Abstract
Gold nanorods (AuNRs) were used as spectroscopic sensing elements to detect specific DNA sequences with a single-base mismatch sensitivity. The assay was based on the observation that the stabilizing repulsive forces between CTA+-coated AuNRs can be removed by citrate ions, which causes aggregation among AuNRs; whereas nucleic acids of different structures[ i.e., peptide nucleic acid (PNA), single-stranded DNA (ssDNA), PNA-DNA complex, and double-stranded DNA (dsDNA)] can retard the aggregation. Moreover, the dsDNA PNA-DNA duplexes provide larger retardation than that by unhybridized ssDNA and PNA probe. This assay can differentiate single-base mismatched targets with base substitution at different locations (center and end) with AuNRs of a larger aspect ratio. Besides ultraviolet–visable spectroscopy measurement of particle assembly-induced plasmonic coupling that in turn provides a spectroscopic detection of the specific DNA, dynamic light scattering and transmission electron microscope (TEM) were used to measure smaller degree of aggregation that can reveal sodium citrate– and dsDNA–AuNRs interactions in fine detail.
Introduction
Noble metal nanoparticles (mNPs) have unique optical properties that arise from their ability to support a localized surface plasmon resonance (LSPR) (Murray and Barnes, 2007; Stewart et al., 2008). The LSPR properties can be adjusted by changing inter-particle distance (i.e., altering the dispersion/aggregation status), and this will result in an observable color change of the mNPs solution or a shift in LSPR spectrum (Zhao et al., 2008). This phenomenon has captured great attention and is widely applied in colorimetric detection of biological molecular interactions and/or biological processes that can alter the particles' aggregation and dispersion status.
Attributed to the ease of synthesis and biofunctionalization, spherical mNPs (e.g., gold and silver nanoparticles) have been widely used as colorimetric sensing probes to detect a large variety of analyses (e.g., nucleic acids, proteins, enzymes etc) (Zhao et al., 2008). Compared with the wide application of spherical mNPs, the use of nonspherical nanoparticles with anisotropic configuration has always been underdeveloped. So far, the colorimetric assays employing nonspherical nanoparticles remain limited except the few examples using gold nanorods (AuNRs) (Dujardin et al., 2001; Pan et al., 2005; Sudeep et al., 2005; Huang et al., 2007; Nakashima et al., 2007; Wang et al., 2007; He et al., 2008; Li et al., 2008).
AuNRs exhibit 2 distinct Plasmon resonance bands arising from their anisotropic configuration, transverse bands and longitudinal bands, which correspond to the oscillation of electrons in the shorter and longer axis respectively. These 2 bands are reflected on the plasmon resonance spectrum as 2 distinct peaks. The longitudinal band is extremely sensitive to changes in local environment and interparticle distance (P`erez-Juste et al., 2005) that depends on the aggregation and dispersion status. The principles used in designing AuNR-based aggregation assays are similar to those using gold nanoparticles (AuNPs) as a probe, that is, a cross-linking mechanism facilitated through formation of chemical bonds between receptor-modified-AuNRs (Sudeep et al., 2005; Nakashima et al., 2007; Wang et al., 2007; Li et al., 2008) and a non-crosslinking mechanism that involves the controlled removal of stabilization forces of unmodified AuNRs (Jain et al., 2006; Huang et al., 2007; Kawamura et al., 2008; Sethi et al., 2009). Attributed to the anisotropic configuration of AuNRs, there are 2 manners through which the nanorods aggregate, end-to-end and side-by-side, whereby the rods join at the tips and sides, respectively. End-to-end aggregation is characterized by a significant red shift of longitudinal peak and retainable transverse peak on LSPR spectrum (Pan et al., 2005; Sudeep et al., 2005; Nakashima et al., 2007; Sethi et al., 2009; Varghese et al., 2008; Zhen et al., 2009). Side-by-side aggregation is signified by a red shift of transverse peak and blue shift of longitudinal peak (Dujardin et al., 2001; Pan et al., 2005; Nakashima et al., 2007; Wang et al., 2007). The distinct fashions through which aggregates are formed imply regions on which aforementioned receptor molecules are coated.
AuNRs are synthesized in a solution of hexadecyltrimethylammonium bromide (CTA+Br−: CTAB) (P`erez-Juste et al., 2005). The resulting AuNRs have a positively charged CTA+ bilayer that provides repulsion forces to stabilize the AuNRs. Introduction of negatively charged species can neutralize the surface charge and thus remove the stabilization forces that in turn cause aggregation through hydrophobic–hydrophobic interaction whereby insoluble AuNRs are grouped together by water molecules (Jain et al., 2006; Huang et al., 2007; Kawamura et al., 2008; Sethi et al., 2009).
