Abstract
IL-17, a major inflammatory cytokine plays a critical role in the pathogenesis of many autoimmune inflammatory diseases. In this study, we report a new function of RNA binding protein HuR in IL-17-induced Act1-mediated chemokine mRNA stabilization. HuR deficiency markedly reduced IL-17-induced chemokine expression due to increased mRNA decay. Act1-mediated HuR polyubiquitination was required for the binding of HuR to CXCL1 mRNA, leading to mRNA stabilization. While IL-17 induced the co-shift of Act1 and HuR to the polysomal fractions in a sucrose gradient, HuR deficiency reduced the ratio of translational-active versus translational-inactive IL-17-induced chemokine mRNAs. Furthermore, HuR deletion in distal lung epithelium attenuated IL-17-induced neutrophilia. In summary, HuR functions to couple receptor proximal signaling to posttranscriptional machinery, contributing to IL-17-induced inflammation.
INTRODUCTION
Interleukin 17 (IL-17, also known as IL-17A) is a key proinflammatory cytokine produced mainly by a distinct subset of CD4 T helper cells called Th17 (1–3). IL-17 is required for host defense against extracellular microorganisms (3–6) and has also has been linked to the development and pathogenesis of many autoimmune disorders, including rheumatoid arthritis, multiple sclerosis, psoriasis and asthma (7–12). Deficiency in IL-17 leads to diminished antigen-specific T cell–mediated immune responses, including allergen-induced pulmonary inflammation and airway hyper-responsiveness (13,14). The major function of IL-17 is to coordinate local tissue inflammation by promoting production of proinflammatory and neutrophil-mobilizing cytokines and chemokines, including IL-6, CSF2, TNFα, IL-1, CXCL1, CCL2, CXCL2, CCL7 and CCL20, resulting in the infiltration of inflammatory cells such as neutrophils, monocytes and lymphocytes.
IL-17 signals through a heterodimeric receptor complex composed of IL-17RA and IL-17RC, members of IL-17R receptor family (15,16). Both IL-17RA and IL-17RC belong to a SEFIR protein family, which is defined by the presence of a conserved cytoplasmic SEFIR domain (17). Act1 (also known as CIKS) is an essential component in IL-17 signaling and required for IL-17-dependent immune responses (15,18,19). Act1 is also a member of the SEFIR protein family, containing a SEFIR domain at its C-terminus (20). Upon IL-17 stimulation, Act1 is recruited to IL-17R through a SEFIR-dependent interaction. Furthermore, Act1 possesses a U-Box domain that is functionally required for its E3-ligase activity. Upon IL-17 stimulation Act1, together with the Ubc13-Uev1A E2 complex, exerts K63-linked polyubiquitination of TRAF6 (21,22). This ubiquitination event is required for TRAF6-mediated activation of TAK1 and the IKK complex, resulting in activation of transcription factor NFkB and subsequent NFkB-dependent transcription of pro-inflammatory and neutrophil-mobilizing cytokines and chemokines. While IL-17 regulates gene transcription, it also induces gene expression by stabilizing otherwise unstable mRNAs of pro-inflammatory genes (23). Recently, we reported that following IL-17 stimulation, a phosphorylated form of Act1 forms a complex with TRAF2 and TRAF5 (24). This phosphorylation event and complex formation are functionally required for the stabilization of CXCL1 and CSF3 mRNA.
Many cytokine and chemokine mRNAs exhibit very short half-lives due to the presence of AU-rich sequence elements (ARE) located within their 3’ untranslated regions (3’UTR) (25–27). Therefore, the regulation of mRNA stability is an important control of inflammatory gene expression (28). The AREs within the 3’ UTR can be recognized by RNA binding proteins that function to mediate the sequential deadenylation, decapping, and ultimately exonucleolytic degradation of the RNA (29–31). These ARE binding proteins include AUF1 (hnRNP D), Tristetraprolin (TTP), butyrate response factors (BRF1 and BRF2), and KH domain containing splicing regulatory factor KSRP (32,33). We recently identified a novel mRNA destabilizing factor, called SF2/ASF, which is linked to IL-17 receptor signaling through its interaction with Act1 and TRAF5 (24). SF2/ASF binds chemokine mRNAs in unstimulated cells, whereas the SF2/ASF-mRNA interaction is markedly diminished following IL-17 stimulation. SF2/ASF promotes chemokine mRNA decay and its depletion results in prolonged mRNA half life.
The important question is what happens to the IL-17-induced chemokine mRNAs after their dissociation from SF2/ASF; how the chemokine mRNAs are actually stabilized and translated. In this study, we report that ARE-binding protein HuR (Human antigen R) plays a critical role in IL-17-induced chemokine mRNA stabilization. IL-17 stimulation induced the interaction of HuR with Act1 and TRAF5, but not SF2/ASF. Whereas Act1 is required for IL-17-induced dissociation of SF2/ASF from mRNA, IL-17 induced the HuR-mRNA interaction in an Act1-dependent manner. HuR depletion indeed substantially impaired IL-17-induced CXCL1 and CXCL5 expression due to increased mRNA decay. Furthermore, upon IL-17 stimulation Act1 mediated K63-linked polyubiquitination of HuR, which is required for HuR-mRNA interaction and HuR-mediated chemokine mRNA stabilization. Moreover, IL-17 stimulation induced the co-shift of Act1 and HuR towards the fractions of polysomes in a sucrose gradient, implicating the importance of Act1-HuR complex in moving the mRNAs to the translational active state. The impact of HuR on IL-17-induced chemokine mRNA stabilization had important biological consequence, since HuR deficiency attenuated IL-17-induced neutrophilia and pulmonary inflammation in vivo.
MATERIALS AND METHODS
Reagents
Antibodies against Act1, GAPDH, HuR, SF2, TRAF2, TRAF5, α-tubulin and β-actin were from Santa Cruz Biotechnology. Anti-HA and anti-Ubiquitin antibody were from Sigma; anti-pro-SP-C antibody from Upstate. Adenoviruses encoding GFP (Ad-GFP) and Cre-GFP (Ad-Cre-GFP) were obtained from Vectorbiolabs. Purified GST-HuR was purchased from Novus Biologicals. Cell culturing of MEFs, HeLa and HeLa Tet-Off cells were as previously described (21,34). Reporter plasmids CXCL1–3’UTR Δ4 and CSF2–3’UTR encoding the 5′ UTR and coding region of mouse CXCL1 and either the truncated 3′ UTR of CXCL1 (GenBank accession No. NM_008176.3; residues 720–950; http://www.ncbi.nlm.nih.gov/nuccore/NM_008176.3) or truncated 3’ UTR of CSF2 (GenBank accession No. NM_009969.4; residues 923–973; http://www.ncbi.nlm.nih.gov/nuccore/NM_009969.4), respectively, were described previously (24). Mouse Act1, its D-Ubox mutant, WT TRAF6 were cloned into pMSCV-IRES-GFP as described previously (21). Wild-type and mutant HA-ubiquitin constructs were described previously (21). SP-C-rtTA/tetO-CRE mice, described previously (35), were kind gift from Dr. Jeffrey Whitsett.
Generation of conditional HuR knockout mice
Conditional HuR knock-out ES cells and mice were generated using gene targeting technology. A targeting vector containing a 5’homology arm (4.5 kb), 3’homology arm (3.5 kb) and conditional region (0.6 kb) was generated by PCR. The targeting construct also contained loxP sequences flanking the conditional KO region, the Neo expression cassette (for positive selection of ES cells) flanked by FRT sequences (for subsequent removal of the Neo cassette) and a DTA expression cassette (for negative selection of the potentially targeted ES cells). The final targeting construct is shown in supple. Fig. 1a. Successfully targeted ES cells were injected into blastocysts and implanted into pseudopregnant females. Chimeric male offspring were mated to WT C57BL/6 female and germline transmission of the mutant HuR allele was confirmed by Southern (Data not shown) and PCR analyses (supple. Fig. 1b); the primers used were: Neo expression cassette forward: 5’-GGTTTCCAAATGTGTCAGTTTCATAGC-3’, intron 3 reverse: 5’-AGAGATAGATGGTTAGGCATAGAGATGCAG-3’ and intron 3 forward: 5’-TATGCTTTAAGAGACCCAGAAGCCAG-3’.
