Abstract
A large-scale sequencing screen of X-linked synaptic genes in individuals with autism spectrum disorder (ASD) or schizophrenia (SCZ), two common neurodevelopmental disorders, identified many variants most of which have no easily predictable effect on gene function. In this report, we evaluated the impact of these rare missense and silent variants on gene splicing. For this purpose, we used complementary in silico analyses, in vitro minigene-based assays and RNA prepared from lymphoblastoid cells derived from patients with these mutations. Our goal was to identify the variants which might either create or disrupt an acceptor splice site, a donor splice site or an exonic splicing enhancer, thus leading to aberrant splicing that could be involved in the pathogenesis of ASD or SCZ. We identified truncating mutations in distinct X-linked gamma-aminobutyric acid A (GABAA) receptor subunit-encoding genes, GABRQ and GABRA3, in two different families. Furthermore, missense and silent variants in nuclear RNA export factor 5 and histone deacetylase 6 were shown to partially disrupt the protein. While genes from the GABAergic pathway have previously been thought to be involved in the pathophysiology of ASD, this is the first report of ASD patients with truncating mutations in GABA receptors genes.
Keywords: autism, schizophrenia, GABRA3, GABRQ, splicing alteration
Introduction
Autism spectrum disorder (ASD) and schizophrenia (SCZ) are two common developmental neuropsychiatric disorders that result from a combination of genetic and environmental factors. A growing number of studies, including ours, suggest that rare and highly penetrant variants, many of which are de novo mutations, have an important role in ASD and SCZ.1, 2, 3, 4, 5 As many arguments suggest a possible role of the X chromosome in these diseases (a high proportion of genes involved in brain development and cognition are located on the X chromosome;6, 7, 8 males are more severely affected with SCZ than females9 and there is a higher prevalence of ASD in males10), our group previously tested a specific subset of X-chromosome genes for rare variants in ASD and SCZ. We identified potentially pathogenic truncating mutations in IL1RAPL1 and in MAOB in addition to multiple rare missense variants in genes encoding proteins of various synaptic functions.1, 11 Prioritization of these variants was on the basis of both familial segregation and bioinformatics analyses (for example, Polyphen and SIFT software) that predicted the deleterious impact of amino acid changes on the resulting protein.12 However, by focusing only on amino acid changes in these X-linked genes, we may have missed other potentially damaging variants. Indeed, some silent and missense nucleotide substitutions can also dramatically alter protein function by impairing the normal splicing process. Splicing can be affected through alteration of normal splice site sequences, creation of new cryptic sites or by the disruption/creation of the internal exonic elements involved in the regulation of splicing. These latter elements are short degenerate sequences (6–8 bp) to which the splicing factors can bind and modulate the recognition of splice sites.13 They can either enhance (exonic splicing enhancers, ESEs) or reduce (exonic splicing silencers, ESSs) splicing at nearby splice sites. The ESEs and ESSs recruit trans-splicing factors, often SR (serine/arginine-rich) proteins and heterogeneous nuclear ribonucleoproteins that either promote or inhibit the spliceosome assembly.
Few studies have focused on the systematic evaluation of the effects of mutations on splicing. In addition to substitutions that affect canonical splice sites, variants are analyzed for effects on splicing only in well-documented disease-causing genes such as BRCA1 (Breast cancer 1)14, 15 or ATM (Ataxia-telangiectasia mutated)16 or when a silent or intronic mutation is identified by familial linkage analysis.17, 18 Cartegni et al19 published an important list of silent mutations associated with altered splicing in different diseases, showing that it is a mechanism to take into account when potential effects of mutations are analyzed. Therefore, in order to assess the contribution of missense and silent variants on splicing alteration in ASD and SCZ, we conducted a systematic analysis (in silico and in vitro) of 253 private variants previously identified from our screening of 111 X-linked synaptic genes in 285 individuals with either ASD (n=142; 122 males and 20 females) or SCZ (n=143; 95 males and 48 females).11 Specifically, we used splice site and ESE prediction programs, followed by the testing of the most promising variants in a minigene-based splicing system and by RNA analysis of lymphoblastoid cell lines. Our study identified ASD-associated protein truncation variants in two genes: gamma-aminobutyric acid (GABA) receptors alpha 3 (GABRA3) and theta (GABRQ). Furthermore, two other variants affecting splicing were identified in the nuclear RNA export factor 5 (NXF5) and histone deacetylase 6 (HDAC6) genes in individuals with SCZ and ASD, respectively. Our results suggest for the first time that GABRA3 and GABRQ may be involved in ASD. This is also the first report to describe the systematic characterization of effect on splicing for rare genetic variants in ASD and SCZ.
Materials and methods
In silico splicing predictions
We compare prediction results with the outcome of four different splice-site prediction algorithms: (1) BDGP Splice Site Prediction by Neural Network (BDGP/NNSplice; http://www.fruitfly.org/seq_tools/splice.html);20 (2) ESEfinder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home)21 and simultaneously, via the Human Splicing Finder (HSF) interface (http://www.umd.be/HSF/):22 (3) MaxEntScan (MES; http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html),23 which is a program based on maximum entropy calculation and (4) HSF,22 which give scores based on matrices derived from Shapiro et al.24 Default parameters are used for ESEfinder and BDGP (threshold 6.67 and 0.4, respectively, for the two programs) to consider the creation of a potential splice acceptor or donor sites. For HSF and MaxEnt, we arbitrarily put a threshold of 80 and 5 to define a strong site, respectively, according to what was already published on normal intron–exon junctions in ATM gene,16 and what is described by Desmet et al.22 The HSF program also permits the search of potential splicing regulatory elements interrogating (1) ESEfinder matrices for SR proteins and (2) HSF matrices for heterogeneous nuclear ribonucleoprotein A1, Tra2-b and 9G8 proteins, defined using systematic evolution of ligands by exponential (SELEX) enrichment,21, 22 (3) RescueESE,25 containing more than two-hundred hexamer sequences, which are potential ESE, found to be significantly more abundant in exons than in introns and (4) putative ESE octamers published by Zhang & Chasin.26 In silico predictions for the higher-ranked variants are shown in Tables 1 and 2.
