Abstract
Absolute quantification of positron emission tomography (PET) data requires invasive blood sampling in order to obtain the arterial input function (AIF). This procedure involves considerable costs and risks. A less invasive approach is to estimate the AIF directly from images, known as an image derived input function (IDIF). One promising method, EPICA, extracts IDIF by applying independent components analysis (ICA) on dynamic PET data from the entire brain. EPICA requires exclusion of non-brain voxels from the PET images, which is achieved by using a brain mask prior to ICA. Including the entire brain in the mask may degrade the performance of ICA due to noise, artifacts and confounding information. We applied EPICA to 3 [18F]FDG and 3 [11C]WAY data sets and investigated if altering the brain mask by including or excluding tissue structures improves EPICA performance. EPICA applied to whole brain data yields poor performance but with the appropriate brain mask IDIF curves approximate the AIF well. Different tissue structures are important for different radiotracers suggesting that the kinetics of the radiotracer and its diffusion characteristics in the brain influence IDIF estimation with ICA.
Keywords: IDIF, AIF, input function, PET, MRI, arterial blood sampling, ICA, independent components analysis
I. INTRODUCTION
Absolute quantification of positron emission tomography (PET) data requires invasive arterial blood sampling though a catheter inserted into the radial artery, in order to obtain the arterial input function (AIF). This procedure involves considerable costs and risks to patients. Less invasive alternative approaches have been proposed for estimating the AIF from image data commonly referred to as an image derived input function (IDIF). IDIF approaches include, extracting blood signal from manually labeled carotid arteries, sinuses or by identifying cranial blood pools using independent components analysis (ICA) [1].
These approaches estimate whole blood and plasma curves, and therefore are useful only for radiotracers that do not form metabolites. Another type of approach, simultaneous estimation (SIME), estimates the metabolite corrected input function from multiple regional time activity curves [2], but cannot estimate the plasma curve needed for calculating the vascular correction (% blood in the brain) parameters in kinetic models. Thus, methods for estimating both the plasma and metabolite corrected input functions are needed for less invasive quantitative PET. To this end it is necessary to improve and optimize existing promising IDIF methods for estimating the plasma curve.
Several studies comparing IDIF approaches have been conducted. The most promising IDIF methods are also the most demanding in terms of processing time and patient discomfort, as they require either manual labeling and several blood samples [3] to correct for spillover effects or enhanced procedures for reconstructing PET data [4]. These methods may not be amenable to many research centers and data sets; especially data acquired on older systems PET systems that are not compatible with newer reconstruction techniques.
One promising method for estimating the plasma curve is the extraction of the plasma time activity curve using ICA (EPICA) [5]. EPICA requires only a single blood sample and implicitly accounts for spillover effects. This method also works with unprocessed PET data, requiring only the exclusion of non-brain voxels, a procedure that can be automated. Despite these advantages, EPICA achieved mediocre scores in evaluation [1]. In our own evaluation of EPICA we noticed that it was highly sensitive to the brain mask used to exclude non-brain voxels. It is known that for IDIF calculations, ICA spatial constraints, defined as the amount of blood vs. tissue voxels included, needs to be evaluated [6]. Since this type of analysis has not yet been reported for EPICA, there may be an opportunity to improve, optimize, and generalize the method, thus enabling its use across different PET data sets and radiotracers.
In this paper, we investigate the use of different spatial constraints on ICA by learning which tissues should be included in the brain mask to optimize EPICA performance.
II. METHODS
A. Subjects
Data from three [18F]FDG and three [11C]WAY-100635 PET and corresponding MRI scans of healthy volunteers were analyzed. All data were previously collected during an Alzheimer’s [7] and depression study [8]. Participants signed informed consent and studies followed institutional review board–approved protocols.
B. Data acquisition
Data collection is described in [7], [8]. PET images were acquired on an ECAT EXACT HR+ scanner (Siemens/CTI). All data were collected after a 10-minute transmission scan. Reconstruction was performed using filtered back-projection with attenuation correction using the transmission data, and scatter correction was performed using a model-based approach. The reconstruction and estimated image filters were Shepp 0.5 (2.5 mm FWHM), the Z filter was all-pass 0.4 (2.0 FWHM) and the zoom factor was 4.0, leading to a final image resolution of 5.1mm FWHM at the center of the field of view. Images were reconstructed to a 128 × 128 matrix (pixel size 2.5 × 2.5 mm).