Intranasal instillation of IL-17
Mice were anesthetized with isoflurane. Carrier-free, murine rIL-17 (R&D Systems) resuspended in sterile saline (0.9%) was instilled into nasal opening in 50 µl (0.5 µg) aliquot per mouse. A total of 0.7 ml of HL-1 medium (BioWhittaker) was used to obtain BAL fluid through trachea using a blunt needle and 1-ml syringe. Lungs were collected and snap frozen immediately in liquid nitrogen container. Total RNA was obtained by using TRIzol (Invitrogen) and OMNI TH tissue homogenizer (Omni International). H&E staining was obtained on lung tissue after fixation in 10% neutral buffered formalin and paraffin embedding. Paraffin-embedded lung sections were stained with hematoxylin and eosin to evaluate inflammation. For frozen sections, lungs were embedded in OCT (Tissue-Tek) and snap frozen in liquid nitrogen. Sections (10µm) were incubated with anti-HuR (1:100) and anti-pro-SP-C (1:100). Antigens were visualized following incubation with fluorescence-conjugated secondary antibodies (Molecular probe). For ELISA assay, CXCL1 production was measured using DuoSet ELISA Development Systems obtained from R&D Systems, following manufacturer’s instruction.
Affymetrix genechip microarray analysis
200ng of RNA was used for target labeling, and the target preparation was done on Biomeck-FXP (Beckman Coulter Inc, Brea, CA, USA) using GeneChip HT 3' IVT Express Kit (Affymetrix, Santa Clara, CA, USA) following manufactures recommendation. Labeled cRNA were hybridized on Affymetrix genechip HT-MG-430PM-96 (Affymetrix, Santa Clara, CA, USA). All array hybridization, washing, and scanning were performed on Genetitan (Affymetrix, Santa Clara, CA, USA) according to the manufacture’s recommendations. Three independent biological replicates were analyzed in each experiment, which yielded very similar results. The t-test was used to assess significance, with P < 0.05 deemed significant.
Immunoprecipiation
Cells were lysed in buffer A (0.5% Triton X-100, 20 mM HEPES, pH 7.4, 150 mM NaCl, 12.5 mMβ-glycerophosphate, 1.5 mM MgCl2, 10 mM NaF, 2 mM dithiothreitol, 1 mM sodium orthovanadate, 2 mM EGTA, 20µM aprotinin, 1 mM phenylmethylsulfonyl fluoride)or, to pevent possible protein-protein interaction, in buffer B (buffer A plus 0.1% SDS and 0.5% deoxycholate). Cell extracts were incubated with 1 µg of different antibodies overnight at 4°C with 20 µl of protein A Sepharose beads. After incubation beads were washed 4 times with lysis buffer, separated by SDS-PAGE and analyzed by Western blot.
In vitro ubiquitination assay
The polyubiquitination assays were perfomed in 10 µl reaction volume in buffer (20 mM Tris HCl, PH7.5, 2 mM ATP, 5 mM MgCl2) at 37°C for 1 h, together with the following components provided accordingly: 100 ng E1, 100 ng E2, 100 ng individual E3 (GST-TRAF6 or GST-Act1), 5 µg ubiquitin. For Act1-mediated polyubiquitination of HuR, in addition to E1, E2 and ubiquitin, 100 ng of GST-HuR was added as substrate, together with Act1 in the same reaction buffer.
Quantitative real-time PCR
Total RNA was isolated with the TRIzol reagent (Invitrogen). Real-time PCR was performed using SYBER Green PCR Master Mix kit (Applied Biosystems). The primers used were β-actin: 5’-GGTCATCACTATTGGCAACG-3’ and 5’-ACGGATGTCAACGTCACACT-3’. CXCL1: 5′-TAGGGTGAGGACATGTGTGG-3′ (forward) and 5′-AAATGTCCAAGGGAAGCGT-3′ (backward); CSF2: 5′-GGCCTTGGAAGCATGTAGAGG-3′ (forward) and 3’-GGAGAACTCGTTAGAGACGACTT-3′ (backward); CXCL5: 5′-GCCCTACGGTGGAAGTCATA-3′ (forward) and 5′-GTGCATTCCGCTTAGCTTTC-3′ (backward); HuR: 5′-CCCAGAGCAGGTCAGCGTCTC-3’(forward) and 5′-GGTGCTACAAGCCCGTCATCA-3′ (backward).
Polysomal fractionation analysis
2 × 108 cells were left untreated or stimulated with IL-17A (50 ng/ml) for 90 minutes. Cytoplasmic exacts were carefully layered over 10–50% linear sucrose gradients in polysome buffer (10 mM HEPES, pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 50 units of recombinant RNasin (Promega), and 0.1% Igepal-CA630 (Sigma) and centrifuged at 17,000 rpm in a Beckman SW32.1 Ti rotor for 4h at 4°C. Gradients were fractioned by using an ISCO gradient fractionation system equipped with a UA-6 detector. Light RNP fractions, 40S, 60S, and 80S and heavy polysome fractions were monitored by the continuous UV absorption profile at A254, and 9 tubes of 750µl fractions were collected. The fractions representing light RNP and free ribosomes were used to prepare the translation-inactive pool of proteins and mRNAs, and the fractions representing heavy polysomes were used to isolate the translation-active proteins and mRNAs. 1/10 of each fraction was used for western analysis; 1/5 of each fraction was used for RNA isolation by extraction with TRIzol.
Subcellular Fractionation
Confluent cells in 15 centimeter plates, untreated or treated with IL-1 (1ng/ml) for various times, were resuspended in 1ml of ice-cold hypotonic buffer (10 mM HEPES, pH 7.4, 1.5 mM MgCl2, 10mM KCl, 0.2 mM phenylmethylsulfonyl fluoride, and 0.5 mM dithiothreitol) and homogenized on ice with 45 strokes of a Dounce homogenizer. Unlysed cells, nuclei, and cell debris were pelleted by centrifugation at 1000 g for 5 min for five times. Soluble (supernatant, S100) and particulate (pellet, P100) fractions were generated by centrifugation at 100,000× g for 1 h.
RNA binding assays
The ability of HuR, SF2 and Act1 to bind to RNA in vivo was conducted as described previously (33). Briefly, cells were transiently transfected with TRE-regulated CXCL1–3’UTR Δ4 and CSF2–3’UTR reporter constructs. Twenty hours after transfection, 2 × 106 cells were trypsynized, washed twice, and resuspended in 10 ml of ice-cold PBS. Cells were fixed in 0.1% formaldehyde for 15 min at room temperature, whereupon the cross-linking reaction was stopped with glycine (pH 7.0, 0.25 M). The cells were then washed twice with ice-cold PBS, resuspended in 2 ml of RIPA buffer (50 mM Tris-HCl (pH 7.5), 1% nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl, and proteinase inhibitors), and sonicated. The lysate was centrifuged (15 min, 4°C, 16,000 × g), and 1 ml of each supernatant was immunoprecipitated overnight at 4°C, using protein G-agarose beads preincubated with 20 µg of anti-HuR or anti-Act1 Ab. The beads were washed 5 times with 1 ml RIPA buffer and resuspended in 150 µl of elution buffer (50 mM Tris-Cl (pH 7.0), 5 mM EDTA, 10 mM DTT, 1% SDS). Cross-linking was reversed by incubation at 70°C for 45 min, and RNA was purified from immunoprecipitates with TRI Reagent, treated with RNase-free DNase, and 10% of the total RNA sample was reverse-transcribed with Moloney murine leukemia virus (M-MLV) reverse transcriptase. Two microliters (10%) of the reverse transcriptase product was subjected to quantitative real-time PCR. The primers for CXCL1 coding region were forward, 5′-CTGGCCACAGGGGCGCCTATC-3’; reverse, 5′-GGACACCTTTTAGCATCTTT-3’ and for GAPDH were forward, 5′-TCACCATCTTCCAGGAGCGAGAT-3”; reverse, 5′-GTTGGTGGTGCAGGAGGCATTGCT-3’.
Adenoviral infection
Primary kidney epithelial cells were subdivided into 60-mm dishes and infected by exposing cells to media containing 2 × 105 IFU/PFU of adenovirus/ml overnight.
Statistical analysis
The data are presented as the mean + standard deviation. The significance of difference between two groups was determined by Student’s t test.
