Abstract
The synthesis and comparison of activities of ‘tag-free’ probes with diazirines in varying positions is described. Remarkable differences in effects on P. aeruginosa and on human bronchial epithelial cells were observed, supporting efforts to isolate and identify receptors for N-acyl homoserine lactones.
Quorum sensing (QS) is a mechanism that enables organisms to regulate gene expression in response to changes in cell population density.1 The main purpose of QS is to coordinate the behavior of single individuals, in order for them to function as one multicellular organism. In bacteria a variety of physiological processes are regulated by QS. Some established phenotypes are the induction of bioluminescence, virulence factor expression, biofilm formation and motility. Common to all these processes is that biological function is attained only upon simultaneous molecular recognition by a large number of individuals.2
Bacterial QS occurs through secretion and recognition of small diffusible molecules, called autoinducers (AI). Upon reaching a certain threshold concentration these AIs diffuse back into the cell where they bind their respective receptor, setting in motion cascades of gene transcription and protein expression. In Gram negative bacteria the main AIs are N-acyl homoserine lactones (AHLs). Our research focuses on a representative member of the AHL family, N-(3-oxododecanoyl) homoserine lactone (3-oxo-C12-HSL, C12, Fig. 1, 1), the primary QS signal of Pseudomonas aeruginosa. This microorganism is ubiquitous and highly adaptable, and can be found in soil, in water, in the skin fauna, but also in most man-made environments, where it can take advantage of pre-existing weaknesses in the host’s immune system.3 The primary receptor for C12 in P. aeruginosa is the transcriptional activator LasR, which is considered a global regulator for P. aeruginosa virulence genes.4 While QS plays an important role in P. aeruginosa pathogenicity, and contributes to its virulence and resistance to host responses, many molecular details regarding its regulation and timing are still underexplored, including for instance the recognition of C12 by more than one regulator.5
Fig 1.
Structures of 3-oxo-C12-HSL (C12, 1) and diazirine alkynyl probes 2, 3 and 4.
In the past decades several lines of research have revealed an exchange of QS related signals between bacteria and higher organisms. Prokaryotes and eukaryotes have co-existed for millions of years and recent studies suggest that the evolution of intricate mechanisms of interaction that are based on chemical signaling events.1, 2
One example of such an interaction can be found in Candida albicans, a pathogenic yeast. C. albicans is part of the natural microfauna in the human organism. However, in immune compromised individuals it can cause serious infections and recent studies show that Candida pathogenicity is modulated by C12.6 Since P. aeruginosa and C. albicans are often found in the same tissues or organs, it is reasonable to assume that some form of cross talk has evolved between these two organisms.
Kolter and co-workers described an effect of C12 on the morphology of C. albicans, which indicates an adaptive defense mechanism of the fungus.6 Moreover, yeast growth is reduced by bacterial phenazines, such as pyocyanin, produced by the bacteria. In turn, farnesol, the designated QS signal of Candida, leads to a decrease in pyocyanin. Thereby, secreted factors from both species have mutually antagonistic effect on each other’s virulence, and eventually on survival.6 Interaction of bacterial QS signals with mammalian cells to date has mainly focused on C12-mediated effects on inflammatory and innate immune responses. However, the observed effects have been shown to vary, probably depending on the concentration of C12. Multiple studies,7–20 conducted with a wide range of C12 concentrations, showed a variety of pro- and anti-inflammatory responses in several different types of cells, such as fibroblasts,7–11 endothelial8 and epithelial7,9 cells, keratinocytes,12 T cells,13–16 macrophages11 and neutrophils.17 In addition, we recently discovered that C12 amplifies LPS-stimulated IL-10 expression in macrophages.21
While a lot of new information has been uncovered in recent years with regard to the effects of C12 on eukaryotes,22,23 so far no bona fide receptor has been found. Several candidate proteins have been identified using a range of methods, but in none of these studies C12 was shown to actually bind the protein, let alone bind it in a specific manner with high affinity.24–27 One candidate for a eukaryotic receptor for C12 is the peroxisome proliferator activated receptor gamma (PPARγ), a transcription factor that participates in the regulation of fatty acid storage and glucose metabolism in mammalian cells. It has been demonstrated that C12 modulates the transcriptional activity of PPARγ,24–26 potentiating the expression of pro-inflammatory response genes. Thereby, additional proteins distinct from PPARγ are likely to be involved in C12-mediated immunosuppressive responses. In a different study, Vikström and co-workers showed that C12 affects migration of human intestinal epithelial Caco-2 cells.27 They moreover showed an interaction between the IQ-motif-containing GTPase-activating protein IQGAP1 and a C12-biotin probe, as well as other fluorescently-tagged probes. However, no specific binding studies with C12 were performed, and given the small size and hydrophobic nature of AHLs, it is expected that even the smallest modification might substantially affect interaction of C12 with its target protein(s).16,22 Therefore, conventional methods of modifying C12 with a bulky tag or conjugation to an affinity matrix28 may cause a significant loss of affinity and prevent detection by its receptor.
