Abstract
Cellular membranes are composed of hundreds of different lipids, ion channels, receptors and scaffolding complexes that act as signalling and trafficking platforms for processes fundamental to life. Cellular signalling and membrane trafficking are often regulated by peripheral proteins, which reversibly interact with lipid molecules in highly regulated spatial and temporal fashions. In most cases, one or more modular lipid-binding domain(s) mediate recruitment of peripheral proteins to specific cellular membranes. These domains, of which more than 10 have been identified since 1989, harbour structurally selective lipid-binding sites. Traditional in vitro and in vivo studies have elucidated how these domains coordinate their cognate lipids and thus how the parent proteins associate with membranes. Cellular activities of peripheral proteins and subsequent physiological processes depend upon lipid binding affinities and selectivity. Thus, the development of novel sensitive and quantitative tools is essential in furthering our understanding of the function and regulation of these proteins. As this field expands into new areas such as computational biology, cellular lipid mapping, single molecule imaging, and lipidomics, there is an urgent need to integrate technologies to detail the molecular architecture and mechanisms of lipid signalling. This review surveys emerging cellular and in vitro approaches for studying protein–lipid interactions and provides perspective on how integration of methodologies directs the future development of the field.
Introduction
Cellular membranes provide dynamic barriers that anchor receptors, ion channels, carriers, and reversibly bound peripheral proteins.1 Binding of numerous peripheral proteins to membranes is essential for cell growth, metabolism, response to exogenous signals, and general homeostasis.2 Dysregulation of lipid binding is associated with various diseases including cancer3 and metabolic syndromes4 and thus there is a pressing need to better understand membrane-mediated processes. A detailed understanding of these processes should help identify new targets for therapeutic intervention.
Biological membranes contain lipids of various size and charge that are organized into a bilayer comprising a hydrophobic core and highly polarized interfacial regions.5 Membranes of cellular organelles are composed of phospholipids, glycerolipids, sphingolipids, sterols, and other lipid species in varying concentrations. This allows for control of signalling events on spatial and temporal levels through specific protein–lipid interactions. In eukaryotes these interactions are mediated by several conserved lipid-binding domains often found in signal transduction and membrane-trafficking proteins.6
Many of these domains have been well characterized for their lipid-binding properties, such as the C1 and C2 domains of Protein Kinase C (PKC), which were discovered in the late 1970’s.7 Despite rigorous biochemical and biophysical analysis of several lipid-binding domains and their host proteins there remains a paucity of predictive lipid-binding data. This necessitates ongoing in vitro and in vivo screening of specific protein–lipid interactions to discover new lipid-binding proteins. Meanwhile, the mechanisms by which these interactions regulate host-protein activity in disease pathologies are being unravelled. Thus, it is essential to advance our understanding of lipid targeting mechanisms through the use of novel technologies.
The aim of this review is to introduce general principles governing the lipid binding and cellular localization of the most well characterized lipid-binding domains and to discuss how emerging methodologies will shape the coming decade of research in this area.
Lipid binding
Since the discovery of the C18 and C29 domains, a number of conserved lipid-binding modules have been identified. The modules have been categorized into families and characterized for their lipid binding specificities. The majority of these domains have been found in trafficking proteins, metabolic enzymes, and proteins involved in growth and regulatory pathways. In general, these domains employ specific lipid head-group recognition complimented by additional hydrogen bonding10 and non-specific electrostatic interactions to reversibly associate with the membrane bilayer. Some lipid-binding domains are involved in additional hydrophobic interactions and partially insert into the membrane interior.11 At least 12 conserved lipid-binding domains have been identified to date, including C1,12 C2,13 PH,14 FYVE,14b,15 PX,14b,15a ENTH,16 ANTH,17 BAR,18 FERM,19 PDZ,20 and TUBBY,21 (See MeTaDoR: http://proteomics.bioengr.uic.edu/metador/MeTaDoR.html)22 or The Pawson Lab: http://pawsonlab.mshri.on.ca/index.php?option=com_content&task=view&id=30&Itemid=63 for more information) domains.23 Recently yeast and mammalian kinase-associated 1 (KA1) have been added to the list of proteins capable of binding to anionic phospholipids in vitro and in vivo.24 This discovery brings to light the exciting possibility that there remain yet-to-be-identified lipid-binding modules.25 Additionally, identification of some lipid-binding domain family members is a challenging task due in part to low sequence homology, particularly the PH26 and C227 domains. To overcome these challenges proteomic methods are being developed to characterize new lipid-binders using mass spectrometry.28 The integration of proteomics with lipid binding assays will be essential to discovering new lipid-binding modules or identifying subfamilies of previously characterized modules.
Biophysical analysis of lipid-binding
A number of well established biochemical and biophysical experiments have been employed to determine the lipid specificity of proteins. Centrifugal liposome assays29 and antibody-based “lipid-blots”30 have been used to rapidly assess selectivity. On the quantitative side, surface plasmon resonance (SPR)31 remains the gold standard for measuring protein affinity for lipids and has been used successfully in determining the Kd values for interactions of C2,32 ENTH,33 FYVE,34 PH,35 and PX36 domains with lipid vesicles. Additionally, this technique has provided a quantitative assessment of lipid-binding proteins such as Protein Kinases C37 and cPLA2α.37b,38 However, it can be difficult to obtain reliable and reproducible binding data using SPR due to nonspecific binding to the sensor chip, mass transport effects, and protein stability.39 Thus, it is essential to carefully optimize conditions to produce reliable SPR measurements.40 Isothermal titration calorimetry using soluble lipid headgroup species has also been utilized, albeit less extensively.41 To determine the ability of peripheral membrane proteins to insert into the hydrophobic core of the membrane the monolayer penetration technique42 has been employed.34b Additionally, the depth of penetration and protein orientation at the membrane interface have been determined by electron paramagnetic resonance spectroscopy.43 Taken together, biophysical assays allow for the thorough characterization of lipid binding specificities and mechanisms of membrane association of peripheral proteins. However, the inability to investigate lipid binding in a high-throughput fashion limits progress toward rapid identification of new lipid-binding proteins. A significant step in this direction was the synthesis of chemically modifiable phosphoinositide (PI) probes that can be used in high-throughput screening.44 Chemically modifiable lipids hold much promise for proteomic studies in which they can be cross-linked to high affinity targets from cell lysates for protein identification.45
Cellular localization
Regulation of the spatial and temporal cellular distribution of lipid-binding domains has become an area of intense study in the past decade. The discovery that the lipid composition of cellular membranes is highly dynamic has aided in understanding the cellular localization and function of peripheral proteins. In addition, temporal modulation of lipids such as PIs and diacylglycerol (DAG) in response to an assortment of signals can dramatically regulate protein–lipid binding.14b,23a Targeting of proteins to a particular organelle can also be regulated by requiring a specific degree of membrane curvature for proper interaction. This property of the BAR domain-containing proteins18c,46 was shown to be essential for endocytosis and recycling of plasma membrane proteins. Membrane targeting can be further mediated by secondary factors such as cytoplasmic calcium, which activates cPLA2α in inflammatory processes47 or by pH. For example, the PI(3)P-binding pocket of the FYVE domain contains conserved histidine residues, protonation of which at a lower pH is necessary for association with early endosomes.48 Furthermore, the pH dependence is not limited to amino acids as protonation of the headgroup of the phosphomonoester phosphatidic acid (PA) is also pH dependent and can regulate effector proteins such as Opi1 in yeast.49
Coincidence detection
Finally, the requirement of two distinct lipid-binding sites within the C2, PH, and PX domains has been demonstrated for robust membrane localization of several proteins. This mode of dual lipid recognition termed “coincidence detection” is seen in the C2 domain of PKCα that associates with PS32,50 and PIs,35d Akt1 and PDK-1 PH domains that bind to PS51 and PI(3,4,5)P3,52 and the p47phox PX domain that recognises PI(3,4)P2 and PA36a (Fig. 1). Several PH domains exhibit coincidence detection through a protein–protein interaction site in addition to a protein–lipid interaction site. Four-phosphate adaptor protein 1 (FAPP1)53 and OSBP54 bind PI(4)P and small GTPases in the Golgi membrane (Fig. 1). As we discuss below, it is possible that some protein–lipid interactions require additional conditions that should be taken into consideration when investigating these specific interactions. For instance, sphingolipids such as ceramide,55 ceramide-1-phosphate (C1P),56 and sphingosine-1-phosphate(S1P)57 have recently been identified to target a number of effector proteins. Because these lipids are often overlooked during screening of lipid-binding proteins there may be a number of unidentified receptors for these sphingolipids. In general, lipid binding screens in search of cognate ligand usually employ anionic lipids such as PIPs, PS and PA, which may limit discovery of new lipid binding properties of peripheral proteins. A prime example of such is C1P,58 which has been shown to associate with a cationic patch in the β-groove13a of the cPLA2α C2 domain.38 This is an example of coincidence detection where the C2 domain detects a glycerophospholipid (PC) through the hydrophobic loop regions11,59 and a sphingolipid (C1P) through the adjacent cationic patch.38 It is important to note the C1P binding properties of the C2 domain were discovered several years after the association with PC had been accepted as the main mode of lipid-binding. This underscores the importance of considering sphingolipid binding to well-known lipid-binding modules.