RESULTS
HuR depletion diminishes IL-17-induced expression of CXCL1 and CXCL5 but not CSF2
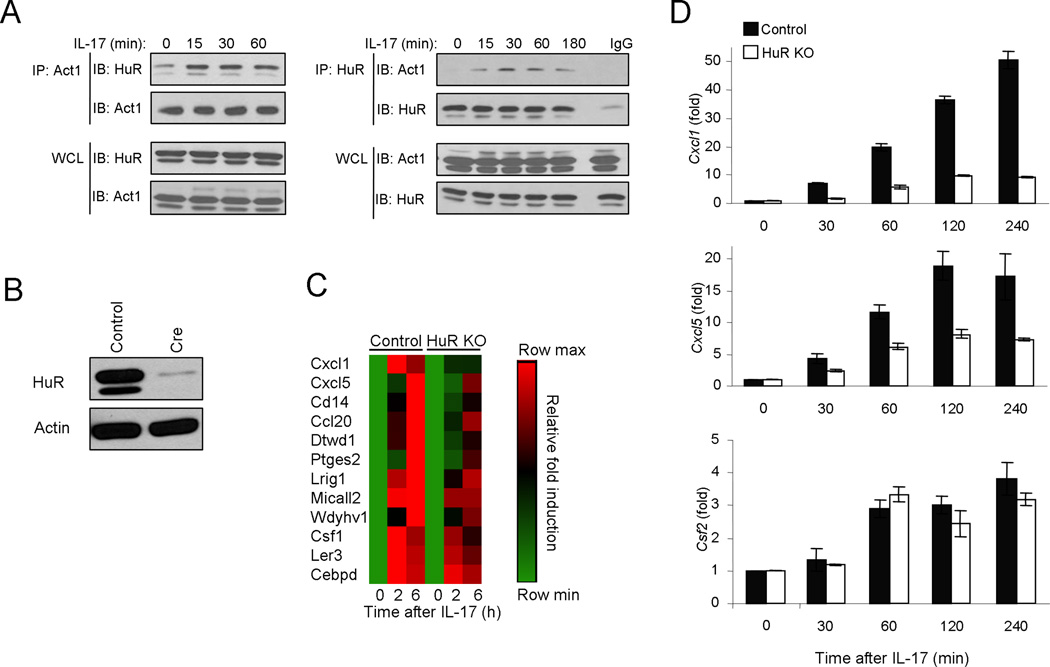
Although IL-17-induced Act1-mediated signaling induces transcriptional activation of genes encoding inflammatory molecules, it also affects their production at the post-transcriptional level. Many cytokine and chemokine mRNAs are known to have half-lives shortened by mechanisms that depend on adenine- and uridine-rich elements (ARE) located in the 3′ untranslated region. To investigate the mechanism for Act1-dependent mRNA stabilization during IL-17 signaling, using co-immunoprecipitation assay we searched for possible interaction of Act1 with ARE-binding proteins that have been implicated in regulation of mRNA stability. Through the search, we found that IL-17 stimulation induced the interaction of endogenous Act1 with mRNA stabilizing protein – HuR (Fig. 1A). HuR was previously shown to inhibit mRNA decay through its cooperation with other ARE-BPs. To identify the potential HuR target mRNAs induced by IL-17, we performed array analysis of kidney epithelial cells from HuR flox/flox mice. Cells were infected with adenovirus encoding GFP or GFP-Cre. Cre-mediated deletion reduced expression of HuR protein by about 95% (Fig. 1B). We found that expression of a subset of IL-17-induced genes was substantially reduced in HuR-deficient cells compared to that in the control cells (Fig 1C). By real-time PCR, we indeed confirmed that CXCL1 and CXCL5 mRNA accumulation was markedly diminished in HuR-deficient cells compared to that in control cells, whereas the accumulation of CSF2 mRNA was unaffected (Fig. 1D).
Figure 1. HuR depletion diminishes IL-17-induced expression of CXCL1 and CXCL5 but not CSF2.
(A) Cell lysates from HeLa cells untreated or treated with IL-17 (50 ng/ml) for the indicated time were immunoprecipitated (IP) with anti-Act1, anti-HuR and IgG control, followed by western analysis (IB) with anti-HuR and anti-Act1. WCL: whole-cell lysates. (B) Western analysis of HuR and β-actin in lysates of primary mouse kidney epithelial cells isolated from HuRflox/flox mice infected with a GFP-encoding adenovirus (Control) or Cre-GFP encoding adenovirus (Cre). (C) Affymetrix microarray analyses of primary kidney epithelial cells infected as in Fig. 1b, left untreated or stimulated with IL-17 (50 ng/ml) for the indicated time. A heat map is shown for the IL-17-induced and HuR-dependent genes. (D) Real-time PCR analysis of Cxcl1, Cxcl5 and Csf2 in mouse primary kidney epithelial cells infected as in fig. 1b, left untreated or stimulated with IL-17A (50 ng/ml) for the indicated time. The results are presented as fold induction of the expression levels in treated samples over the untreated. Data are representative of three independent experiments (error bars (d), s.d.).
HuR binds and stabilizes CXCL1 and CXCL5 mRNAs in response to IL-17 stimulation
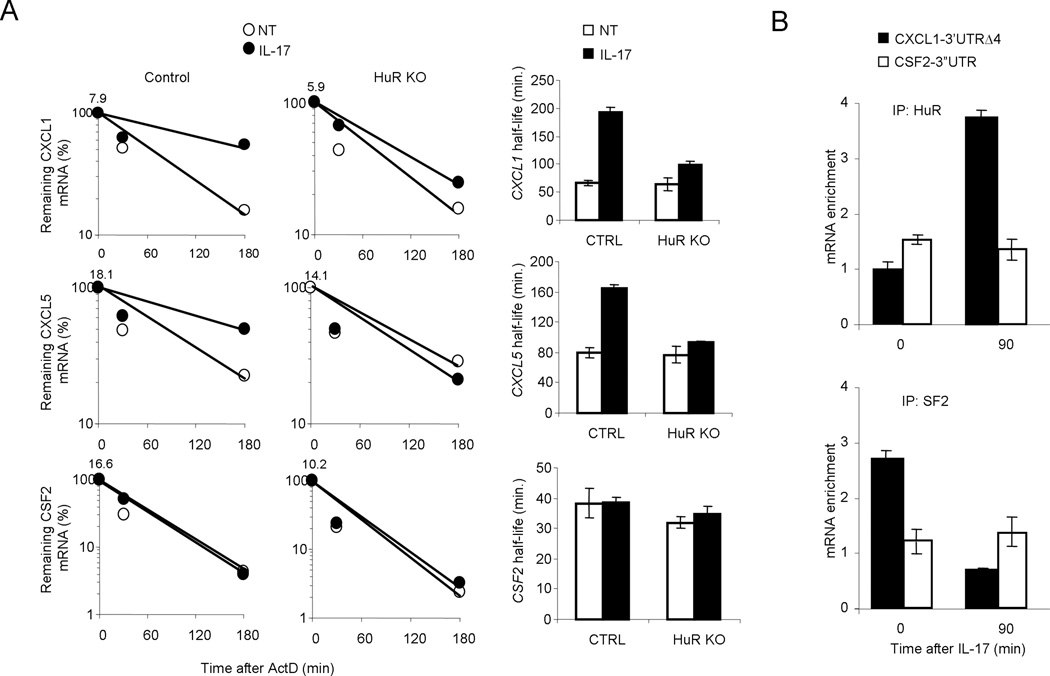
We next measured the impact of HuR deficiency on IL-17-induced mRNA stabilization. Wild-type and HuR-deficient kidney cells were pretreated for 0.5 h with TNFα to promote inflammatory gene transcription, then treated them for 0.5–3 h with actinomycin D (to block transcription) along with IL-17 (for mRNA stabilization). Although CXCL1 and CXCL5 mRNA was induced to similar extent in wild-type and HuR-deficient kidney cells after the initial treatment with TNFα, the mRNAs decayed more rapidly in HuR-deficient kidney epithelial cells than that in wild-type cells (Fig. 2A). But stability CSF2 mRNA was unaffected by HuR deficiency. These results indicated that HuR is necessary for IL-17-mediated stabilization of CXCL1 and CXCL2 mRNAs.
Figure 2. HuR binds and stabilizes CXCL1 and CXCL5 mRNAs in response to IL-17 stimulation.
(A) Real-time PCR analysis of Cxcl1, Cxcl5 and Csf2 in primary kidney epithelial cells infected as in fig. 1b, pre-treated for 2 h with TNF (10 ng/ml) followed by actinomycin D (5 µg/ml) alone (NT) or in combination with IL-17 (50 ng/ml) for the indicated time. The results are presented as decay over time (left panel) and as half-life (right panel). (B) HeLa Tet-Off cells transfected with the CXCL1 (KCΔ4) and CSF2–3’UTR reporter constructs, were untreated (0) or treated for 90 min with IL-17, followed by RNA-immunoprecipitation with anti-HuR or anti-SF2 (ASF) and real-time RT-PCR analyses of KCΔ4 and KC-GM-CSF mRNAs; results are presented as relative to results obtained by immunoprecipitation with nonspecific IgG. Data are representative of three independent experiments (error bars (a,b), s.d.).
