Abstract
Deletions of the chromosomal region 2q37 cause brachydactyly-mental retardation syndrome (BDMR), also known as Albright hereditary osteodystrophy-like syndrome. Recently, histone deacetylase 4 (HDAC4) haploinsufficiency has been postulated to be the critical genetic mechanism responsible for the main clinical characteristics of the BDMR syndrome like developmental delay and behavioural abnormalities in combination with brachydactyly type E (BDE). We report here on the first three generation familial case of BDMR syndrome with inheritance of an interstitial microdeletion of chromosome 2q37.3. The deletion was detected by array comparative genomic hybridization and comprises the HDAC4 gene and two other genes. The patients of this pedigree show a variable severity of psychomotor and behavioural abnormalities in combination with a specific facial dysmorphism but without BDE. Given that only about half of the patients with 2q37 deletions have BDE; we compared our patients with other patients carrying 2q37.3 deletions or HDAC4 mutations known from the literature to discuss the diagnostic relevance of the facial dysmorphism pattern in 2q37.3 deletion cases involving the HDAC4 gene. We conclude that HDAC4 haploinsufficiency is responsible for psychomotor and behavioural abnormalities in combination with the BDMR syndrome-specific facial dysmorphism pattern and that these clinical features have a central diagnostic relevance.
Keywords: 2q37, HDAC4, BDMR, Albright hereditary osteodystrophy-like syndrome, brachydactyly
Introduction
Brachydactyly-mental retardation syndrome (BDMR, MIM 600430), synonymous with Albright hereditary osteodystrophy-like syndrome, is caused by chromosomal 2q37 deletions ranging from small submicroscopic interstitial deletions to large terminal deletions that are detectable by routine cytogenetics.1 Major features of BDMR are mild to moderate developmental delay/intellectual disability, and behaviour disorders.
Brachydactyly type E (BDE) is a variable clinical sign and only documented in about half of the patients with 2q37 deletions. Further clinical findings in these patients include obesity, short stature, seizures, hypotonia, and structural anomalies of the CNS, heart, trachea and the gastrointestinal/genitourinary tract. In rare cases, Wilms tumours have been observed.1, 2 By array comparative genomic hybridization (array CGH), the minimal deletion interval responsible for the phenotypical features of BDMR has been refined.3, 4 Recently, histone deacetylase 4 (HDAC4, MIM 605314) haploinsufficiency has been identified to be the critical genetic mechanism responsible for the major BDMR features in patients with 2q37 deletions or intragenic HDAC4 mutations.5 HDAC4 acts as a transcription repressor by altering chromatin structure and influences a broad transcriptional network that is essential for brain, muscle and bone development, as well as function.6, 7, 8 In the present study, we show that a heterozygous 2q37.3 microdeletion involving the genes HDAC4, TWIST2 and FLJ43879 is inherited in an autosomal-dominant manner and is associated with psychomotor and behavioural abnormalities in combination with the BDMR-specific facial dysmorphism pattern.
Clinical description
Patient 1
This is the female index patient who is the only child of non-consanguineous parents (Figure 1a and b). She was born at 38-weeks' gestation by spontaneous delivery after an uncomplicated pregnancy. During her first weeks of life, constipation and a diminished motor activity were noticed. At age of 6 months the girl was referred to our clinical genetics unit because of motor developmental and growth delays. Upon clinical examination, the patient showed a borderline short stature with a height of 62 cm (-2 SD), while her occipitofrontal head circumference (41.8 cm; −0.9 SD) and weight (6.4 kg; −1.3 SD) were in the normal range. At this age, she was not able to grasp nor to roll from back to ventral position. The patient presented a short neck, widely placed hypoplastic nipples, a four finger crease on the left hand and clinodactyly of both fifth fingers. In the follow-up examination at the age of 11 months unattended sitting was not possible. Her height was in the lower normal range and primary dentition had started meanwhile at the age of 10 months. At the age of 2 years and 8 months, physical examination revealed midface hypoplasia, mild ptosis, deeply set eyes, posteriorly rotated and low-set ears, thin upper lip and pointed chin (Table 1). Motor skills now were according for age and her speech development was unremarkable being able to communicate with short sentences of three to four words. Behavioural problems in the form of aggressive tantrum-like behaviour and sleep abnormalities became progressively disturbing. Clinically and radiologically, BDE was not present and was also excluded by a metacarpophalangeal pattern profile (Supplementary figure 1). The middle finger length was at 4.6 cm (3rd–25th centile) and the total hand length was at 10.9 cm (3rd–25th centile).
