA-mCherry-carrying endosomes fell in this group for cells singly transfected with TrkA-mCherry (Fig. 5C). Abnormally enlarged TrkA-mCherry endosomes in the cytoplasm could be the result of defective TrkA degradation or defective endocytic trafficking of TrkA.
We next performed experiments to define the nature of the enlarged vesicles that contained TrkA-mCherry in Rab7-knockdown cells by siRNA. Surface receptor-mediated endocytosis proceeds from early endosomes (marked by Rab5) to late endosomes/lysosomes (marked by Rab7) (Rink et al., 2005). This is also true for the NGF/TrkA signals (Saxena et al., 2005; Deinhardt et al., 2006; Wu et al., 2007, 2009). Because Rab7 plays a key role in regulating the biogenesis of lysosomes and autophagosomes (Bucci et al., 2000; Cogli et al., 2009), we predicted that reduction of Rab7 would result in accumulation of TrkA in early endosomes marked by Rab5. For this purpose, we cotransfected PC12M cells with EGFP-Rab5wt, TrkA-mCherry, and siRNA against Rab7 and performed live cell imaging. As shown in Figure 5D, we indeed observed excellent colocalization of EGFP-Rab5wt with enlarged vesicles containing TrkA-mCherry (Fig. 5D, arrowheads). These results are evidence that TrkA-mCherry is stalled in Rab5-early endosomes, and its degradation is prevented when Rab7 is reduced by siRNA knockdown.
Expression of CMT2B Rab7 mutants selectively impairs NGF-induced Erk1/2 activation but not activation of the PI3K/Akt pathway
To investigate the effect of CMT2B Rab7 mutants on NGF-TrkA downstream signaling pathways, we transfected these constructs into PC12M cells and examined the Erk1/2 and PI3K/Akt signaling pathways. In general, we achieved ∼30% transfection efficiencies. By immunoblotting analysis, we found that the expression levels of these constructs in PC12M cells were similar (Fig. 6A) and the level of endogenous Rab7 was not markedly affected (Fig. 6C).
We then examined whether CMT2B Rab7 mutants exhibited elevated levels of activation, (i.e., GTP-bound form) by performing the GTP-agarose pulldown assay. After transfection, PC12M cells were rinsed and lysed as described in the Materials and Methods. The lysates were incubated with GTP-agarose beads to pull down the GTP-bound forms that were detected by SDS-PAGE/immunoblotting (Fig. 6B). We indeed observed that the constitutively active mutant (EGFP-Rab7Q67L) and the CMT2B EGFP-Rab7L129F and EGFP-Rab7K157N mutants had significantly more Rab7 in the GTP-bound state compared with the controls (EGFP alone, EGFP-Rab7wt, EGFP-Rab7T22N). The CMT2B EGFP-Rab7N161T and EGFP-Rab7V162 mutants were also present in their GTP-bound state more than the controls but less pronounced than the constitutively active mutant (Q67L) and the CMT2B EGFP-Rab7L129F and EGFPRab7K157N mutants. Consistent with previously published reports (Spinosa et al., 2008; McCray et al., 2010), these results are evidence that CMT2B Rab7 mutants exhibit enhanced levels of activation.
In a parallel set of experiments, we treated transfected cells with NGF (final concentration, 50 ng/ml for 30 min), analyzed the cell lysates by SDS-PAGE/immunoblotting, and probed for both pErk1/2 and pAkt. Nontransfected cells either treated with NGF (+) or with NGF vehicle (−) were used as controls (Fig. 6D,E). Similar to a recent report (BasuRay et al., 2010), the activation of Akt was not markedly affected by the expression of these various Rab7 constructs (Fig. 6D). The effects of Rab7 expression on the Erk1/2 pathway appeared controversial; either an enhancement (BasuRay et al., 2010) or a suppression (Saxena et al., 2005) of the Erk1/2 pathway by Rab7 has been reported. We observed that NGF-induced activation of Erk1/2 appeared to be suppressed in CMT2B Rab7 mutant-expressing cells, but statistical analysis showed that this reduction was not significant (Fig. 6E). Because activation of the Erk1/2 pathway requires internalization of NGF/TrkA (Zhang et al., 2000), our findings suggest that internalization and endocytic trafficking of NGF/TrkA may be impaired in cells expressing CMT2B Rab7 mutants.