We have recently reported that SF2/ASF bound CXCL1 chemokine mRNA in unstimulated cells to mediate mRNA decay, but the SF2 (ASF)-mRNA interaction was much lower after stimulation with IL-17. To determine whether HuR directly mediates IL-17-induced mRNA stabilization, we examined the binding of HuR to CXCL1 mRNA with and without IL-17 stimulation. We used reporter constructs containing the coding region of CXCL1 with truncated 3’ UTR of either CXCL1 (CXCL1–3’UTR Δ4) or CSF2 (CSF2–3’UTR) under the control of a tetracycline-response element (TRE). These constructs were transfected into HeLa Tet-Off cells stably expressing the tetracycline-controlled trans activator24. Cells extracts of the transfected cells untreated or treated with IL-17 for 90 minutes, were immunoprecipitated with anti-HuR, anti-SF2 or IgG. The amounts of HuR- and SF2-bound KCΔ4 and KC-GM-CSF mRNAs, relative to abundance of GAPDH mRNA, were determined by real-time PCR. IL-17 treatment resulted in 4-fold increase in abundance of HuR-bound CXCL1–3’UTR Δ4 mRNA, whereas SF2 bound CXCL1–3’UTR Δ4 mRNA in untreated cells and was dissociated from the transcript in IL-17-treated cells (Fig 2B). Neither HuR- or SF2-bound CSF2–3’UTR was altered in response to IL-17 stimulation, which is consistent with the fact that HuR and SF2 deficiency did not have much impact on CSF2 mRNA stability.
HuR is ubiquitinated upon IL-17 stimulation
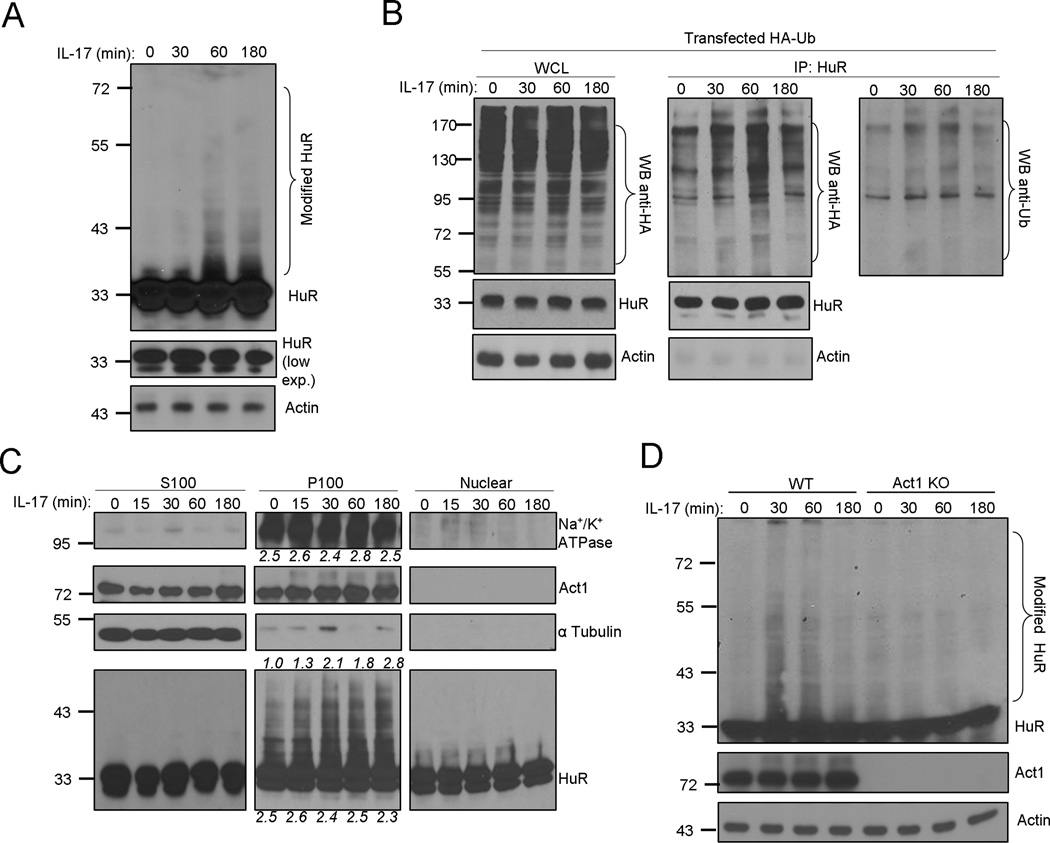
It was noted that IL-17 stimulation induced slower migrating forms of HuR, implicating IL-17-dependent HuR posttranslational modification (Fig. 3A). The laddering feature of the modified HuR suggests possible ubiquitiantion of HuR. To test this, HeLa cells were transfected with HA-tagged ubiquitin, followed by treatment with IL-17. The cell lysates were immunoprecipitated with anti-HuR antibody and immunobloted with anti-HA, anti-HuR and anti-Ubiquitin antibodies. These results showed that IL-17 indeed induced ubiquitination of HuR (Fig. 3B). Previous studies have suggested that while HuR is mostly localized in the nucleus, extracellular stimuli can promote the translocation of HuR from the nucleus to the cytosol. We wondered where HuR is modified and whether IL-17 stimulation has any impact on HuR’s subcellular localization. HeLa cells untreated or treated with IL-17, were fractionated into membranes and cytosol, which were then analyzed with antibodies against HuR and Act1. The Na+/K+ ATPase and α-tubulin were used as markers for the membrane and cytosol fractions, respectively. Whereas unmodified HuR was found in both membrane and nuclear fractions before IL-17 treatment, the modified HuR induced by stimulation with IL-17 was found only in the membrane fraction, but not in the cytosol or nucleus (Fig. 3C). On the other hand, we have previously reported that IL-17 induces Act1 phosphorylation through the activation of IKKi. Interestingly, the IL-17-induced phosphorylated Act1 (slower migration) was co-localized with the modified HuR in the membrane fraction, whereas unmodified Act1 was found in both membrane and cytosol fractions before IL-17 treatment (Fig. 3C). The co-localization of Act1 with HuR is consistent with the IL-17-induced complex formation between Act1 and HuR (Fig. 1A), implicating the possible role of Act1 in HuR modification. To determine the importance of Act1 in IL-17-induced polyubiquitination of HuR, we examined several primary cell types from wild-type and Act1-deficient mice, including MEFs, kidney epithelial cells and bone marrow macrophages. We found that while IL-17 induced HuR polyubiquitination in wild-type cells, Act1 deficiency abolished IL-17-dependent HuR modification (Fig. 3D and data not shown)
Figure 3. HuR is ubiquitinated upon IL-17 stimulation.
(A) HeLa cells untreated or treated with IL-17 (50 ng/ml) for the indicated time, followed by western analysis with anti-HuR and anti-β-actin. (B) HeLa cells transfected with HA-ubiquitin untreated or treated with IL-17 (50 ng/ml) for the indicated time, followed by immunoprecipitated (IP) with anti-HuR and western analysis with anti-HA, anti-Ub, anti-HuR and anti-β-actin. WCL: whole-cell lysates. (C) Cell lysates from HeLa cells untreated or treated with IL-17 (50 ng/ml) for the indicated time were fractionated into cytosolic (S100), membrane (P100) and nuclear fractions as described in Methods, followed by western analyses of α-Tubulin, Act-1, HuR and Na+/K+ ATPase. The levels of Na+/K+ ATPase, modified and unmodified HuR in P100 fraction were quantified by densitometry. (D) Bone marrow macrophages from wild-type and Act1-deficient mice untreated or treated with IL-17 (50 ng/ml) for the indicated time were subjected to western analysis with anti-HuR. Data are representative of three independent experiments.
