Abstract
The major active retinoid, all-trans retinoic acid, has long been recognized as critical for the development of several organs, including the eye. Mutations in STRA6, the gene encoding the cellular receptor for vitamin A, in patients with Matthew–Wood syndrome and anophthalmia/microphthalmia (A/M), have previously demonstrated the importance of retinol metabolism in human eye disease. We used homozygosity mapping combined with next-generation sequencing to interrogate patients with anophthalmia and microphthalmia for new causative genes. We used whole-exome and whole-genome sequencing to study a family with two affected brothers with bilateral A/M and a simplex case with bilateral anophthalmia and hypoplasia of the optic nerve and optic chiasm. Analysis of novel sequence variants revealed homozygosity for two nonsense mutations in ALDH1A3, c.568A>G, predicting p.Lys190*, in the familial cases, and c.1165A>T, predicting p.Lys389*, in the simplex case. Both mutations predict nonsense-mediated decay and complete loss of function. We performed antisense morpholino (MO) studies in Danio rerio to characterize the developmental effects of loss of Aldh1a3 function. MO-injected larvae showed a significant reduction in eye size, and aberrant axonal projections to the tectum were noted. We conclude that ALDH1A3 loss of function causes anophthalmia and aberrant eye development in humans and in animal model systems.
INTRODUCTION
Retinoic acid (RA) metabolism is vital for normal morphogenesis (reviewed in 1,2). In mammals, retinol is bound to Retinol-binding protein 4 (RBP4) and enters the cell by binding to Stimulated by Retinoic Acid Gene 6 homologue (STRA6) transmembrane protein (3). After entry into the cytoplasm, retinol is oxidized to retinaldehyde by alcohol dehydrogenases and then converted to RA by three retinaldehyde dehydrogenases—Aldehyde dehydrogenase 1 family, member A1 (ALDH1A1), Aldehyde dehydrogenase 1 family, member A2 (ALDH1A2) and Aldehyde dehydrogenase 1 family, member A3 (ALDH1A3) (1,2,4). RA can then bind to two classes of nuclear receptors—the RA receptors (RARs) and the retinoid X receptors (RXRs), to effect downstream gene signaling. RA binds as all-trans RA (atRA) and 9cis-RA to the RAR α, β and γ receptors and as 9cis-RA to the RXR α, β and γ receptors (5).
Compelling evidence that RA signaling is critical for eye development has been gained from perturbations of most of the steps of retinol metabolism outlined earlier (2). Deficiencies of vitamin A have been associated with coloboma in humans and mice (6,7). Mutations in STRA6 have been demonstrated in patients with isolated anophthalmia (8,9) and in patients with Matthew–Wood syndrome (MIM 601186), also known as Pulmonary hypoplasia-Diaphragmatic hernia-Anophthalmia-Cardiac defects (PDAC) syndrome (10–12), and in patients with intermediate phenotypes (13). Mice with deficiencies of RARs and RXRs studied by crossing double-homozygous null mutants show variable ocular defects similar to those observed with vitamin A or RA deficiency, including coloboma, absence of the optic nerve, shortening of the ventral retina and malformation of the anterior segment of the eye (5).
Anophthalmia (absent eyes), microphthalmia (small eyes) and coloboma (defective closure of the optic fissure) are birth defects that can be devastating because of the resultant lack of vision. Currently, <50% of patients with anophthalmia/microphthalmia (A/M) and coloboma receive a molecular genetic diagnosis after available investigations to determine the cause of their birth defect. The major causative gene to date, SOX2, is mutated in 10–20% of patients (14,15), and the remaining known pathogenic genes are mutated in only a small number of individuals (16–20). Chromosome aberrations are identified in as many as 25–30% of individuals with eye malformations (21,22), but are less commonly observed in A/M. As part of ongoing research to determine new causative genes in patients with A/M, we performed whole-exome and whole-genome sequencing in three patients with bilateral A/M, two from the same family and one isolated case. Our results implicate ALDH1A3 in the pathogenesis of the eye malformations, with further implications for the role of RA signaling in human eye development.