Expression of CMT2B Rab7 mutants inhibits NGF-induced differentiation in PC12M cells and causes axonal degeneration in DRG neurons
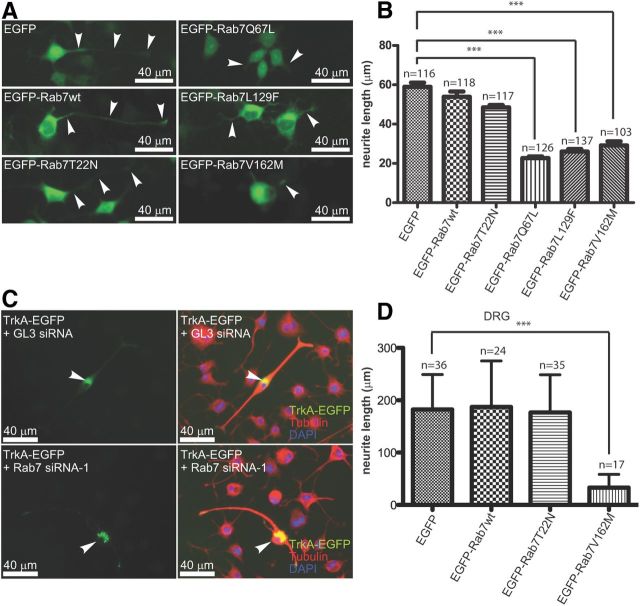
Based on the results that CMT2B Rab7 mutants impaired NGF-induced Erk1/2 activation, we expected that expression of CMT2B Rab7 mutants would block or decrease NGF-induced neurite outgrowth. We transfected the various Rab7 constructs into PC12M cells and treated the cells with 50 ng/ml NGF to induce differentiation. We then measured the length of neurites after 3 d of NGF treatment. Representative images are shown in Figure 7A, and quantitative results are shown in Figure 7B. PC12M cells transfected with EGFP alone, the wild-type (EGFP-Rab7wt), and the dominant-negative (EGFP-Rab7T22N) all showed similar mean neurite lengths of 58.97, 53.89, and 48.47 μm, respectively. However, expression of the constitutively active mutant (EGFP-Rab7Q67L) and the two CMT2B Rab7 mutants (EGFP-Rab7L129F and EGFP-Rab7V162M) all resulted in a significant reduction in neurite length (22.69, 26.20, and 27.36 μm, respectively). These results demonstrate that hyperactivated CMT2B Rab7 mutants inhibit NGF-induced differentiation in PC12M cells.
Figure 7.
Expression of CMT2B Rab7 Mutants inhibits NGF-induced differentiation in PC12M cells and causes axonal degeneration in DRG neurons. A, B, PC12M cells were transfected, treated with 50 ng/ml NGF for 3 d, and then the length of neurites was measured and quantitated. A, Representative images. B, Quantitation. Similar to EGFP-Rab7Q67L, expression of EGFP-Rab7L129L and EGFP-Rab7V162M resulted in significant inhibition of NGF-induced differentiation in these cells. C, The TrkA-GFP construct was cotransfected into PC12M cells with either Rab7 siRNA-1 (bottom) or GL3 control siRNA (top). Cells were then treated with NGF (50 ng/ml) for 3 d to induce differentiation. Cells were fixed and stained with an antibody against tubulin and DAPI to stain for the nuclei. Left, GFP images. Right, Images of tubulin and DAPI for revealing the cell morphology. Knockdown of Rab7 did not affect cell differentiation. D, Rat E15/16 DRG neurons were cultured for 3 d and then transfected using Magnefect-NanoII (Nanotherics), a technique that allowed us to transfect adhered neurons and preserve the existing axons/neurites. After 72 h, the length of the axons was measured using ImageJ and the quantitative results are shown. Expression of EGFP-Rab7V162M causes shortening of axons. *p < 0.001.