To overcome this challenge, we have recently developed an approach to label and isolate receptors for C12 via a two-step tag free photoaffinity purification procedure.29 This approach is based on a powerful methodological approach termed activity based protein profiling (ABPP), which was developed by the groups of Cravatt and Bogyo.30–32
In our previous study we described the design and synthesis of a diazirine containing C12 mimic with an alkyne as a strategic group for reaction with a fluorescent or biotinylated azide (Fig. 1). The choice of the diazirine as a reactive group was motivated by its small size, effecting minimal structural changes to the C12 mimic (allowing maximal receptor recognition) and relatively mild irradiation conditions.33
Here, we describe the synthesis and evaluation of C12 based probes with the diazirine moiety at different positions along the alkyl chain (Fig. 1). The rationale behind this exploration of the potential impact of the position of the diazirine on biological function is based on our hypothesis that the microenvironment of the bound probe strongly affects the probability to successfully target true receptors among the many weaker binders of small hydrophobic ligands.
Both diazirine alkynyl probes 3 and 4 were synthesized based on procedures that are similar to our published route29 to probe 2 – with some clear distinctions (see ESI for details). The synthesis of probe 3 started from commercially available (5-iodo-1-pentynyl) trimethylsilyl-silane (5), to which lithiated 3,4-dihydro-2H-pyran was added, followed by oxidation with Jones reagent (Scheme 2S, ESI). The alkylated product 6 was then treated with liquid ammonia to afford the diaziridine intermediate, which was oxidized with iodine to afford the desired diazirine 7. Coupling of 7 with Meldrum's acid in the presence of DCC and DMAP, followed by treatment with homoserine lactone yielded diazirine alkynyl probe 3.
Probe 4 was synthesized in a similar fashion (Scheme 3S, ESI). Homologation of THP-protected hydroxycaproic acid 9 with mono tert-butyl malonate resulted in 10, which was reacted with propargyl bromide to afford 11. Deprotection, followed by Jones oxidation and TFA mediated dealkylation yielded the desired 6-oxodec-9-ynoic acid, 14. The final coupling to prepare probe 4 was performed in good yield, following the previously described procedure for probe 3.
In order to analyze binding of the probes to the native Pseudomonas receptor, LasR, we overexpressed the ligand binding domain (LBD) of LasR in E. coli, in the presence of probes 2–4, followed by UV irradiation of the cells (at 365 nm), resulting in a covalent bond between the probes and their receptor. The cells were then lysed, and the LasR-LBD was purified by affinity chromatography, after which the protein was irradiated again for 10 min and directly analysed by LC/MS (Fig. 2). The additional UV irradiation improved the labeling, indicating that part of the diazirine probe undergoes slow activation and that the probes in general have high affinity for LasR, remaining in its binding pocket throughout purification. Cells that were incubated with C12 alone (20 µM) yielded a single LasR-LBD peak (22,427 Da (calc. 22,430 Da), Fig. 3A). An additional peak with a mass difference of 291 Da appeared upon addition of probe 3 (20 µM, ESI, Fig. S1A, corresponding to the expected addition of 291 Da upon reaction with the activated diazirine probe; probe 4 yielded a similar result (+289 Da). In both cases the additional peak was absent when C12 (20 µM) was added to the reaction (ESI, S1A and B), demonstrating the specificity of the probes for the LasR binding pocket, similar to probe 2.29
Fig. 2.
Deconvoluted ESI mass spectra of crude LasR-LBD: (A) overexpressed in E. coli in the presence of C12 (20 µM). Calculated mass: 22,430 Da. Observed: 22,427 Da. (B) overexpressed in the presence of diazirine probe 4 (20 µM). Calculated mass increase upon labeling with 4: 291 Da. Observed increase: 289 Da.
Fig. 3.