RESULTS
Case reports
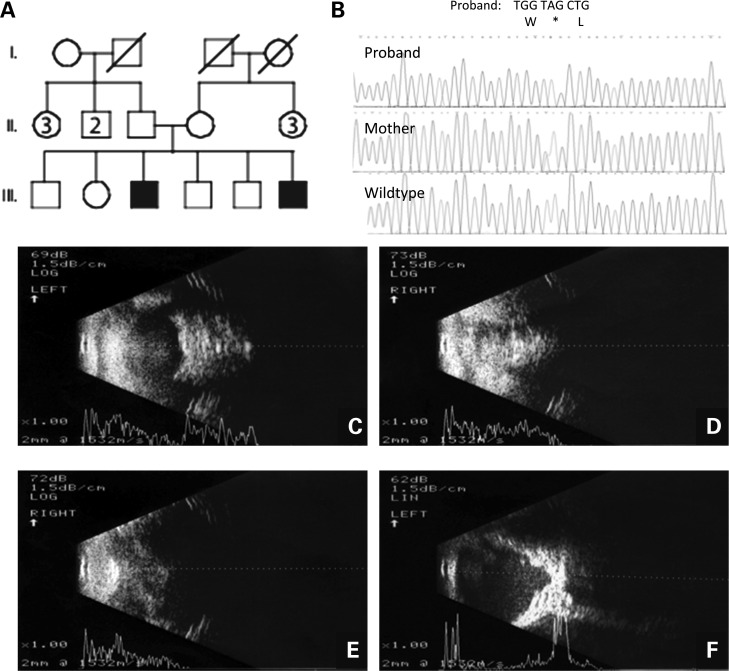
Patients were diagnosed with A/M according to published clinical criteria (17). We report clinical features of three children with A/M, comprising two brothers (for pedigree, see Fig. 1A) from an Egyptian cohort of anophthalmia patients and one simplex case ascertained from the International Children's Anophthalmia Network (http://www.anophthalmia.org/general_information.shtml). The first affected boy, III-3, was an 11-year old male who presented with bilateral anophthalmia. His examination showed absent eyes with whitened conjunctivae, short palpebral fissures and bilateral entropion. Transpalpebral ultrasonography confirmed total anophthalmia of both eyes and no cysts were observed (Figs 1C and D). He had normal development and attended a regular school. His affected younger brother, III-6, was 1 year of age and had right-sided anophthalmia and severe microphthalmia of the left eye. Physical examination showed an absent right eye, very small and barely visible left eye and short palpebral fissures with bilateral entropion similar to his brother. Transpalpebral ultrasonography showed total absence of the right eye (Fig. 1E) and a very small globe with posterior coloboma, detached retina and the presence of the optic nerve in the left orbit (Fig. 1F). Both boys were born full term after uneventful pregnancies and uncomplicated deliveries and neither had additional phenotypic abnormalities or health problems. Their parents and grandparents were unaffected and the brothers had four siblings who were normal on complete eye examination, but the parents refused venipuncture for the unaffected children. Ethnicity was Egyptian and although the family had no known history of consanguinity, both parents originated from a rural region southwest of Cairo.
Figure 1.
Pedigree, chromatograms and transpalpebral ultrasonography from family with ALDH1A3 loss-of-function mutation p.Lys190*. (A) Pedigree of family with two sons affected by A/M. The oldest, III-3, has bilateral anophthalmia and the youngest, III-6, has anophthalmia of the right eye and severe microphthalmia of the left eye. (B) The chromatogram of the oldest son showing c.568A>T, predicting p.Lys190* and premature protein truncation of ALDH1A3. (C) Transpalpebral ultrasonography of the right eye of III-3 showed total anophthalmia without cyst. (D) Transpalpebral ultrasonography of the left eye of III-3 showed total anophthalmia without cyst. (E) Transpalpebral ultrasonography of the right eye of III-6 showed total absence of the eye. (F) Transpalpebral ultrasonography of the left eye of III-6 showed a very small globe with posterior coloboma, detached retina and the presence of the optic nerve in the left orbit.