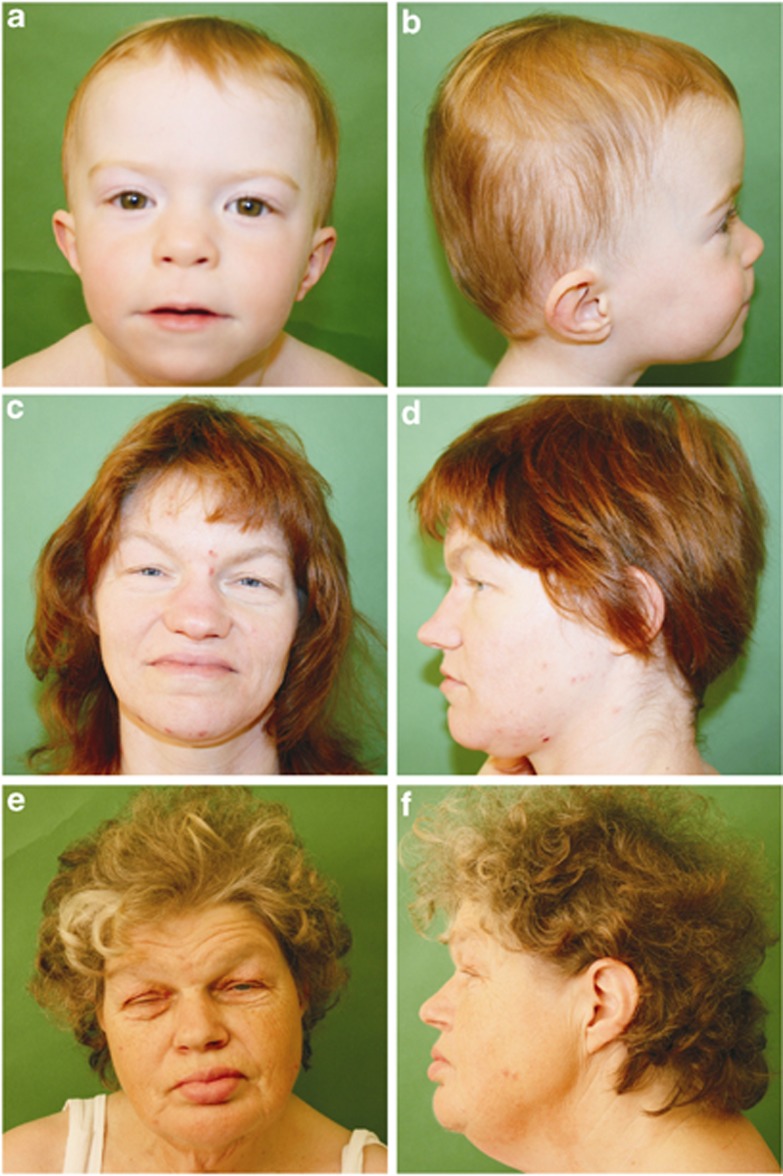
Figure 1.
Facial features of patients with 2q37.3 deletion including HDAC4. Patient 1 (a, b), a female at age of 2 years and 8 months, shows features like midface hypoplasia, and mild ptosis. Note in patient 2 (c, d) highly arched eyebrows, narrow palpebral fissures, deep set eyes and broad nasal bridge, as well as tip that are more pronounced in her mother (e, f).