Table 1. Variants predicted to affect/create a splice acceptor/donor site (ASS/DSS).
|
Variant |
Splicing predictions |
Minigene assay |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Position on cDNA | AA change | position on Hg18 | Flanking sequences | Change | HSF (>80) | MaxEnt (>5) | BDGP (>0,4) | ESE Finder (>6,67) | Exon Length | Effect on splicing |
| PLXNB3 | NM_005393 (c.464 C->G) | A155G | chrX:152685940 | 5′-CTGTACCAGG-3′ [C/G] 5′-TGAGGACCCT-3′ | new DSS | 91 | 7,1 | 0,99 | 9,8 | exon 4 (1041+262 =1303bp) | unknown |
| NXF5 | NM_032946 (c.287A->G) | N96S | chrX:100983140 | 5′-TCTGTGAAGA-3′ [A/G] 5′-TAAGTTGAAG-3′ | new DSS | 84 | 9,3 | 0,98 (normal 0,96) | — | exon 6 (85+262 =347 bp) | see table 2 |
| CNKSR2 | NM_014927 (c.2199 G->A) | T733T | chrX:21537163 | 5′-TTTCCTCCAC-3′ [G/A] 5′-GAGACTTCTC-3′ | new ASS | 93 | 7,4 | 0,8 | 9,16 | exon 20 (547+262 =809bp) | no effect |
| GABRQ | NM_018558 (c.306 G->C) | M102I | chrX:151564711 | 5′-AAATGAATAT-3′ [G/C] 5′-GTAAGTGTGT-3′ | close to DSS | Normal 94 to 83 | normal 11 to 10,4 | normal 1 to 0,99 | normal 10,05 to 0 | exon 3 (68+262 =330bp) | Diminution of the use of the normal DSS; use of an alternative DSS in intron |
Abbreviations: AA, amino acids; cDNA, complementary DNA, DSS, donor splice site; ESE, exonic splicing enhancer; HSF, Human Splicing Finder.
Information about each variant predicted to affect an acceptor or donor site by at least three prediction programs is given in this table: the gene in which the variant is located, the position of the variant in cDNA and in NCBI2006/Hg18, the amino acid change and the flanking sequence. Splicing predictions are given for the new site (or the disrupted normal site), according to four different programs: HSF, MaxEnt, BDGP and ESEfinder, and threshold parameters used to define splice sites are shown in parentheses. The exon included in the minigene system is indicated, as well as the size expected for the RT-PCR products and the results from the minigene analyses.
Table 2. Variants predicted to disrupt an Exonic Splicing Enhancer (ESE).
|
Variant |
Splicing predictions |
Minigene assay |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | AA change | Position on cDNA | Position on Hg18 | Flanking sequences | ESE finder | Rescue ESE | Zhang octamers | HSF ESE | Nber (pg) | Nber ESE lost | Exon length | Effect on splicing |
| GABRA3 | T12S | NM_000808.3 (c.34 A->T) | chrX:151283665 | 5′-CTGTTACATG-3′ [A/T] 5′-CCAGCCTTGG-3′ | Srp40 lost, Srp55 new | 1 lost | 3 lost | 2 lost | 4 | 7 | exon 2 (166+262=428 bp) | skipping of exon 2 |
| CDKL5 | S426S | NM_003159.2 (c.1278 A->C) | chrX:18532243 | 5′-CAAAGCCTTC-3′ [A/C] 5′-GAAGGCCCAG-3′ | Srp40 lost, SF2 new | 5 lost | 3 lost | — | 3 | 9 | exon 12 (967+262=1229 bp) | no effect |
| GRIPAP1 | K428K | NM_020137 (c.1284 G->A) | chrX:48724785 | 5′-GTGCGGAGAA-3′ [G/A] 5′-CGGAAGGCCA-3′ | SF2 lost, Srp55 lost, SF2 new | 1 new | 2 lost | 2 lost | 3 | 6 | exon 16 (213+262=475 bp) | no effect |
| MAOB | G335S | NM_000898 (c.1003 G->A) | chrX:43525661 | 5′-CAAACCTGAA-3′ [G/A] 5′-GCAACTATGC-3′ | SF2 lost, SC35 lost, Srp40 new | 3 new | 3 lost, 1 new | 1 lost | 3 | 6 | exon 9 (93+262=355 bp) | no effect |
| HCFC1* | T241M | NM_005334.2 (c.722 C->T) | chrX:152880942 | 5′-GACACCCTGA-3′ [C/T] 5′-GTGGAATAAG-3′ | Srp40 lost Srp55 new | 1 new | 3 lost | 1 lost | 3 | 5 | exon 5 (85+262=347 bp) | no effect |
| ATP2B3 | E314E | NM_021949 (c.942 G->A) | chrX:152464765 | 5′-GGGCCATGGA-3′ [G/A] 5′-AGTAGCCAGA-3′ | SF2 lost | 2 new | 1 lost | 2 lost | 3 | 4 | exon 6 (42+262=304 bp) | no effect |
| MAOB | P109L | NM_000898 (c. 326 C->T) | chrX:43547549 | 5′-GTATGGAATC-3′ [C/T] 5′-AATTACCTAC-3′ | SC35 lost | 2 lost | — | 1 lost | 3 | 4 | exon 4 (105+262=367 bp) | no effect |
| NXF5 | N96S | NM_032946 (c. 287 A->G) | chrX:100983140 | 5′-TCTGTGAAGA-3′ [A/G] 5′-TAAGTTGAAG-3′ | — | 1 lost | 2 lost | 1 lost | 3 | 4 | exon 6 (81+262=343 bp) | skipping of exon 6 |
| KCND1 | E462K | NM_004979.4 (c.1384 G->A) | chrX:48708012 | 5′-CGGCAGTGGC-3′ [G/A] 5′-AGGAACAGGC-3′ | SF2 lost | — | 1 lost | 1 lost | 3 | 3 | exon 2+3 (84+99+262=445 bp) | no effect |
| DRP2 | E176K | NM_001939 (c.428 G->A) | chrX:100380713 | 5′-GCACCCATTT-3′ [G/A] 5′-AGGAGTTAGA-3′ | Srp55 lost | — | 1 lost | 1 lost | 3 | 3 | exon 6 (121+262=383 bp) | no effect |
| HDAC6 | A39A | NM_006044 (c. 117 C->T) | chrX:48546245 | 5′-AAAAGGGAGC-3′ [C/T] 5′-GTTCCCCGCT-3′ | SF2 lost | — | 3 new, 1 lost | 1 lost | 3 | 3 | exon 3 (129+262=391 bp) | partial skipping of exon 3 |
Abbreviations: AA, amino acids; cDNA, complementary DNA; ESE, exonic splicing enhancer; HSF, Human Splicing Finder.