Fig. 1.
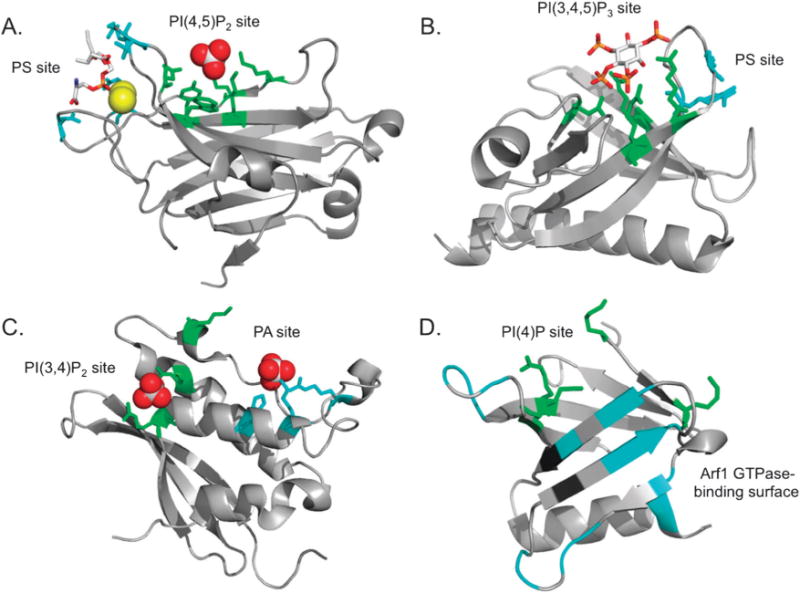
Lipid-binding domains termed “coincidence detectors” harbour two distinct binding sites. Residues involved in lipid-head group coordination or protein binding are highlighted. (A) Protein Kinase Cα (PKCα) C2 domain bound to phosphatidylserine (PS) (red and gray sticks) and two calcium ions (yellow) with PS-coordinating residues shown in cyan (Asn-189, Arg-216, Arg-249, and Thr-251). The PtdIns(4,5)P2 headgroup is coordinated by Tyr-195, Lys-197, Lys-209, Lys-211, Trp-245 and Asn-253 shown in green. The sulfate group (red and gray) was bound in the inositol-binding site (Protein Data Bank (PDB) Code 1DSP). (B) The structure of the Akt1 PH domain (PDB 1UNQ) was determined in complex with the PI(3,4,5)P3 headgroup, Ins(1,3,4,5)P4 (red and gray). PI(3,4,5)P3 is bound by Arg-23, Arg-25, Asn-52, and Arg-86 shown in green. Akt1-PH additionally recognizes PS through Arg-15 and Lys-20 (cyan) adjacent to the PI(3,4,5)P3 pocket. (C) The p47phox PX domain structure was obtained with two sulfate molecules bound in each lipid-binding site (PDB 1O7K). This domain coordinates PA by His-51, Lys-55, and Arg-70 (cyan), and PI(3,4)P2 by Arg-43, Lys-79, and Arg-90 (green). (D) FAPP1-PH coordinates the PI(4)P headgroup at the bottom of the β-barrel with Lys-7, Arg-18, Lys-41 and Lys-45 (green) and co-localizes with Arf1 GTPase at the Golgi membrane (PDB 3RCP). The Arf1 binding surface, as determined by NMR resonance perturbation analysis, includes the β1–β2 and β5–β6 loops as well as the β5,6 and 7 strands (cyan).
Lipid-dependent drug targets
Association with specific membranes is required for the proper biological function of various enzymes and non-catalytic components of cellular signalling. Dysregulation of these signalling pathways can lead to adverse consequences manifested in metabolic disorders and cancers.60 For instance, an oncogenic E17K mutation in the PH domain of Akt1 has been found in human cancer tissue.3a This mutation appears to abolish specificity for PI(3,4,5)P3 and promotes constitutive association of Akt1 with the PI(4,5)P2-enriched plasma membrane.61 Additionally, mutations in the PH domain of Dynamin that compromise GTPase activity result in Centronuclear Myopathy.62 Clearly, peripheral proteins are potent drug targets for which therapeutic intervention at the level of protein–lipid binding can be invaluable.63 Recently, seminal studies have demonstrated the feasibility of such an approach in which small molecules that inhibit lipid binding of C264 and PH domains65 were generated and tested. Progress has been made in understanding the role of protein–lipid interactions in cellular processes such as endo- and exocytosis, cellular lipid trafficking and disease pathologies. However, there remains a great need for more detailed structural, biophysical and system-wide lipidomic analyses. Computational biology, theoretical and “omic” studies have already begun to play a leading role in the field and should be bolstered by a wealth of information gained in the next decade. In the following sections we evaluate recent technological advances in probing protein–lipid interactions.
Structural biology
The three dimensional structures of lipid-bound proteins provide invaluable information regarding the molecular mechanisms underlying the biological activities of these proteins. In the last three decades the atomic-resolution crystal and solution structures of various water-soluble domains have been determined; however, structural details of membrane-associated modules are challenging to obtain.66 A number of relatively soluble PI-recognizing domains have been studied using conventional X-ray crystallographic and NMR spectroscopic approaches.14b,23a The mechanistic basis of the association with membranes has been elucidated from the structures of these domains in complex with lipid head groups or intact lipids containing short acyl chains. However, biochemical and structural characterization of less soluble peripheral membrane proteins requires additional manipulation and often necessitates the use of detergents during purification. In addition, the lipid system in which the protein can be reconstituted in a form that retains its biological function must also be identified and optimised.
Cryomicroscopy (CryoEM) techniques, i.e. electron crystallography of two-dimensional crystals and single particle analysis of detergent-solubilized protein complexes, provide an alternative method of structural characterization.67 As with many other structural tools, both crystallographic approaches have limitations. The major concern is the crowding of protein molecules, particularly in two dimensional crystals, and a limited amount of lipids present and/or bound to the protein. Therefore, additional analysis is always necessary to confirm the lipid binding sites and the relative orientation of the protein molecules. Nevertheless, despite the common view of X-ray crystallography as a technique that produces a static scaffold, it provides the most desirable insight into protein–ligand interactions and allows for determining the structures of large proteins and complexes. Recent notable advances in obtaining well-diffracting crystals of membrane proteins for X-ray crystallographic analysis and the electron crystallography structure of aquaporin determined at 1.9 Å resolution68 reveal the great potential of these approaches in studying lipid-bound proteins.