To investigate whether siRNA knockdown of Rab7 affected NGF-induced differentiation in PC12M cells, TrkA-EGFP was cotransfected with either Rab7 siRNA-1 or GL3 siRNA into Neuroscreen-1, a subcell line of PC12M cells. Cells were then treated with NGF (50 ng/ml) for 3 d to induce neurite outgrowth. As shown in Figure 7C, compared with cells transfected with GL3-negative control siRNA, Rab7 knock-down caused enlarged TrkA-GFP vesicles, consistent with the results for TrkA-mCherry (Fig. 5B,D). However, NGF-induced differentiation was not apparently impacted when Rab7 was knocked down by siRNA (Fig. 7C). This observation is consistent with the results obtained from the dominant-negative mutant, EGFP-Rab7T22N (Fig. 7A,B), which also did not appear to affect NGF-induced Erk1/2 activation (Fig. 6E). These results suggest that neither the cellular level of Rab7 nor its activation is required for NGF-induced differentiation in PC12 cells.
To investigate whether CMT2B Rab7 mutants cause axonal degeneration in DRG neurons, we grew E15–E16 rat DRG neurons on glass coverslips for 3 d, at which point the neuron's axons/neurites were well extended. We then transfected the neurons with the various Rab7 constructs using Magnefect-NanoII (Nanotherics), a technique that allowed us to transfect adhered neurons preserving the existing axons/neurites. After 72 h, we measured the length of the axons/neurites using ImageJ; the results are shown in Figure 7D. DRG neurons transfected with EGFP had a mean neurite length of 182.3 μm. Similarly, DRG neurons transfected with the wild-type (EGFP-Rab7wt) and the dominant-negative (EGFP-Rab7T22N) constructs showed a mean neurite length of 187.0 μm and 176.7 μm, respectively. However, the DRG neurons transfected with the CMT2B mutant (EGFP-Rab7V162M) showed a significant reduction in neurite length with a mean value of 32.79 μm. We thus conclude that the CMT2B Rab7 mutant causes axonal degeneration in DRG neurons.
Discussion
Genomic analyses of various forms of CMT diseases have identified many new genes, whose protein products are involved in axonal transport (Yoshioka et al., 1996; Saito et al., 1997; Keller and Chance, 1999a, b; Dubourg et al., 2000; Chance, 2001; Gemignani and Marbini, 2001; Georgiou et al., 2002; Lus et al., 2003; Chance, 2004; Rotthier et al., 2009). It is thus conceivable that dysfunction of axonal transport contributes prominently to these various forms of CMT diseases. Using cellular and biochemical analysis and live cell imaging techniques, we investigated how signaling and trafficking of NGF/TrkA were affected by CMT2B Rab7 mutations (L129F, K157N, N161T, and V162M) in DRG neurons as well as PC12 cells.
We found that all CMT2B Rab7 mutant-containing vesicles moved faster within DRG axons in the anterograde direction. The speed of TrkA transport was significantly decreased once Rab7 mutants were present in the same vesicles. We also showed that expression of the activated form of Rab7 appeared to reduce the level of surface binding of NGF to its TrkA receptors, likely the result of premature degradation of TrkA receptors in lysosomes/autophagosomes, whereas siRNA-mediated knockdown of Rab7 caused accumulation of TrkA in Rab5-early endosomes within the cytoplasm of PC12 cells. In addition, NGF-induced Erk1/2 activation and cell differentiation were suppressed by expression of CMT2B Rab7 mutants. We further showed that CMT2B Rab7 mutants resulted in axonal degeneration in DRG neurons. These findings suggest that CMT2B Rab7 mutants disrupt axonal trafficking and signaling of NGF/TrkA, which, in turn, contributes to CMT2B disease (Bucci et al., 2000; Cogli et al., 2009; Cogli et al., 2010; McCray et al., 2010).