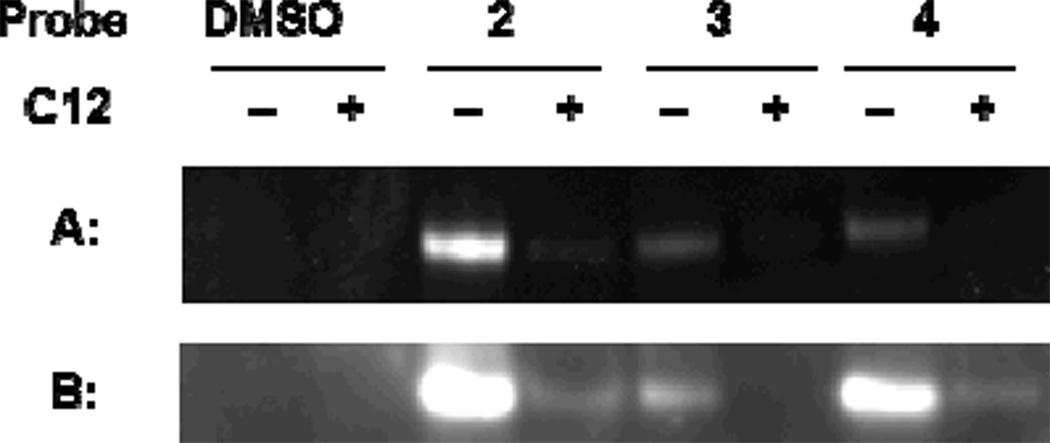
Live cell photolabeling of LasR in using probes 2–4, followed by lysis and CuAAC: A) fluorescent labeling of tagged LasR-LBD by rhodamine azide; (B) Western blot analysis biotinylated proteins, using streptavidin-HRP.
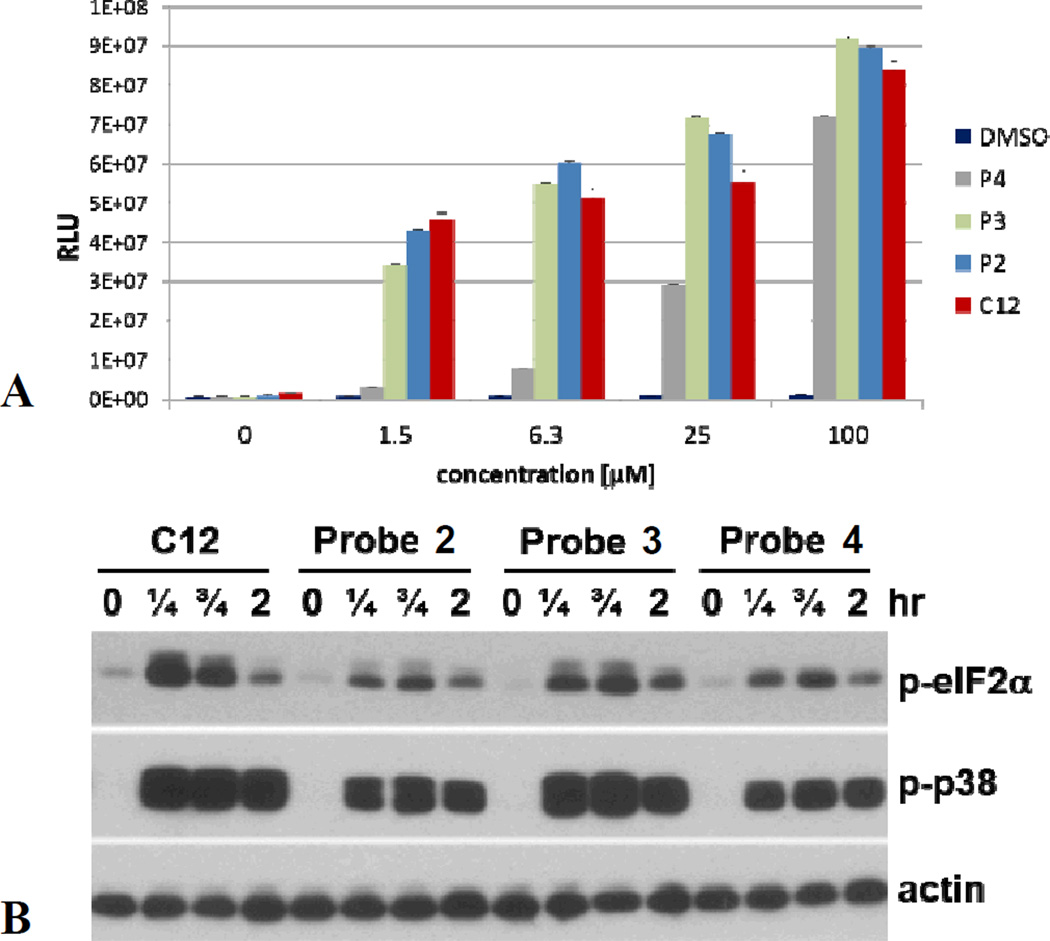
In addition, we performed in vivo labeling experiments with LasR-LBD overexpressed in E. coli, using biotin/rhodamine-azide as tags. Binding of the probes to LasR-LDB was validated using LC/MS. After cell lysis, employing the ligand’s alkyne moiety, a bioorthogonal copper catalyzed azide alkyne cycloaddition (CuAAC) reaction was performed, yielding fluorescent or biotinylated proteins. Analysis of the lysed proteome (Fig. 3), following SDS-PAGE and Western blotting, shows fluorescent bands at the expected molecular mass for LasR-LBD, while for cells that were treated with probes 2–4 in the presence of C12 (2-fold excess), the intensity of the bands was much lower, again demonstrating competition for the same binding site. We also evaluated the ability of probes 2–4 to induce bioluminescence in the QS reporter strain PAO-JP2-luxCDABE (Fig. 4A), a P. aeruginosa strain that has the complete QS machinery, but cannot synthesize C12. We found that at low micromolar concentrations probes 2 and 3 were good mimics of C12, while probe 4 performed surprisingly poorly, attesting to the importance of the position of the diazirine moiety in different species, not only with regard to binding but also in terms of receptor activation.