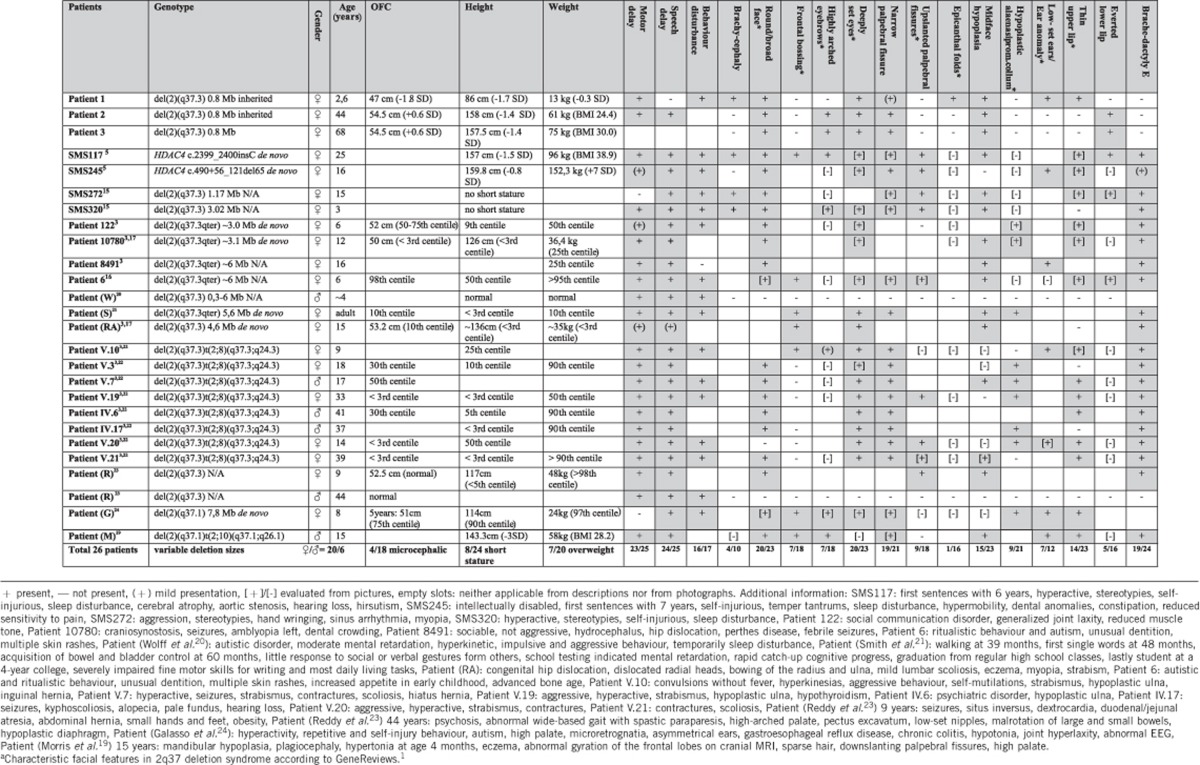
Table 1. Comparison of genotypes and clinical features in patients of the current study and patients known from literature.
Patient 2
The 45-year-old mother of the index patient had a history of general developmental and growth delays during childhood (Figure 1c,d). However, later the patient was able to attend normal school. In adulthood she noticed a reduced spatial orientation and memory deficits. On clinical examination, a coarse facial appearance with a broad and depressed nasal bridge, highly arched eyebrows, deep set eyes and narrow palpebral fissures were observed (Table 1). Her growth parameters were all in the normal range: height of 158 cm (−1.4 SD), weight of 61 kg (BMI 24.4), and OFC of 54.5 cm (+0.6 SD). Clinically her hands and feet appeared to be normal, BDE was excluded radiologically and by a metacarpophalangeal pattern profile (Supplementary figure 1). The middle finger length was at 8 cm (50th centile) and the total hand length was at 17.6 cm (25th centile).
Patient 3
The 68-year-old grandmother of the index patient is the mother of patient 2 (Figure 1e and f), her only child. Her sisteŕs daughter had suffered from intellectual disability in combination with hydrocephalus and paraplegia of unknown cause and died at the age of 28 years.