During the [18F]FDG scan, emission data were acquired in 3D mode for 60 minutes as 26 frames of increasing duration (8×15 s, 6×30 s, 5×1 min, 4×5 min, and 3×10 min). Thirteen arterial blood samples (each 0.3 ml) were drawn by pump and manually during the scan (at 10, 20, 30, 40, 50, 60, 70, 80, 90 sec, and 1.5, 5, and 20 min after injection). The last sample was drawn 30 sec after the final acquisition. During the [11C]WAY scan, emission data were collected in 3D mode for 110 min as 20 frames of increasing duration (3×20 s, 3×1 min, 3×2 min, 2×5 min, 9×10 min). Following radiotracer injection, 30 arterial samples were collected, 24 samples automatically every 5 seconds for the first 2 min, and 6 samples manually thereafter at longer intervals. Following centrifugation, plasma was collected in 200 ml aliquots and radioactivity was counted in a gamma counter (Wallac 1480 Wizard 3M Automatic Gamma Counter).
MRI images were acquired using a 1.5T (Signa Advantage, GE Healthcare). All scans from the 1.5T camera were acquired in the coronal plane (orthogonal to the anterior- commissure posterior-commissure plane over the whole brain) with the following parameters: 3-dimensional spoiled gradient recalled acquisition in the steady state; repetition time, 34 ms; echo time, 5 ms; flip angle, 45°; 1.5-mm slice thickness (zero gap); 124 slices; and field of view, 220 × 160 mm. All images from the 1.5T camera were reconstructed to a size of 256 × 256, with a resolution of 1.5 × 0.86 × 0.86 mm.
C. Image Processing
Image analysis was performed using Matlab 2011a (The Mathworks), with extensions to the following open source packages: Functional MRI of the Brain’s Linear Image Registration Tool (FLIRT), version 5.2; Brain Extraction Tool (BET) version 1.2; and University College of London’s Statistical Parametric Mapping (SPM5) segmentation routines, as previously described [9].
The PET scan was motion corrected utilizing de-noising techniques to improve intra-frame co-registration. Any significant residual motion was assessed visually and manually corrected. The mean of motion corrected frames was co-registered to the MRI and the resulting transformation was applied to all PET frames. Blood flow (BF) images (where brain arteries and veins are visible) were made by averaging the first 60 seconds of data, approximately the time at which the radiotracer reaches blood vessels after injection. To standardize image orientation, using the MRI, BF images were aligned to the orientation of the 2mm single subject MNI template (Collins et al, 1998) using a rigid body transform. MRI images were segmented using SPM5 into gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) maps and transformed into PET space using inverse MRI-PET transform from above.
C. Blood and metabolite analysis
Plasma radioactivity curves derived from arterial sampling were fit with a model in order to interpolate the activity at times corresponding to the binning of the PET data. The model used is a straight line from time zero to the peak, followed by a sum of three decreasing exponentials after the peak:
| (1) |
Since [11C]WAY metabolizes in the body, its parent fractions are measured in order to correct the plasma radioactivity curve. Hills Function was used to model the metabolite curve, given by the equation (2).
| (2) |
The metabolite corrected AIF is therefore calculated using equation (3):
| (3) |
D. Blood vessel labeling
ITK-SNAP1 was used to delineate the internal carotid arteries and the superior, transverse, and sigmoid sinus on the aligned BF PET image. An anatomical protocol was not followed in this study because the only goal was to highlight cranial vessels that may aid the ICA classification. Nevertheless the manual segmentation procedure is described here for reference. The entire span of the internal carotid, cervical (C1) to communicating (C7) sections, are typically not visible on the PET image due to patient position, motion, and/or low field of view (FOV). Therefore, carotid arteries were labeled from petrous (C2) to C7. The sinuses were traced from the superior aspect of the superior sagittal sinus to the superior aspect of the jugular bulb. The superior sagittal sinus was traced in the sagittal view, excluding the smaller less visible superior-anterior sections. The jugular bulb was excluded from carotids and sinuses because it is difficult to differentiate at that level. A representative tracing of left and right carotid arteries and sinus sections are shown in Fig. 1.
Figure 1.

Carotid and sinus labeling. Left: transverse view of left/right carotid arteries (red) and left/right transverse sinuses (green). Middle: sagittal view, medial slice. Right: 3D rendering of fully labeled subject, noting the exclusion of the jugular bulb where the carotid and sinuses meet.