Information about each variant predicted to affect an ESE by at least three different programs is given in this table: the gene in which the variant is located, the position of the variant in cDNA and in NCBI2006/Hg18, the amino acid change, the flanking sequence (mutation between brackets). Predictions of ESE alterations caused by each variant are given by four different programs (ESEfinder, Rescue ESE, Zhang octamers and HSF ESE). The number of programs predicting an ESE loss and the total number of ESE disrupted are also indicated. The exon included in the minigene system, as well as the size expected for RT-PCR products and results from the minigene analyses are indicated. Asterix indicates that this variant was also found in the proband's SCZ affective sister. Variants that affect splicing in vitro minigene assay are in bold.
Minigene vector constructs
The pSPL3B vector was obtained from Life Technologies Inc., Burlington, Ontario, Canada. Exons of interest were amplified by PCR, with 150–200 bp of 5′ and 3′ flanking intronic sequences, from corresponding patient/control DNA blood samples and using forward and reverse primers carrying 5′ tails that contained sequence homologous to pSPL3B sequence around the multiple cloning site. Primer sequences are available on request. The PCR products were cloned into the pSPL3 vector, between two exons of rabbit β-globin, using the SLIC method described by Li et al27 or using BamH1 digestion/ligation. All constructs were verified by sequencing. Minigenes carrying the wild-type and the variant allele were then transfected separately into COS-7 cells during the same experiment. The splicing patterns corresponding to the wild-type and to the mutant allele were then compared by reverse transcriptase-PCR analysis of the RNA from transfected cells.
Cell culture and transfections
COS-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 4,500 mg l−1 glucose supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml at 5% CO2 and 37 °C. One day before transfection, cells were seeded in six-well culture dishes at a density of 4 × 105 cells per well. Cells were transfected with 2 μg pSPL3B using lipofectamine 2000 transfection reagent (Life Technologies Inc.), according to the manufacturer's instructions. The next day, the media was changed and after another 24 h, the cells were harvested and total RNA was isolated using TRIzol reagent (Life Technologies Inc.).
Exon-trapping analysis
First strand complementary DNA was synthesized using 1 μg RNA, 1 μg random hexamers and MMLV reverse transcriptase (Life Technologies Inc.). The Complementary DNAs were amplified with rabbit β-globin-specific primers (5′-CTGAGTCACCTGGACAACC-3′ and 5′-ATCTCAGTGGTATTTGTGAGC-3′) and the PCR products were resolved on a 2% agarose gel and sequenced using the rabbit β-globin-specific primers. The results presented are representative of three independent tranfection assays when a splicing effect was observed. Both expected bands obtained with the wild-type construct and the alternative product were purified from gel and subcloned in TA-plasmid (TA-Cloning kit, Life Technologies Inc.). Plasmids were amplified in bacteria and purified before being sent to sequencing using universal T7 and Sp6 primers.
RNA analysis
Total cellular RNA from patients was isolated from lymphoblastoid cell lines using the TRIzol reagent, and reverse-transcribed using MMLV reverse transcriptase (Life Technologies Inc.). Complementary DNA was PCR-amplified with specific primers (Supplementary Table S1). PCR products obtained from patients and an unaffected control were sent to sequencing. PCR products were resolved on a 2% agarose gel.
X inactivation HUMARA assay
The X Chromosome Inactivation assay was performed on genomic DNA extracted from peripheral blood, as described elsewhere.28
GABRA3 and GABRQ mutation screening in control individuals
Patient and control cohorts have been described in Piton et al,11 and primers for the amplification and the sequencing of coding regions of the GABRA3 and GABRQ genes were those already used in that study. The sequencing was performed at the Genome Quebec Innovation Center (Montreal, Quebec, Canada) and genomic variations were identified using Mutation Surveyor v3.10 software (SoftGenetics, State College, PA, USA).