In a previous work (Kanjanawarut and Su, 2010), we reported that the trivalent sodium citrate ion can largely aggregate AuNRs and double-stranded DNA (dsDNA) or DNA-peptide nucleic acid (DNA-PNA) hybrids can retard sodium citrate-induced AuNRs aggregation better than a single-stranded DNA or PNA probe. Using ultraviolet-visiable (UV-vis) spectroscopy, we detected DNA hybridization based on the spectrum shift. In this current paper, we extended the study designed DNA assay with the following extensions. Firstly, not only UV-vis spectroscopy but also dynamic light scattering (DLS) was used to reveal the fine details of AuNRs aggregation under different sampling conditions (i. e., sodium citrate or dsDNA of low to high concentration). DLS is one of the most popular methods used to determine particle size. Shining a monochromatic light beam, such as a laser, onto a solution with particles in Brownian motion caused a Doppler shift when the light hits the moving particle, changing the wavelength of the incoming light. This change is related to the size of the particle (Urban and Schurtenberger, 1998; Murdock et al., 2008). We have previously proved that with spherical gold nanoparticles, DLS can observe a smaller degree of aggregation by direct measurement of particle size than UV-vis spectroscopy, which measures size change based on plasmon coupling (Murdock et al., 2008). In this study using DLS we have observed a smaller degree of AuNR aggregation as seen under transmission electron microscope image (TEM), but which is not detectable under UV-vis measurement. Secondly, we compared different nucleic acids molecules (dsDNA, single-stranded DNA [ssDNA], PNA-DNA complex, and PNA) in their ability to retard the citrate ion-induced aggregation, and discussed their distinct behavior with the support of i TEM measurement. Thirdly, we evaluated the sensitivity of the assay using AuNRs of different aspect ratios (ARs) and found that AuNRs of a higher aspect ratio are more sensitive than those with lower aspect ratios in response to aggregation forces. Lastly, we have demonstrated that this assay is highly feasible, being able to detect single-base mismatches at different positions (i.e., having a mismatch-position selectivity), which is impossible by the solid-liquid phase method, like surface plasmon resonance spectroscopy. The development of rapid methods for detecting single-nucleotide polymorphisms (SNP) and identifying specific SNP positions in short DNA bases is practical for diagnostic and nucleic acid therapy research (July et al., 2004; Zienolddiny et al., 2012).
Methods and Materials
Reagent
Supernatant of gold nanorods (deionized water with <0.1% ascorbic acid and <0.1% CTAB surfactant capping agent) of 25-nm radius and aspect ratio of 2.5 (62 nm length), 30 (73 nm length), and 3.5 (86 nm length) were purchased from the Nanopartz™ Inc. The trisodium citrate dihydrate (99.9%) was obtained from Aldrich. Ultrapure water (18MΩ, prepared with Millipore Elix 3 purification system) was used as solvent for nucleic acid and sodium citrate. Single-stranded DNA probe (13-mer and 20-mer) and their fully complementary target and 1-base mismatch targets for the 13-mer probe (with the base substitutes at either center or end) were purchased from Research Biolabs Pte Ltd. Thirteen-mer and twenty-mer PNA probe was synthesized by the Eurogentec S.A. Leige. (See Table 1 for sequences of nucleic acids used).To form DNA-DNA and PNA-DNA duplexes, DNA target was annealed with the DNA or PNA probes at a 1:1 molar ratio in phosphate buffered saline buffer (pH 7.2) containing 0.1M NaCl, 0.001M EDTA for 5 minutes at 92°C.
Table 1.
Oligonucleotide Sequences
| Oligonucleotides (denoted name) | Sequences | |
|---|---|---|
| 13-mer PNA (PNA13) | N′-TTCCCCTTCCCAA-C′ | |
| 13-mer ssDNA (ssDNA13) | 5′-TTCCCCTTCCCAA-3′ | |
| 13-mer target DNA | Fully complementary (fc13) | 5′-TTGGGAAGGGGAA-3′ |
| Single-base mismatch at center (m113c) | 5′- TTGGGAGGGGGAA-3′ | |
| Single-base mismatch at end (m113e) | 5′- TTGGGAAGGGGAC-3′ | |
| 20-mer PNA (PNA20) | N′-TTGCACTGTCCTCCTCTTGA-C′ | |
| 20-mer ssDNA (ssDNA20) | 5′-TTGCACTGTACTCCTCTTGA-3′ | |
| 20-mer fully complementary DNA (fc20) | 5′-TCAAGAGGAGTACAGTACAA-3′ | |
PNA, peptide nucleic acid; ssDNA, single-stranded DNA.
Apparatus and characterization
UV-vis absorption spectra were measured using TECAN infinite M200 spectrophotometer (Tecan Trading). Ninety-six-well clear flat bottom UV-transparent microplates (Corning Incorporated) were used as reaction carriers. Zeta potential measurements were carried out using a Zeta Plus zeta potential analyzer (Brookhaven Instruments). Zeta Plus dynamic light scattering particle size analyzer (Brookhaven Instruments) was used to measure the size distribution and effective hydrodynamic diameter of the AuNRs and their aggregates.
Sodium citrate- and dsDNA-induced AuNRs aggregation
For UV-vis spectroscopy measurement of AuNRs aggregation induced by sodium citrate, 150 μL AuNRs in a 96-well plate was mixed with sodium citrate (final concentration of sodium citrate was 0.5–2.5 mM) for 1 minute, and UV-vis spectra was taken from 400 nm to 900 nm wavelengths. The same aggregation experiments were performed for DLS measurement with 2.5 mL of AuNRs solution and the same concentration of sodium citrate.
To study the behavior of AuNRs in dsDNA, 150 μL AuNRs (for UV-vis measurement) and 2.5 mL of AuNRs (for DLS measurement) was mixed with 13-mer dsDNA (final concentration 0.5–3.0 μM) prior to the UV-vis spectrum scan and DLS measurement. For DLS, two sets of measurements consisting of 10 runs each were conducted.
Nucleic acids retarded aggregation and DNA detection
In 150 μL of AuNRs solution, 1.875 μL of 40 μM nucleic acids (i.e., ssDNA, PNA, or preannealed dsDNA and PNA-DNA complex) were added, followed by addition of 15 μL of 10 mM sodium citrate. The final concentration of the nucleic acids and sodium citrate are 0.5 μM and 1.0 mM, respectively. After 1 minute of incubation, UV-vis spectra were scanned from 400 nm to 900 nm wavelengths.