Act1-mediated HuR polyubiqutiantion is required for HuR-mRNA interaction
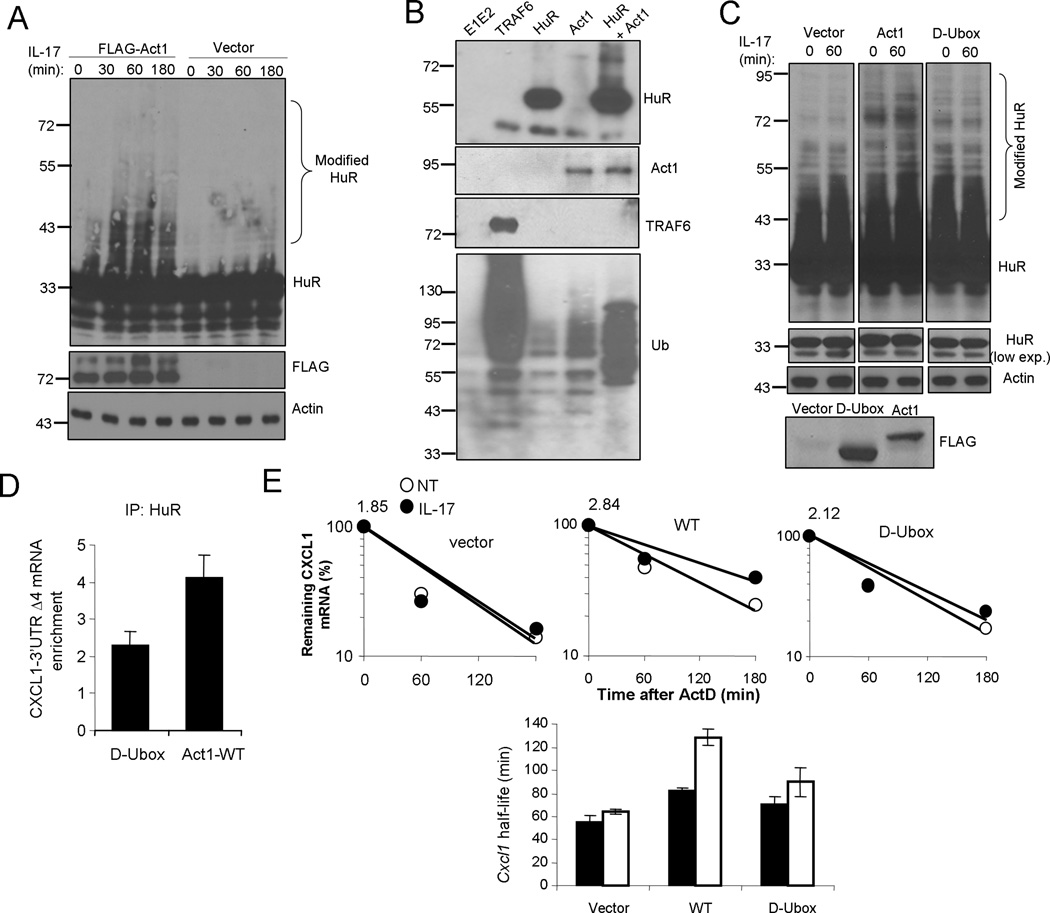
We have previously reported the E3 ligase activity of Act1 that exerts K63-linked polyubiquitination of TRAF6 to mediate NFκB activation (20). Since Act1 was required for IL-17-induced HuR polyubiquitiantion, we hypothesize that Act1 might also function as an E3 ligase for HuR. We found that overexpression of Act1 in HeLa cells induced polyubiquitination of HuR in unstimulated cells and further enhanced IL-17-induced HuR modification, suggesting the ability of Act1 in mediating HuR ubiquitination (Fig. 4A). To test whether Act1 can directly ubiquitinate HuR, we performed in vitro polyubiquitination assay. Recombinant Act1 was able to use Ubc13/Uev1A E2 complex to specifically catalyze polyubiquitination on HuR in this in vitro assay (Fig. 4B), suggesting that Act1 is likely an E3 ubiquitin ligase for HuR. While TRAF6 was used as a positive control, the specificity of the assay was also confirmed by different control reactions (Fig 4B). Moreover we found that wild-type ubiquitin and ubiquitin mutant K48R (Lys 48 mutated to Arg) but not K63R (Lys 63 mutated to Arg), were able to mediate HuR polyubiquitiantion in vivo upon Act1 overexpression (Supple. Fig. 2). These results indicate that Act1 is a bona fide E3 ligase for HuR.
Figure 4. Act1 is E3 ligase for HuR.
(A) HeLa cells transfected with either empty vector (Vector) or flag-tagged Act1(Act1), were untreated or treated with IL-17 (50 ng/ml) for the indicated time, followed by western analysis with anti-HuR, anti-Flag and anti-β-actin. (B) In vitro ubiquitination assays of HuR using GST-Act1 as E3 ligase in the presence of E1 and E2, as described in Material and Methods. The reaction of E1+E2+ubiquitin was used as negative control. E1+E2+ubiquitin were added to all the reactions, including TRAF6 (positive control), HuR, Act1 and HuR+Act1. The samples from these reactions were subjected western analyses with anti-HuR, anti-Act1, anti-TRAF6 and anti-Ub. (C) Act1-deficient MEFs transfected with either empty vector (Vector), flag-tagged wild-type Act1(Act1) or flag-tagged Act1-D-Ubox mutant (D-box), were untreated or treated with IL-17 (50 ng/ml) for the indicated time, followed by western analysis with anti-HuR and anti-β-actin. (D) HeLa Tet-Off cells were co-transfected with the CXCL1–3’UTR Δ4 reporter construct and flag-mouse Act1-D-Ubox mutant or flag-mouse Act1-WT, followed by RNA-immunoprecipitation with anti-HuR and real-time RT-PCR analysis of KCΔ4 mRNA. Results are presented as fold induction relative to that of IgG control. (E) Act1−/− MEFs reconstituted with either empty vector, flag-tagged Act1-WT or flag-tagged Act1-D-Ubox mutant, were pre-treated for 2 h with TNF (10 ng/ml) followed by actinomycin D (5 µg/ml) alone (NT) or in combination with IL-17 (50 ng/ml) for 0, 60 and 180 min. RNAs from these cells were subjected to real-time PCR analysis for CXCL1 mRNA; the results are presented as decay over time, with the quantity of mRNA at time point zero indicated above graph (top panel) and as half-life (bottom panel). Data are representative of two (a,b,c) or three independent experiments (error bars (d,e), s.d.).
We have previously shown that Act1 contains a U-box-like region and is a member of the U-box type E3 ubiquitin ligase family. Therefore, it is important to determine whether the U-box of Act1 is required for IL-17-induced HuR polyubiquitination. While restoration of Act1-deficient MEFs with retroviral Act1 induced HuR polyubiqutination and rendered IL-17-induced HuR modification, U-box deletion mutant of Act1 failed to mediate HuR ubiquitination (Fig 4C). Overexpression of Act1 wild-type, but not Ubox mutant of Act1, induced HuR–mRNA interaction (Fig. 4D). Furthermore, CXCL1 mRNA stability was reduced in Act1-deficient MEFs reconstituted with the D-Ubox mutant of Act1 compared with that of wild-type Act1 (Fig. 4E). Taken together, these results indicate the importance of Act1-mediated HuR polyubiquitination for HuR’s RNA binding and stabilizing activity.
Act1-TRAF2/5-SF2 and Act1-TRAF2/5-HuR are two independent complexes
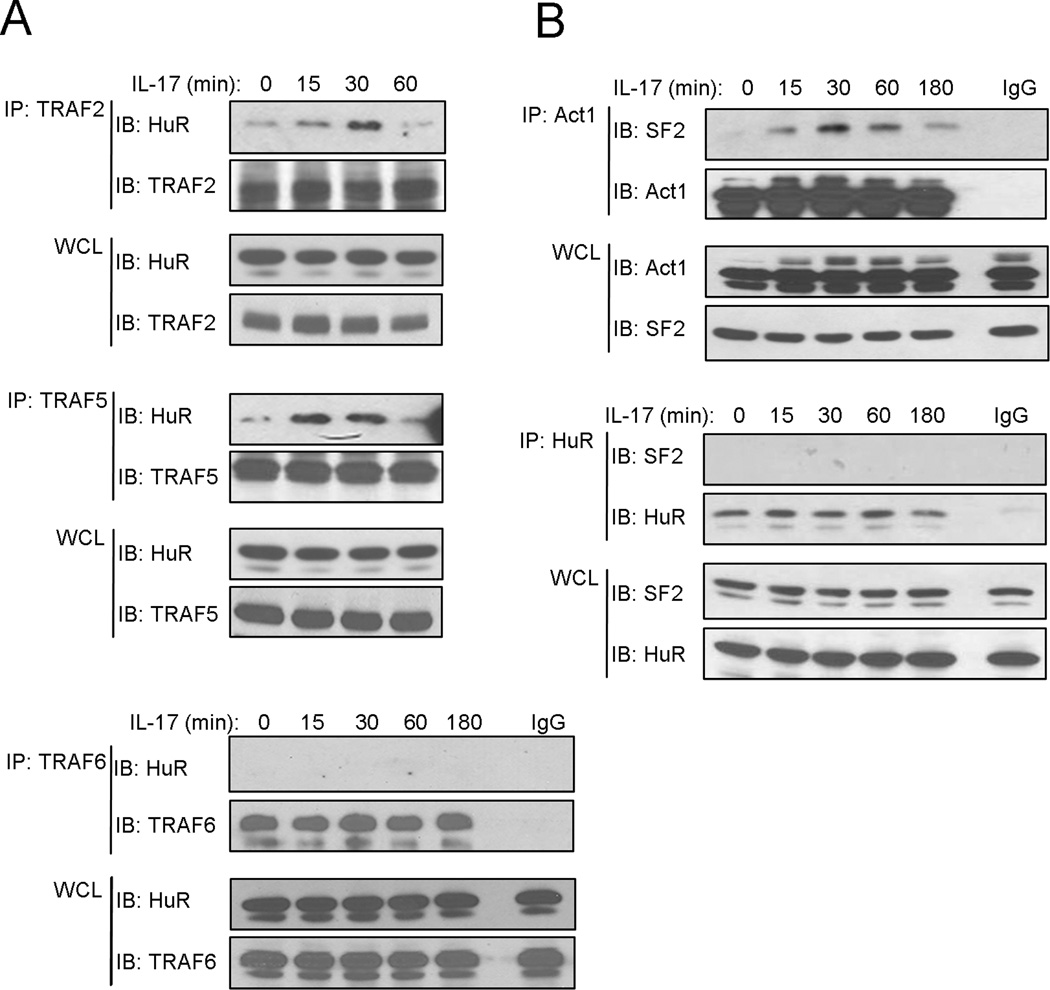
We recently reported that IKKi-mediated Act1 phosphorylation plays a critical role in directing the interaction of Act1 with TRAF2/5, which is required for the formation of Act1-TRAF2/5-SF2 complex leading to dissociation of SF2 from mRNA. Here we show that the interaction of Act1 with HuR is essential for the binding of HuR to mRNA. We wondered whether and what TRAF molecules are involved in this process. Through co-immunoprecipitation experiments, we found that IL-17 induced the interaction of HuR with TRAF2 and TRAF5, but not TRAF6 (Fig. 5A and data not shown). Since both SF2 and HuR interact with Act1-TRAF2/5, the important question is whether SF2 and HuR are in the same complex. It is interesting to note that we failed to co-immunoprecipitate HuR with SF2 (Fig. 5B), suggesting that these two RNA binding proteins are not in the same complex. These results suggest that Act1-TRAF2/5-SF2 and Act1-TRAF2/5-HuR are probably two independent complexes. While Act1-TRAF2/5 facilitates the dissociation of SF2 from mRNA, Act1-TRAF2/5 enhances the interaction of HuR with mRNA, thereby stabilizing mRNA.