The past medical history of patient 3 is positive for severe osteoarthritis, most accentuated in both knee joints. She underwent multiple surgeries and an extensive orthopaedic treatment. On examination, she presented a dysmorphic facial aspect similar to that of her daughter with highly arched eyebrows, markedly narrow palpebral fissures and everted as well as full lips (Table 1). Her growth parameters were all in the normal range: height −157.5 cm (−1.4 SD), weight −75 kg (BMI 30.0), and OFC −54.5 cm (+0.6 SD). She was able to communicate in simple sentences and her intellectual skills appeared to be lower than normal. Clinically her hands were normal.
Methods
Informed consent
Written informed consent was given by the patients or the legal guardian for genetic testing and publication of images.
Conventional cytogenetics
Standard cytogenetic analysis with a high resolution 550 GTG-banding was performed according to standard procedures using a lithium-heparin peripheral blood sample from patient 1.
Array CGH
Genomic DNA samples from patients 1 and 2 were extracted from EDTA peripheral blood samples. The DNA sample from patient 1 was analysed by whole genome 244 K oligonucleotide array according to the manufacturer's protocol (Agilent Technologies, Santa Clara, CA, USA). Image data were analysed using Feature Extraction 9.5.3.1 and CGH Analytics 3.4.40 software (Agilent Technologies) with the following analysis settings: aberration algorithm ADM-2; threshold: 6.0; window size: 0.2 Mb; filter: five probes, log2ratio=0.29. Genome coordinates are shown according to the human genome build hg18 (NCBI 36.1). The Array-Format BlueGenome CytoChip ISCA 4 × 180K v1.0 (Bluegenome, Cambridge, UK) was used to compare deletion sizes between patients 1 and 2.
Fluorescence in situ hybridization
To confirm the array CGH result of patient 1, a fluorescence in situ hybridization (FISH) analysis was performed on metaphase chromosomes from peripheral blood lymphocytes using the BAC clone RP11-546M8 (2q37.3) labelled in red. BAC clone RP11-27O22 probe (2p16.1) labelled in green served as control probe. The same procedure was performed for patients 2 and 3.
Real-time quantitative PCR
To clarify whether TWIST2 is also affected by the microdeletion 2q37.3 we performed real-time quantitative PCR (qPCR) for patients 1 and 2 measuring four amplicons covering the TWIST2 gene. Genomic DNA samples from the patients were extracted from EDTA peripheral blood samples. For comparison the qPCR analysis included quantification of HDAC4 exon 12 and FVIII exon 8. The primer sequences are annotated in the Supplementary table 1. The qPCR analysis was performed on ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) as described previously.9
Results
While the standard cytogenetic analysis showed a normal female karyotype in patient 1, we identified by array CGH a heterozygous interstitial microdeletion 2q37.3 of about 800 kb, according to ISCN 2009 arr 2q37.3q37.3(239,395,957-240,154,599)x1 (Figure 2a). The following FISH analysis confirmed this chromosomal aberration and revealed that the deletion was maternally inherited from the grandmother via the mother to the index patient (Figure 2b). To clarify that the deletion is of the same size in patient 1 and at least her mother (patient 2), we performed a second array CGH analysis hybridizing the DNA samples from patient 1 and 2 against each other. The array CGH shows now a balanced profile on chromosome 2 indicating that both patients (1 and 2) carry exactly the same deletion. (Supplementary figure 2).
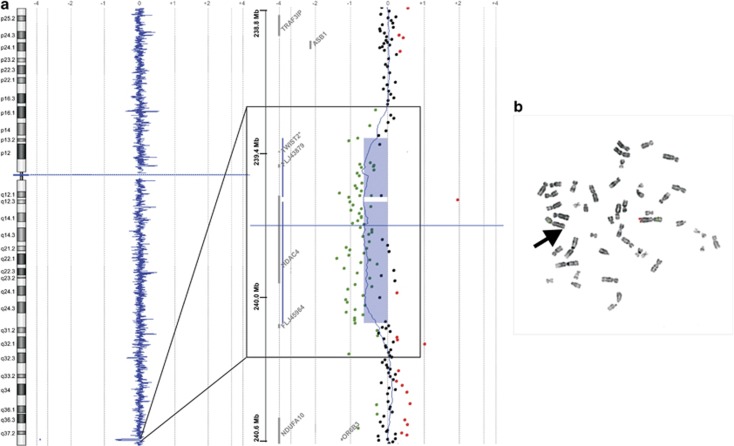
Figure 2.