Results and Discussions
UV-vis and DLS measurement of AuNRs aggregation induced by sodium citrate and short dsDNA
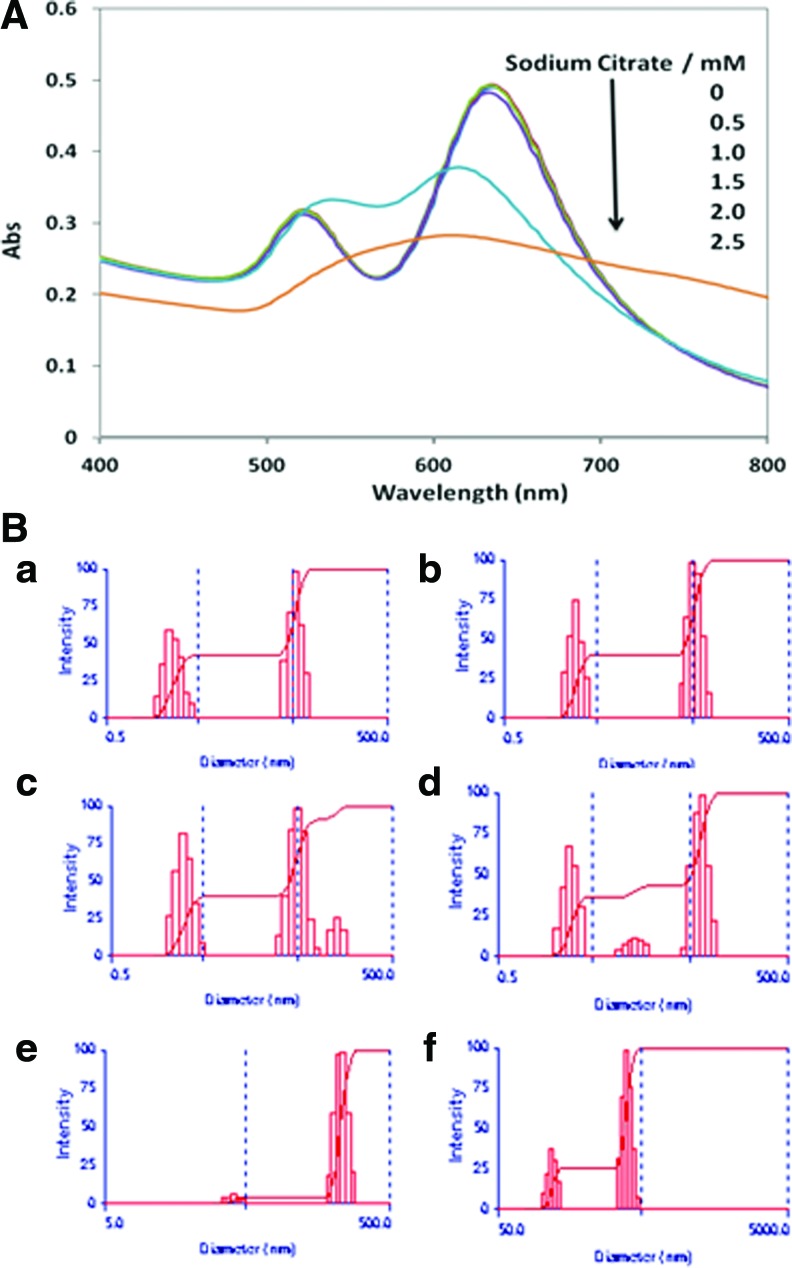
Figure 1A is the UV-vis spectra of AuNRs (AR 2.5) in the presence of sodium citrate of different concentration. A concentration of 2 mM of citrate ion is required to aggregate the AuNRs (437 pM) to the degree that is detectable under UV-vis spectrum. We believe the aggregation is caused by the neutralization of positive charges by the negatively charged citrate ions that remove the stabilizing repulsive forces between AuNRs. Based on the UV-vis adsorption spectrum, we inter that the aggregates formed mainly adopt a side-by-side arrangement as shown by the red shift and the blue shift of the transverse and longitudinal peaks, respectively. The reason for this observation is presumably that the CTA+ coating density (i.e., charge density) is higher along the longitudinal rather than the tip (Sudeep et al., 2005). Therefore, more citrate ions will be attracted electrostatically to the longitudinal, resulting in greater charge neutralization and larger longitudinal (side-by-side) aggregation. In addition, we believe the more preferable formation of side-by-side aggregation rather than end-to-end is a result of greater surface area of contact at the side, which confers stronger van der Waals force between AuNRs.
FIG. 1.
Gold nanorod (AuNR; AR 2.5) aggregation induced by sodium citrate (SC) at concentrations of 0, 0.5, 1.0, 1.5, 2.0, and 2.5 mM, measured using (A) UV-vis spectroscopy and (B) dynamic light scattering. In (B), from (a) to (f), the final concentration of sodium citrate is 0, 0.5, 1.0, 1.5, 2.0, and 2.5 mM. The corresponding effective particle sizes (deff) are 22.2±0.2, 22.4±.2, 24.7±0.5, 24.9±0.4, 196.2±1.7, and 252.4±4.5 nm, respectively. AR, aspect ratio. Color images available online at www.liebertpub.com/nat
The sodium citrate-induced AuNRs aggregation is further evidenced by dynamic light scattering (DLS) characterization that measures the size distribution and effective particle size. It is notable that DLS characterization can measure AuNP aggregation at a smaller degree under a lower concentration of sodium citrate than UV-vis spectrophotometery, which is an indirect measurement of size through particle size/distance-determined plasmon coupling. In DLS size distribution profiles (Fig. 1B), when sodium citrate concentration is as low as 1.0 mM, a small but noticeable population of particles of larger size was observed. On the other hand, only a large degree of aggregation is measured by UV-vis at 2.0 mM sodium citrate, whereby the effective diameter increases significantly to 196.2 nm.