NMR spectroscopy has developed as a powerful tool to determine the three-dimensional structures of membrane-associated proteins and to investigate protein dynamics and lipid binding properties.66a,69 One of the major requirements, however, is that an NMR sample must contain large amounts of soluble protein and interacting lipid. This can be achieved by using water-soluble short chain lipids in the case where the protein is soluble, or alternatively a membrane mimetic system that solubilizes the hydrophobic protein and the lipid in an aqueous environment. A number of membrane mimetics have been developed for this purpose, including detergent-based and lipid-based spherical micelles, disk-like bicelles, amphipols and organic solvents.70 The lipid-binding site can be identified from intermolecular nuclear Overhauser enhancements (NOEs) observed between the protein and lipid protons or through resonance perturbations occurred in the protein upon binding of the lipid. The 1H, 15N heteronuclear single quantum coherence (HSQC) technique is particularly useful in monitoring chemical shift changes in the 15N-labeled protein as an unlabeled lipid is titrated in.35b,c,71 These experiments not only allow for identification of residues that are most likely involved in the interaction and/or accompanying conformational changes but also provide an effective method for examining the stability and folding of the protein and assessing the strength of the interaction. In addition, the internal motions can be investigated through measuring relaxation times T1 and T2 and heteronuclear NOEs.72 The depth of penetration and topology of the peripheral proteins can be measured by adding lipophilic or soluble paramagnetic probes and spin-labelled lipids.73
The size of a protein or a protein–lipid complex remains a major limitation of solution NMR, and it becomes increasingly difficult to obtain the atomic-resolution structure of a macromolecule of over 30–40 kDa. Another concern is that micelles and bicelles represent modest mimetics of lipid bilayers because of pronounced curvature and different surface tension. Therefore, data produced in these systems need to be thoroughly validated by lipid binding assays such as SPR or vesicle sedimentation assays. Furthermore, formation of a tight complex between the protein and membrane-mimetic particle significantly increases the effective size, which can easily reach the limit for NMR structure determination (for example, the size of commonly used dodecylphosphocholine (DPC) micelles is ~20 kDa). This results in slower tumbling and broadening of NMR resonances beyond detection. Nevertheless, remarkable breakthroughs recently seen in the application of NMR spectroscopy to structural analysis of biological macromolecules offer new tools and strategies that could be applicable in probing membrane proteins. These include measurements of residual dipolar couplings (RDCs) in bicelles, polyacrylamide gels and other aligned media, aiding in a more accurate structure refinement and assessment of relative orientation of multiple linked domains.74 Paramagnetic relaxation enhancement (PRE) of NMR relaxation rates have been used as another long-range restraint.74b,75 In these experiments, a paramagnetic spin-label is attached to the thiol group of a cysteine residue leading to distant-dependent line broadening of NMR resonances of the protein. Additionally, development of transverse relaxation optimized spectroscopy (TROSY)-based pulse sequences, deuteration of proteins, application of high dimensionality experiments and advanced labelling strategies allow for determination of structures of macromolecules as large as 100 kDa.75b,76 The use of cryoprobes and ultra-high field solution NMR spectrometers as well as recent innovations of solid state NMR should significantly increase sensitivity and resolution and further broaden the application of NMR to the structural characterization of lipid-binding proteins.
Computational biology
While significant progress has been made in characterizing protein–lipid interactions experimentally a number of limiting factors remain. Generally membrane proteins are difficult to express in bacterial systems or to obtain in a soluble form in amounts sufficient for biochemical experiments. On the other hand, design and expression of fluorescently labelled constructs in vivo is laborious and concerns remain regarding the physiological relevance of the data obtained with over-expressed probes. Some of these obstacles can be overcome using in silico approaches. Computational biology was first used as a tool to predict structures of membrane proteins based on previously determined structures77 or to assess lipid binding properties as was shown for the PH and FYVE domains.78 Additionally, electrostatic potential calculations complemented by an array of biophysical lipid-binding assays helped to elucidate the PI-induced docking mechanisms of the ENTH, FYVE, and PX domains.33a,34b,36b
Molecular dynamics
Molecular dynamics (MD) simulations have shed light on the molecular mechanisms of membrane anchoring. In silico studies of the FYVE, PH and PX domains have demonstrated how non-specific interactions of the protein residues with the hydrophobic core of the bilayers enhance binding to PI-containing membranes.79 MD simulations of the C2 domain of PKCα combined with experimental lipid-binding studies, cell translocation assays, and mutagenesis suggested that the Ca2+-induced binding to PS is augmented by PI(4,5)P2 due to the increased membrane residence time of the domain.35d Subsequent biochemical studies coupled with MD simulations revealed the PI(4,5)P2 stoichiometry.80
MD simulations have also contributed to a better understanding of the mechanisms underlying the protein-induced vesicle tubulation that has been observed experimentally with some lipid-binding domains. Studies have shown how amphyphysin N-BAR domains can induce local curvature upon association with a membrane, and do so with greater affinity when negatively-charged PI(4,5)P2 is added to the membrane.81 It has also been shown that when amphipathic α-helices adjacent to the N-BAR domain are removed this strong membrane association is lost. Interestingly, high concentrations of the amphipathic α-helices alone can drive membrane curvature independent of the BAR domain at higher concentrations.82 MD studies have uncovered how the complementarity of N-BAR concentration, oligomerization, and membrane composition affect the level and type of curvature, tubulation, or vesiculation that can occur in different lipid models.81b
Bioinformatics
In addition to in silico modelling techniques, recently developed databases, algorithms, and statistical methods have broadened the toolkit for analysis of protein–lipid interactions. Computational approaches including sequence alignments and charge mapping also serve as an important platform to design cellular and biochemical assays. For instance, a recursive-learning algorithm was used to identify PI(3,4,5)P3-specific PH domains, and these findings were confirmed experimentally through observation of the translocation of fluorescently labelled proteins to the plasma membrane upon PI(3,4,5)P3 stimulation.83 Recently, a new combinatorial computational approach has been applied in a genome-wide scale identification of ANTH domains.84 This study not only defined the ANTH domain family but also annotated its members based upon structural features and biophysical properties including membrane deforming activity. Additionally, a machine-learning algorithm has been used to predict lipid-binding abilities of a subset of C2 domains.85
Integration of computational work with genomic, proteomic, and lipidomic studies will be essential to rapidly identify novel lipid-binding proteins and may reveal new mechanisms of lipid recognition. Furthermore, linking identification of new lipid binding proteins86 with lipid metabolic pathways87 could uncover additional roles of lipid-binding proteins in health and disease. However, computational biology still has limitations as demonstrated by association of peripheral proteins with PA. Despite the experimental characterization of more than 22 PA binding proteins88 bioinformatic approaches have not been able to identify a common motif or protein module that is able to specifically recognize PA.
Cellular imaging
Imaging of fluorescently labelled proteins including the C2, FYVE, PH, and PX domains have been used to estimate the subcellular distribution of PS, PI(3)P, PI(4)P, PI(4,5)P2 and PI(3,4,5)P3. (See Fig. 2).89 However, there are concerns regarding the ability of these lipid-binding probes to accurately reach all organelles. These concerns were recently highlighted in a study where cellular PS distribution was assessed using confocal and electron microscopy. Electron microscopy detected PS which was not visible with a fluorescent C2 domain probe in subcellular compartments such as caveolae and the inner mitochondrial membranes.90 Similar rigorous analysis will be essential for mapping the compartmentalization and distribution of other signalling lipids. In addition to conventional lipid-binding probes, unique sensors have been developed to estimate lipid concentrations. A C1 domain DAG sensor has been constructed, which undergoes a change in FRET upon binding DAG while an ENTH domain with an environmentally sensitive fluorophore that detects PI(4,5)P2 was engineered.91 The new strategy applied to the ENTH domain allows for quantification of PI(4,5)P2 in different cellular compartments under physiological conditions. This sensor methodology should also be applicable to other specific lipid-binding domains to quantify the native state of lipids such as PI(3)P, PI(4)P and PI(3,4,5)P3 in cells. Although monitoring dynamics of some lipids with fluorescently tagged lipid-binding domains has become more or less routine it is important to note that a significant gap remains for sensing glycerophospholipids and sphingolipids due to a lack of corresponding lipid-binding probes.
Fig. 2.
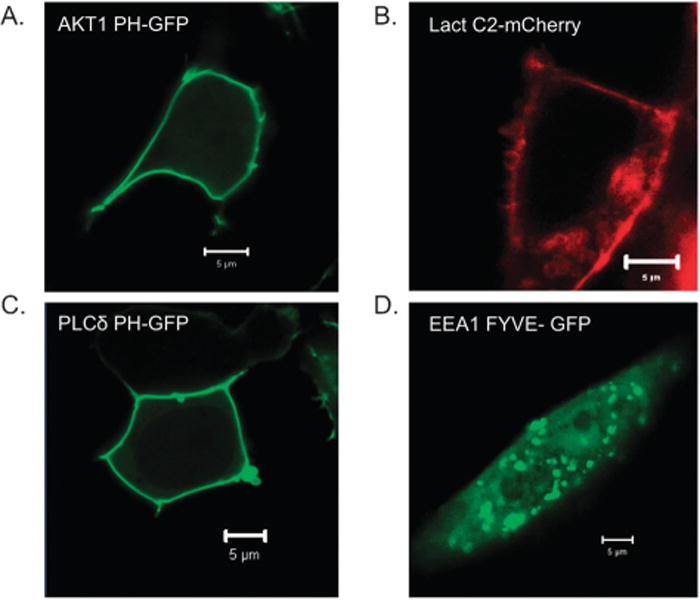
GFP- and mCherry-labelled lipid-binding domains serve as markers of cellular lipids. (A) Akt1 PH-GFP localizes to the plasma membrane in response to PI(3,4)P2 or PI(3,4,5)P3, (B) Lactadherin C2-mCherry marks PS-enriched membranes including the cytoplasmic leaflet of the plasma membrane, (C) PLCδ PH-GFP binds PI(4,5)P2 most prominently in the plasma membrane and (D) EEA1 FYVE-GFP localizes to PI(3)P-containing early endosomes.