Results
Identification of rare missense/silent variants predicted to induce splicing alteration
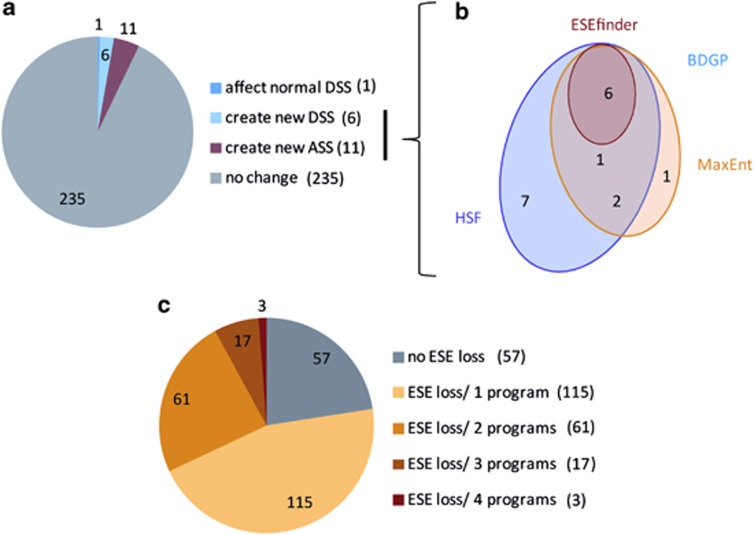
To identify which of the 253 previously identified private variants (see the list in Piton et al, Supplementary Table 411) affect normal splicing, we first performed in silico analysis using different programs able to predict acceptor (ASS) and donor splice sites (DSS) used in mammalian cells (see Materials and methods). Only one variant was predicted to affect a regular DSS (M102I in GABRQ gene, Table 1). On the basis of at least one of the programs (Figure 1a), 17 variants were predicted to create a new DSS or a new ASS. The algorithms used by BDGP and the ESEfinder seemed the most stringent, as these predicted splice alterations only when both MaxEntScan and HSF did (Figure 1b). Altogether, we show that the overlap between the different prediction programs is very high (Figure 1b).
Figure 1.
Proportion of variants that alter/create a DSS, an ASS or disrupt an ESE among the 253 unique variants identified during the resequencing of 111 X-linked synaptic genes in 285 individuals with ASD or SCZ. (a) Number of variants predicted to affect a normal DSS or to create new cryptic DSS and ASS, according to at least one splice prediction program (BDGP/NNSplice: http://www.fruitfly.org/seq_tools/splice.html,20 ESEfinder: http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home,21 HSF interface: http://www.umd.be/HSF/22) and MaxEntScan (http://genes.mit.edu/burgelab/maxent/ Xmaxentscan_scoreseq.html23). (b) Overlap between these different programs for the variants predicted to create new cryptic DSS and ASS sites. (c) Number of variants predicted to disrupt one or more exonic splicing elements (ESEs), according to ESEfinder, RescueESE,25 HSF internal program for ESE motif experimental predictions and PESE octamers published by Zhang & Chasin.26
We also made a parallel analysis of the same 253 variants to identify those that could alter splicing-regulatory elements. We focused on ESEs, as their disruption due to a nucleotide substitution could lead to the skipping of the corresponding exon in the mature mRNA. As for splice site predictions, we used multiple functional prediction programs (see Materials and methods). Over 75% of the variants were predicted to disrupt an ESE by at least one program (Figure 1c); and in contrast to our observed splice site predictions, the overlap between the different programs was poor, making the interpretation of this in silico analysis difficult.
Before the in vitro validation experiments, we selected a subset of 14 missense and silent variants predicted to affect a DSS (1 variant, Table 1), to create a cryptic DSS or ASS (3 variants, Table 1) or to affect an ESE (11 variants, Table 2, variant in NXF5 is common between two lists) using the following restrictive criteria: the variants must be predicted to have an effect on splicing by at least three programs and, for the ESE, not be located in the first or the last exon (11/20 variants). Variants which did not co-segregate with the disease were also filtered out. These 14 variants comprise nine missense and five silent variants located in 13 genes. They were examined using a complementary in vitro approach based on the use of a reporter minigene system with exon-trapping model.29 The exons to be tested were subcloned in SPL3B vector, between two exons of the rabbit β-globin gene. For each variant, wild-type and mutant splicing patterns were compared, as described in Materials and methods.
Altered splicing process in two GABA receptor subunit genes in patients with ASD
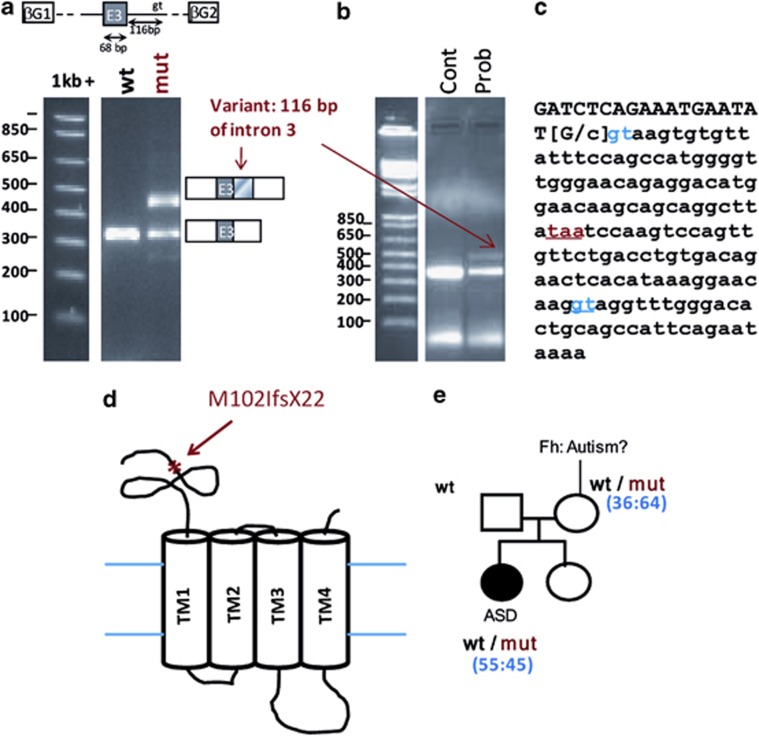
The missense M102I (c.306G>C) in the GABRQ gene, which encodes for the GABA-A receptor theta subunit, identified in one ASD female patient, is located just upstream to the normal DSS and predicted to affect this splice site, either mildly (MaxEnt and BDGP), significantly (HSF) or very drastically (ESEfinder) (Supplementary Figure S3A). The minigene assay shows that the band corresponding to the inclusion of the entire exon 3 (band at 330 bp) is weaker in the mutant than the wild-type, and at least one upper band (446 bp) is exclusively observed in the mutant (Figure 2a). This latter represents a splicing event corresponding to the use of an alternative DSS in intron 3, located 116-bp downstream of the normal site. This result shows that normal DSS is less used with the C