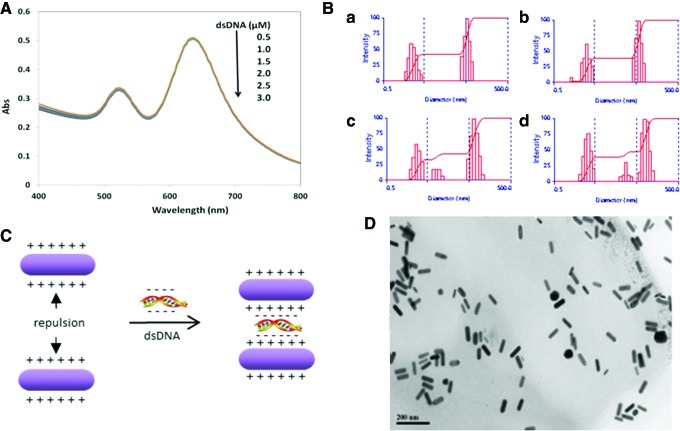
Besides citrate anions, other negatively charged species such as dsDNA should be able to induce aggregation among AuNRs, and this has been proven previously by using large genome DNA (Dujardin, 2001; Pan et al., 2007). For the 13-bp dsDNA oligos, we didn't observe detectable AuNRs aggregation under UV-vis spectroscopy for the tested DNA concentration up to 3 μM (Fig. 2A). However, under DLS, we detected the increase of effective size at 0.5 μM of 13-bp dsDNA (Fig. 2B). Zeta potential measurement confirmed the coating of dsDNA oligo to AuNRs by showing a surface charge decrease from +53.3±1.8 mV to +49.1±1.6 mV. The decreased charge density due to the neutralization is then responsible for the slight aggregation only measured using DLS. The exceptional coagulating effect of dsDNA (as at concentration as low as 0.5 μM) relative to sodium citrate (mM range) can be explained by the presence of two strands of negatively charged phosphate backbones. Electrostatic attraction between the anionic phosphate backbone and positively charged CTA+ coating on one AuNRs can bridge two AuNRs together as illustrated by Fig. 2C. Additional attraction between AuNRs in addition to van der Waals forces when dsDNA is used as coagulant that results in high degree of aggregation even at low concentration. TEM image (Fig. 2D) confirmed the small degree of aggregation (i.e., the appearance of small population of dimer or trimer with side-by-side arrangement).
FIG. 2.
(A) Ultraviolet/visible (UV-vis) spectroscopy and (B) Dynamic light scattering of AuNR solutions (AR 2.5) in the presence of 13-bp double-stranded DNA (dsDNA) at concentrations of (a) 0; (b) 0.5; (c) 2.0; and (d) 3.0 μM. The corresponding deff for each is 21.8±0.6, 25.2±0.4, 28.4±0.7, and 30.3±0.9 nm, respectively. (C) Schematic illustration of coagulation of AuNRs induced by a short oligo dsDNA and the (D) corresponding TEM image of AuNRs (25 nm, AR 3.5) in the presence of 0.5 μM of 13-bp dsDNA. AR, aspect ratio; TEM, transmission electron microscope. Color images available online at www.liebertpub.com/nat
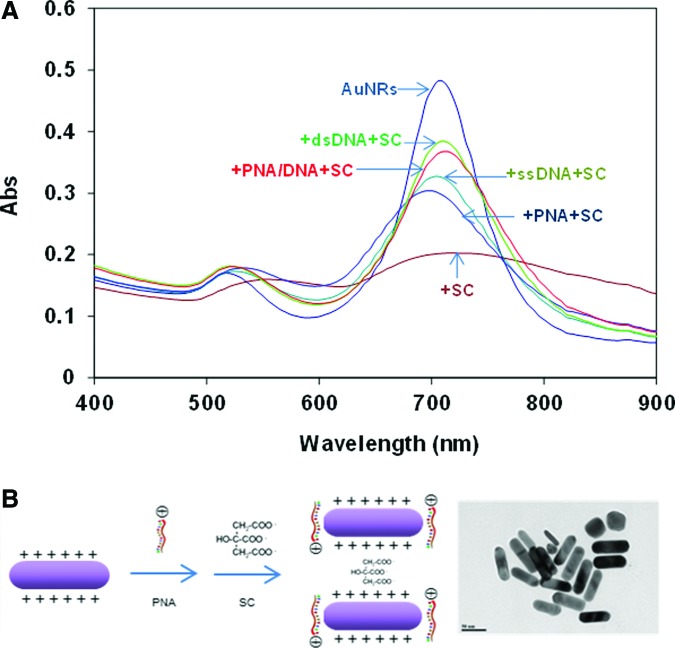
Oligonucleotides can protect AuNRs from citrate-induced aggregation measured as retarded UV-vis spectrum shift
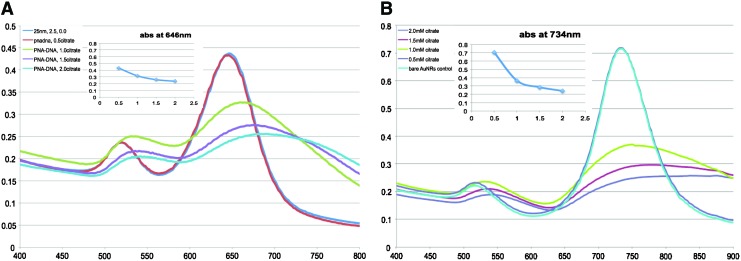
In the previous session we have shown that dsDNA can coat on AuNRs and induce minor degree of nanrods assembly detectable under DLS, but not under UV-vis spectroscopy. In this session we further demonstrate that dsDNA and nucleic acids of other forms (i.e., ssDNA, PNA probe, and PNA-DNA hybrids) have apparent protection of AuNRs from sodium citrate-induced aggregation under UV-vis measurement. Furthermore, the protection effect is of different degree depending on