Tools to manipulate cellular lipid compositions such as reagents to deplete plasma membrane PI(4,5)P292 allow for analysis of lipid-mediated signalling pathways. These types of assays are agonist-inducible where binding of a drug triggers association of cytosolic bait (the enzyme) with a protein receptor in a target membrane. Recently, this strategy has been applied to monitor the modulation of PI(4)P levels in the Golgi.93 Such an approach holds promise for engineering targeted enzymes essential in sphingolipid metabolism, which is an underexplored area of lipid biology. Despite the progress in monitoring lipid dynamics, there remains a concern that the lipid probes act as dominant negative effectors when over-expressed. This overexpression may block downstream signalling events due to sequestration of target lipids. Small molecule sensors or chemically modified lipid-binding reporters that can sense lipids with subnanomolar affinities,94 would alleviate these issues. This is admittedly a difficult problem but a step in the right direction has been the development of small fluorescent molecules that can recognize PS exposed on apoptotic cells95 as well as small molecules that act as PI-binding mimetics of PH domains.96 Lipid biosensors that could easily enter cells and readily map available pools of different lipids would be a great boon to the lipid community.
Single molecule imaging
Single molecule imaging enables highly sensitive detection of transient interactions between labelled proteins and lipid vesicles or cellular membranes.97 One such example is total internal reflection fluorescence (TIRF) imaging, which has become a useful tool for measuring protein–lipid interactions with giant unilamellar vesicles or at the plasma membrane of live cells.97,98 TIRF generates an evanescent wave focused approximately in the 100 nm between the cover slip and the sample; exciting fluorophores near the plasma membrane or vesicle-media interface. This technique significantly eliminates background noise from other fluorophores to allow accurate interpretation of single molecule binding events.99 TIRF has been used to characterize the membrane binding affinity of peptides97a as well as the kinetics of PH domain association with PI(3,4,5)P3-containing membranes.99 Because TIRF can be combined with single molecule detection and analysis methods it is possible to monitor absolute protein concentrations with high sensitivity per unit area of membrane. TIRF is also robust when integrated with software that measures the number and brightness of the fluorescent molecules. This allows properties such as protein oligomerization to be observed in real time. However, the bleaching of fluorophores can complicate interpretation of TIRF results, as labelled proteins may be bound and photobleached simultaneously. To minimize this scenario, fluorophores with high quantum yield are often used and the laser power is carefully controlled.97a This technique has proved useful in monitoring the kinetics of assembly and egress of fluorescently labelled HIV-1,100 which binds PI(4,5)P2 and PS in the cytoplasmic leaflet of the plasma membrane.101
Lipidomics
Lipidomics has emerged as a platform to integrate system-wide cellular information. Enhanced extraction and separation protocols, improved analysis software, and the availability of integrated databases have led to a wealth of new information on cellular lipid compositions.102 It is our hope that technological advances will help to elucidate the composition of cellular organelles and enable comparision of lipids between healthy and diseased state models. One can envision an integrated approach to build a cellular map of lipid distribution using lipidomics combined with lipid-binding probe studies that can resolve lipid compartmentalization.90 The availability of lipidomic data will enhance our understanding of lipid–protein interactions and the biophysical characteristics of cellular membranes required to recruit effector proteins. While the basics of formation of lipid domains are understood to some extent, recent data demonstrate that lipid domains are heterogeneous in nature and can differentially attract specific lipid-binding proteins.103 Robust analysis of lipid domain formation and composition of cell membranes will be indispensable for studying signalling platforms and trafficking pathways.
Lipidomics and disease
Several diseases are known to be linked to lipid imbalances including Alzheimer’s,104 diabetes,105 atherosclerosis,106 and lipid storage disorders.107 Lipid metabolic changes can rapidly alter peripheral protein localization and activity, thus information regarding the temporal and spatial composition of organelle lipids in healthy and pathological states is invaluable. Lipidomic analysis can detect small but physiologically significant changes of signalling lipid concentrations and lead to a targeted search for enzymes responsible for these fluctuations. In addition, lipidomic studies of bacterial and viral pathogens can provide metabolic profiles of infection and replication of these organisms in human hosts.108 In the last year lipidomic analysis of diseased tissue has emerged with the analysis of mitochondrial cardiolipins linked to diet and disease,109 oxidative lipidomic analysis in traumatic brain injury,110 analysis of human and mouse brain with Alzheimer’s disease,111 and eicosanoid release in inflamed synovial joints.112 Taken together, these seminal studies provide a template to build targeted approaches for specific lipid analysis of pathological conditions.
Lipidomics and systems biology
With the ‘omics’ revolution have come efforts to identify lipid-binding domains in a system-wide fashion. Using PI(3,4,5)P3-coated beads and mass spectrometry, two new PI(3,4,5)P3-binding PH domains were recently discovered.28 Additionally, novel lipid-binding patterns were identified in yeast by a custom-made lipid array containing 56 lipid metabolites. Using this approach, 530 lipid–protein associations were detected, most of which were novel.87 These examples illustrate significant progress in a field filled with challenges such as organelle separations, resolution, bilayer asymmetry, and lipid metabolism during purification. Additionally, three main databases have been assembled to collect lipidomic information including the Lipid Metabolite and Pathways Strategy (LIPID MAPS, http://www.lipidmaps.org/), METLIN (Metabolite and Tandem MS Database, http://metlin.scripps.edu/) and the Human Metabolome Database (http://www.hmdb.ca/). Integrating information through databases and cross-disciplinary research to rapidly discover druggable targets will be essential to moving the field forward. This is well exemplified in a study that mined the ovarian cancer literature integrating various lipid studies to identify potential drug targets.113 Once again, this emphasizes the importance of cross-disciplinary research to expedite drug discovery.
Conclusion and perspectives
The lipid-binding field has undergone a technological revolution in the last decade and is well primed with available and evolving methodologies to fill in the aforementioned gaps. The implication of lipid–protein interactions in diverse disorders will continue to prompt intense investigation of specific protein targets. Lipidomics and bioinformatics should enable researchers to evaluate protein–lipid interactions in relation to cellular lipid profiles and irregularities in disease states. System-wide studies will unveil new lipid-binding proteins and uncover unknown intricacies of those already under study. Researchers are encouraged to partake in cross-disciplinary studies that will surely underlie future breakthroughs in the field.
Insight, innovation, integration.
In the last two decades significant progress has been made towards understanding the molecular mechanisms by which lipid-binding proteins are recruited to membranes. While investigating the most common biochemical properties of individual lipid-binding domains has become more or less routine, studies of intact proteins within membranes can be limited by the available technology. Lipid-binding proteins have been implicated in a number of diseases so as this field expands there is an urgent need for integration of data from newly emerging technologies into quantitative databases that will direct translational studies. This article presents emerging technologies in the field and provides perspectives on how these methodologies will address critical questions regarding cellular lipid-binding events.
Acknowledgments
Research in the Stahelin lab is supported by grants from the NIH (AI081077) and the NSF (1122068) and research in the Kutateladze lab is supported by the NIH (GM096863 and CA113472) and the AHA (10GRNT2600175) grants. J.L.S. is supported by a CBBI NIH training fellowship (T32GM075762), E.A.G. is supported by a Notre Dame Eck Institute for Global Health graduate training fellowship and C.A.M. is an NIH NRSA postdoctoral fellow (F32HL096399).
Biographies

Robert V. Stahelin, PhD, is an Assistant Professor in the Department of Biochemistry and Molecular Biology at the Indiana University School of Medicine-South Bend and an Adjunct Assistant Professor in the Department of Chemistry and Biochemistry at the University of Notre Dame. His research group investigates the molecular basis of lipid–protein interactions in viral assembly, inflammation, cancer and other human diseases. Stahelin’s laboratory applies biochemical, biophysical, and cellular approaches to better understand lipid-mediated events in the context of disease.