allele, and therefore confirms the in silico analysis predictions. We performed mRNA analysis on lymphoblastoid cell lines from the ASD patient and observed the same pattern of bands thus validating the minigene assay result (Figure 2b). The retention of intron 3 causes a frameshift and introduces a premature stop codon (Figure 2c) presumed to lead to a very early-truncated protein (M102IfsX22) (Figure 2d). As the M102I missense was inherited from an healthy mother (Figure 2e), we performed the analysis of the X inactivation pattern in blood DNA of the ASD female and her mother; however, this analysis failed to explain the phenotype difference (Figure 2e). We screened 180 ethnically matched control individuals for mutations in the GABRQ gene and no M102I or any other truncating mutations were found. A missense, M102V (c.304A>G), affecting the same amino acid as variant M102I (c.306G>C) was found in the control cohort. However, the M102V variant was not predicted to significantly affect the use of the normal DSS (Supplementary Figure S3A), and this was confirmed by the minigene assay that shows no effect on splicing (Supplementary Figure S3B). In addition, three unique missense variants (I401T, V486G and S506L) were identified in the control cohort but none of them were predicted to alter the protein function, according to Polyphen2 or to affect the splice site or disrupt ESE sequences.
Figure 2.
In vitro analysis of GABRQ c.306 G->C (M102I) variant identified in one ASD female. (a) In vitro analysis of the effect of GABRQ c.306 G->C variant using a minigene assay. The genomic DNA segment tested is shown schematically above the panel: white boxes represent β-globin exons of the minigene vector and gray box represents exon 3 of GABRQ cloned into the vector. Migration patterns of RT-PCR products from COS-7 cells transfected with SPL3B_GABRQ_exon3_wild-type (wt) or SPL3B_GABRQ_exon3_ c.306 G->C (mut) constructs. The splicing patterns are shown schematically alongside the right-hand margin. (b) RT-PCR products obtained from patient (Prob) and control individual (Cont) lymphoblastoid cells RNA. The sequences of primers used are given in the Materials and methods section. (c) Genomic sequence of the end of Exon 3 and the beginning of intron 3 of the GABRQ gene. The variant c.306 G->C is indicated by brackets, the normal donor site ‘gt' is annotated in blue and the intronic gt sequence with the best prediction scores (ESEfinder: 10.6; Nnsplice:0.99; SpliceView:0.88) in blue and underlined. The premature stop codon taa is indicated in red and is underlined. (d) Schematic view of GABRQ protein: the asterix indicates the localization of the truncation caused by the mutation c.306 G->C p. M102IfsX22. (e) Pedigree of the family with the mutation c.306 G->C. Fh: familial history of autism in the maternal side (the son of the mother's sister was diagnosed with ASD but his DNA was unavailable for testing). In blue, between brackets, are indicated the results from the X inactivation profile analyses performed on blood DNA (% of cells expressing the wild-type allele: % of cells expressing the mutant allele).
The T12S (c.34A>T) missense variant, identified in one ASD male in GABRA3, another GABA receptor subunit gene, is located ∼30-bp downstream from the beginning of exon 2 and predicted to disrupt ESE by the four different prediction programs (Supplementary Table S2). Using the minigene system, we observed an expected product at 428 bp from the wild-type allele but not from the variant allele (Figure 3a). This result shows that the variant induces the skipping of the entire GABRA3 exon 2. Unfortunately, we were not able to validate our result using patient RNA, as the expression level of GABRA3 was far below a reliable detection threshold in lymphoblastoid cells (data not shown). Interestingly, as exon 2 contains the initiation codon, this skipping is predicted to prevent GABRA3 protein expression in this male. The variant, inherited from a healthy heterozygote mother (Figure 3b) was recently reported in dbSNP (rs61734348). It has been identified at the heterozygote state in one female out of 66 apparently healthy individuals from the Coriell collection. This variant was also found as a rare variant in the Exome Variant Server, with a minor allele frequency of 0.16%. No truncating mutation in the GABRA3 gene was identified in 180 ethnically matched control individuals or in 285 additional ASD patients.
Figure 3.