the molecular form (Fig. 3A). For the dsDNA, ssDNA, and PNA-DNA complex, they contain phosphate backbones that are overall negatively charged. They can thus coat onto AuNRs by electrostatic attraction between CAT+ and negatively charged phosphate groups of the nucleic acids and then create locally negatively charged regions on the rods. This provides repulsion against citrate ions (negatively charged) and retards the neutralization of positive charges on AuNRs surfaces by citrate ions. Hence, the aggregation is slowed down. The degree of protection with various oligonucleotides can be explained by the amount of electrostatic charges carried. Given that the number of bases is constant, assuming that the ssDNA molecule carries 1 whole unit of negative charge, the corresponding PNA molecule therefore carries less than 1 unit of charge. Hence, the amount of charges carried by oligonucleotides of the same base number is: PNA (TM−)<ssDNA (−1 unit)<PNA-DNA complex (TM− and −1 unit)<dsDNA(−2 units due to double helix structure). The repulsion against citrate ions increases across the series, which connotes greater protection.
FIG. 3.
(A) Differential aggregation of AuNRs (AR 3.0) induced by sodium citrate (0.5 mM) in the presence of 20-mer PNA, PNA-DNA complex, dsDNA, and single-stranded (ss) DNA. Oligonucleotide concentration is 1 μM. Sodium citrate was added 1 minute after oligonucleotides were mixed with AuNRs. The mixture was scanned after 1 minute of incubation. (B) Schematic illustration of formation of side-by-side aggregate of PNA-coated AuNRs and corresponding TEM image. Color images available online at www.liebertpub.com/nat
Since these negatively charged nucleic acids are mainly attracted electrostatically toward the longitudinal axis where the CTA+ density is higher, citrate ions will likely experience greater charge repulsion if they approach the longitudinal, leading to them coating mainly on the tips (transverse) where there is less ssDNA, dsDNA or PNA-DNA protection. Hence, the aggregate will be built up by end-to-end AuNRs aggregation. This hypothesis can be confirmed by the slight red shift of the longitudinal peak of ∼5 nm for dsDNA and PNA-DNA cases. In the case of PNA molecules, the fact that slightly positively charged PNA molecules (a lysine residue at one end) are also able to retard the aggregation (to a small extent) due presumably to the repulsion between the electronegative oxygen and nitrogen atoms, which bear TM– charges, and the negatively charged citrate ions. Since PNA are slightly positively charged, it would approach AuNRs at the tips (less CTA+ density), resulting in side-by-side AuNRs aggregation when citrate ions are added (Fig. 3B).
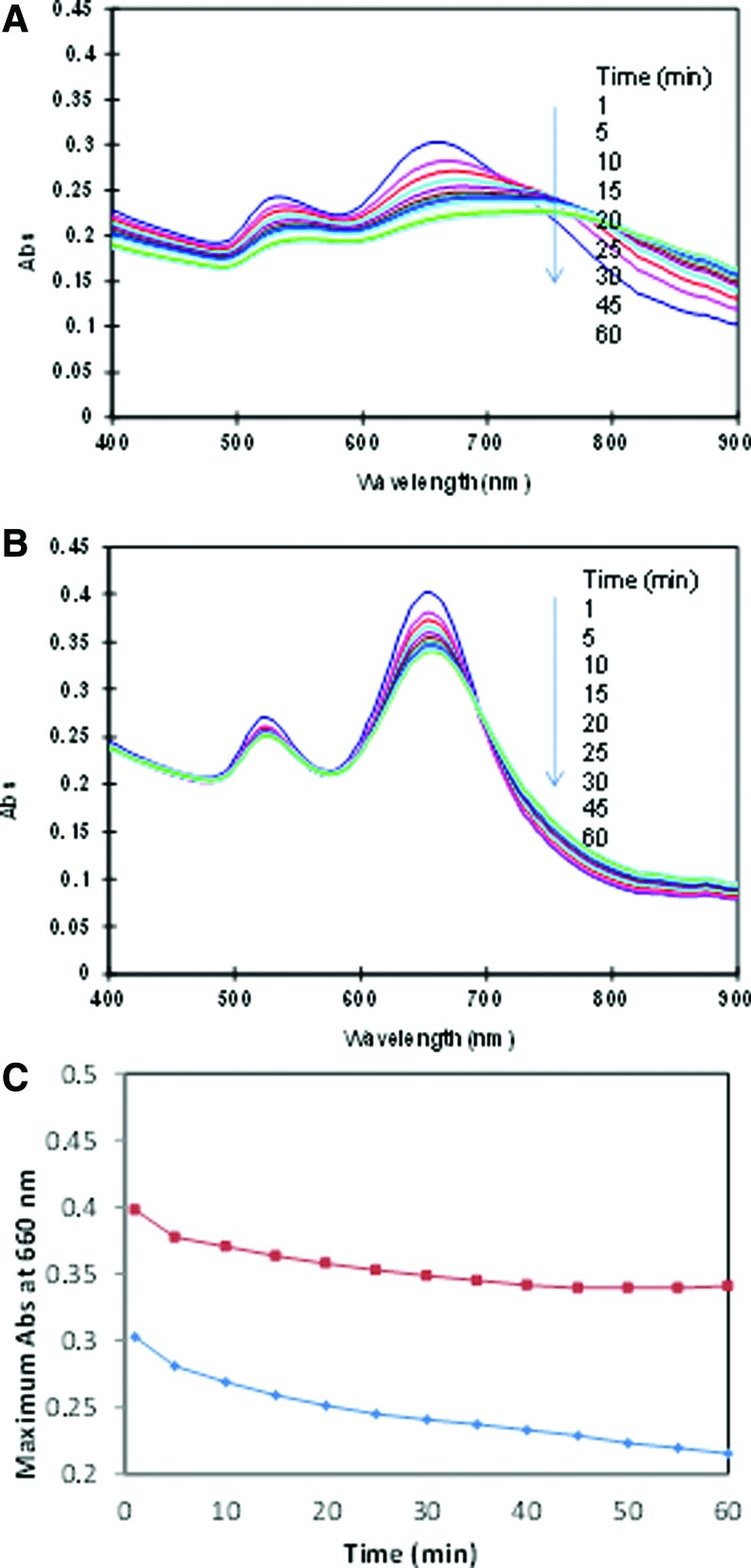
Retarted aggregation by ssDNA and dsDNA over time