Rab7 resides primarily in late endosomes/lysosomes (Bucci et al., 2000). Our results showed that Rab7 travels only at approximately half the speed of that of TrkA within axons of DRG neurons, suggesting that under normal conditions the majority of TrkA-containing signaling endosomes that move within axons are able to evade fusion with Rab7 vesicles and avoid their premature degradation during axonal transport. One possible explanation is that activation of TrkA leads to activation of Rab5GAP, which in turn suppresses activation of Rab5 (Liu et al., 2007). Activated Rab5 is a prerequisite for Rab7 activation that promotes endocytic progression from Rab5 early endosomes to Rab7-late endosomes/lysosomes (Rink et al., 2005). By suppressing Rab5 activation, TrkA-signaling endosomes manage to prevent the Rab5 to Rab7 conversion. However, such tight regulation is lost in CMT2B because of hyperactivation of Rab7 mutants. TrkA-signaling endosomes are prematurely fused with and converted into Rab7-late endosomes/lysosomes/autophagosomes within axons. Lysosomes and autophagosomes show slow movement in axons with an average transport speed of ∼0.3 μm/s (Lee et al., 2011). Consistent with these results, the transport speed for TrkA-mCherry that overlapped with Rab7 is reduced to ∼0.36 μm/s, suggesting that these vesicles are likely lysosomes/autophagosomes. Conversely, axonal movement of TrkA-mCherry signals was not affected when the dominant-negative Rab7 mutant (Rab7N125I) was coexpressed in the same neuron. Therefore, hyperactivated CMT2B Rab7 mutants are responsible for the observed defects in transport of NGF/TrkA trophic signals.
Defects in axonal transport can be catastrophic to neuronal survival. Such defects, however, are not restricted to blockage or slowing down of axonal transport. As revealed by our studies, CMT2B Rab7 mutants actually travel faster than wild-type in the anterograde direction, indicating that not only the absolute speed but also the balance of axonal transport flux may play a significant role in CMT2B diseases. Consistent with this notion, the autophagosomes are initiated at the distal terminals and become mature during transport toward the cell body in DRG neurons (Maday et al., 2012). Acceleration of anterograde transport in CMT2B Rab7 mutants can potentially promote early maturation of autophagosomes and thus affect endosome-mediated NGF/TrkA trophic signaling.
Based on our results, we propose the following model to describe how NGF-TrkA signaling is regulated in normal neurons, and how such regulation is lost in CMT2B neurons. In normal neurons, NGF binds to and activates TrkA at the axonal terminals. A portion of NGF-TrkA complex undergoes endocytosis and is internalized into the cell cytoplasm to form signaling endosomes, which are retrogradely transported along the microtubule toward the cell body to propagate the Erk1/2 signals to the nucleus. The NGF/TrkA machinery is likely degraded in the cell body (Fig. 8A). In CMT2B neurons, the population of active Rab7 is significantly increased (Cogli et al., 2010; McCray et al., 2010). The increased Rab7 activity and the increased anterograde axonal transport push the degradation gradient, for instance, the autophagosomal machinery (Maday et al., 2012), toward the axonal terminus (Fig. 8B). CMT2B Rab7 mutants may well promote the maturation of these autophagosomes in axons to cause a slowdown of axonal transport and premature degradation of NGF/TrkA signals, preventing the trophic signal from reaching the cell nucleus. As a result, retrograde transmission of trophic signals is disrupted leading to axonal degeneration and neuronal atrophy.
Figure 8.