Figure 5. IL-17 induces the formation of a complex containing Act1, TRAF2, TRAF5 and HuR.
(A) Cell lysates from HeLa cells untreated or treated with IL-17 (50 ng/ml) for the indicated time were immunoprecipitated (IP) with anti-TRAF2, anti-TRAF5, anti-TRAF6 and IgG, followed by western analysis (IB) with anti-HuR, anti-TRAF2 anti-TRAF5 and anti-TRAF6. WCL: whole-cell lysates. (B) Cell lysates from HeLa cells untreated or treated with IL-17 (50 ng/ml) for the indicated time were immunoprecipitated (IP) with anti-Act1 and anti-HuR and IgG, followed by western analysis (IB) with anti-SF2, anti-Act1 and anti-HuR. WCL: whole-cell lysates. Data are representative of three independent experiments.
IL-17-induced shift of Act1-HuR to polysomes facilitates the translation of ARE-mRNAs
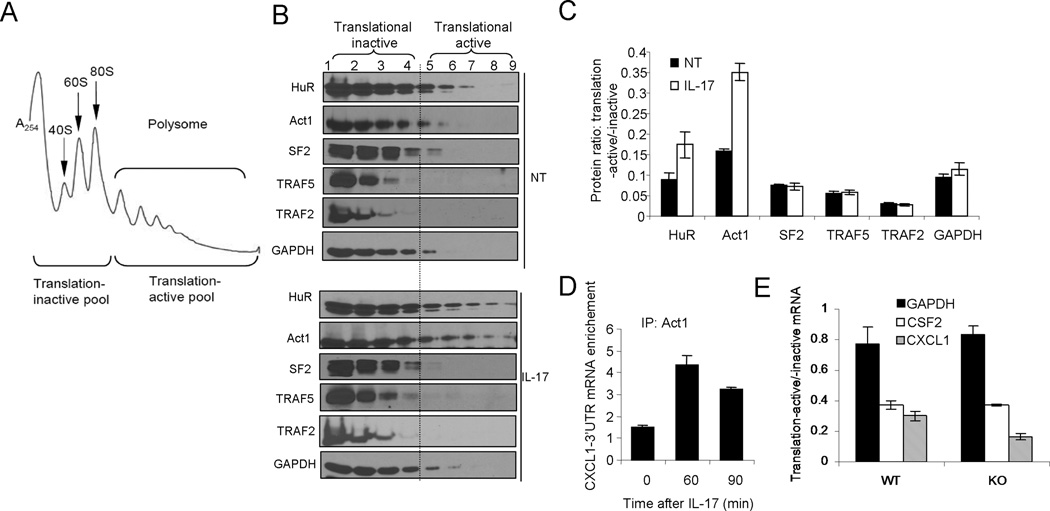
It is well known that ARE-containing mRNAs (ARE-mRNAs) follow HuR trafficking to polysomes for the transcripts to be translated. Since IL-17 induced the interaction of Act1 with HuR to promote HuR-ARE-mRNAs interaction, we hypothesize that IL-17 stimulation might induce the shift of HuR-ARE-mRNAs to the polysomes to facilitate the translation. Therefore, we examined the distribution of HuR in a sucrose gradient, which was used to separate translation-inactive free ribosomes from translation-active polysomes (Fig. 6B). Interestingly, increased amounts of HuR were indeed detected in the fractions of translation-active polysomes from lysates of IL-17 treated than that of untreated cells. Surprisingly, Act1 was also detected in the same polysomal fractions as HuR, suggesting that the Act1-HuR complex is associated with translational active polysomes (Fig. 6C). In support of this, IL-17 treatment resulted in more than 2-fold increase in abundance of Act1-bound CXCL1 mRNA (Fig. 6D). We then wondered whether the IL-17-induced association of Act1-HuR with the polysomal fractions has any impact on the translation of IL-17-induced chemokines. Compared with wild-type kidney epithelial cells, the HuR-deficient cells had more CXCL1 mRNAs in the translation-inactive pool than that in the translation-active pool. As a result, the ratio of IL-17-induced CXCL1 mRNAs in translation-active versus translation-inactive pools was reduced in HuR-deficient cells compared with wild-type cells (Fig. 6E). These results suggest that HuR is not only important for mRNA stabilization of a subset of IL-17-induced ARE-mRNAs, but also essential for their translation.
Figure 6. IL-17-induced shift of Act1-HuR to polysomes facilitates the translation of ARE-mRNAs.
(A) UV absorbance profile of RNP and polysome complexes separated on a sucrose density gradient. (B–C) Cytoplasmic extracts of HeLa cells untreated (NT) or treated with IL-17 (50 ng/ml) for 90 min were fractionated through a 10% to 50% sucrose gradient as described in Methods. (B) Western analyses of HuR, Act1, SF2, TRAF2, TRAF5 and GAPDH in sucrose gradient fractions (for corresponding UV absorbance profiles see supple. Fig. 3; the fraction numbers correspond to lanes in the Western blots). (C)The ratios of HuR, Act1 and GAPDH from translation-active (fractions 4–9) and -inactive (1–4) pools. (D) HeLa Tet-Off cells transfected with the CXCL1–3’UTR Δ4 reporter plasmid, were untreated or treated with IL-17 for the indicated time, followed by RNA-immunoprecipitation with anti-Act1 and real-time RT-PCR analyses of KCΔ4 mRNA. Results are presented relative to results obtained by immunoprecipitation with nonspecific IgG. (E) Primary kidney epithelial cells isolated from HuRflox/flox mice, infected as in Fig. 1b, were treated with IL-17 (50 ng/ml) for 90 min and fractionated as in Fig. 6a. CXCL1, CSF2 and GAPDH mRNAs from translation-active pools and translation-inactive pools were analyzed by quantitative RT-PCR and normalized to β-actin. The ratios of mRNAs from in translation-active and inactive pools are shown. Data are representative of three independent experiments (error bars (b, c, d), s.d.).