In previous session, we scanned UV-vis spectrum upon addition of citrate ions to show the aggregation and retarded AuNRs aggregation by DNA. To better understand the aggregation kinetics, we perfromed UV-vis scan over 60 minutes, for cases where dsDNA and ssDNA are present (Fig. 4). It is interesting to see that the aggregation become more and more over time, for both the cases. The summary of aggregation degree measured using A600 (Fig. 4C) shows that the difference between dsDNA and ssDNA cases increase with prolonged assay time, indicating that detection of DNA hybridization or formation of dsDNA, prolonged assay time may introduce higher sensitivtiy.
FIG. 4.
Time-dependent AuNRs (AR 2.5) aggregation induced by sodium citrate (0.5 mM) in the presence of (A) ssDNA and (B) dsDNA. DNA concentration is 0.5μM. (C) A summary of aggregation degree (measured as A600 drop) over 60 minutes. Color images available online at www.liebertpub.com/nat
Since this assay detects DNA hybridization based on the distinct structural properties between dsDNA (rigid and heavy negative charges) and ssDNA (flexible and coiled structure), it would work for longer sequences (>20-mer). This is because the longer the sequence, the more significant is of the structural difference between double helix and ssDNA (longer ssDNA is more flexible and adopts a largely coiled structure) (Zhang et al., 2001). Saying so, we must acknowledge that for DNA of longer sequences the assay conditions (SC concentration and DNA-AuNRs incubation time, etc.) would be re–determined.
Aspect ratio dependence of AuNRs aggregation
The maximum absorption of the longitudinal plasmon resonance peak of AuNRs shifts to a longer wavelength when aspect ratio is increased (Tao et al., 2008). Previous study has shown that AuNRs of larger AR ratio are more responsive to aggregation factors. We investigated this phenomenon by using 25 nm AuNRs of 2.5 and 3.5 AR. Figure 5 shows the UV-vis spectra of these AuNRs containing PNA-DNA complex (20 bp) with sodium citrate of 0.5, .0, 1.5, and 2.0 mM. Results show that for the same diameter, AuNRs of smaller aspect ratio (shorter length) are more stable than those of larger aspect ratio (longer length). This is because the smaller aspect ratio has a shorter longitudinal axis so that less citrate ion is required in the presence of PNA-DNA complex-coated AuNRs. However, the trend of stability of different aspect ratios is the converse of sensitivity effect; the sensitivity increases with the increase in aspect ratio of the nanorods, as previously reported (Perez et al., 2004; Hu et al., 2006). This result guided us to use AuNRs of 3.5 AR to detect DNA hybridization and especially a single-base mismatch at different location.
FIG. 5.
