Abstract
We conducted a genome-wide association study of male breast cancer using 823 cases and 2,795 controls of European ancestry with validation in independent sample sets totalling 438 cases and 474 controls. A novel variant in RAD51B (14q24.1) was significantly associated with male breast cancer risk (P = 3.02 ×10−13, odds ratio (OR) = 1.57). TOX3 (16q12.1) was also a susceptibility locus (P = 3.87 ×10−15, OR = 1.50).
Male breast cancer accounts for 1% of all breast cancer diagnoses. Family history is a significant risk factor for male breast cancer; the relative risk of breast cancer for a female with an affected brother is approximately 30% higher than for a female with an affected sister1. Approximately 10% of male breast cancer cases are BRCA2 mutation carriers while BRCA1 mutation carriers are reported less frequently2. Predicated on the assumption that common variation contributes appreciably to the heritable risk of male breast cancer, and since investigation of risk alleles for breast cancer in men may provide novel insight into genetic susceptibility for the disease in females, we performed a genome-wide association study (GWAS).
Using Illumina OmniExpress arrays (Illumina, San Diego CA) we genotyped 920 male breast cancer cases ascertained from the UK (n = 805) and US (n = 115) (Supplementary Methods; Supplementary Table 1). For controls we used publicly available data on 2,912 individuals from the 1958 British Birth Cohort, genotyped on Illumina 1.2M DuoCustom arrays. After applying pre-specified quality control measures (Supplementary Methods; Supplementary Figure 1; Supplementary Tables 2a and 2b), we estimated odds ratios (ORs) and 95% confidence intervals (CI) for 447,760 autosomal SNPs with minor allele frequencies (MAF) ≥ 5% in 823 cases and 2,795 controls. Quantile-quantile plots of P-values showed minimal inflation of test statistics, indicating that there was no substantial cryptic population substructure or differential genotyping between cases and controls (genomic control inflation factor λ = 1.05; Supplementary Figure 2).
A total of 17 SNPs, mapping to six independent genomic regions, showed evidence of association with male breast cancer at P ≤ 5.0 × 10−7(Supplementary Figure 3). We attempted to validate the most significantly associated SNP mapping to each of the six regions in 438 cases and 474 controls recruited from 12 case-control series (Supplementary Methods; Supplementary Table 1). In a combined analysis the associations of two SNPs, rs1314913 (P = 3.02 × 10−13, OR = 1.57) and rs3803662 (P = 3.87 × 10−15, OR = 1.50) attained genome-wide significance (Table 1; Supplementary Tables 3 & 4).
Table 1.
Summary data for the 14q21.1 SNP rs1314913 and 16q12.1 SNP rs3803662 associated with risk of male breast cancer.
| Locus | Control MAF | Control Genotype Counts | Case MAF | Case Genotype Counts | P-value | ORtrend | 95% CI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1314913 | GG | GA | AA | GG | GA | AA | ||||||
| 14q24.1 | GWAS | 0.14 | 2047 | 675 | 65 | 0.21 | 520 | 261 | 42 | 4.09 × 10−10 | 1.55 | 1.35–1.78 |
| RAD51B | Replication | 0.15 | 333 | 117 | 12 | 0.22 | 258 | 155 | 16 | 1.71 × 10−04 | 1.61 | 1.25–2.07 |
| Combined | 0.15 | 2380 | 782 | 77 | 0.21 | 778 | 416 | 58 | 3.02 × 10−13 | 1.57 | 1.39–1.77 | |
|
| ||||||||||||
| rs3803662 | GG | GA | AA | GG | GA | AA | ||||||
| 16q12.1 | GWAS | 0.26 | 1540 | 1046 | 205 | 0.34 | 356 | 372 | 95 | 2.51 × 10−10 | 1.46 | 1.30–1.64 |
| TOX3 | Replication | 0.27 | 257 | 181 | 36 | 0.37 | 173 | 204 | 59 | 2.38 × 10−06 | 1.62 | 1.32–1.99 |
| Combined | 0.26 | 1797 | 1227 | 241 | 0.35 | 529 | 576 | 154 | 3.87 × 10−15 | 1.50 | 1.35–1.66 | |
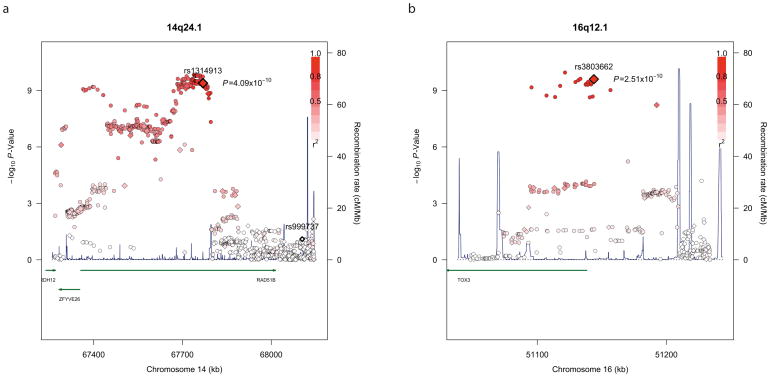