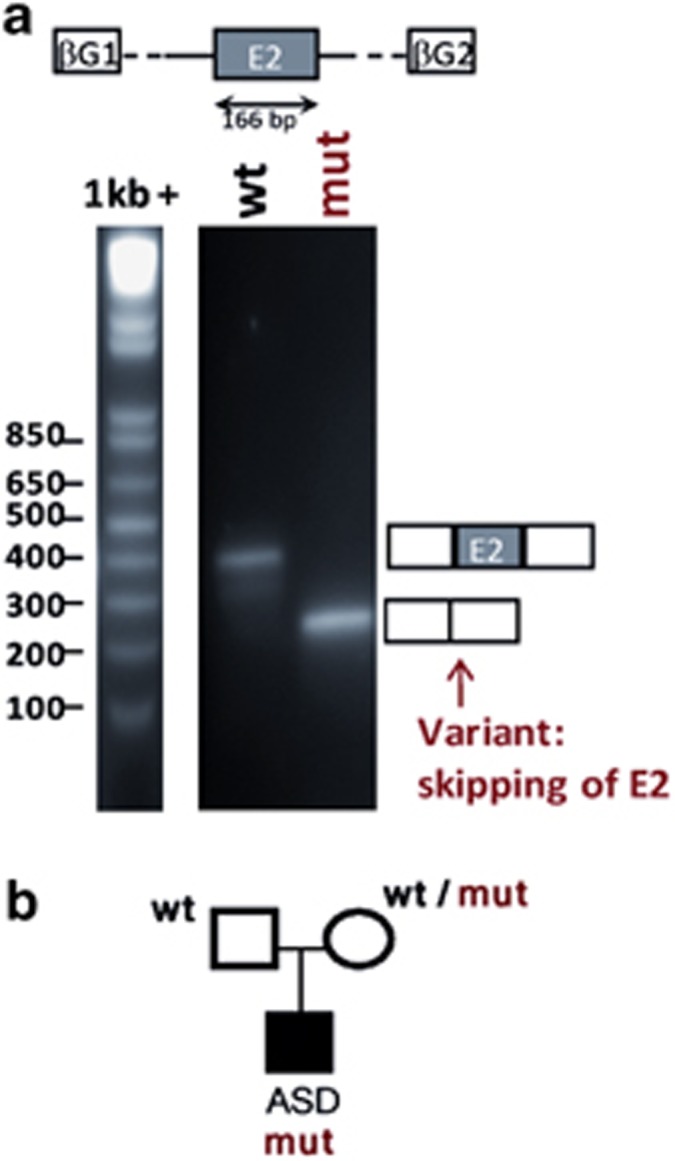
In vitro analysis of GABRA3 c.34A->T variant identified in one ASD male. (a) In vitro analysis of the effect of GABRA3 c.34 A->T variant using a minigene assay. The genomic DNA segment tested is shown schematically above the panel: white boxes represent β-globin exons of the minigene vector and gray box represents exon 2 of GABRA3 cloned into the vector. Migration patterns of RT-PCR products from COS-7 cells transfected with SPL3B_GABRA3_exon2_wild-type (wt) or SPL3B_GABRA3_exon2_ c.34 A->T (mut) constructs. The splicing patterns are shown schematically alongside the right-hand margin. (b) Pedigree of the family of the ASD male carrying the mutation c.34 A->T.
NXF5 and HDAC6 splicing alterations in SCZ and ASD patients
Among the remaining twelve variants that were tested with the minigene-based assay, two displayed a distinct gel migration pattern different from the wild-type allele (Supplementary Figure S1). The variant N96S (c.287A>G) in the NXF5 gene, found in a SCZ female, was predicted to create a cryptic DSS in the exon and to disrupt ESE sequences (Supplementary Table S2). Using the minigene-based analysis, we validated that the newly created cryptic DSS was not functional but that the loss of ESE sequences led to the skipping of the entire exon 6 (band at 262 bp in Supplementary Figure S1), causing an in-frame loss of 27 amino acids. We also observed a natural skipping of this exon in more than half of the transcripts with the wild-type sequence, but it is not known if this phenomenon occurs normally in cells or if it is just because of the minigene context. Our mRNA lymphoblastoid cell line-based analysis was inconclusive because of the weak expression level of NXF5 transcript in this cell type (data not shown). The variant was found in a SCZ girl from a large Pakistani family, as well as in 3 out of 5 other available affected individuals in this pedigree (Supplementary Figure S2). This variant was recently reported in dbSNP (rs142009656) and in Exome Variant Server, with a low minor allele frequency (0.12%).
Another variant A39A (c.117C>T) in the HDAC6 gene, predicted to disrupt ESE sequences (Supplementary Table S2), affects the normal splicing in the minigene system (Supplementary Figure S1). We found that this variant induces exon 3 to be skipped in less than one-third of the transcripts (Supplementary Figure S1). This skipping is predicted to cause an in-frame deletion of amino acids 32–74 of the HDAC6 protein, removing a functional nuclear export signal (NES1, AA 67–76)30 that could affect normal import/export regulation of HDAC6. This variant, identified at the hemizygote state in one ASD male and transmitted by his asymptomatic mother was recently added to dbSNP (rs147420530) and has a very low frequency in Exome Variant Server (minor allele frequency=0.02%). RNA analysis of lymphoblastoid cells from the patient failed to detect the alternative spliced product observed in the minigene assay.
For one variant in the PLXNB3 gene, predicted to create a strong DSS, the wild-type exon, as well as the mutated one were not retained (Supplementary Figure S1), making impossible to evaluate the effect of the variant. The nine remaining variants tested did not affect splicing, according to the minigene results (Supplementary Figure S1).
Discussion
We carried out a large-scale and systematic bioinformatical study to characterize genetic variants that were previously identified in ASD and SCZ patients11 and investigate those predicted to affect normal splicing. Prediction programs used for splicing analysis, such as ESEfinder and NNsplice/BDGP, can be easily run in a systematic manner, allowing the prioritization of a large number of variants, according to their potential effect on splicing. These programs have been extensively used by other groups and proved their usefulness for the predictions of splicing defects.14, 15, 31 To make in vivo/in vitro assessments of a variant true impact on splicing, the analysis of patient RNA is ideal but the availability of disease relevant material is often limited, especially for neurological disorders like ASD and SCZ for which brain biopsies are rare. The minigene approach allows the rapid testing of any variant and it circumvents the problems associated with the difficult access to patient RNA (from patients' derived biopsies, immortalized cell lines or blood sample recollection several years after the original recruitment). Furthermore, it also helps to overcome challenges like low level of expression in blood cells of genes involved in neurodevelopmental diseases, and the masking effect of preferential amplification of the wild-type allele. As an alternative, we and others showed that using monoallelic minigene is a powerful tool to assess the impact of sequence variants on splicing in vitro,14, 15, 32 as high correlation is observed between minigene results and patient RNA analysis. Among the 14 variants predicted to affect splicing by different programs and thus tested through the functional splicing assay, 29% (4/14) had an actual effect on splicing. For some variants, in CDKL5 and CNKSR2 for instance, unexpected patterns of bands were observed in the minigene assay both in the mutant and the wild type (Supplementary Figure S1). If the shorter bands correspond to the absence of the exon, the other bands may represent alternative splice events naturally occurring in these genes or could be owing to the heterologous cellular system used in the minigene assay. Indeed, the exon of interest is cloned into an intronic sequence that significantly differs from its normal genomic environment. The overexpression of the minigene in COS-7 cells could also induce alternative splicing events. However, to control for such a bias, we analyze in parallel both the variant and the mutant alleles in an identical cellular background. It is noteworthy that one of the limitation of the minigene system is the size of the exon to be tested; for instance in the case of the PLXNB3 gene, the variant was in an exon over 1 kb in size, which was not retained during the splicing in the minigene system.