Differential aggregation of AuNRs containing 20-mer PNA-DNA complex induced by various concentrations of sodium citrate in different aspect ratios of AuNRs (A) AR 2.5 (25 nm and 62 nm length) and (B) AR 3.5 (25 nm and 87 nm length). Color images available online at www.liebertpub.com/nat
Detecting specific locations of mismatch substitution in single-base mismatch target
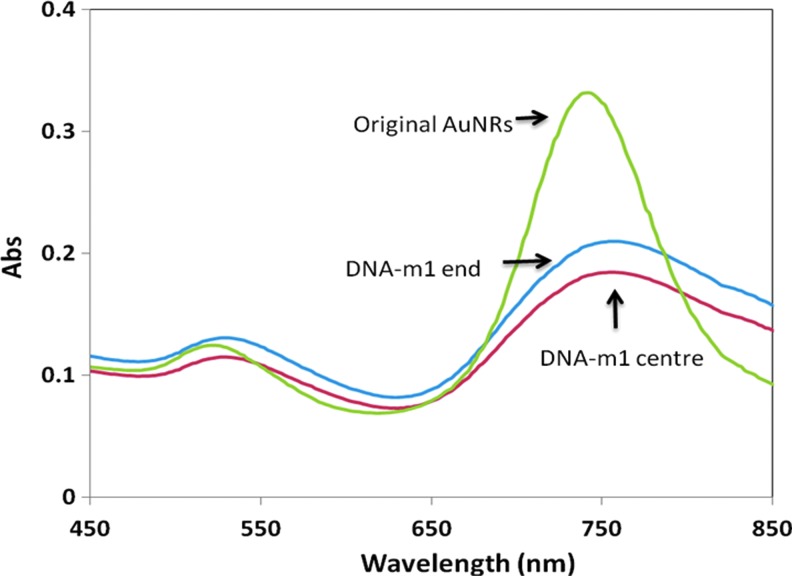
Mismatch targets having the substituted base at different locations (center vs. terminus) can be differentiated using the assay developed. After hybridizing 13-mer DNA probe with its mismatch target, we found that the probe hybridized with a single base mismatch (m1) target with its mismatch at the center was less effective in protecting AuNRs from aggregation (Fig. 6) than target with the mismatch at the end. This result concurs with that obtained by others using oligonucleotide microarrays and neural network analyses (Hidetoshi et al., 2002), in which a mismatch at the center was found to have greater destabilizing effect on dsDNA than when it is at the end. Hence, equilibrium between DNA probe and m1 target lies more to the dissociated side when the mismatch is at the center. In other words, for the center mismatch target, there will be a smaller amount of dsDNA in the solution to exert the protection effect, causing the particles aggregate more than in the case with the mismatch at the end.
FIG. 6.
UV-vis spectra of AuNRs (aspect ratio 3.5 and axial diameter 25 nm) with 13-mer DNA probe being hybridized with its 1-base mismatch targets having a A→G substitution at the center or an A→C substitution at end, followed by the addition of citrate (1.0 mM). The mixture was incubated for 1 minute before scanning the UV-vis spectrum. DNA concentration is 0.5 μM. Color images available online at www.liebertpub.com/nat
Conclusion
We have demonstrated that negatively charged species (citrate ions and dsDNA oligos) can induce aggregation among AuNRs, and DLS can observe a much smaller degree of AuNR aggregation than UV-vis spectroscopy. We have shown that oligonucleotides of different geometries (PNA, ssDNA, PNA-DNA complex, and dsDNA) and base number (13-mer, 20-mer) can adsorb onto CTA+-coated AuNRs and protect AuNRs from citrate-induced aggregation. Furthermore, the dsDNA and PNA-DNA complex have more significant effect than their respective probes (i.e., ssDNA and PNA). By employing this principle, we have developed UV-vis spectroscopy based assay using AuNRs as a probe for detecting nucleic acid hybridization DNA. This assay confers high efficiency with no tedious on-particle hybridization and can produce colorimetric response quickly. Moreover, this assay also demonstrates high sensitivity and selectivity towards single-base mismatch at different location.
Acknowledgments
The authors would like to acknowledge the Agency for Science, Technology, and Research (A*STAR), Singapore for the financial support under grant no. CCOG01_005_2008 and JCOAG03_FG02_2009.
Author Disclosure Statement
No competing financial interests exist.
References
- Dujardin E. Hsin L-B. Wang C.R.C. Mann S. DNA-driven self assembly of gold nanorods. Chem. Commun. 2001:1264–1265. [Google Scholar]
- He W. Huang Z. C. Li Y. F. Xie J. P. Yang R. G. Zhou P. F. Wang J. One-step label-free optical genosensing system for sequence-specific DNA related to the human immunodeficiency virus based on the measurements of light scattering signals of gold nanorods. Anal. Chem. 2008;80:8424–8430. doi: 10.1021/ac801005d. [DOI] [PubMed] [Google Scholar]
- Hidetoshi U. Peter A. N. Said E. F. John J. K. David A. S. Single-base-pair discrimination of terminal mismatches by using oligonucleotide microarrays and neural network analyses. Appl. Environ. Microbiol. 2002;68:235–244. doi: 10.1128/AEM.68.1.235-244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M. Chen J. Li Z. Au L. Hartland V. G. V. Li X. Marquez M. Xia Y. John J. K. Gold nanostructures: engineering their plasmonic properties for biomedical applications. Chem. Soc. Rev. 2006;35:1084–1094. doi: 10.1039/b517615h. [DOI] [PubMed] [Google Scholar]
- Huang Y. F. Lin Y. W. Chang H. T. Control of the surface charges of Au-Ag nanorods: selective detection of iron in the presence of poly(sodium 4-stryenesulfonate) Langmuir. 2007;23:12777–12781. doi: 10.1021/la701668e. [DOI] [PubMed] [Google Scholar]
- Jain P. K. Eustis S. Ei-Sayed M. A. Plasmon coupling in nanorod assemblies: Optical absorption, discrete dipole approximation simulation, and exciton-coupling model. J. Phys. Chem. B. 2006;110:18243–18253. doi: 10.1021/jp063879z. [DOI] [PubMed] [Google Scholar]