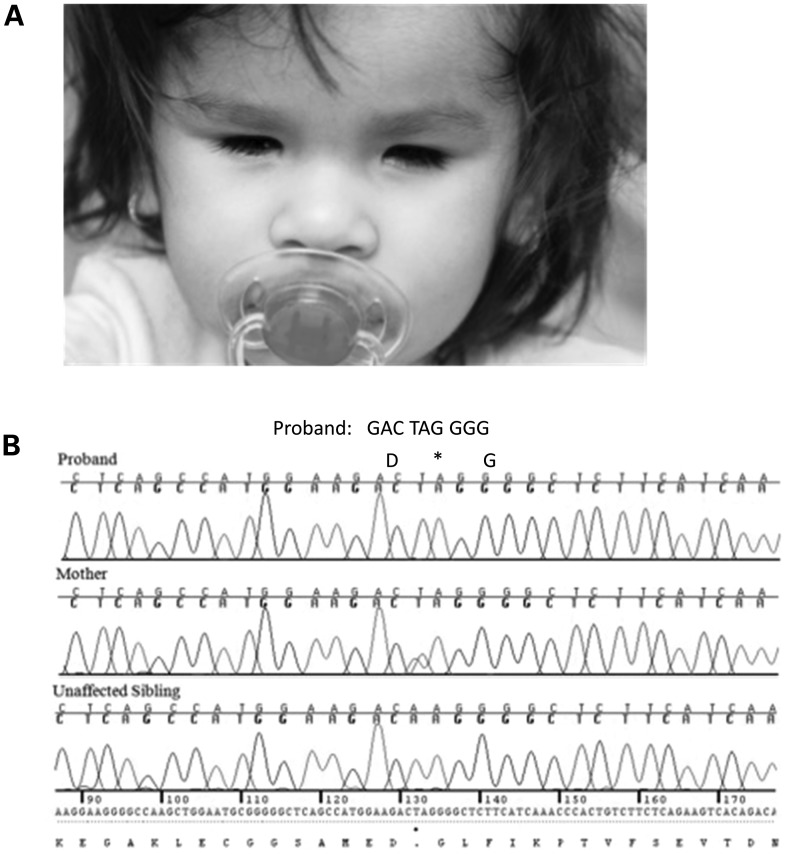
The third patient was a 4-year-and-6-month-old female of Hispanic ethnicity (Fig. 2A). She was born by vertex vaginal delivery at full term with a birthweight of 3030 g (25–50th centile). At birth, she was noted to have anophthalmia that resulted in the placement of conformers for orbital expansion. She passed her newborn hearing screen. A cranial magnetic resonance imaging scan of the head and orbits at 6 months of age identified bilateral agenesis of the ocular globes with small optic nerves and a small optic chiasm, but was otherwise unremarkable. TORCH titers were normal. A real-time ultrasound scan of the abdomen was reported as normal, although the kidneys showed slight echogenicity bilaterally. At 5 months, her weight was 7.14 kg (50–75th centile), height 63.75 cm (50–75th centile) and head circumference 40 cm (10th centile). Her development and general health were normal and there were no significant examination findings apart from the eye abnormalities and hyperkeratosis of the skin. She had a full brother and a paternal half-brother, who were both healthy and there was no history of ocular abnormalities in other family members. Her parents were first cousins. Previous genetic testing included a bacterial artificial chromosome microarray, clinical testing for SOX2 mutations and research testing for OTX2, BMP4, CHX10, PITX2 and FOXE3 mutations (14–19), all of which were unrevealing.
Figure 2.
Chromatograms and the phenotype from a simplex case with ALDH1A3 loss-of-function mutation p.Lys389*. (A) Facial view of the patient (simplex case) at 4 years and 6 months of age, showing bilateral anophthalmia. (B) The chromatogram of the patient (simplex case) showing c.1165A>T, predicting p.Lys389* and premature protein truncation of ALDH1A3. Both parents are heterozygous for the mutation, and an unaffected sibling was wild-type.
Exome sequencing
Exome sequencing was performed in the two brothers (Supplementary Material, Table S1). A total of 190 973 and 181 233 sequence variants were identified in the older and younger boy, respectively (Supplementary Table S1). Examination of homozygous, coding, non-synonymous or frameshift mutations in homozygous regions shared by the affected brothers that were not present in their parents showed only one common mutation that was validated by Sanger sequencing: c.568A>G, predicting p.Lys190*, a nonsense mutation in ALDH1A3 (Fig. 1B; transcript NM_000693.2). This gene was located in the longest, 4.5 Mb region of homozygosity on chromosome 15 (Supplementary Table S2). The p.Lys190* mutation was not observed on Sanger sequencing of 92 chromosomes from healthy individuals stated to be of Egyptian origin and 384 chromosomes from healthy individuals stated to be of European origin (data not shown).