D. IDIF algorithm
A version of EPICA (v1.0) is freely available2 and uses a pow5exp ICA cost function, which suits irreversible radiotracers such as [18F]FDG. In order to implement EPICA for [11C]WAY, the author kindly provided a revised version (v1.3) that includes a kurtosis cost function more appropriate for neuroreceptor studies. Although the core EPICA algorithm remained untouched, some changes to the software were made for the purposes of batch processing. EPICA separates the PET signal into blood and tissue components. It cannot differentiate the two automatically. Instead, two plots show ICA components representing blood and the tissue curves and the user must select the one that best resembles the shape of a typical AIF curve. For our purposes this process was automated by selecting the curve with the lowest error sum of squares (SSE) relative to the measured AIF.
Brain Masks
One of the pre-processing steps for EPICA is to remove non-brain regions from the PET image by using a brain mask derived from an MRI or by segmenting the PET image [5]. We noticed that this typically yields poor results with our data. In the original EPICA studies, the FOV and resolution of PET data were significantly lower than our scans, forcing the authors to use a narrower brain mask with superior and inferior regions of the PET cutoff. In fact, this is evident in the demo images provided on the EPICA website. We hypothesized that optimizing the brain mask might improve EPICA performance for our data. To find the optimal brain mask we created brain masks consisting of various combinations of tissues: GM, WM, CSF, carotid (CA) and sinuses (SV) from the MRI and manual labeling, respectively. For example, a brain mask could consist of GM+WM, GM+CA or GM+WM+SV. All n-choose-k combinations were created yielding 186 different brain mask types. Furthermore, we evaluated 11 different superior and inferior cutoffs: (1) slice 5 held constant, superior slice gradually increased in steps of 10 slices from the 10th to 60th. (2) superior slice 60 held constant, while inferior slice was increased in steps of 10 slices from the 10th to the 50th. Considering all brain mask types and cutoffs, a total of 2046 different brain masks were evaluated. Representative brain masks are shown in Fig. 2.
Figure 2.

Example brain mask types. From left to right: whole brain mask, GM+WM, WM+CSF and GM only, respectively, overlaid on the blood flow PET image from a representative subject. The whole brain mask is the type recommended for use with EPICA while the tissue combinations are examples of what is used in this study.
E. Evaluation
In order to determine the optimal brain mask for [18F]FDG and [11C]WAY data EPICA was run with all brain mask types and SSE was calculated between all IDIFs and corresponding AIFs. The brain mask that yielded the lowest SSE for a particular subject was considered optimal for that subject. In an attempt to generalize the approach across subjects, we then searched for a single (common) brain mask that gave the lowest average SSE across all subjects. For each, bth, brain mask we averaged SSE across all, sth, subjects using equation (4), where, N = total number of subjects and T = total scan time. The IDIF curves that correspond to the lowest across subject average SSE are reported.
| (4) |
III RESULTS
A. EPPICA without modifications
EPICA was applied to our data set using a whole brain mask as previously recommended [5]. For FDG, the IDIF underestimated the AIF for four subjects. In all four subjects the IDIF contained negative values in the tail. In only one subject did the IDIF closely approximate AIF curve. For WAY data, the IDIF overestimated the peak in two cases and the tail slope was positive in two cases, which is physiologically impossible. We also see that for WAY, the IDIF approximates the plasma radioactivity levels and not the AIF due to the presence of metabolites. EPICA results using a whole brain mask are shown in Fig. 3.
Figure 3.

IDIF results obtained with EPICA applied to FDG (a,b,c) and WAY (d,e,f) data from 6 subjects, using default settings. Each graph shows results for a single subject. For FDG data the AIF is the measured plasma radioactivity. For WAY, the AIF is the measured plasma radioactivity corrected for metabolites. Significant underestimation of tail for FDG, overestimation of the peak and increasing slope in WAY, are evident with EPICA calculated IDIF curves.