SNP rs1314913 localises to intron seven of the RAD51B gene (RAD51 homolog B) on chromosome 14q24.1 at 67,769,347 bp (NCBI build 36). It maps to the distal end of a linkage disequilibrium block of approximately 52 kb (Supplementary Figure 4). RAD51 family members function in both mitotic and meiotic homologous recombination and in DNA double-strand break repair. rs999737, located in intron 10 of RAD51B, has previously been shown to be associated with risk of female breast cancer3. This SNP maps approximately 335 kb telomeric to rs1314913 and is separated from it by strong recombination hotspots (Figure 1a; Supplementary Figure 4). rs999737 and rs1314913 are only weakly correlated in the male breast cases (r2 = 0.02) and the HapMap CEU population (r2 = 0.006). To test formally for independence between rs1314913 and rs999737 we fitted a logistic regression model, using the discovery phase samples, adjusted for rs999737 in which the OR for rs1314913 was 1.54 (P = 1.04 × 10−9). Conversely the OR for rs999737, adjusted for rs1314913, was 0.93 (P = 0.25).
Figure 1. Association and recombination plots for the 14q24.1 and 16q12.1 loci.
Directly genotyped SNPs from the discovery phase are represented as diamonds and imputed SNPs as circles. A larger diamond indicates the GWAS “hit” in each region. The strength of linkage disequilibrium between each SNP and the GWAS hit is indicated by the colour intensity of the symbol, from white (r2 = 0) to dark red (r2 = 1). Recombination rates, plotted in dark blue, are based on the HapMap CEU samples and genomic coordinates are based on NCBI build 36 of the human genome. Results are shown for the (a) 14q24.1 and (b) 16q12.1 loci. The location of rs999737 is indicated in bold at the distal end of RAD51B in the 14q24.1 plot.
To provide further insight into the association at 14q24.1 we imputed genotypes in cases and controls using data from the 1,000 Genomes Project. Fifty-two imputed SNPs were more strongly associated with male breast cancer than rs1314913 and delineated an 85 kb cluster from 67.68 Mb to 67.77 Mb (Figure 1a; Supplementary Table 5). To examine if any directly genotyped or imputed SNPs annotated a putative transcription factor binding site or enhancer element we conducted a bioinformatic search of the region (Supplementary Methods). Seven associated SNPs, including rs1314913, were highly evolutionarily conserved (Supplementary Table 6). Analysis of ENCODE project data, including the Broad histone modification datasets for human mammary epithelial cells (HMECs), showed that two conserved SNPs, rs1314913 and an adjacent SNP, rs1316014, were located in a transcription factor-binding site lying within a DNAse hypersensitive site flanked by regions of high H3K4 mono/di-methylation and low tri-methylation, features that are characteristic of enhancer elements (Supplementary Figures 5 and 6). In silico predictions are compatible with the minor alleles of both rs1314913 and rs1316014 abrogating the DNA binding sites of AP-1 and related transcription factors (Supplementary Figure 7). It is possible that the role of AP-1 in modulating estrogen signalling and transcription4 might explain the association between rs1314913 and male breast cancer.
We have previously shown in a much smaller study that rs3803662, a synonymous SNP in LOC643714 mapping to chromosome 16q12.1 at 51,143,812 bp, was associated with male breast cancer risk, albeit not at genome-wide levels of significance5. Here we provide robust confirmatory evidence of that association (Table 1). Examination of imputed data suggests that the association spans a 61 kb region from 51.09 Mb to 51.16 Mb and is proximal to LOC643174(Figure 1b), localising to the gene TOX3 (TOX high mobility group family member three).