In the simplex case, genotyping showed several large regions of loss of heterozygosity (assumed to be regions of autozygosity; data not shown) and we analyzed novel sequence variants detected by whole-genome sequencing in these regions. Coverage was excellent (average 50–60× across the genome; data not shown) and sequence variants were filtered out using the following scheme:
variants in regions of homozygosity—1038 novel sequence variants;
filter for and remove synonymous variants—497 novel sequence variants remaining;
filter for homozygous sequence variants without known minor allele frequency and not present in the Database of Single Nucleotide Polymorphisms (dbSNP; http://www.ncbi.nlm.nih.gov/projects/SNP/)—45 novel sequence variants remaining (Supplementary Table S3). Of these 45 variants, there was only one nonsense mutation—c.1165A>T predicting p.Lys389* in ALDH1A3 (transcript NM_000693.2)—that was verified by Sanger sequencing (Fig. 2B) and predicted to cause nonsense-mediated decay and complete absence of protein function. The nonsense mutation was absent from the Exome Variant Server (http://evs.gs.washington.edu/EVS/). One hundred and twenty control chromosomes of diverse ethnicity were sequenced in view of the Hispanic ethnicity of the patient, but did not show the nonsense mutation (Coriell Institute for Medical Research, Multi-Ethnic Panels, DNA Polymorphism Discovery Resource, http://www.coriell.org; data not shown). Both parents were heterozygous for the same mutation and an unaffected sibling was wild-type (Fig. 2B).
Both mutations were predicted to be disease-causing by MutationTaster (P = 0.99 and P = 0.99 for p.Lys190* and p.Lys389*, respectively) and to be deleterious by the Protein Variation Effect Analyzer (PROVEAN), with scores of −1149.97 and −430.49, respectively (http://provean.jcvi.org/) (23,24). The nucleotides at position c.568 and c.1165 were conserved in chimp, rhesus, cat, mouse and chicken (http://www.mutationtaster.org/).
Antisense morpholino studies targeting the Aldh1a3 ortholog in Danio rerio
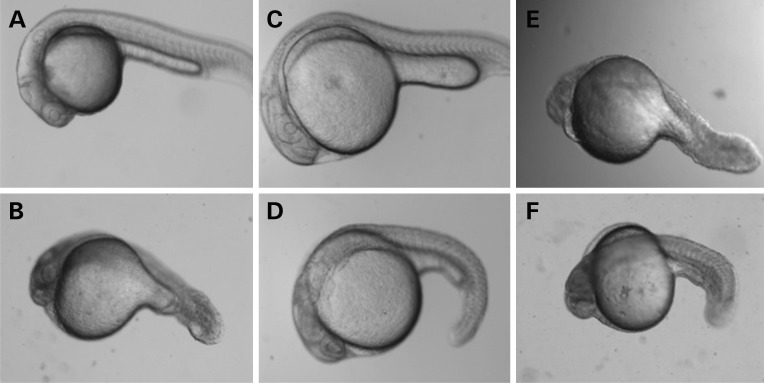
We used Danio rerio and antisense morpholinos (MOs) as a model system to study the effects of loss of function of the orthologous Aldh1a3 gene on eye development. Mutant embryos injected with 1.5 to 6 ng translational MO or splice MO targeting the intron 2 exon 2 boundary of D. rerio Aldh1a3 showed a significant reduction in eye size relative to wild-type larvae and control-injected larvae at 48–72 h post-fertilization (h.p.f.; Supplementary Fig. S1), as previously observed by others (25). Additional phenotypes seen with variable penetrance in MO-injected larvae included delayed closure of the optic fissure, coloboma-like lesions, cardiac edema and kinking of the tail. We verified the specificity of the phenotype observed in morphant larvae and performed rescue experiments by injecting sense mRNA for the wild-type human ALDH1A3 gene together with translational MO targeting the D. rerio Aldh1a3 gene (Fig. 3A–D). Wild-type human ALDH1A3 sense mRNA was able to rescue the morphant phenotype, but attempted rescue with mutant human ALDH1A3 sense mRNA containing either the p.Lys190* or the p.Lys389* mutations was unsuccessful, as would be predicted from the likely loss of function caused by these mutations (Fig. 3E and F). As the clinical phenotype associated with ALDH1A3 loss of function includes optic nerve hypoplasia and ALDH1A3 is known to contribute to the axonal projections of retinal cells into the brain (26,27), we used Pou4f3 (previously known as Brn3c) transgenic fish to study the retinotectal projections of morphant larvae. This transgenic zebrafish line expresses membrane-targeted green fluorescent protein (GFP) under the control of the Pou4f3 promoter/enhancer, labeling a subset of retinal ganglion cells that project into one of the layers of the tectum and into a subset of the extratectal arborization fields (28). Confocal images of Pou4f3:mGFP-labeled retinotectal projections of 5 day post-fertilization (d.p.f.) tecta in live larvae showed that the tectum from wild-type larvae was filled with retinal axons and the optic tract had branched into stereotyped fascicles (Fig. 4A), whereas the tectum from MO-treated larvae appeared less innervated (Fig. 4B).