Jordan L. Scott received her BS in Biology from Calvin College where she studied the activation of the glucose transporter GLUT1 with Professor Larry Louters. She is currently a third year PhD student in the Stahelin lab in the Department of Chemistry and Biochemistry at the University of Notre Dame. At Notre Dame, she has focused her molecular mechanistic studies on the PI4P-binding protein FAPP1 and other phosphoinositide effectors.

Catherine A. Musselman, PhD, graduated from the University of Michigan. She is currently a postdoctoral fellow in Tatiana Kutateladze’s laboratory in the Department of Pharmacology at the University of Colorado Denver School of Medicine. Catherine investigates the molecular mechanisms of histone binding by PHD fingers and other epigenetic readers.

Emmanuel Adu-Gyamfi received his BS in Biochemistry from the University of Ghana and is currently a fourth year PhD candidate in the Stahelin lab in the Department of Chemistry and Biochemistry at the University of Notre Dame. Adu-Gyamfi is applying state-of-the-art approaches including high resolution TEM and single molecule imaging to understand the molecular basis of Ebola virus assembly and spreading.

Tatiana G. Kutateladze, PhD, is an Associate Professor in the Department of Pharmacology at the University of Colorado Denver School of Medicine. The research in her laboratory focuses on epigenetic biology and phosphoinositide signaling. Her group applies NMR spectroscopy, X-ray crystallography and biochemical approaches to study the three-dimensional structures and functions of chromatin- and lipid-binding proteins implicated in cancer and other human diseases.
Notes and References
- 1.(a) Cho W. Building signaling complexes at the membrane. Science’s STKE. 2006;2006:pe7. doi: 10.1126/stke.3212006pe7. [DOI] [PubMed] [Google Scholar]; (b) Glatz JF, Luiken JJ, van Bilsen M, van der Vusse GJ. Cellular lipid binding proteins as facilitators and regulators of lipid metabolism. Mol Cell Biochem. 2002;239:3–7. [PubMed] [Google Scholar]
- 2.(a) Zwaal RF, Comfurius P, Bevers EM. Lipid–protein interactions in blood coagulation. Biochimica et biophysica acta. 1998;1376:433–453. doi: 10.1016/s0304-4157(98)00018-5. [DOI] [PubMed] [Google Scholar]; (b) Clarke SD, Armstrong MK. Cellular lipid binding proteins: expression, function, and nutritional regulation. FASEB. 1989;3:2480–2487. doi: 10.1096/fasebj.3.13.2680704. [DOI] [PubMed] [Google Scholar]; (c) Antonescu CN, Aguet F, Danuser G, Schmid SL. Phosphatidylinositol-(4,5)-bisphosphate regulates clathrin-coated pit initiation, stabilization, and size. Mol Biol Cell. 2011;22:2588–2600. doi: 10.1091/mbc.E11-04-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) LeBlanc MA, McMaster CR. Surprising roles for phospholipid binding proteins revealed by high throughput genetics. Biochem Cell Biol. 2010;88:565–574. doi: 10.1139/O09-171. [DOI] [PubMed] [Google Scholar]; (e) Smathers RL, Petersen DR. The human fatty acid-binding protein family: evolutionary divergences and functions. Human genomics. 2011;5:170–191. doi: 10.1186/1479-7364-5-3-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]; (b) Falke JJ. Membrane Recruitment as a Cancer Mechanism: A Case Study of Akt PH Domain. Cellscience. 2007;4:25–30. [PMC free article] [PubMed] [Google Scholar]; (c) Nardella C, Carracedo A, Salmena L, Pandolfi PP. Faithfull modeling of PTEN loss driven diseases in the mouse. Curr Top Microbiol Immunol. 2010;347:135–168. doi: 10.1007/82_2010_62. [DOI] [PubMed] [Google Scholar]
- 4.Nagao K, Yanagita T. Bioactive lipids in metabolic syndrome. Prog Lipid Res. 2008;47:127–146. doi: 10.1016/j.plipres.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 5.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 7.Takai Y, Kishimoto A, Iwasa Y, Kawahara Y, Mori T, Nishizuka Y. Calcium-dependent activation of a multifunctional protein kinase by membrane phospholipids. The Journal of biological chemistry. 1979;254:3692–3695. [PubMed] [Google Scholar]
- 8.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 9.Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 10.(a) Boggs JM, Chia LS, Rangaraj G, Moscarello MA. Interaction of myelin basic protein with different ionization states of phosphatidic acid and phosphatidylserine. Chem Phys Lipids. 1986;39:165–184. doi: 10.1016/0009-3084(86)90110-6. [DOI] [PubMed] [Google Scholar]; (b) Kooijman EE, Tieleman DP, Testerink C, Munnik T, Rijkers DT, Burger KN, de Kruijff B. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J Biol Chem. 2007;282:11356–11364. doi: 10.1074/jbc.M609737200. [DOI] [PubMed] [Google Scholar]; (c) Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Mollica L, Fraternali F, Musco G. Interactions of the C2 domain of human factor V with a model membrane. Proteins: Struct, Funct, Bioinf. 2006;64:363–375. doi: 10.1002/prot.20986. [DOI] [PubMed] [Google Scholar]
- 11.Bittova L, Sumandea M, Cho W. A structure-function study of the C2 domain of cytosolic phospholipase A2. Identification of essential calcium ligands and hydrophobic membrane binding residues. J Biol Chem. 1999;274:9665–9672. doi: 10.1074/jbc.274.14.9665. [DOI] [PubMed] [Google Scholar]
- 12.Blumberg PM, Kedei N, Lewin NE, Yang D, Czifra G, Pu Y, Peach ML, Marquez VE. Wealth of opportunity –the C1 domain as a target for drug development. Curr Drug Targets. 2008;9:641–652. doi: 10.2174/138945008785132376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta, Mol Cell Biol Lipids. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]; (b) Corbalan-Garcia S, Gomez-Fernandez JC. The C2 domains of classical and novel PKCs as versatile decoders of membrane signals. BioFactors. 2010;36:1–7. doi: 10.1002/biof.68. [DOI] [PubMed] [Google Scholar]
- 14.(a) Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp. 2007:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nat Chem Biol. 2010;6:507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Kutateladze TG. Mechanistic similarities in docking of the FYVE and PX domains to phosphatidylinositol 3-phosphate containing membranes. Prog Lipid Res. 2007;46:315–327. doi: 10.1016/j.plipres.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kutateladze TG. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim Biophys Acta, Mol Cell Biol Lipids. 2006;1761:868–877. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]; (b) Horvath CAJ, Vanden Broeck D, Boulet GAV, Bogers J, De Wolf MJS. Epsin: Inducing membrane curvature. Int J Biochem Cell Biol. 2007;39:1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Duncan MC, Payne GS. ENTH/ANTH domains expand to the Golgi. Trends Cell Biol. 2003;13:211–215. doi: 10.1016/s0962-8924(03)00076-x. [DOI] [PubMed] [Google Scholar]
- 18.(a) Rao Y, Haucke V. Membrane shaping by the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Cell Mol Life Sci. 2011:1–11. doi: 10.1007/s00018-011-0768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2010;21:340–349. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]; (c) Qualmann B, Koch D, Kessels MM. Let’s go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. The FERM domain: organizing the structure and function of FAK. Nature reviews. Nat Rev Mol Cell Biol. 2010;11:802–814. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- 20.Gallardo R, Ivarsson Y, Schymkowitz J, Rousseau F, Zimmermann P. Structural diversity of PDZ-lipid interactions. ChemBioChem. 2010;11:456–467. doi: 10.1002/cbic.200900616. [DOI] [PubMed] [Google Scholar]
- 21.Mukhopadhyay S, Jackson PK. The tubby family proteins. GenomeBiology. 2011;12:225. doi: 10.1186/gb-2011-12-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhardwaj N, Stahelin RV, Zhao G, Cho W, Lu H. MeTaDoR: a comprehensive resource for membrane targeting domains and their host proteins. Bioinformatics. 2007;23:3110–3112. doi: 10.1093/bioinformatics/btm395. [DOI] [PubMed] [Google Scholar]
- 23.(a) Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature reviews. Molecular cell biology. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]; (b) Stahelin RV. Lipid binding domains: more than simple lipid effectors. J Lipid Res. 2009;50(Suppl):S299–304. doi: 10.1194/jlr.R800078-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191[pii]. [DOI] [PubMed] [Google Scholar]