B. EPICA with modified brain masks
Brain masks selective of different tissue classes were applied to WAY and FDG data. For each subject individually, we searched for an optimal brain mask that gave the lowest SSE between IDIF and measured AIF. For FDG, the optimal brain masks yielded IDIFs that closely approximated the tail and peak shape, with the exception of a slight time delay. Optimal brain masks were composed of different tissue classes, GM (n=2), WM (n=2), CSF (n=2), CA (n=2), SV was not present in any optimal brain mask, n = # of subjects. Cutoff ranged from slice 5-20 to slice 60, essentially most of the brain was included. For WAY, optimal brain masks improved peak estimation and the positive tail slope was reduced to better match the tail of the measured plasma curve. Again, brain masks were composed of different tissue classes, CSF (n=3), CA (n=2), SV (n=3), WM (n=1), GM was not present in any of the masks. EPICA results using optimal brain masks are shown in Fig. 4.
Figure 4.
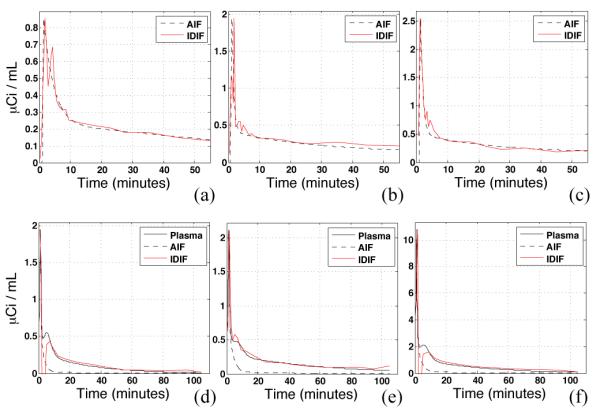
IDIF results obtained with EPICA applied to FDG (a,b,c) and WAY (d,e,f) data from 6 subjects, using an optimal brain mask for each subject. AIF is the measured plasma radioactivity for FDG and measured plasma radioactivity corrected for metabolites for WAY. The IDIF curves closely approximate true plasma radioactivity for both WAY and FDG. Discrepancies, evident around the 5-minute time point for 1 FDG and 2 WAY subjects.
A common brain mask was selected for each radiotracer based on SSE criteria in equation (4). Slight under- and over-estimation of the tail was present in FDG IDIFs, and the peak in WAY IDIFs. The IDIF for both radiotracers using the common brain mask were closer to the AIF than when using the default whole brain mask. As expected however, the results were worse than when the optimal brain mask was selected for each subject individually. EPICA results using common brain masks are shown in Fig. 5.
Figure 5.
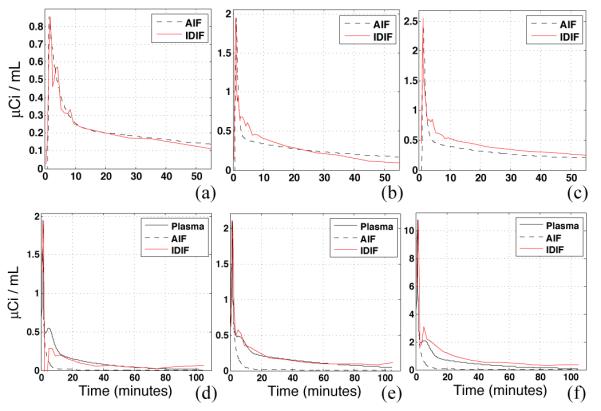
IDIF results obtained with EPICA applied to FDG (a,b,c) and WAY (d,e,f) data from 6 subjects, using a common brain mask. The common brain mask yielded the lowest average SSE across subjects between IDIF and AIF curves. The FDG brain mask was composed of GM, WM and SV tissues with a range of slices 20-60, bottom 19 slices cutoff. The WAY brain mask was composed of CSF and SV tissues with a range of slices 5-40, top 19 slices cutoff. Under- and over- estimation of the tail is evident with FDG while the peak is under or overestimated with WAY.
IV. CONCLUSION
We introduced a procedure for optimizing the brain mask used for IDIF estimation with ICA. This technique was demonstrated using the EPICA IDIF estimation software. It was shown that there exists a brain mask in each subject that can yield IDIFs that approximate the AIF very well, which suggests that imposing spatial constraints on ICA can improve results. In practice however, identifying a brain mask type for each subject is not practical so we attempted to find common brain mask that works well across subjects, one for each radiotracer. The results were less impressive than the individual masks but a great improvement over the default whole brain mask recommended for EPICA.