Figure 3.
Phenotype in D. rerio larvae injected with antisense MO for Aldh1a3 with rescue experiments. (A) Uninjected D. rerio larva at 24 h.p.f. (B) Representative D. rerio larva injected with translational antisense MO (ATG-MO) for Aldh1a3 at 24 h.p.f. The eye is reduced in size, edema is present and the tail is shortened. (C) Representative D. rerio larva injected with control at 24 h.p.f. (D) Representative D. rerio larva injected with translational antisense morpholino (ATG-MO) for Aldh1a3 together with wild-type human mRNA for ALDH1A3 at 24 h.p.f. Rescue of the morphant phenotype can be observed. (E) Representative D. rerio larva injected with translational antisense morpholino (ATG-MO) for Aldh1a3 and mutant human mRNA for ALDH1A3/p.Lys190* at 24 h.p.f. Rescue of the morphant phenotype is not observed. (F) Representative D. rerio larva injected with translational antisense morpholino for Aldh1a3 and mutant human mRNA for ALDH1A3/p.Lys190* at 24 h.p.f. Rescue of the morphant phenotype is not observed.
Figure 4.

Aldh1a3 morphant D. rerio larvae show reduced innervation of the tectum. (A and B) Confocal images of Pou4f3:mGFP-labeled retinotectal projections at 5 d.p.f. tecta in live larvae. The wild-type tectum (A) is filled with retinal axons, and the optic tract has branched into stereotyped fascicles. The MO-treated tectum (B) appears less innervated. Figures are shown with the anterior aspect of the larvae superiorly. Dotted lines outline the tectal neuropil.
DISCUSSION
We have demonstrated homozygosity for two nonsense mutations in ALDH1A3, p.Lys190* and p.Lys389*, predicted to be associated with complete loss of function, in three patients with bilateral A/M. The absence of ALDH1A3 is predicted to reduce cellular RA synthesis and thus diminish downstream signaling. Homozygosity for two missense mutations and one splice site mutation in ALDH1A3 were previously described in three patients with A/M (29; for summary of clinical features in all affected patients to date, see Table 1). Both missense mutations were shown to result in the loss of ALDH1A3 function (29), similar to the deficiency of ALDH1A3 predicted by the two nonsense mutations in this report. It is worth noting that two of the previously reported patients had optic nerve hypoplasia (29), as found in the female child in this report. Optic nerve hypoplasia has been observed in D. rerio models of Aldh1a3 loss of function (25), and may prove to be a characteristic phenotypic feature associated with ALDH1A3 mutations, although it is not specific for this gene. To date, there is insufficient data to determine the frequency of ALDH1A3 mutations in human patients with A/M and whether cardiac defects (for example, the pulmonary valve stenosis and atrial septal defect seen in one child) (29) will be observed in other affected individuals. The three patients described here have no known cardiac defects or neurological findings, although echocardiograms have not been performed.
Table 1.