Fig. 3.
(A) Changes in P. aeruginosa bioluminescence as a function of C12 and probe 2–4 concentration. (B) Biological activity of C12 and its diazirine alkynyl analogs 2–4 in normal human bronchial epithelial cells. Cells were treated with C12 (25 µM) or its derivatives (Probe 2, 3 or 4, 25 µM) as indicated, and samples were analyzed by Western blot for phosphorylated forms of eIF2a and p38, and actin as a loading control.
Next, we analysed stimulus-induced signaling events in human bronchial epithelial cells (Fig. 4B), representing a cell type in lung tissue exposed to P. aeruginosa at sites of infection. The phosphorylation of eukaryotic initiation factor 2α (eIF2α) and protein kinase p38 were used as biochemical markers of the biological activity of AHLs in human and murine cells.22 The results of these experiments revealed that all three probes possess the ability to activate eIF2α and p38 in human bronchial epithelial cells, and, in contrast with the P. aeruginosa assays, probe 3 appeared to be a superior mimic of C12.
Taken together, the functional and binding assays show that while all three probes are good mimics for C12 in both bacterial and mammalian systems, significant differences in target binding and / or function demonstrate that the exact position of the diazirine moiety - even between adjacent positions on along the lipid chain - may be a crucial factor in photoaffinity labeling and informs on the importance of a precise design of such small photoaffinity probes. We are currently using the three probes to identify putative and unknown receptors for C12 in different species.
Supplementary Material
Acknowledgments
This work was supported by the European Research Council (Starting Grant 240356, MMM), the National Institutes of Health (AI094348, VVK), and by the US–Israel Binational Science Foundation (Startup Grant 2006287, MMM). We thank R. J. Ulevitch for valuable insight and support, and M. J. Bottomley and M. G. Surette for bacterial strains and plasmids.
Footnotes
Electronic Supplementary Information (ESI) available: synthetic procedures, spectral characterization and analytical protocols.
References
- 1.Engebrecht J, Nealson K, Silverman M. Cell. 1983;32:773. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 2.Hastings JW, Nealson KH. Annu. Rev. Microbiol. 1977;31:549. doi: 10.1146/annurev.mi.31.100177.003001. [DOI] [PubMed] [Google Scholar]
- 3.Lyczak JB, Cannon CL, Pier GB. Microb. Infect. 2000;2:1051. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 4.Pearson JP, Gray KM, Passador L, Tucker K, Eberhard A, Iglewski BH, Greenberg EP. PNAS. 1994;91:197. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winzer K, Williams P. Int. J. Med. Microbiol. 2001;291:131. doi: 10.1078/1438-4221-00110. [DOI] [PubMed] [Google Scholar]
- 6.Hogan DA, Vik A, Kolter R. Mol. Microbiol. 2004;54:1212. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 7.Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP. J. Bacteriol. 2008;190:4408. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiner EK, Terentyev D, Bryan A, Sennoune S, Martinez-Zaguilan R, Li G, Gyorke S, Williams SC, Rumbaugh KP. Cell. Microbiol. 2006;8:1601. doi: 10.1111/j.1462-5822.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 9.Smith RS, Fedyk ER, Springer TA, Mukaida N, Iglewski BH, Phipps RP. J. Immunol. 2001;167:366. doi: 10.4049/jimmunol.167.1.366. [DOI] [PubMed] [Google Scholar]
- 10.Smith RS, Kelly R, Iglewski BH, Phipps RP. J. Immunol. 2002;169:2636. doi: 10.4049/jimmunol.169.5.2636. [DOI] [PubMed] [Google Scholar]