HuR depletion reduces IL-17-induced pulmonary inflammation
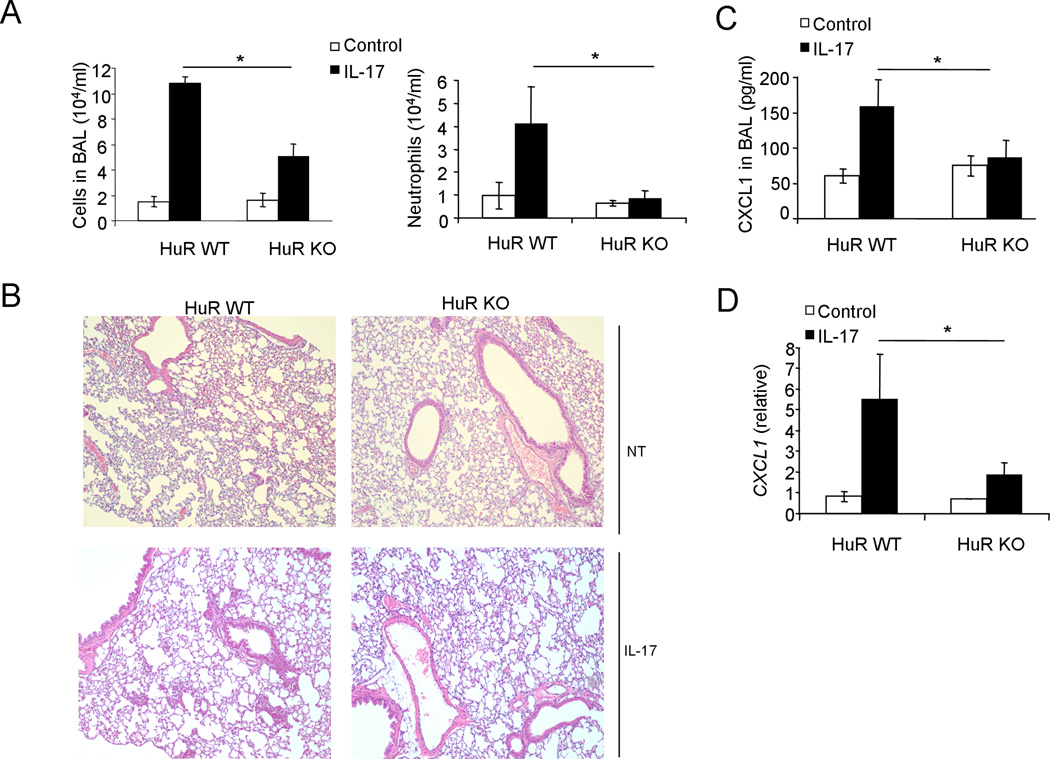
The main function of IL-17 is to coordinate local tissue inflammation via the up-regulation of proinflammatory and neutrophil-mobilizing cytokines and chemokines. We have previously reported that epithelial-derived Act1 is required for IL-17-induced neutrophilia in the airway and for allergic pulmonary inflammation (36). Furthermore, IKKi, the kinase that mediates IL-17-induced mRNA stabilization, is also required for IL-17-induced neutrophilia and pulmonary inflammation. Since the study here showed that HuR functions downstream of IKKi mediating chemokine mRNA stabilization ex vivo, we examined the importance of HuR in this in vivo inflammation model. The SFTPC (Surfactant Protein C or SP-C) gene promoter was used to generate the SP-C-rtTA/(tetO)7CMV-Cre transgenic mice that allow doxycycline-regulated expression of Cre recombinase in the distal-lung respiratory epithelium. HuRflox/flox mice were bred onto SP-C-rtTA/(tetO)7CMV-Cre to generate conditional distal-lung-specific HuR-deficient mice [SP-C-rtTA(tetO)7CMV-Cre-HuRflox/flox] and control mice ([SP-C-rtTA(tetO)7CMV-Cre-HuRflox/wt], (Supple. Fig. 4). These HuR conditional and control mice were administrated with doxycycline for one week, followed by treatment with recombinant IL-17 via intranasal injection. Twenty-four hours after challenge, the mice were analyzed for Bronchalveolar lavage (BAL) cells and lung inflammation. Infiltration of cells, especially neutrophils, was significantly reduced in conditional distal-lung-specific HuR-deficient mice [SP-C-rtTA(tetO)7CMV-Cre-HuRflox/flox] compared to that in control mice [SP-C-rtTA(tetO)7CMV-Cre-HuRflox/wt] (Fig. 7A). Histological analysis of lung tissue showed that HuR deficiency in distal-lung epithelium led to decreased lung inflammation (Fig. 7B). The decreased inflammation phenotype correlated with decreased levels of CXCL1 (a potent neutrophil chemokine) in the BAL and lung (Fig. 7C and D). Taken together, these results indicate that HuR-mediated chemokine mRNA stability has an important impact on IL-17-induced neutrophilia and pulmonary inflammation.
Figure 7. HuR regulates IL-17-induced gene expression in vivo.
(A) Total BAL and differential cell counts in samples from HuRflox/wt (HuR WT) and HuRflox/flox (HuR KO) mice expressing SP-CrtTA/tetO-CRE (n = 6 per group) fed with doxycycline for one week and then left unchallenged (Control) or challenged for 24 h by intranasal injection of IL-17 (0.5 µg). *P < 0.05 (two-tailed t-test). (B) Lung sections of saline (NT) and IL-17-challenged mice (as described in Fig. 7a), stained with hematoxylin and eosin. Magnification ×100. (C) Enzyme-linked immunosorbent assay of CXCL1 in BAL fluid from control or IL-17-challenged mice (as described in Fig. 7a). (D) Real-time PCR analysis of Cxcl1 in lung tissue from control or IL-17-treated mice (as described in Fig. 7a). Results are presented in arbitrary units relative to the expression of β-actin mRNA. *P < 0.05 (two-tailed t-test). Data are representative of two independent experiments (mean and s.d. in a, c, d).
DISCUSSION
IL-17, a signature cytokine produced by pathogenic Th17 cells plays a critical role in the pathogenesis of many autoimmune inflammatory diseases. While IL-17 induces transcriptional activation of inflammatory genes, it also impacts their production at the post-transcriptional level (23). Elucidation of signaling mechanisms that govern IL-17-induced gene expression and protein production is crucial for the development of new therapeutic strategies to attenuate this major pro-inflammatory pathway. In this study, we report a new function of RNA binding protein HuR in IL-17-induced Act1-mediated chemokine mRNA stabilization. HuR depletion substantially impaired IL-17-induced CXCL1 and CXCL5 expression due to increased mRNA decay, in contrast to the prolonged mRNA half-life in cells deficient in SF/ASF, a destabilizing factor in IL-17 pathway. While the interaction of Act1 with SF2 results in dissociation of SF2 from CXCL1 mRNA, IL-17-induced Act1-HuR interaction and subsequent Act1-mediated HuR ubiquitination promote the binding of HuR to CXCL1 mRNA, leading to mRNA stabilization. Although Act1 utilized the same TRAFs (TRAF2/5) to form the Act1-TRAF2/5-SF2 and Act1-TRAF2/5-HuR complexes, HuR and SF2/ASF interaction was not detected, indicating the action of two distinct complexes in the IL-17-induced mRNA stabilization process. Furthermore, IL-17 stimulation induced the co-shift of Act1 and HuR towards the polysomal fractions in a sucrose gradient, implicating the importance of Act1-HuR complex in moving the mRNAs to the translational active state. Moreover, HuR deficiency in distal lung epithelium attenuated IL-17-induced neutrophilia and pulmonary inflammation in vivo, demonstrating the biological importance of HuR.
We have previously shown that IKKi is activated upon IL-17 stimulation and acts as a critical upstream kinase of Act1 (34). The serine in the Act1 molecule targeted by IKKi is located adjacent to a putative TRAF-interacting consensus motif (P/S/T/A-X-Q/E-acidic/polar) on Act1. IKKi deficiency and mutation of Act1-Ser 311 abolished IL-17-induced Act1-TRAF2/5 complex formation and chemokine mRNA stabilization, whereas the Act1-TRAF6-NF-κB axis was retained. Consistent with this, while IL-17 induced the interaction of HuR with Act1, TRAF2 and TRAF5, HuR-TRAF6 interaction was not detected. It is possible that when HuR is recruited to the Act1-TRAF2/5 complex, a conformational change might take place, allowing direct contact between Act1 and HuR for Act1 to mediate HuR polyubiquitination.
While polyubiquitination with the ubiquitin linked through Lys 48 targets a protein for proteasomal degradation, polyubiquitination chains linked through Lys 63 of ubiquitin mediates protein-protein interactions and cell signaling (37). We have previously shown that Act1 utilizes Ubc13-Uev1A E2 complex to exert K63-linked polyubiquitination of TRAF6, which is required for TRAF6-mediated activation of TAK1 and the IKK complex, resulting in activation of transcription factor NFκB21. In this study, we found that Act1, together with Ubc13-Uev1A E2 complex ubiquitinates HuR, which is critical for HuR to bind and stabilize chemokine mRNAs. Interestingly, previous study has shown that heat shock (HS) potently induces HuR ubiquitination followed by proteasome-mediated HuR protein degradation, implicating Lys 48-linked HuR polyubiquitination (38). It is important to note that IL-17 stimulation does not induce HuR degradation, consistent with the fact that Act1 mediates K63-linked polyubiquitination through the Ubc13-Uev1A E2 complex. Future studies are required to investigate additional IL-17-induced HuR modification, especially phosphorylation, since phosphorylation often proceeds ubiquitination.
The important question is how HuR stabilizes mRNAs. HuR has been shown to bind and increase stability of a large number of functionally different ARE-containing transcripts including c-Fos, c-Myc, cyclooxygenase-2, TNF-α, and IL-3 (39–42). HuR, a member of the embryonic lethal abnormal vision (ELAV) family of RNA-binding proteins, contains three classical RNA-binding domains [RNA recognition motifs (RRMs), implicated in ARE recognition (43). It was proposed that HuR stabilizes ARE mRNAs by competing with destabilizing proteins for common binding sites thus preventing the recruitment of deadenylases and exonucleases and the degradation of the transcripts (44,45). In this study, we show that IL-17 stimulation induced the dissociation of destabilizing factor SF2 from CXCL1, whereas association of HuR with the transcript. Importantly, HuR and SF2 were not simultaneously present in the same complex, suggesting that HuR may compete with SF2 for the binding to the ARE mRNAs in response to IL-17 stimulation. One novel finding is that Act1-mediated HuR ubiquitination facilitated the RNA binding of HuR. Future studies are required to elucidate the molecular mechanism for how the ubiquitination of HuR might affect its RNA binding.