Rare variants in two breast cancer susceptibility genes, BRCA2 and CHEK2, have larger ORs in males compared with females2,6 and we show here that this is also true for two common susceptibility alleles. Both rs1314913 and rs3083662 are striking in the magnitude of their effects. Comparing the breast cancer OR for rs3803662 in our data with the published estimate for females (OR = 1.20 [1.16–1.24])7, the effect was significantly greater in males (P = 7.76 × 10−5). Since rs1314913 is a novel breast cancer susceptibility variant there are no equivalent female estimates for comparison.
Variants at 24 loci have so far been shown to influence female breast cancer risk3,7–14. Their associations with male breast cancer are shown in Supplementary Table 7. In addition to rs3803662, SNPs at 2q35, 6q25.1, 10q21.2, 11q13.3, 12p11.22 were significantly associated at P< 0.05. Loci at 3p24.1, 9p21.3 and 14q24.1 showed borderline associations at P ≤ 0.1. There was no significant association, however, between variants at the FGFR2 locus on chromosome 10q26.13 and male breast cancer risk (rs2981582; OR = 1.07 [0.96–1.20]; P = 0.21). This observation is surprising since male breast cancer is almost entirely estrogen receptor (ER) positive. rs2981582 is the SNP with the strongest known association with ER positive breast cancer in females15 and the power of our study to detect an allele with the same effect size as for female breast cancer at P ≤ 0.05 is close to 100%. rs3803662, however, is strongly associated with both ER negative and ER positive breast cancer in females15. Therefore the ER status of male breast cancers does not obviously explain the SNP associations.
These data provide evidence for low penetrance susceptibility to male breast cancer. Given the modest size of our study it is likely that additional risk variants can be identified by future GWAS.
Supplementary Material
Acknowledgments
We thank the men who participated in the study. For the ICR sample collection we are grateful to Beverley Smith, Deborah Hogben, Robert McCann, Jane Melia, Sharon Squires, Margaret Snigorska, Joanne Micic, and Kim Sibley for administrative help and advice; to the research nurses Alison Butlin, Margo Pelerin, Jill Wood, and Tracy Gardener for collection of blood samples and questionnaire data from participants; to Dawn Thomas and her team from the BGS Study for coordinating collection of controls; and to the cancer registries of England and Wales for providing us with information on eligible participants. SEARCH thanks the SEARCH team. We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (funded 2001–2009 by NHMRC and currently by the National Breast Cancer Foundation and Cancer Australia #628333) for their contributions to this resource, and the many families who contribute to kConFab. We thank Greg Wilhoite for preparing the US samples for this study. We thank Dr Arja Jukkola-Vuorinen, Dr Vesa Kataja and Dr Päivi Auvinen as well as Sanna Hallamies and Salla Ranta for their contribution in the Finnish Male Breast Cancer Study. We thank Jeronimo Forteza, Maximo Fraga, Catuxa Celeiro and the staff of the Department of Pathology of CHUS Santiago; and Angel Carracedo and Carmen M Redondo for the contributions to the Spanish study. Finally, we thank the consultants under whose care the patients were for their advice and help.
This work was supported by Breakthrough Breast Cancer, Movember and the Institute of Cancer Research, who acknowledge NHS funding to the NIHR Biomedical Research Centre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The High Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics is funded by Wellcome Trust grant 090532/Z/09/Z and MRC Hub grant G0900747 91070.
This work made use of data and samples generated by the 1958 Birth Cohort under grant G0000934 from the Medical Research Council, and grant 068545/Z/02 from the Wellcome Trust.
The Finnish Male Breast Cancer Study acknowledges funding from the Finnish Breast Cancer Group, Finnish Oncology Association and the Salonoja Foundation.
L.O. acknowledges funding from the Italian Association for Cancer Research (AIRC, IG 8713).
D.P. acknowledges funding from the Istituto Toscano Tumori, Florence, Italy.
SEARCH is funded by a grant from Cancer Research UK (C490/A10124) and is supported by the University of Cambridge NIHR Biomedical Research Centre. A.M.D. has been funded by Cancer Research UK grant CA8197/A10865 and by the Joseph Mitchell Fund.