- 11.Vikstrom E, Magnusson KE, Pivoriunas A. Microbes Infect. 2005;7:1512. doi: 10.1016/j.micinf.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Paes C, Nakagami G, Minematsu T, Nagase T, Huang L, Sari Y, Sanada H. Biochem. Biophys. Res. Commun. 2012;427(2):273. doi: 10.1016/j.bbrc.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 13.Telford G, Wheeler D, Williams P, Tomkins PT, Appleby P, Sewell H, Stewart GS, Bycroft BW, Pritchard DI. Infect. Immun. 1998;66:36. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie AJ, Jansson A, Stallberg J, Nilsson P, Lysaght P, Cooley MA. Infect. Immun. 2005;73:1648. doi: 10.1128/IAI.73.3.1648-1655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie AJ, Yam AO, Tanabe KM, Rice SA, Cooley MA. Infect. Immun. 2003;71:4421. doi: 10.1128/IAI.71.8.4421-4431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chhabra SR, Harty C, Hooi DS, Daykin M, Williams P, Telford G, Pritchard DI, Bycroft BW. J. Med. Chem. 2003;46:97. doi: 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- 17.Tateda K, Ishii Y, Horikawa M, Matsumoto T, Miyairi S, Pechere JC, Standiford TJ, Ishiguro M, Yamaguchi K. Infect. Immun. 2003;71:5785. doi: 10.1128/IAI.71.10.5785-5793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner C, Zimmermann S, Brenner-Weiss G, Hug F, Prior B, Obst U, Hansch GM. Anal. Bioanal. Chem. 2006;387:481. doi: 10.1007/s00216-006-0698-5. [DOI] [PubMed] [Google Scholar]
- 19.Thomas GL, Bohner CM, Williams HE, Walsh CM, Ladlow M, Welch M, Bryant CE, Spring DR. Mol. BioSyst. 2006;2:132. doi: 10.1039/b517248a. [DOI] [PubMed] [Google Scholar]
- 20.Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Grauer DC, Lehmann M, Meijler MM, Janda KD, Ulevitch RJ. Science. 2008;321:259. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]
- 21.Glucksam-Galnoy Y, Sananes R, Silberstein N, Krief P, Kravchenko VV, Meijler MM, Zor T. J. Immunol. 2013 doi: 10.4049/jimmunol.1300368. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Wood MR, Brogan AP, Lehmann M, Mee JM, Iwata K, Pan Q, Fearns C, Knaus UG, Meijler MM, Janda KD, Ulevitch RJ. J. Biol. Chem. 2006;281:28822. doi: 10.1074/jbc.M606613200. [DOI] [PubMed] [Google Scholar]
- 23.Teplitski M, Mathesius U, Rumbaugh KP. Chem. Rev. 2011;111(1):100. doi: 10.1021/cr100045m. [DOI] [PubMed] [Google Scholar]
- 24.Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP. J. Bacteriol. 2008;190:4408. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooley MA, Whittall C, Rolph MS. Microbes and Infect. 2010;12:231. doi: 10.1016/j.micinf.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Griffin PE, Roddam LF, Belessis YC, Strachan R, Beggs S, Jaffe A, Cooley MA. PLOS One. 2012;7(7):e42241. doi: 10.1371/journal.pone.0042241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson T, Turkina MV, Yakymenko O, Magnusson K-E, Vikström E. PLOS Path. 2012;8(10):e1002953. doi: 10.1371/journal.ppat.1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Praneenararat T, Beary TM, Breitbach AS, Blackwell HE. Bioorg. Med. Chem. Lett. 2011;21:5054. doi: 10.1016/j.bmcl.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubinsky L, Jarosz LM, Amara N, Krief P, Kravchenko VV, Krom BP, Meijler MM. Chem. Commun. 2009;47:7378. doi: 10.1039/b917507e. [DOI] [PubMed] [Google Scholar]
- 30.Adam GC, Sorensen EJ, Cravatt BF. Mol. Cell. Proteomics. 2002;1:781. doi: 10.1074/mcp.r200006-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Nat. Chem. Biol. 2005;1:33. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 32.Speers AE, Adam GC, Cravatt BF. J. Am. Chem. Soc. 2003;125:4686. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 33.Dubinsky L, Krom BP, Meijler MM. Bioorg. Med. Chem. 2012;20:554. doi: 10.1016/j.bmc.2011.06.066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.