- July L. V. Beraldi E. So A. Fazli L. Evans K. English J. C. Gleave M. E. Nucleotide-based therapies targeting clusterin chemosensitize human lung adenocarcinoma cells both in vitro and in vivo. Mol. Cancer Ther. 2004;3:223–232. [PubMed] [Google Scholar]
- Kanjanawarut R. Su X. D. Study of nucleic acid-gold nanorod interactions and detecting nucleic acid hybridization using gold nanorod solutions in the presence of sodium citrate. Biointerphases. 2010;5:1–7. doi: 10.1116/1.3496962. [DOI] [PubMed] [Google Scholar]
- Kawamura G. Yang Y. Nogami M. End-to-end assembly of CTAB-stabilized gold nanorods by citrate anions. J. Phys. Chem. C. 2008;112:10632–10636. [Google Scholar]
- Li X. Qian J. He S. Impact of the self-assembly of multilayer polyelectrolyte functionalized gold nanorods and its application to biosensing. Nanotechnology. 2008;19:355501–355508. doi: 10.1088/0957-4484/19/35/355501. [DOI] [PubMed] [Google Scholar]
- Murdock R. C. Braydich-Stolle L. Schrand A. M. Schlager J. J. Hussain S. M. Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol Sci. 2008;101:239–253. doi: 10.1093/toxsci/kfm240. [DOI] [PubMed] [Google Scholar]
- Murray W.A. Barnes W.L. Plasmonic materials. Adv. Mater. 2007;19:3771–3782. [Google Scholar]
- Nakashima H. Kashimura K. Torimitsu K. Anisotropic assrmbly of gold nanorods assisted by selective ion recognition of surface-anchored crown ether derivatives. Chem. Commun. 2007:1080–1082. doi: 10.1039/b609162h. [DOI] [PubMed] [Google Scholar]
- P`erez-Juste J. Pastoriza-Santos I. Liz-Marz`an L. M. Mulvaney P. Gold nanorods: Synthesis, characterization and applications. Coord. Chem. Rev. 2005;249:1870–1901. [Google Scholar]
- Pan B. Cui D. Ozkan C. Xu P. Huang T. Li Q. Chen H. Liu F. Gao F. He R. DNA-templated ordered array of gold nanorods in one and two dimensions. J. Phy. Chem. C. 2007;111:12572–12576. [Google Scholar]
- Pan B.F. Ao L.M. Gao F. Tian H.Y. Cui R. He R. Daxiang C. End-to-end self-assembly and colorimetric characterization of gold nanorods and nanospheres via oligonucleotide hybridization. Nanotechnology. 2005;16:1776–1780. [Google Scholar]
- Perez J. Liz L. M. Carnie S. Chan D. Y. C. Mulvaney P. Electric-Field-Directed Growth of Gold Nanorods in Aqueous Surfactant Solutions. Adv. Funct. Mater. 2004;14:571–579. [Google Scholar]
- Sethi M. Joung G. Knecht M. R. Stability and electrostatic assembly of au nanorods for use in biological assays. Langmuir. 2009;25:317–325. doi: 10.1021/la802096v. [DOI] [PubMed] [Google Scholar]
- Stewart M.E. Anderton C. R. Thompson L.B. Maria J. Gray S.K. Rogers J. A. Nuzzo R.G. Nanostructured plasmonic sensors. Chem. Rev. 2008;108:494–521. doi: 10.1021/cr068126n. [DOI] [PubMed] [Google Scholar]
- Sudeep P.K. Joseph S.T. Thomas G.K. Selective detection of cysteine and glutathione using gold nanorods. J. Am. Chem. Soc. 2005;127:6516–6517. doi: 10.1021/ja051145e. [DOI] [PubMed] [Google Scholar]
- Tao J. Lu Y. Zheng R. Lin K. Xie Z. Luo Z. Li S. Wang P. Ming H. Effect of Aspect Ratio Distribution on Localized Surface Plasmon Resonance Extinction Spectrum of Gold Nanorods. Chin. Phys. Lett. 2008;25:4459. [Google Scholar]
- Urban C. Schurtenberger P. Characterization of Turbid colloidal Suspensions Using Light Scattering Techniques Combined with Cross-Correlation Methods. Journal of colloid and interface science. 1998;207(1):150–158. doi: 10.1006/jcis.1998.5769. [DOI] [PubMed] [Google Scholar]
- Varghese N. Vivekchand S.R.C. Rao C.N.R. A calorimetric investigation of the assembly of gold nanorods to form necklaces. Chem. Phys. Lett. 2008;450:340–344. [Google Scholar]
- Wang C. Chen Y. Wang T. Ma Z. Su Z. Biorecognition-driven self-assembly of gold nanorods: a rapid and sensitive approach toward antibody sensing. Chem. Mater. 2007;19:5809–5811. [Google Scholar]
- Zhang Y. Zhou H. Ou-Yang ZC. Stretching single-stranded DNA: interplay of elestatic, base-pairing, and base-pair stacking interactions. Biophys. J. 2001;81:1133–1143. doi: 10.1016/S0006-3495(01)75770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. A. Brook M. A. Li Y. Design of gold nanoparticle-based colorimetric biosensing assays. ChemBioChem. 2008;9:2363–2371. doi: 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- Zhen S. J. Huang C. Z. Wang J. Li F. End-to End Assembly of Gold Nanorods on the Basis of Aptamer-Protein Recognition. J. Phys. Chem. C. 2009;113:21543–21547. [Google Scholar]
- Zienolddiny S. Skaug V. Single nucleotide polymorphisms as susceptibility, prognostic, and therapeutic markers of nonsmall cell lung cancer. Lung Cancer: Targets Ther. 2012;3:1–14. doi: 10.2147/LCTT.S13256. [DOI] [PMC free article] [PubMed] [Google Scholar]