We identified one potential pathological missense, T12S (c.34A>T) in the GABRA3 gene encoding the α3 subunit of GABA receptor, which induces a total skipping of the first coding exon of the gene and therefore presumably leads to an absence of the GABRA3 protein. Interestingly, knockout mice for GABRα3 receptor have a sensorimotor gating deficit,33, 34 and in human, GABRA3 expression was found to be significantly reduced in parietal cortex of ASD individuals.35 We also report a splicing mutation M102I (c.306G>C) in the GABRQ gene (not retrieved in Exome Variant Server and dbSNP) that affect the splicing and is predicted to cause a premature truncation in the θ-subunit of GABA receptors in one ASD female. This subunit has been poorly studied, although it is known that it produces a functional receptor when co-expressed with α, β and γ-subunits together.36 The θ-subunit is significantly expressed in anterior cingulate cortex, in hypothalamic regions, in the amygdala and in the hippocampus, regions known to be involved in autism pathology.37 In rat, the α3- and θ-subunits show striking overlapping expression patterns throughout the adult brain, suggesting that these subunits may assemble to form novel GABAA receptors.38 They are also expressed early in the central nervous system, thus suggesting a role during brain development.
It has been proposed that ASD may result from an alteration in the homeostasis between inhibitory (GABAergic) and excitatory synapses (glutamatergic).39, 40, 41 Numerous studies, which have employed a diverse set of approaches including autoradiography, molecular biology, genetics, cytogenetics or knock-out mice models, strongly suggest that the GABAR subunit genes may have an important role in the etiology of ASD.39, 40, 41 A recent study described a mouse model lacking Mecp2 specifically in GABA-releasing neurons that recapitulates numerous autistic features, including repetitive behaviors.42 Given the putative role of the GABAergic system in ASD etiology, the identification, for the first time, of two truncating rare variants in the X-linked GABRQ and GABRA3 genes in ASD probands is a promising result. However, considering the mother/daughter transmission of the GABRQ variant and the minor allele frequency reported for GABRA3 T12S variant (0.16%), a highly penetrant disease causing role for these variants can be ruled out and it may be better to consider these truncating mutations in light of the ‘second/multiple hits' model recently put forward for ASD.43 These mutations in GABA receptor subunits might indeed affect synaptic excitation/inhibition balance and this could, in interaction with rare variants in other genes also involved in the synaptic homeostasis, lead to ASD. In any case, further studies are needed to confirm if GABRA3 and GABRQ genes are involved in the disease susceptibility.
Atypical splicing has already been observed in ASD patients for NLGN3 and NLGN4X, two X-linked genes,44 and for CADPS2 or SHANK3 gene.45, 46 It is therefore of great interest to show that additional splicing variants in synapse-related genes could also have a pathogenic role in ASD.
Another splicing alteration identified in our study is the skipping of an entire exon of the NXF5 gene causing an in-frame loss of 27 amino acids. NXF5 was previously reported to be disrupted in patients with intellectual disability, ASD and other behavioral diseases.47 This gene is involved in mRNA nuclear transportation and is specifically expressed in neurites of hippocampal neurons.48 Although the minigene assay showed an altered splicing effect of the N96S variant, our familial segregation analysis does not support a major role of this variant in SCZ.
We also report one variant in HDAC6 in one ASD male that partially affects splicing in the minigene system. Histone deacetylases (HDACs) are involved in neuronal dendritic growth regulation and regulate transcription factors activity associated to ASD or intellectual disability, such as MECP249 or MEF2C.50, 51 One may speculate that the alternative transcribed protein, missing the nuclear export signal, may have a dominant negative effect. However, we failed to demonstrate any altered splicing in mRNA extracted from the patient lymphoblastoid cells. This may be because of rapid RNA degradation of the alternative form or to another mechanism.
In conclusion, we showed that combining an in silico analysis, using splicing prediction programs, to an in vitro splicing minigene assay is a reasonable approach to identify variants that affect splicing. We report the identification of two truncating mutations in GABA receptor genes in ASD patients, supporting the notion that alterations in GABAergic pathway may have a role in the etiology of ASD.