- 26.DiNitto JP, Lambright DG. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim Biophys Acta, Mol Cell Biol Lipids. 2006;1761:850–867. doi: 10.1016/j.bbalip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Zhang D, Aravind L. Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene. 2010;469:18–30. doi: 10.1016/j.gene.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci U S A. 2010;107:11829–11834. doi: 10.1073/pnas.1006153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho W, Bittova L, Stahelin RV. Membrane binding assays for peripheral proteins. Anal Biochem. 2001;296:153–161. doi: 10.1006/abio.2001.5225. [DOI] [PubMed] [Google Scholar]
- 30.Dowler S, Kular G, Alessi DR. Protein lipid overlay assay. Science’s STKE. 2002;2002:pl6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- 31.Mozsolits H, Aguilar MI. Surface plasmon resonance spectroscopy: an emerging tool for the study of peptide-membrane interactions. Biopolymers. 2002;66:3–18. doi: 10.1002/bip.10200. [DOI] [PubMed] [Google Scholar]
- 32.Stahelin RV, Rafter JD, Das S, Cho W. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J Biol Chem. 2003;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]
- 33.(a) Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J Biol Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]; (b) Yoon Y, Tong J, Lee PJ, Albanese A, Bhardwaj N, Kallberg M, Digman MA, Lu H, Gratton E, Shin YK, Cho W. Molecular basis of the potent membrane-remodeling activity of the epsin 1 N-terminal homology domain. J Biol Chem. 2010;285:531–540. doi: 10.1074/jbc.M109.068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.(a) Blatner NR, Stahelin RV, Diraviyam K, Hawkins PT, Hong W, Murray D, Cho W. The molecular basis of the differential subcellular localization of FYVE domains. J Biol Chem. 2004;279:53818–53827. doi: 10.1074/jbc.M408408200. [DOI] [PubMed] [Google Scholar]; (b) Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 35.(a) Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinosi-tide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG. Molecular mechanism of membrane targeting by the GRP1 PH domain. J Lipid Res. 2008;49:1807–1815. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) He J, Scott JL, Heroux A, Roy S, Lenoir M, Overduin M, Stahelin RV, Kutateladze TG. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J Biol Chem. 2011;286:18650–18657. doi: 10.1074/jbc.M111.233015. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Manna D, Bhardwaj N, Vora MS, Stahelin RV, Lu H, Cho W. Differential roles of phosphatidylserine, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in plasma membrane targeting of C2 domains. Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKCalpha C2 domain. J Biol Chem. 2008;283:26047–26058. doi: 10.1074/jbc.M802617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.(a) Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J Biol Chem. 2003;278:14469–14479. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- 37.(a) Bittova L, Stahelin RV, Cho W. Roles of ionic residues of the C1 domain in protein kinase C-alpha activation and the origin of phosphatidylserine specificity. J Biol Chem. 2001;276:4218–4246. doi: 10.1074/jbc.M008491200. [DOI] [PubMed] [Google Scholar]; (b) Stahelin RV, Cho W. Roles of calcium ions in the membrane binding of C2 domains. Biochem J. 2001;359:679–685. doi: 10.1042/0264-6021:3590679. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Melowic HR, Rafter JD, Cho W. Diacylglycerol-induced membrane targeting and activation of protein kinase Cepsilon: mechanistic differences between protein kinases Cdelta and Cepsilon. J Biol Chem. 2005;280:19784–19793. doi: 10.1074/jbc.M411285200. [DOI] [PubMed] [Google Scholar]
- 38.Stahelin RV, Subramanian P, Vora M, Cho W, Chalfant CE. Ceramide-1-phosphate binds group IVA cytosolic phospholipase a2 via a novel site in the C2 domain. J Biol Chem. 2007;282:20467–20474. doi: 10.1074/jbc.M701396200. [DOI] [PubMed] [Google Scholar]
- 39.(a) Rich RL, Myszka DG. Grading the commercial optical biosensor literature-Class of 2008: ’The Mighty Binders’. J Mol Recognit. 2010;23:1–64. doi: 10.1002/jmr.1004. [DOI] [PubMed] [Google Scholar]; (b) Rich RL, Myszka DG. Survey of the 2009 commercial optical biosensor literature. J Mol Recognit. 2011;24:892–914. doi: 10.1002/jmr.1138. [DOI] [PubMed] [Google Scholar]
- 40.Besenicar M, Macek P, Lakey JH, Anderluh G. Surface plasmon resonance in protein–membrane interactions. Chem Phys Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Falconer RJ, Collins BM. Survey of the year 2009: applications of isothermal titration calorimetry. J Mol Recognit. 2011;24:1–16. doi: 10.1002/jmr.1073. [DOI] [PubMed] [Google Scholar]
- 42.Maget-Dana R. The monolayer technique: a potent tool for studying the interfacial properties of antimicrobial and membrane-lyticpeptides and their interactions with lipid membranes. Biochim Biophys Acta, Biomembr. 1999;1462:109–140. doi: 10.1016/s0005-2736(99)00203-5. [DOI] [PubMed] [Google Scholar]
- 43.(a) Malmberg NJ, Falke JJ. Use of EPR power saturation to analyze the membrane-docking geometries of peripheral proteins: applications to C2 domains. Annu Rev Biophys Biomol Struct. 2005;34:71–90. doi: 10.1146/annurev.biophys.34.040204.144534. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huang H, Cafiso DS. Conformation and membrane position of the region linking the two C2 domains in synaptotagmin 1 by site-directed spin labeling. Biochemistry. 2008;47:12380–12388. doi: 10.1021/bi801470m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.(a) Gong D, Smith MD, Manna D, Bostic HE, Cho W, Best MD. Microplate-based characterization of protein–phosphoinositide binding interactions using a synthetic biotinylated headgroup analogue. Bioconjugate Chem. 2009;20:310–316. doi: 10.1021/bc8004107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Smith MD, Best MD. Characterization of protein–membrane binding interactions via a microplate assay employing whole liposome Immobilization. Methods Mol Biol. 2011;751:477–489. doi: 10.1007/978-1-61779-151-2_30. [DOI] [PubMed] [Google Scholar]
- 45.Rowland MM, Bostic HE, Gong D, Speers AE, Lucas N, Cho W, Cravatt BF, Best MD. Phosphatidylinositol 3,4,5-trisphosphate activity probes for the labeling and proteomic characterization of protein binding partners. Biochemistry. 2011;50:11143–11161. doi: 10.1021/bi201636s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 47.Leslie CC, Gangelhoff TA, Gelb MH. Localization and function of cytosolic phospholipase A2alpha at the Golgi. Biochimie. 2010;92:620–626. doi: 10.1016/j.biochi.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He J, Vora M, Haney RM, Filonov GS, Musselman CA, Burd CG, Kutateladze AG, Verkhusha VV, Stahelin RV, Kutateladze TG. Membrane insertion of the FYVE domain is modulated by pH. Proteins: Struct, Funct, Bioinf. 2009;76:852–860. doi: 10.1002/prot.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.(a) Shin JJ, Loewen CJ. Putting the pH into phosphatidic acid signaling. BMC Biol. 2011;9:85. doi: 10.1186/1741-7007-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, Smits GJ, Loewen CJ. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- 50.Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang BX, Akbar M, Kevala K, Kim HY. Phosphatidylserine is a critical modulator for Akt activation. J Cell Biol. 2011;192:979–992. doi: 10.1083/jcb.201005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milburn CC, Deak M, Kelly SM, Price NC, Alessi DR, Van Aalten DM. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem J. 2003;375:531–538. doi: 10.1042/BJ20031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Matteis MA, Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–397. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 54.Levine CG, Mitra D, Sharma A, Smith CL, Hegde RS. The efficiency of protein compartmentalization into the secretory pathway. Mol Biol Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanada K, Kumagai K, Tomishige N, Yamaji T. CERT-mediated trafficking of ceramide. Biochim Biophys Acta, Mol Cell Biol Lipids. 2009;1791:684–691. doi: 10.1016/j.bbalip.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 56.(a) Arana L, Gangoiti P, Ouro A, Trueba M, Gomez-Munoz A. Ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 2010;9:15. doi: 10.1186/1476-511X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lamour NF, Chalfant CE. Ceramide kinase and the ceramide-1-phosphate/cPLA2alpha interaction as a therapeutic target. Curr Drug Targets. 2008;9:674–682. doi: 10.2174/138945008785132349. [DOI] [PubMed] [Google Scholar]