(a) Array CGH profile of chromosome 2 indicating a heterozygous interstitial deletion in 2q37.3 (hg18 position chr2:239,395,957-240,154,599 bp) as shown by CGH Analytics 3.4.40 software. Note the magnification box, the deleted region (blue bars) encompasses HDAC4, FLJ43879 and TWIST2 (*gene symbol added according to UCSC hg18 NCBI36/Version Mar 2006). (b) FISH analysis confirming the 2q37.3 deletion in lymphocytic chromosomes of the index patient's grandmother. Note, there is only one hybridization signal of the 2q37.3 locus-specific RP11-546M8 probe (red) on one of the chromosomes 2 (control probe RP11-27O22 labelled in green). The aberrant chromosome 2 is marked by an arrow.
The genome coordinates chr2:239,395,957-240,154,599 in the UCSC human genome build 18 (NCBI36/ Version Mar 2006) indicated that the deletion included HDAC4 and two additional genes FLJ43879 and TWIST2 (MIM 607556) (Figure 3). For confirmation we used target-specific qPCR analysis and confirmed that both genes with known function HDAC4 and TWIST2 are affected by the microdeletion 2q37.3 in patients 1 and 2, and, therefore, most probably also in patient 3 (Supplementary figure 3). The coordinates for the minimum and maximum deletion intervals are chr2:239,395,957-240,154,599 and chr2:239,385,056-240,165,585, respectively.
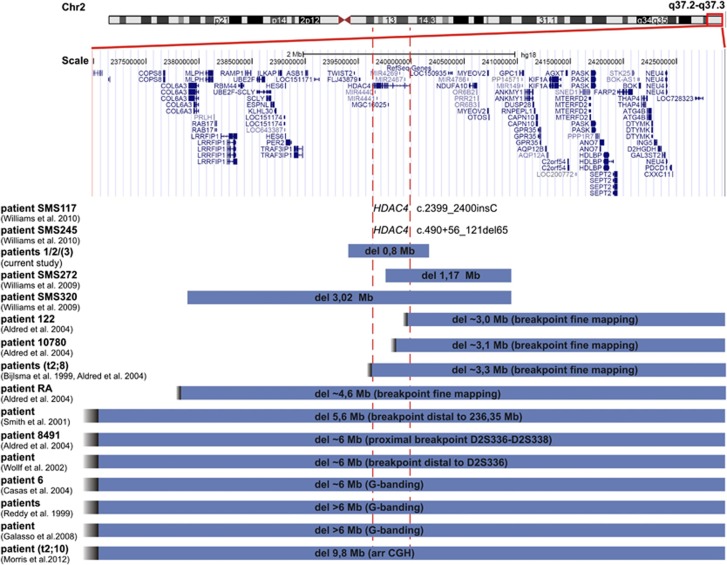
Figure 3.
Schematic representation of the 2q37.3 deletions in the family of the current study relative to recently reported HDAC4 mutations (SMS117, SMS245) or 2q37.3 deletions involving HDAC4. The 2q37.3 deletions of SMS272 (hg18, chr2:239,764,593-240,938,545), SMS320 (hg18, chr2:237,920,956-240,938,547) and the patient described by Morris et al.19 were detected by array CGH, while the deletions of the other patients were located by break point mapping using microsatellite analysis or by cytogenetic analysis with G-banding. RefSeq Gene positions are according to UCSC hg18 (NCBI36/ Version Mar 2006).
Discussion
Currently, about 100 patients with 2q37 deletions have been reported in the literature and 10 reports of patients with 2q37.3 microdeletions of different sizes are annotated in the DECIPHER database. Most of them have emerged de novo. Studies on parent-of-origin of de novo deletions revealed no preferential maternal or paternal transmission. Only a minority of 2q37.3 deletions recur in families with structural chromosomal abnormalities.3 Most of these are derivatives from a balanced parental chromosomal translocation. The current study is to our knowledge the first report on a three generation familial case with an inherited interstitial 2q37.3 microdeletion comprising HDAC4, TWIST2 and FLJ43879. There is so far only one report on parent to child transmission of an apparently pure terminal 2q37.3 deletion from an unaffected parent which is distally to the HDAC4 gene and does not overlap with the interstitial 2q37.3 deletion reported here.9
Recently, HDAC4 haploinsufficiency has been identified as the critical genetic mechanism responsible for developmental delay, behavioural abnormalities, and BDE in BDMR patients with 2q37 deletion.5 This was supported by the detection of intragenic de novo HDAC4 mutations in two patients with core findings of BDMR syndrome.