A proposed model for the pathogenic mechanism of CMT2B Rab7 mutations. NGF binds to and activates TrkA at the surface leading to activation of the PI3K/Akt pathways. However, activation of the Erk1/2 signaling cascade requires endocytosis of the NGF/TrkA signaling complex into Rab5-early endosomes (i.e., signaling endosomes). The signaling endosomes transmit the NGF/TrkA/pErk1/2 trophic signal to the soma via retrograde axonal transport. Under normal circumstances, upon completion of the delivery, the trophic signaling endosomes are downregulated after fusion with the Rab7-late endosomes/lysosomes/autophagosomes (A). However, in CMT2B, the mutations cause acceleration in anterograde axonal transport of Rab7-late endosomes/lysosomes/autophagosomes, which likely leads to hyperactivation of these degradation organelles within axons. Endocytosed NGF/TrkA signals encounter and fuse with these Rab7-late endosomes/lysosomes/autophagosomes prematurely. The low pH value and the abundant presence of degrading proteases within Rab7-vesicles will result in premature termination of the NGF/TrkA signal (e.g., Erk1/2). As a result, retrograde transmission of trophic signals is disrupted leading to axonal degeneration and neuronal atrophy (B).
In conclusion, our present work suggests that dysregulation of axonal transport of Rab7-late endosomes/lysosomes may play a critical role in the pathogenesis of CMT2B neuropathies. Understanding the transport and signaling properties of CMT2B Rab7 mutants-containing endosomes is crucial for developing prevention and treatment strategies.
Footnotes
This work was supported by National Institutes of Health Grants ADRC P50, EY016525-08, and NS082125, Down Syndrome Research and Treatment Foundation, Larry L. Hillblom Foundation, University of California, San Diego Neurosciences Startup, K. C. Wong Education Foundation Hong Kong (C.W.), the Searle Scholar Award, the Packard Science and Engineering Fellowship, a Postdoctoral Fellowship from the American Cancer Society (K.Z.), and the Dreyfus New Faculty Award (B.C.). We thank Sadie Bartholomew, Renae Shibata, Pauline Yue Hu, Josiah To, and Benz Chaijarasphong for technical assistance; Prof. Bao Lan of SIBCB for constructive suggestions; and Dr. H. Brown for constructing and providing the expression vector TrkA-mCherry.
The authors declare no competing financial interests.
References
- Asbury AK. Sensory neuronopathy. Semin Neurol. 1987;7:58–66. doi: 10.1055/s-2008-1041406. [DOI] [PubMed] [Google Scholar]
- Auer-Grumbach M. Hereditary sensory neuropathies. Drugs Today (Barc) 2004;40:385–394. doi: 10.1358/dot.2004.40.5.850487. [DOI] [PubMed] [Google Scholar]
- Auer-Grumbach M, De Jonghe P, Wagner K, Verhoeven K, Hartung HP, Timmerman V. Phenotype-genotype correlations in a CMT2B family with refined 3q13–q22 locus. Neurology. 2000;55:1552–1557. doi: 10.1212/WNL.55.10.1552. [DOI] [PubMed] [Google Scholar]
- Auer-Grumbach M, Mauko B, Auer-Grumbach P, Pieber TR. Molecular genetics of hereditary sensory neuropathies. Neuromolecular Med. 2006;8:147–158. doi: 10.1385/NMM:8:1-2:147. [DOI] [PubMed] [Google Scholar]