D.T.B. acknowledges funding by Cancer Research UK (Programme Award C588/A10589)
M.G.D. and J.E.C. acknowledge the support of the Department of Pathology of the CHUS Santiago Hospital, the Galician Foundation of Genomic Medicine, and the Program Grupos Emergentes, Cancer Genetics Unit of CHUVI, Instituto de Salud Carlos III, Spain.
H.O. and I.H. acknowledge funding from the Swedish Cancer Society and The Swedish Medical Research Council.
The collection of samples and epidemiologic data for the US cases was supported by U.S. ARMY Grant DAMD-17-96-I-6266 and NIH R01CA74415. S.L.N. was partially supported by the Morris and Horowitz Families Endowed Professorship at the City of Hope.
Footnotes
URLs
Genotype Libraries and Utilities (GLU): http://code.google.com/p/glu-genetics; R: http://cran.r-project.org; UCSC Genome Browser: http://genome.ucsc.edu/; 1,000 Genomes Project: http://www.1000genomes.org/; HapMap: http://hapmap.ncbi.nlm.nih.gov/; SNAP: http://www.broadinstitute.org/mpg/snap/; IMPUTE2: http://mathgen.stats.ox.ac.uk/impute/impute_v2.html; SNPTESTv2: https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html; SHAPEIT: http://www.shapeit.fr/; HaploView: http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview; EigenSoft v3: http://genepath.med.harvard.edu/~reich/Software.htm; TRANSFAC Matrix Database: http://www.biobase-international.com/pages/index.php?id=transfac; 1958 British Birth Cohort: http://www2.le.ac.uk/projects/birthcohort; http://www.bristol.ac.uk/alspac/; http://www.cls.ioe.ac.uk/ncds; http://www.esds.ac.uk/findingData/ncds.asp
Author Contributions
A.A. and A.J.S. conceived the Breakthrough Breast Cancer Male Breast Cancer Study and obtained financial support; N.O. and A.J.S. designed the GWAS; N.O. drafted the manuscript with substantial input from O.F., M.J., C.J.L., R.S.H., M.G.C., A.A., and A.J.S; N.O., R.C., S.S. McD. C.M. and M.Z. performed statistical and bioinformatic analyses; K.T. managed sample handling and DNA extraction; A.L., N.J., G.B., and C.B. performed genotyping. A.H.T. and P.A.J. coordinated sample collection at the Peter MacCallum Cancer Centre, East Melbourne, Australia; S.E.B. and S.B. coordinated sample collection at Copenhagen University Hospital; H.N. and J.M. coordinated sample collection for the Finnish Male Breast Cancer Study; E.F. and Y.L. coordinate sample collection at Sheba Medical Centre and the Sackler School of Medicine; D.P., G.M. and I.Z. coordinated sample collection at ISPO Cancer Research and Prevention Institute, Florence; L.O. and G.G. coordinated sample collection at “Sapienza” University of Rome; A.H. and A.M.W.O. coordinated sample collection at Erasmus University Medical Center, Rotterdam; S.N. and M.K. coordinated sample collection from Institute of Oncology Ljubljana; M.G.D. and J.E.C. coordinated sample collection from Galicia, Spain; H.O. and I.H. coordinated sample collection from Lund University; D.F.E., P.D.P.P. and A.M.D. coordinated sample collection from the SEARCH/Cambridge studies; D.T.B. coordinated sample collection at Leeds IMM; S.L.N. and L.S. coordinated collection of US samples. All authors contributed to the final paper.
References
- 1.Bevier M, Sundquist K, Hemminki K. Breast Cancer Res Treat. 2012;132:723–728. doi: 10.1007/s10549-011-1915-2. [DOI] [PubMed] [Google Scholar]
- 2.Basham VM, et al. Breast Cancer Res. 2002;4:R2. doi: 10.1186/bcr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas G, et al. Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, et al. Mol Endocrinol. 2011;25:1527–1538. doi: 10.1210/me.2011-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr N, et al. PLoS Genet. 2011;7:e1002290. doi: 10.1371/journal.pgen.1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meijers-Heijboer H, et al. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 7.Easton DF, et al. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haiman CA, et al. Nat Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens KN, et al. Cancer Res. 2012;72:1795–1803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher O, et al. J Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 11.Stacey SN, et al. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 12.Stacey SN, et al. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 13.Turnbull C, et al. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghoussaini M, et al. Nat Genet. 2012;44:312–318. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Closas M, et al. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.