Acknowledgments
We would like to thank the families involved in our study. We also thank Dr Lan Xiong and Sirui Zhou for providing DNA and information for the SCZ family pedigree, as well as Dr Hussein Daoud, Dr Helene Catoire and Dr Olga Kotsopoulos for their scientific advices.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Piton A, Michaud JL, Peng H, et al. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum Mol Genet. 2008;17:3965–3974. doi: 10.1093/hmg/ddn300. [DOI] [PubMed] [Google Scholar]
- Awadalla P, Gauthier J, Myers RA, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafrenière RG, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH. X-linked genes and mental functioning. Hum Mol Genet. 2005;14 Spec No 1:R27–R32. doi: 10.1093/hmg/ddi112. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Cuthbert PC, Grant SG. The role of neuronal complexes in human X-linked brain diseases. Am J Hum Genet. 2007;80:205–220. doi: 10.1086/511441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse D. Genetic influences on the neural basis of social cognition. Philos Trans R Soc Lond B Biol Sci. 2006;361:2129–2141. doi: 10.1098/rstb.2006.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JM, Seidman LJ, Goodman JM, et al. Are there sex differences in neuropsychological functions among patients with schizophrenia. Am J Psychiatry. 1998;155:1358–1364. doi: 10.1176/ajp.155.10.1358. [DOI] [PubMed] [Google Scholar]
- Fombonne E. Epidemiological trends in rates of autism. Mol Psychiatry. 2002;7 (Suppl 2:S4–S6. doi: 10.1038/sj.mp.4001162. [DOI] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF, et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2010. [DOI] [PMC free article] [PubMed]
- Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2010;14:533–537. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Bonnet C, Krieger S, Vezain M, et al. Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet. 2008;45:438–446. doi: 10.1136/jmg.2007.056895. [DOI] [PubMed] [Google Scholar]
- Thery JC, Krieger S, Gaildrat P, et al. Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. Eur J Hum Genet. 2011;19:1052–1058. doi: 10.1038/ejhg.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng L, Coutinho G, Nahas S, et al. Nonclassical splicing mutations in the coding and noncoding regions of the ATM gene: maximum entropy estimates of splice junction strengths. Hum Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- Collin RW, de Heer AM, Oostrik J, et al. Mid-frequency DFNA8/12 hearing loss caused by a synonymous TECTA mutation that affects an exonic splice enhancer. Eur J Hum Genet. 2008;16:1430–1436. doi: 10.1038/ejhg.2008.110. [DOI] [PubMed] [Google Scholar]
- Fukao T, Boneh A, Aoki Y, Kondo N. A novel single-base substitution (c.1124A>G) that activates a 5-base upstream cryptic splice donor site within exon 11 in the human mitochondrial acetoacetyl-CoA thiolase gene. Mol Genet Metab. 2008;94:417–421. doi: 10.1016/j.ymgme.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Shapiro BA, Nussinov R, Lipkin LE, Maizel JV. An interactive dot matrix system for locating potentially significant features in nucleic acid molecules. J Biomol Struct Dyn. 1987;4:697–706. doi: 10.1080/07391102.1987.10507673. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Burn TC, Connors TD, Klinger KW, Landes GM. Increased exon-trapping efficiency through modifications to the pSPL3 splicing vector. Gene. 1995;161:183–187. doi: 10.1016/0378-1119(95)00223-s. [DOI] [PubMed] [Google Scholar]
- Bertos NR, Gilquin B, Chan GK, Yen TJ, Khochbin S, Yang XJ. Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. J Biol Chem. 2004;279:48246–48254. doi: 10.1074/jbc.M408583200. [DOI] [PubMed] [Google Scholar]
- Vreeswijk MP, Kraan JN, van der Klift HM, et al. Intronic variants in BRCA1 and BRCA2 that affect RNA splicing can be reliably selected by splice-site prediction programs. Hum Mutat. 2009;30:107–114. doi: 10.1002/humu.20811. [DOI] [PubMed] [Google Scholar]
- Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Fiorelli R, Rudolph U, Straub CJ, Feldon J, Yee BK. Affective and cognitive effects of global deletion of alpha3-containing gamma-aminobutyric acid-A receptors. Behav Pharmacol. 2008;19:582–596. doi: 10.1097/FBP.0b013e32830dc0c7. [DOI] [PubMed] [Google Scholar]
- Yee BK, Keist R, von Boehmer L, et al. A schizophrenia-related sensorimotor deficit links alpha 3-containing GABAA receptors to a dopamine hyperfunction. Proc Natl Acad Sci USA. 2005;102:17154–17159. doi: 10.1073/pnas.0508752102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD, Thuras PD. GABA(A) receptor downregulation in brains of subjects with autism. J Autism Dev Disord. 2009;39:223–230. doi: 10.1007/s10803-008-0646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnert TP, McKernan RM, Farrar S, et al. Theta, a novel gamma-aminobutyric acid type A receptor subunit. Proc Natl Acad Sci USA. 1999;96:9891–9896. doi: 10.1073/pnas.96.17.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Pape JR, Bertrand SS, Lafon P, et al. Expression of GABA(A) receptor alpha3-, theta-, and epsilon-subunit mRNAs during rat CNS development and immunolocalization of the epsilon subunit in developing postnatal spinal cord. Neuroscience. 2009;160:85–96. doi: 10.1016/j.neuroscience.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Hussman JP. Suppressed GABAergic inhibition as a common factor in suspected etiologies of autism. J Autism Dev Disord. 2001;31:247–248. doi: 10.1023/a:1010715619091. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Heinrich J, Delorme R, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, Butler MG. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. J Med Genet. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakata T, Washida M, Iwayama Y, et al. Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients. J Clin Invest. 2007;117:931–943. doi: 10.1172/JCI29031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- Grillo L, Reitano S, Belfiore G, et al. Familial 1.1 Mb deletion in chromosome Xq22.1 associated with mental retardation and behavioural disorders in female patients. Eur J Med Genet. 2010;53:113–116. doi: 10.1016/j.ejmg.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Jun L, Frints S, Duhamel H, et al. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr Biol. 2001;11:1381–1391. doi: 10.1016/s0960-9822(01)00419-5. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Le Meur N, Holder-Espinasse M, Jaillard S, et al. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet. 2010;47:22–29. doi: 10.1136/jmg.2009.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier M, Gregor A, Zweier C, et al. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat. 2010;31:722–733. doi: 10.1002/humu.21253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.