Here, we show that a heterozygous 2q37.3 microdeletion involving HDAC4 is associated with psychomotor and behavioural abnormalities in combination with the BDMR-specific facial dysmorphism pattern (Figure 1). The missing BDE in the patients reported here matches to observations that BDE is a variable clinical feature in BDMR patients. Despite the positive history of growth delay in infancy of patients 1 and 2, upon examination, all the patients of the current study presented a height in the lower normal range (Table 1). Interestingly, HDAC4 exerts its inhibitory activity on key players of skeletogenesis like RUNX2 and MEF2C by binding them with its N-terminal part. Therefore, a loss of the N-terminal function of HDAC4 leads to a premature ossification resulting in shortness of bones in mice. In contrast the loss of the enzymatic activity in the C-terminus of HDAC4 allows normal bone development in mice.8, 10 Thus, other RUNX2 or MEF2C-binding factors compensating the effect of the HDAC4 N-terminal part might have taken effect in the patients reported here and led to a normal metacarpal bone development. For instance, TWIST2 which is also heterozygously deleted in these subjects is known to be involved in skeletogenesis.11, 12 However, as TWIST2 maintains cells in a preosteoblast phenotype its deletion should have forced the manifestation of bone shortness. On the other hand, neither mutations in TWIST2 nor in FLJ43879 are described in patients with skeletal abnormalities. Instead, homozygous nonsense mutations in TWIST2 are reported to segregate in families with Setleis syndrome (MIM 227260), which is characterized by thin skin and sparse hair with a bi-temporal forceps marks-like pattern.13, 14 No function of the FLJ43879 gene is known yet.
A comparison of the spectrum of the clinical findings of previously reported patients with 2q37.1, 2q37.2 and 2q37.3 terminal deletions has been performed by Galasso et al.24 The distribution of malformations or anomalies of the central nervous system, gastrointestinal, cardiac, oral, tracheal, bony and genitourinary systems was different between these subgroups, however, more frequent in patients with break points in 2q37.1. To evaluate further diagnostic criteria for the 2q37 deletion syndrome, we compared the clinical findings of our patients and a representative cohort of other published patients carrying HDAC4 mutations or overlapping interstitial or terminal 2q37 deletions (Figure 3, Table 1).3, 5,15, 16, 17, 18, 19, 20, 21, 22, 23, 24 The female to male ratio was 21/6. Regarding the body measurements, 4/18 of the patients were microcephalic, 8/24 revealed a short stature and 7/20 an overweight. Twenty-three out of 24patients had a history of motor delay. Except for patient 1, all patients for whom data were available showed speech delay. Altogether, the developmental delay/intellectual disability of the presented individuals (patients 1-3) was of only mild degree compared with the other here discussed patients. Behavioural problems were reported in 16/17 affected individuals. The reported behavioural abnormalities include hyperactivity, sleep disturbance, auto-/aggression or temper tantrums and stereotypic/ritualistic or autistic behaviour. Only patient 8491 is described as sociable and not aggressive.3 As only the child (patient 1) of the current study reveals behavioural problems, and no obvious behavioural abnormalities are present in the adult patients, it is supposable that these behavioural problems though progressively disturbing during childhood will disappear with age. Overall, these behavioural problems remind that of Smith–Magenis syndrome (SMS; MIM 182290). Accordingly, a regulatory connexion between HDAC4 and RAI1 the major gene for the phenotypic features of SMS has been proven.5 Interestingly, with regard to the memory deficits claimed by the index patient́s mother, HDACs seem to be related to memory functions of the brain as shown in an Alzheimeŕs disease mouse model.18