It is known that HuR binding to its target mRNAs not only contributes to their protection from degradation but also favors their translation (46). We indeed found that IL-17 stimulation induced the increased association of HuR with polysomal fractions promoting protein translation. It is intriguing that IL-17 treatment also induced the shift of Act1 to the polysomal fractions and formation of Act1-mRNA complex. Since there is no detectable RNA binding domain in Act1, it is likely that the association of Act1 with RNA is through HuR or other RNA binding proteins. Additionally, it is intriguing that the IL-17-dependent modified forms of Act1 and HuR were co-localized in the membrane (P100) fraction, which could represent polysomal containing rough ER. Future studies are required to investigate the detailed mechanism by which Act1-HuR associates with polysomes and promotes protein translation of ARE mRNAs.
It is important to note that HuR deficiency in distal-lung epithelium led to decreased IL-17-induced neutrophilia and lung inflammation. The decreased inflammation phenotype correlated with decreased levels of CXCL1 (a potent neutrophil chemokine) in the BAL and lung. These results are consistent with our previous findings about the critical role of epithelial-specific Act1 signaling in IL-17-mediated lung inflammation (36). Taken together, these results implicate that HuR-mediated chemokine mRNA stability has an important impact on IL-17-induced neutrophilia and pulmonary inflammation. As a future study, it will be critical to evaluate the importance of HuR in more physiologically relevant airway inflammation model. Since HuR is also a downstream molecule for other inflammatory pathways including TLR signaling, it would be important to identify pathway specific HuR-targets to evaluate the impact of HuR in a pathway-specific manner on the pathogenesis of airway inflammation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Jeffrey Whitsett for the generous gift of SP-C-rtTA/tetO-CRE mice.
This work was supported by NIH grants (1R01NS071996 and 1P01 HL103453) and the Sandler Award for Asthma Research to X.L.
Reference List
- 1.Fossiez F, Djossou O, Chomarat P, Flores-Romo L, Ait-Yahia S, Maat C, Pin JJ, Garrone P, Garcia E, Saeland S, Blanchard D, Gaillard C, Das MB, Rouvier E, Golstein P, Banchereau J, Lebecque S. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J Exp. Med. 1996;183:2593–2603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp. Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp.Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, Magorien JE, Blauvelt A, Kolls JK, Cheung AL, Cheng G, Modlin RL, Miller LS. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin. Invest. 2010;120:1762–1773. doi: 10.1172/JCI40891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp. Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat. Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- 8.Blauvelt A. T-helper 17 cells in psoriatic plaques and additional genetic links between IL-23 and psoriasis. J Invest Dermatol. 2008;128:1064–1067. doi: 10.1038/jid.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirota K, Hashimoto M, Yoshitomi H, Tanaka S, Nomura T, Yamaguchi T, Iwakura Y, Sakaguchi N, Sakaguchi S. T cell self-reactivity forms a cytokine milieu for spontaneous development of IL-17+ Th cells that cause autoimmune arthritis. J Exp. Med. 2007;204:41–47. doi: 10.1084/jem.20062259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Z, Altuntas CZ, Gulen MF, Liu C, Giltiay N, Qin H, Liu L, Qian W, Ransohoff RM, Bergmann C, Stohlman S, Tuohy VK, Li X. Astrocyte-restricted ablation of interleukin-17-induced Act1-mediated signaling ameliorates autoimmune encephalomyelitis. Immunity. 2010;32:414–425. doi: 10.1016/j.immuni.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molet S, Hamid Q, Davoine F, Nutku E, Taha R, Page N, Olivenstein R, Elias J, Chakir J. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J Allergy Clin. Immunol. 2001;108:430–438. doi: 10.1067/mai.2001.117929. [DOI] [PubMed] [Google Scholar]
- 13.Iwakura Y, Ishigame H. The IL-23/IL-17 axis in inflammation. Journal of Clinical Investigation. 2006;116:1218–1222. doi: 10.1172/JCI28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakae S, Komiyama Y, Nambu A, Sudo K, Iwase M, Homma I, Sekikawa K, Asano M, Iwakura Y. Antigen-specific T cell Sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity. 2002;17:375–387. doi: 10.1016/s1074-7613(02)00391-6. [DOI] [PubMed] [Google Scholar]
- 15.Shen F, Gaffen SL. Structure-function relationships in the IL-17 receptor: implications for signal transduction and therapy. Cytokine. 2008;41:92–104. doi: 10.1016/j.cyto.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toy D, Kugler D, Wolfson M, Vanden Bos T, Gurgel J, Derry J, Tocker J, Peschon J. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J. Immunol. 2006;177:36–39. doi: 10.4049/jimmunol.177.1.36. [DOI] [PubMed] [Google Scholar]
- 17.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A, Eisenhaber F. The STIR-domain superfamily in signal transduction, development and immunity. Trends in Biochemical Sciences. 2003;28:226–229. doi: 10.1016/S0968-0004(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 18.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol. Chem. 2006;281:35603–35607. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 19.Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, Kordula T, Zhang QW, Vallance B, Swaidani S, Aronica M, Tuohy VK, Hamilton T, Li X. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Commane M, Nie H, Hua X, Chatterjee-Kishore M, Wald D, Haag M, Stark GR. Act1, an NF-kappa B-activating protein. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10489–10493. doi: 10.1073/pnas.160265197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, Qian W, Qian Y, Giltiay NV, Lu Y, Swaidani S, Misra S, Deng L, Chen ZJ, Li X. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci. Signal. 2009;2:ra63. doi: 10.1126/scisignal.2000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J Exp. Med. 2000;191:1233–1240. doi: 10.1084/jem.191.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartupee J, Liu C, Novotny M, Li X, Hamilton T. IL-17 enhances chemokine gene expression through mRNA stabilization. J. Immunol. 2007;179:4135–4141. doi: 10.4049/jimmunol.179.6.4135. [DOI] [PubMed] [Google Scholar]
- 24.Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF) Nat. Immunol. 2011;12:853–860. doi: 10.1038/ni.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw, Kamen GR. Pillars article: a conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 2012;46:659–667. doi: 10.1016/0092-8674(86)90341-7. 1986. J Immunol. 189: 5–13. [DOI] [PubMed] [Google Scholar]
- 27.Wilson GM, Sun Y, Sellers J, Lu H, Penkar N, Dillard G, Brewer G. Regulation of AUF1 expression via conserved alternatively spliced elements in the 3' untranslated region. Mol. Cell Biol. 1999;19:4056–4064. doi: 10.1128/mcb.19.6.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson P. Post-transcriptional control of cytokine production. Nat. Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 29.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 30.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5'-3' decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta S, Biswas R, Novotny M, Pavicic PG, Jr, Herjan T, Mandal P, Hamilton TA. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J Immunol. 2008;180:2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 34.Bulek K, Liu C, Swaidani S, Wang L, Page RC, Gulen MF, Herjan T, Abbadi A, Qian W, Sun D, Lauer M, Hascall V, Misra S, Chance MR, Aronica M, Hamilton T, Li X. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat. Immunol. 2011;12:844–852. doi: 10.1038/ni.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perl AK, Zhang L, Whitsett JA. Conditional expression of genes in the respiratory epithelium in transgenic mice: cautionary notes and toward building a better mouse trap. Am. J Respir. Cell Mol. Biol. 2009;40:1–3. doi: 10.1165/rcmb.2008-0011ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swaidani S, Bulek K, Kang ZZ, Liu CN, Lu Y, Yin WG, Aronica M, Li XX. The Critical Role of Epithelial-Derived Act1 in IL-17-and IL-25-Mediated Pulmonary Inflammation. Journal of Immunology. 2009;182:1631–1640. doi: 10.4049/jimmunol.182.3.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen ZJJ, Sun LJJ. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Molecular Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, Gorospe M. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengupta S, Jang BC, Wu MT, Paik JH, Furneaux H, Hla T. The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J Biol.Chem. 2003;278:25227–25233. doi: 10.1074/jbc.M301813200. [DOI] [PubMed] [Google Scholar]
- 42.Wang JG, Collinge M, Ramgolam V, Ayalon O, Fan XC, Pardi R, Bender JR. LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J Immunol. 2006;176:2105–2113. doi: 10.4049/jimmunol.176.4.2105. [DOI] [PubMed] [Google Scholar]
- 43.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol.Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cellular and Molecular Life Sciences. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.