- Axelrod FB, Gold-von Simson G. Hereditary sensory and autonomic neuropathies: types II, III, and IV. Orphanet J Rare Dis. 2007;2:39. doi: 10.1186/1750-1172-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic N, Claeys KG, Sirotkoviæ-Skerlev M, Löfgren A, Nelis E, De Jonghe P, Timmerman V. Charcot-Marie-Tooth disease: a clinico-genetic confrontation. Ann Hum Genet. 2008;72:416–441. doi: 10.1111/j.1469-1809.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- BasuRay S, Mukherjee S, Romero E, Wilson MC, Wandinger-Ness A. Rab7 mutants associated with Charcot-Marie-Tooth disease exhibit enhanced NGF-stimulated signaling. PLoS One. 2010;5:e15351. doi: 10.1371/journal.pone.0015351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronfman FC, Escudero CA, Weis J, Kruttgen A. Endosomal transport of neurotrophins: roles in signaling and neurodegenerative diseases. Dev Neurobiol. 2007;67:1183–1203. doi: 10.1002/dneu.20513. [DOI] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JR, Rodriguez-Waitkus PM, Ng BK, Liang P, Glaser M. Progesterone synthesized by Schwann cells during myelin formation regulates neuronal gene expression. Mol Biol Cell. 2000;11:2283–2295. doi: 10.1091/mbc.11.7.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance PF. Molecular basis of hereditary neuropathies. Phys Med Rehabil Clin North Am. 2001;12:277–291. [PubMed] [Google Scholar]
- Chance PF. Genetic evaluation of inherited motor/sensory neuropathy. Suppl Clin Neurophysiol. 2004;57:228–242. doi: 10.1016/S1567-424X(09)70360-5. [DOI] [PubMed] [Google Scholar]
- Chen JY, Lin JR, Cimprich KA, Meyer T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol Cell. 2012;45:196–209. doi: 10.1016/j.molcel.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogli L, Piro F, Bucci C. Rab7 and the CMT2B disease. Biochem Soc Trans. 2009;37:1027–1031. doi: 10.1042/BST0371027. [DOI] [PubMed] [Google Scholar]
- Cogli L, Progida C, Lecci R, Bramato R, Krüttgen A, Bucci C. CMT2B-associated Rab7 mutants inhibit neurite outgrowth. Acta Neuropathol. 2010;120:491–501. doi: 10.1007/s00401-010-0696-8. [DOI] [PubMed] [Google Scholar]
- Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. One at a time, live tracking of NGF axonal transport using quantum dots. Proc Natl Acad Sci U S A. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinhardt K, Salinas S, Verastegui C, Watson R, Worth D, Hanrahan S, Bucci C, Schiavo G. Rab5 and Rab7 control endocytic sorting along the axonal retrograde transport pathway. Neuron. 2006;52:293–305. doi: 10.1016/j.neuron.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron. 2003;39:69–84. doi: 10.1016/S0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- Dubourg O, Barhoumi C, Azzedine H, Birouk N, Brice A, Bouche P, Leguern E. Phenotypic and genetic study of a family with hereditary sensory neuropathy and prominent weakness. Muscle Nerve. 2000;23:1508–1514. doi: 10.1002/1097-4598(200010)23:10<1508::AID-MUS6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Kaplan DR, Price DL, Koliatsos VE. NGF-stimulated retrograde transport of trkA in the mammalian nervous system. J Cell Biol. 1995;130:149–156. doi: 10.1083/jcb.130.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemignani F, Marbini A. Charcot-Marie-Tooth disease (CMT): distinctive phenotypic and genotypic features in CMT type 2. J Neurol Sci. 2001;184:1–9. doi: 10.1016/S0022-510X(00)00497-4. [DOI] [PubMed] [Google Scholar]
- Georgiou DM, Zidar J, Korosec M, Middleton LT, Kyriakides T, Christodoulou K. A novel NF-L mutation Pro22Ser is associated with CMT2 in a large Slovenian family. Neurogenetics. 2002;4:93–96. doi: 10.1007/s10048-002-0138-4. [DOI] [PubMed] [Google Scholar]