Our results show that more work needs to be done understanding and characterizing the locations of blood pools within PET brain images. An interesting unexpected finding was the difference in tissues present in both the optimal and common brain masks. In optimal masks, SV was important for WAY but not for FDG. In common masks, GM and WM seemed to be more important for FDG while CSF was more important to WAY data. It is unclear why this would be since CSF does not contain brain tissue. A possible explanation is that CSF contains useful signal from meninges, surface blood vessels and/or smoothing artifact of nearby gray matter. More work is needed with additional subjects in order to evaluate whether our tissue selection can be used to improve ICA based IDIF methods.
Footnotes
Contributor Information
Arthur Mikhno, Columbia University, NY 10027 USA.
Francesca Zanderigo, Columbia University, NY 10027 USA.
Mika Naganawa, Yale University School of Medicine, New Haven, CT 06520 USA.
Andrew F. Laine, Columbia University, NY 10027 USA
Ramin V. Parsey, Columbia University, NY 10027 USA
REFERENCES
- [1].Zanotti-Fregonara P, El Mostafa Fadaili RM, Comtat C, Souloumiac A, Jan S, Ribeiro MJ, Gaura V, Bar-Hen A, Trébossen R. Comparison of eight methods for the estimation of the image-derived input function in dynamic [ 18F]-FDG PET human brain studies. Journal of Cerebral Blood Flow & Metabolism. 2009;vol. 29(no. 11):1825–1835. doi: 10.1038/jcbfm.2009.93. others. [DOI] [PubMed] [Google Scholar]
- [2].Ogden RT, Zanderigo F, Choy S, Mann JJ, Parsey RV. Simultaneous estimation of input functions: an empirical study. Journal of Cerebral Blood Flow & Metabolism. 2009 Dec.vol. 30(no. 4):816–826. doi: 10.1038/jcbfm.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen K, Bandy D, Reiman E, Huang S-C, Lawson M, Feng D, Yun L, Palant A. Noninvasive Quantification of the Cerebral Metabolic Rate for Glucose Using Positron Emission Tomography, 18F-Fluoro-2-Deoxyglucose, the Patlak Method, and an Image-Derived Input Function. J Cereb Blood Flow Metab. 1998 Jul.vol. 18(no. 7):716–723. doi: 10.1097/00004647-199807000-00002. [DOI] [PubMed] [Google Scholar]
- [4].Mourik JEM, Lubberink M, Klumpers UMH, Comans EF, Lammertsma AA, Boellaard R. Partial volume corrected image derived input functions for dynamic PET brain studies: Methodology and validation for [11C]flumazenil. NeuroImage. 2008 Feb.vol. 39(no. 3):1041–1050. doi: 10.1016/j.neuroimage.2007.10.022. [DOI] [PubMed] [Google Scholar]
- [5].Naganawa M, Kimura Y, Ishii K, Oda K, Ishiwata K, Matani A. Extraction of a plasma time-activity curve from dynamic brain PET images based on independent component analysis. IEEE Transactions on Biomedical Engineering. 2005 Feb.vol. 52(no. 2):201–210. doi: 10.1109/TBME.2004.840193. [DOI] [PubMed] [Google Scholar]
- [6].Zanotti-Fregonara P, Liow J-S, Fujita M, Dusch E, Zoghbi SS, Luong E, Boellaard R, Pike VW, Comtat C, Innis RB. Image-Derived Input Function for Human Brain Using High Resolution PET Imaging with [11C](R)-rolipram and [11C]PBR28. PLoS ONE. 2011 Feb.vol. 6(no. 2):e17056. doi: 10.1371/journal.pone.0017056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen K, Chen X, Renaut R, Alexander GE, Bandy D, Guo H, Reiman EM. Characterization of the image-derived carotid artery input function using independent component analysis for the quantitation of [18F] fluorodeoxyglucose positron emission tomography images. Physics in medicine and biology. 2007;vol. 52:7055. doi: 10.1088/0031-9155/52/23/019. [DOI] [PubMed] [Google Scholar]
- [8].Devanand DP, et al. Pittsburgh compound B (11C-PIB) and fluorodeoxyglucose (18 F-FDG) PET in patients with Alzheimer disease, mild cognitive impairment, and healthy controls. J Geriatr Psychiatry Neurol. 2010 Sep.vol. 23(no. 3):185–198. doi: 10.1177/0891988710363715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Parsey RV, et al. Higher Serotonin 1A Binding in a Second Major Depression Cohort: Modeling and Reference Region Considerations. Biological Psychiatry. 2010 Jul.vol. 68(no. 2):170–178. doi: 10.1016/j.biopsych.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]