With regard to the BDMR-associated facial dysmorphism we compared the facial characteristics of our patients and the other patients reported in the literature with the features previously described in GeneReviews (see features marked with an asterisk in Table 1).1 Sorted by frequency, joint facial signs were narrow palpebral fissures in 91% (19/21), round or broad face in 87% (20/23), deeply set eyes in 87% (20/23), midface hypoplasia in 65% (15/23), thin upper lip in 61% (14/23), upslanted palpebral fissures in 50% (9/18), hypoplastic alae nasi with prominent columnella in 43% (9/21), frontal bossing in 39% (7/18), highly arched eyebrows in 39% (7/18), low-set ears or ear anomalies in 33% (7/21), everted lower lip in 31% (5/16) and epicanthal folds in 6% (1/16) of the patients. From the listed features, particularly our patients did not show frontal bossing, upslanted palpebral fissures nor hypoplastic alae nasi with prominent columnella. Additional remarkable facial features not described by GeneReviews are the markedly narrow palpebral fissures and the everted lower lip, which seems to aggrevate with age, as this could be observed especially in the adult patients of our current study and was described for the patients harbouring HDAC4 mutations. Comparing the photograph of patient 2 to that of patient SMS117 (see Williams et al., 20105) the facial gestalt of both patients is of striking similarity. As shown recently, different levels of HDAC4 gene expression levels from the remaining intact HDAC4 alleles could be one explanation for the differing phenotypic severity in BDMR syndrome.19
In summary, our study shows that a heterozygous 2q37.3 microdeletion involving the genes HDAC4, TWIST2 and FLJ43879 is inherited in an autosomal-dominant manner in a three generation familial case and is associated with psychomotor and behavioural abnormalities in combination with the BDMR-specific facial dysmorphism pattern. We provide another example that HDAC4 haploinsufficiency is not fully penetrant with regard to the BDE phenotype.3, 16 Furthermore, we assume that it is also the critical genetic mechanism for the BDMR-specific facial pattern besides the BDMR-associated moderate developmental delay and behavioural problems. As patients with 2q37.3 microdeletions including HDAC4 show similar behavioural problems as the ones known for SMS HDAC4 deletions and mutations should be considered in patients with BDMR-specific facial dysmorphism pattern, and a phenotypic spectrum of SMS who are negative for 17p11.2 deletions and mutations of RAI1.
Acknowledgments
We would like to thank the family for their collaboration and contribution to this project. We acknowledge F Trotier for excellent technical assistance.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Doherty ES, Solomon BD, Lacbawan F.2q37 Deletion Syndromein Pagon RA, Bird TD, Dolan CR, Stephens K, (eds): GeneReviews Seattle WA; 1993–2011 [Google Scholar]
- Falk RE, Casas KA. Chromosome 2q37 deletion: clinical and molecular aspects. Am J Med Genet C Semin Med Genet. 2007;145C:357–371. doi: 10.1002/ajmg.c.30153. [DOI] [PubMed] [Google Scholar]
- Aldred MA, Sanford RO, Thomas NS, et al. Molecular analysis of 20 patients with 2q37.3 monosomy: definition of minimum deletion intervals for key phenotypes. J Med Genet. 2004;41:433–439. doi: 10.1136/jmg.2003.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni M, Le Merrer M, Raoul O, et al. Molecular cytogenetic analysis of five 2q37 deletions: refining the brachydactyly candidate region. Eur J Med Genet. 2006;49:255–263. doi: 10.1016/j.ejmg.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Williams SR, Aldred MA, Der Kaloustian VM, et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdzadeh N, Wang L, Morrison BE, Bassel-Duby R, Olson EN, D'Mello SR. HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev Neurobiol. 2008;68:1076–1092. doi: 10.1002/dneu.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Langley E, Wolf D, Karlsson C, Pines J, Kouzarides T. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 2001;29:3439–3447. doi: 10.1093/nar/29.16.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega RB, Matsuda K, Oh J, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- van Karnebeek CDM, Koevoets C, Sluijter S, et al. Prospective screening for subtelomeric rearrangements in children with mental retardation of unknown aetiology: the Amsterdam experience. J Med Genet. 2002;39:546–553. doi: 10.1136/jmg.39.8.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I, Savelieva KV, Ye GL, et al. Loss of the putative catalytic domain of HDAC4 leads to reduced thermal nociception and seizures while allowing normal bone development. PLoS One. 2009;4:e6612. doi: 10.1371/journal.pone.0006612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Toita H, Yoshimoto A, et al. Potential involvement of Twist2 and Erk in the regulation of osteoblastogenesis by HB-EGF-EGFR signaling. Cell Struct Funct. 2010;35:53–61. doi: 10.1247/csf.10001. [DOI] [PubMed] [Google Scholar]
- Tukel T, Sosic D, Al-Gazali LI, et al. Homozygous nonsense mutations in TWIST2 cause Setleis syndrome. Am J Hum Genet. 2010;87:289–296. doi: 10.1016/j.ajhg.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan DE, Villarroel CE, Medrano-Hernandez A, et al. Setleis syndrome in Mexican-Nahua sibs due to a homozygous TWIST2 frameshift mutation and partial expression in heterozygotes: review of the focal facial dermal dysplasias and subtype reclassification. J Med Genet. 2010;48:716–720. doi: 10.1136/jmedgenet-2011-100251. [DOI] [PubMed] [Google Scholar]
- Williams SR, Girirajan S, Tegay D, Nowak N, Hatchwell E, Elsea SH, et al. Array comparative genomic hybridisation of 52 subjects with a Smith–Magenis-like phenotype: identification of dosage sensitive loci also associated with schizophrenia, autism, and developmental delay. J Med Genet. 2009;47:223–229. doi: 10.1136/jmg.2009.068072. [DOI] [PubMed] [Google Scholar]
- Casas KA, Mononen TK, Mikail CN, et al. Chromosome 2q terminal deletion: report of 6 new patients and review of phenotype-breakpoint correlations in 66 individuals. Am J Med Genet A. 2004;130A:331–339. doi: 10.1002/ajmg.a.30156. [DOI] [PubMed] [Google Scholar]
- Wilson LC, Leverton K, Oude Luttikhuis MEM, et al. Brachydactyly and mental retardation: an albright hereditary osteodystrophy-like syndrome localized to 2q37. Am J Hum Genet. 1995;56:400–407. [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris B, Etoubleau C, Bourthoumieu S, et al. Dose dependent expression of HDAC4 causes variable expressivity in a novel inherited case of brachydactyly mental retardation syndrome. Am J Med Genet Part A. 2012;158A:2015–2020. doi: 10.1002/ajmg.a.35463. [DOI] [PubMed] [Google Scholar]
- Wolff DJ, Clifton K, Karr C, Charles J. Pilot assessment of the subtelomeric regions of children with autism: detection of a 2q deletion. Genet Med. 2002;4:10–14. doi: 10.1097/00125817-200201000-00002. [DOI] [PubMed] [Google Scholar]
- Smith M, Escamilla JR, Filipek P, et al. Molecular genetic delineation of 2q37.3 deletion in autism and osteodystrophy: report of a case and of new markers for deletion screening by PCR. Cytogenet Cell Genet. 2001;94:15–22. doi: 10.1159/000048775. [DOI] [PubMed] [Google Scholar]
- Bijlsma EK, Aalfs CM, Sluitjer S, et al. Familial cryptic translocation between chromosomes 2qter and 8qter: further delineation of the Albright hereditary osteodystrophy-like phenotype. J Med Genet. 1999;36:604–609. [PMC free article] [PubMed] [Google Scholar]
- Reddy KS, Flannery D, Farrer RJ. Microdeletion of chromosome sub-band 2q37.3 in two patients with abnormal situs viscerum. Am J Med Genet. 1999;84:460–468. doi: 10.1002/(sici)1096-8628(19990611)84:5<460::aid-ajmg10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Galasso C, Lo-Castro A, Lalli C, Nardone AM, Gullotta F, Curatolo P. Deletion 2q37: an identifiable clinical syndrome with mental retardation and autism. J Child Neurol. 2008;23:802–806. doi: 10.1177/0883073808314150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.