- Grimes ML, Zhou J, Beattie EC, Yuen EC, Hall DE, Valletta JS, Topp KS, LaVail JH, Bunnett NW, Mobley WC. Endocytosis of activated TrkA: evidence that nerve growth factor induces formation of signaling endosomes. J Neurosci. 1996;16:7950–7964. doi: 10.1523/JNEUROSCI.16-24-07950.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H, King RH, Muddle JR, Warner TT, Reilly MM, Orrell RW, Ginsberg L. A novel RAB7 mutation associated with ulcero-mutilating neuropathy. Ann Neurol. 2004;56:586–590. doi: 10.1002/ana.20281. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Jullien J, Guili V, Derrington EA, Darlix JL, Reichardt LF, Rudkin BB. Trafficking of TrkA-green fluorescent protein chimerae during nerve growth factor-induced differentiation. J Biol Chem. 2003;278:8706–8716. doi: 10.1074/jbc.M202401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MP, Chance PF. Inherited peripheral neuropathy. Semin Neurol. 1999a;19:353–362. doi: 10.1055/s-2008-1040850. [DOI] [PubMed] [Google Scholar]
- Keller MP, Chance PF. Inherited neuropathies: from gene to disease. Brain Pathol. 1999b;9:327–341. doi: 10.1111/j.1750-3639.1999.tb00230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlenbäumer G, Young P, Hünermund G, Ringelstein B, Stögbauer F. Clinical features and molecular genetics of hereditary peripheral neuropathies. J Neurol. 2002;249:1629–1650. doi: 10.1007/s00415-002-0946-3. [DOI] [PubMed] [Google Scholar]
- Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J Neurosci. 2011;31:7817–7830. doi: 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lamb D, Chou MM, Liu YJ, Li G. Nerve growth factor-mediated neurite outgrowth via regulation of Rab5. Mol Biol Cell. 2007;18:1375–1384. doi: 10.1091/mbc.E06-08-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lus G, Nelis E, Jordanova A, Löfgren A, Cavallaro T, Ammendola A, Melone MA, Rizzuto N, Timmerman V, Cotrufo R, De Jonghe P. Charcot-Marie-Tooth disease with giant axons: a clinicopathological and genetic entity. Neurology. 2003;61:988–990. doi: 10.1212/WNL.61.7.988. [DOI] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray BA, Skordalakes E, Taylor JP. Disease mutations in Rab7 result in unregulated nucleotide exchange and inappropriate activation. Hum Mol Genet. 2010;19:1033–1047. doi: 10.1093/hmg/ddp567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggouh F, Bienfait HM, Weterman MA, de Visser M, Baas F. Charcot-Marie-Tooth disease due to a de novo mutation of the RAB7 gene. Neurology. 2006;67:1476–1478. doi: 10.1212/01.wnl.0000240068.21499.f5. [DOI] [PubMed] [Google Scholar]
- Mersiyanova IV, Perepelov AV, Polyakov AV, Sitnikov VF, Dadali EL, Oparin RB, Petrin AN, Evgrafov OV. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley WC, Rutkowski JL, Tennekoon GI, Gemski J, Buchanan K, Johnston MV. Nerve growth factor increases choline acetyltransferase activity in developing basal forebrain neurons. Brain Res. 1986;387:53–62. doi: 10.1016/0169-328x(86)90020-3. [DOI] [PubMed] [Google Scholar]
- Moises T, Dreier A, Flohr S, Esser M, Brauers E, Reiss K, Merken D, Weis J, Krüttgen A. Tracking TrkA's trafficking: NGF receptor trafficking controls NGF receptor signaling. Mol Neurobiol. 2007;35:151–159. doi: 10.1007/s12035-007-8000-1. [DOI] [PubMed] [Google Scholar]