- 57.Spiegel S, Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol. 2011;11:403–415. doi: 10.1038/nri2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kooijman EE, Sot J, Montes LR, Alonso A, Gericke A, de Kruijff B, Kumar S, Goni FM. Membrane organization and ionization behavior of the minor but crucial lipid ceramide-1-phosphate. Biophys J. 2008;94:4320–4330. doi: 10.1529/biophysj.107.121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.(a) Malkova S, Long F, Stahelin RV, Pingali SV, Murray D, Cho W, Schlossman ML. X-ray reflectivity studies of cPLA2{alpha}-C2 domains adsorbed onto Langmuir monolayers of SOPC. Biophys J. 2005;89:1861–1873. doi: 10.1529/biophysj.105.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Malmberg NJ, Van Buskirk DR, Falke JJ. Membrane-docking loops of the cPLA2 C2 domain: detailed structural analysis of the protein–membrane interface via site-directed spin-labeling. Biochemistry. 2003;42:13227–13240. doi: 10.1021/bi035119+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 61.Landgraf KE, Pilling C, Falke JJ. Molecular mechanism of an oncogenic mutation that alters membrane targeting: Glu17Lys modifies the PIP lipid specificity of the AKT1 PH domain. Biochemistry. 2008;47:12260–12269. doi: 10.1021/bi801683k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.(a) Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology. 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sudhahar CG, Haney RM, Xue Y, Stahelin RV. Cellular membranes and lipid-binding domains as attractive targets for drug development. Curr Drug Targets. 2008;9:603–613. doi: 10.2174/138945008785132420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.(a) Liu Z, Lin L, Yuan C, Nicolaes GA, Chen L, Meehan EJ, Furie B, Huang M. Trp2313-His2315 of factor VIII C2 domain is involved in membrane binding: structure of a complex between the C2 domain and an inhibitor of membrane binding. J Biol Chem. 2010;285:8824–8829. doi: 10.1074/jbc.M109.080168. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Segers K, Sperandio O, Sack M, Fischer R, Miteva MA, Rosing J, Nicolaes GA, Villoutreix BO. Design of protein membrane interaction inhibitors by virtual ligand screening, proof of concept with the C2 domain of factor V. Proc Natl Acad Sci U S A. 2007;104:12697–12702. doi: 10.1073/pnas.0701051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.(a) Jo H, Lo PK, Li Y, Loison F, Green S, Wang J, Silberstein LE, Ye K, Chen H, Luo HR. Deactivation of Akt by a small molecule inhibitor targeting pleckstrinhomology domain and facilitating Akt ubiquitination. Proc Natl Acad Sci U S A. 2011;108:6486–6491. doi: 10.1073/pnas.1019062108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kim D, Sun M, He L, Zhou QH, Chen J, Sun XM, Bepler G, Sebti SM, Cheng JQ. A small molecule inhibits Akt through direct binding to Akt and preventing Akt membrane translocation. J Biol Chem. 2010;285:8383–8394. doi: 10.1074/jbc.M109.094060. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; (c) Meuillet EJ, Zuohe S, Lemos R, Ihle N, Kingston J, Watkins R, Moses SA, Zhang S, Du-Cuny L, Herbst R, Jacoby JJ, Zhou LL, Ahad AM, Mash EA, Kirkpatrick DL, Powis G. Molecular pharmacology and antitumor activity of PHT-427, a novel Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin homology domain inhibitor. Mol Cancer Ther. 2010;9:706–717. doi: 10.1158/1535-7163.MCT-09-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Miao B, Skidan I, Yang J, Lugovskoy A, Reibarkh M, Long K, Brazell T, Durugkar KA, Maki J, Ramana CV, Schaffhausen B, Wagner G, Torchilin V, Yuan J, Degterev A. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proc Natl Acad Sci U S A. 2010;107:20126–20131. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.(a) Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent Advances in the Application of Solution NMR Spectroscopy to Multi-Span Integral Membrane Proteins. Prog Nucl Magn Reson Spectrosc. 2009;55:335–360. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Muller DJ, Wu N, Palczewski K. Vertebrate membrane proteins: structure, function, and insights from biophysical approaches. Pharmacol Rev. 2008;60:43–78. doi: 10.1124/pr.107.07111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidt-Krey I, Rubinstein JL. Electron cryomicroscopy of membrane proteins: specimen preparation for two-dimensional crystals and single particles. Micron. 2011;42:107–116. doi: 10.1016/j.micron.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.(a) Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid–protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Reichow SL, Gonen T. Lipid–protein interactions probed by electron crystallography. Curr Opin Struct Biol. 2009;19:560–565. doi: 10.1016/j.sbi.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.(a) Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang G. NMR of membrane-associated peptides and proteins. Curr Protein Pept Sci. 2008;9:50–69. doi: 10.2174/138920308783565714. [DOI] [PubMed] [Google Scholar]
- 70.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44(Spec No):S24–40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 71.Xu RX, Pawelczyk T, Xia TH, Brown SC. NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein–lipid micelle interactions. Biochemistry. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]
- 72.Chill JH, Louis JM, Baber JL, Bax A. Measurement of 15N relaxation in the detergent-solubilized tetrameric KcsA potassium channel. J Biomol NMR. 2006;36:123–136. doi: 10.1007/s10858-006-9071-4. [DOI] [PubMed] [Google Scholar]
- 73.(a) Brunecky R, Lee S, Rzepecki PW, Overduin M, Prestwich GD, Kutateladze AG, Kutateladze TG. Investigation of the Binding Geometry of a Peripheral Membrane Protein. Biochemistry. 2005;44:16064–16071. doi: 10.1021/bi051127+. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hilty C, Wider G, Fernandez C, Wuthrich K. Membrane protein–lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. ChemBioChem. 2004;5:467–473. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]; (c) Kutateladze TG, Capelluto DGS, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. Multivalent mechanism of membrane insertion by the FYVE domain. J Biol Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]; (d) Papavoine CH, Konings RN, Hilbers CW, van de Ven FJ. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry. 1994;33:12990–12997. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]; (e) Shenkarev ZO, Nadezhdin KD, Sobol VA, Sobol AG, Skjeldal L, Arseniev AS. Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle. FEBS J. 2006;273:2658–2672. doi: 10.1111/j.1742-4658.2006.05282.x. [DOI] [PubMed] [Google Scholar]
- 74.(a) Chill JH, Louis JM, Delaglio F, Bax A. Local and global structure of the monomeric subunit of the potassium channel KcsA probed by NMR. Biochim Biophys Acta, Biomembr. 2007;1768:3260–3270. doi: 10.1016/j.bbamem.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhou Y, Cierpicki T, Jimenez RH, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.(a) Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]; (b) Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 76.(a) Frueh DP, Sun ZY, Vosburg DA, Walsh CT, Hoch JC, Wagner G. Non-uniformly sampled double-TROSY hNcaNH experiments for NMR sequential assignments of large proteins. J Am Chem Soc. 2006;128:5757–5763. doi: 10.1021/ja0584222. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tugarinov V, Choy WY, Orekhov VY, Kay LE. Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc Natl Acad Sci U S A. 2005;102:622–627. doi: 10.1073/pnas.0407792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 78.(a) Diraviyam K, Stahelin RV, Cho W, Murray D. Computer modeling of the membrane interaction of FYVE domains. J Mol Biol. 2003;328:721–736. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]; (b) Singh SM, Murray D. Molecular modeling of the membrane targeting of phospholipase C pleckstrin homology domains. Protein Sci. 2003;12:1934–1953. doi: 10.1110/ps.0358803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.(a) Lumb CN, He J, Xue Y, Stansfeld PJ, Stahelin RV, Kutateladze TG, Sansom MS. Biophysical and Computational Studies of Membrane Penetration by the GRP1 Pleckstrin Homology Domain. Structure. 2011;19:1338–1346. doi: 10.1016/j.str.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Psachoulia E, Sansom MS. Interactions of the pleckstrin homologydomain with phosphatidylinositol phosphate and membranes: characterization via molecular dynamics simulations. Biochemistry. 2008;47:4211–4220. doi: 10.1021/bi702319k. [DOI] [PubMed] [Google Scholar]; (c) Psachoulia E, Sansom MS. PX- and FYVE-mediated interactions with membranes: simulation studies. Biochemistry. 2009;48:5090–5095. doi: 10.1021/bi900435m. [DOI] [PubMed] [Google Scholar]