- Riccio A, Pierchala BA, Ciarallo CL, Ginty DD. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science. 1997;277:1097–1100. doi: 10.1126/science.277.5329.1097. [DOI] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Romero Rosales K, Peralta ER, Guenther GG, Wong SY, Edinger AL. Rab7 activation by growth factor withdrawal contributes to the induction of apoptosis. Mol Biol Cell. 2009;20:2831–2840. doi: 10.1091/mbc.E08-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthier A, Baets J, De Vriendt E, Jacobs A, Auer-Grumbach M, Lévy N, Bonello-Palot N, Kilic SS, Weis J, Nascimento A, Swinkels M, Kruyt MC, Jordanova A, De Jonghe P, Timmerman V. Genes for hereditary sensory and autonomic neuropathies: a genotype-phenotype correlation. Brain. 2009;132:2699–2711. doi: 10.1093/brain/awp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Hayashi Y, Suzuki T, Tanaka H, Hozumi I, Tsuji S. Linkage mapping of the gene for Charcot-Marie-Tooth disease type 2 to chromosome 1p (CMT2A) and the clinical features of CMT2A. Neurology. 1997;49:1630–1635. doi: 10.1212/WNL.49.6.1630. [DOI] [PubMed] [Google Scholar]
- Saxena S, Bucci C, Weis J, Kruttgen A. The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J Neurosci. 2005;25:10930–10940. doi: 10.1523/JNEUROSCI.2029-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A. TRKing signals through the Golgi. Sci STKE. 2005;2005:pe1. doi: 10.1126/stke.2672005pe1. [DOI] [PubMed] [Google Scholar]
- Spinosa MR, Progida C, De Luca A, Colucci AM, Alifano P, Bucci C. Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease. J Neurosci. 2008;28:1640–1648. doi: 10.1523/JNEUROSCI.3677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Gillooly DJ. Intracellular trafficking and turnover of phosphatidylinositol 3-phosphate. Semin Cell Dev Biol. 2001;12:193–199. doi: 10.1006/scdb.2000.0236. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-5-reviews3007. REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui-Pierchala BA, Ginty DD. Characterization of an NGF-P-TrkA retrograde-signaling complex and age-dependent regulation of TrkA phosphorylation in sympathetic neurons. J Neurosci. 1999;19:8207–8218. doi: 10.1523/JNEUROSCI.19-19-08207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A, Van Gerwen V, Wagner K, Hartung HP, Timmerman V. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:722–727. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Lai CF, Mobley WC. Nerve growth factor activates persistent Rap1 signaling in endosomes. J Neurosci. 2001;21:5406–5416. doi: 10.1523/JNEUROSCI.21-15-05406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ramirez A, Cui B, Ding J, Delcroix JD, Valletta JS, Liu JJ, Yang Y, Chu S, Mobley WC. A functional dynein-microtubule network is required for NGF signaling through the Rap1/MAPK pathway. Traffic. 2007;8:1503–1520. doi: 10.1111/j.1600-0854.2007.00636.x. [DOI] [PubMed] [Google Scholar]
- Wu C, Cui B, He L, Chen L, Mobley WC. The coming of age of axonal neurotrophin signaling endosomes. J Proteomics. 2009;72:46–55. doi: 10.1016/j.jprot.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, Torii T, Kusakawa S, Sanbe A, Nakamura K, Takashima S, Hamasaki H, Kawaguchi S, Miyamoto Y, Tanoue A. The mood stabilizer valproic acid improves defective neurite formation caused by Charcot-Marie-Tooth disease-associated mutant Rab7 through the JNK signaling pathway. J Neurosci Res. 2010;88:3189–3197. doi: 10.1002/jnr.22460. [DOI] [PubMed] [Google Scholar]
- Yoshioka R, Dyck PJ, Chance PF. Genetic heterogeneity in Charcot-Marie-Tooth neuropathy type 2. Neurology. 1996;46:569–571. doi: 10.1212/WNL.46.2.569. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Moheban DB, Conway BR, Bhattacharyya A, Segal RA. Cell surface Trk receptors mediate NGF-induced survival while internalized receptors regulate NGF-induced differentiation. J Neurosci. 2000;20:5671–5678. doi: 10.1523/JNEUROSCI.20-15-05671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]