- 80.Lai CL, Landgraf KE, Voth GA, Falke JJ. Membrane docking geometry and target lipid stoichiometry of membrane-bound PKCalpha C2 domain: a combined molecular dynamics and experimental study. J Mol Biol. 2010;402:301–310. doi: 10.1016/j.jmb.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.(a) Ayton GS, Blood PD, Voth GA. Membrane remodeling from N-BAR domain interactions: insights from multi-scale simulation. Biophys J. 2007;92:3595–3602. doi: 10.1529/biophysj.106.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ayton GS, Lyman E, Krishna V, Swenson RD, Mim C, Unger VM, Voth GA. New insights into BAR domain-induced membrane remodeling. Biophys J. 2009;97:1616–1625. doi: 10.1016/j.bpj.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Blood PD, Voth GA. Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations. Proc Natl Acad Sci U S A. 2006;103:15068–15072. doi: 10.1073/pnas.0603917103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Arkhipov A, Yin Y, Schulten K. Membrane-bending mechanism of amphiphysin N-BAR domains. Biophys J. 2009;97:2727–2735. doi: 10.1016/j.bpj.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blood PD, Swenson RD, Voth GA. Factors influencing local membrane curvature induction by N-BAR domains as revealed by molecular dynamics simulations. Biophys J. 2008;95:1866–1876. doi: 10.1529/biophysj.107.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park WS, Heo WD, Whalen JH, O’Rourke NA, Bryan HM, Meyer T, Teruel MN. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Silkov A, Yoon Y, Lee H, Gokhale N, Adu-Gyamfi E, Stahelin RV, Cho W, Murray D. Genome-wide structural analysis reveals novel membrane-binding properties of ANTH domains. J Biol Chem. 2011 doi: 10.1074/jbc.M111.265611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhardwaj N, Stahelin RV, Langlois RE, Cho W, Lu H. Structural Bioinformatics Prediction of Membrane-binding Proteins. J Mol Biol. 2006;359:486–495. doi: 10.1016/j.jmb.2006.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.(a) Catimel B, Schieber C, Condron M, Patsiouras H, Connolly L, Catimel J, Nice EC, Burgess AW, Holmes AB. The PI(3,5)P2 and PI(4,5)P2 interactomes. J Proteome Res. 2008;7:5295–5313. doi: 10.1021/pr800540h. [DOI] [PubMed] [Google Scholar]; (b) Catimel B, Yin MX, Schieber C, Condron M, Patsiouras H, Catimel J, Robinson DE, Wong LS, Nice EC, Holmes AB, Burgess AW. PI(3,4,5)P3 Interactome. J Proteome Res. 2009;8:3712–3726. doi: 10.1021/pr900320a. [DOI] [PubMed] [Google Scholar]
- 87.Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernandez-Tornero C, Jensen LJ, Kuhn M, Trott J, Rybin V, Muller CW, Bork P, Kaksonen M, Russell RB, Gavin AC. A systematic screen for protein–lipid interactions in Saccharomyces cerevisiae. Mol Syst Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.(a) Kooijman EE, Burger KN. Biophysics and function of phosphatidic acid: a molecular perspective. Biochim Biophys Acta, Mol Cell Biol Lipids. 2009;1791:881–888. doi: 10.1016/j.bbalip.2009.04.001. [DOI] [PubMed] [Google Scholar]; (b) Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta, Mol Cell Biol Lipids. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 89.Varnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim Biophys Acta, Mol Cell Biol Lipids. 2006;1761:957–967. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 90.Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sato M, Ueda Y, Umezawa Y. Imaging diacylglycerol dynamics at organelle membranes. Nat Methods. 2006;3:797–799. doi: 10.1038/nmeth930. [DOI] [PubMed] [Google Scholar]
- 92.Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Szentpetery Z, Varnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci U S A. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.(a) Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. The Journal of biological chemistry. 1985;260:3440–3450. [PubMed] [Google Scholar]; (b) Paredes RM, Etzler JC, Watts LT, Zheng W, Lechleiter JD. Chemical calcium indicators. Methods. 2008;46:143–151. doi: 10.1016/j.ymeth.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.(a) Hanshaw RG, Smith BD. New reagents for phosphatidylserine recognition and detection of apoptosis. Bioorg Med Chem. 2005;13:5035–5042. doi: 10.1016/j.bmc.2005.04.071. [DOI] [PubMed] [Google Scholar]; (b) Lampkins AJ, O’Neil EJ, Smith BD. Bio-orthogonal phosphatidylserine conjugates for delivery and imaging applications. J Org Chem. 2008;73:6053–6058. doi: 10.1021/jo8011336. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Smith BA, Xiao S, Wolter W, Wheeler J, Suckow MA, Smith BD. In vivo targeting of cell death using a synthetic fluorescent molecular probe. Apoptosis. 2011;16:722–731. doi: 10.1007/s10495-011-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mak LH, Georgiades SN, Rosivatz E, Whyte GF, Mirabelli M, Vilar R, Woscholski R. A Small Molecule Mimicking a Phosphatidylinositol (4,5)-Bisphosphate Binding Pleckstrin Homology Domain. ACS Chem Biol. 2011 doi: 10.1021/cb2003187. [DOI] [PubMed] [Google Scholar]
- 97.(a) Fox CB, Wayment JR, Myers GA, Endicott SK, Harris JM. Single-molecule fluorescence imaging of peptide binding to supported lipid bilayers. Anal Chem. 2009;81:5130–5138. doi: 10.1021/ac9007682. [DOI] [PubMed] [Google Scholar]; (b) Ge S, Koseoglu S, Haynes CL. Bioanalytical tools for single-cell study of exocytosis. Anal Bioanal Chem. 2010;397:3281–3304. doi: 10.1007/s00216-010-3843-0. [DOI] [PubMed] [Google Scholar]; (c) Toomre D, Manstein DJ. Lighting up the cell surface with evanescent wave microscopy. Trends Cell Biol. 2001;11:298–303. doi: 10.1016/s0962-8924(01)02027-x. [DOI] [PubMed] [Google Scholar]
- 98.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89:141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Knight JD, Falke JJ. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.(a) Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]; (c) Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hamard-Peron E, Muriaux D. Retroviral matrix and lipids, the intimate interaction. Retrovirology. 2011;8:15. doi: 10.1186/1742-4690-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.(a) Gross RW, Han X. Lipidomics at the interface of structure and function in systems biology. Chem Biol. 2011;18:284–291. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 103.(a) Harder T, van Meer G. Lipid-based membrane domains: physics meets immunology. Traffic. 2003;4:812–820. doi: 10.1034/j.1600-0854.2003.00131.x. [DOI] [PubMed] [Google Scholar]; (b) van Meer G, de Kroon AI. Lipid map of the mammalian cell. J Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]; (c) van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Di Paolo G, Kim TW. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fox TE, Kester M. Therapeutic strategies for diabetes and complications: a role for sphingolipids? Adv Exp Med Biol. 2010;688:206–216. doi: 10.1007/978-1-4419-6741-1_14. [DOI] [PubMed] [Google Scholar]
- 106.Obinata H, Hla T. Sphingosine 1-phosphate in coagulation and inflammation. Semin Immunopathol. 2011 doi: 10.1007/s00281-011-0287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.(a) Vieira FS, Correa G, Einicker-Lamas M, Coutinho-Silva R. Host-cell lipid rafts: a safe door for micro-organisms? Biol Cell. 2010;102:391–407. doi: 10.1042/BC20090138. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal Chem. 2011;83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sparvero LJ, Amoscato AA, Kochanek PM, Pitt BR, Kagan VE, Bayir H. Mass-spectrometry based oxidative lipidomics and lipid imaging: applications in traumatic brain injury. J Neurochem. 2010;115:1322–1336. doi: 10.1111/j.1471-4159.2010.07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chan RB, Oliveira TG, Cortes EP, Honig LS, Duff KE, Small SA, Wenk MR, Shui G, Di Paolo G. Comparative lipidomic analysis of mouse and human brain with Alzheimer’s disease. J Biol Chem. 2011 doi: 10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.de Grauw JC, van de Lest CH, van Weeren PR. A targeted lipidomics approach to the study of eicosanoid release in synovial joints. Arthritis Res Ther. 2011;13:R123. doi: 10.1186/ar3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kanagasabai R, Narasimhan K, Low HS, Ang WT, Fernandis AZ, Wenk MR, Choolani MA, Baker CJ. Mining to find the lipid interaction networks involved in Ovarian Cancers. Summit on translational bioinformatics. 2009;2009:61–65. [PMC free article] [PubMed] [Google Scholar]