Phenotypic features of patients with ALDH1A3 loss-of-function mutations
| Phenotypic feature | Patient IV-1, Family 1 (29) | Patient IV-5, Family 1 (29) | Patient IV-1, Family 2 (29) | Patient 1, Family 3 (29) | Patient III-3, Family 1 (this report) | Patient III-6, Family 1 (this report) | Patient 3 (this report) |
|---|---|---|---|---|---|---|---|
| Eye findings | |||||||
| Anophthalmia | +; bilat. | +; bilat. | +; right eye | +; bilat. | |||
| Microphthalmia | +; bilat. | +; bilat. | +; bilat. | +; left eye | |||
| Cysts | + | + | + | − | − | − | |
| ON hypoplasia | + | + | + | ||||
| OC hypoplasia | + | + | + | ||||
| Cardiac defect | + | ||||||
| Intelligence | Autism 3 y | ?Autism 4 y | Normal | Normal | Normal | Normal | |
| Growth | 2nd–10th centile | 2nd–9th centile | Normal | Normal | Normal | ||
| ALDH1A3 mutationa | p.Arg89Cys | p.Arg89Cys | p.Ala493Pro | c.475+1G>T | p.Lys190* | p.Lys190* | p.Lys389* |
ON, optic nerve; OC, optic chiasm; bilat., bilateral; y, years.
aAll mutations are homozygous.
The implication of ALDH1A3 loss of function, and thus diminished RA signaling, in the etiology of human anophthalmia has been supported by papers on STRA6 loss of function and animal model studies in mouse and fish. In D. rerio, the orthologous Aldh1a3 protein shares ∼70% identity with human Aldh1a3 protein and 69% nucleotide identity with the human gene (30). Aldh1a3 expression was detected in the eye from the 10-somite stage (14 h.p.f.) and is present in the ventral retina at 24 h.p.f. (30). The high similarity between the human and D. rerio genes prompted us to use MOs to target the ortholog of Aldh1a3, and our experiments revealed that the MO-treated fish had a smaller eye size together with other malformations, including cardiac edema and a kinked tail. Similar results, including a reduction in eye size, have previously been demonstrated with MO knockdown of other genes involved in RA metabolism, including Stra6 (31) and Bcox, an enzyme involved in vitamin A synthesis (32). Treatment with a RA inhibitor, citral, at 10–11 h.p.f., resulted in the absence of the ventral retina in D. rerio (33). The ventral retinal defects could be partially rescued by RA administration (33). Treatment of D. rerio with another chemical RA inhibitor, diethylaminobenzaldehyde (DEAB), at 9 h.p.f. resulted in larvae with microphthalmia, abnormal retinal development and a reduction in the cell population and variation in retinal cell-layer lamination (25). The visual background adaptation and optokinetic responses were also absent in DEAB-treated embryos (25). The timing of treatment with RA inhibitors was critical, and paradoxically, provision of atRA to murine embryos early in development can result in A/M (33). More widespread phenotypic effects, including an increase in mortality and cardiac edema, were also observed in the DEAB-treated fish, thus resembling the combination of eye and cardiac malformations seen with STRA6 deficiency in humans and with MO knockdown for Stra6 in D. rerio (31). Aldh1a3 homozygous null mice have a lethal phenotype caused by choanal atresia, but also demonstrate eye defects (26). At E14.5, shortening of the ventral retina, lens rotation and persistence of the primary vitreous body were observed, similar to the eye abnormalities in murine embryos with deficiencies of vitamin A or RA signaling (26,34–36).
The exact pathway in which Aldh1a3 exerts its effects on eye development has not been fully determined, but aberrant Wnt signaling has been implicated by several studies. In Aldh1a1−/−/Aldh1a3−/− double-mutant mice, the expression of Dkk2, a downstream effector of Pitx2 and an antagonist of canonical Wnt signaling, was markedly reduced at E12.5 in the perioptic mesenchyme and cornea, suggesting that upregulation of the Wnt/β-catenin signaling pathway contributed to the eye defects seen in the double-mutant mice (4). Additionally, the expression of Axin2, a gene induced by the Wnt/β-catenin pathway (37), was increased in the perioptic mesenchyme of the cornea and the ventral eyelid fold of Aldh1a1−/−/Aldh1a3−/− double-mutants at E12.5, also indicative of the upregulation of Wnt/β-catenin signaling (4). Homozygous null mice for Lrp6, a Wnt co-receptor, exhibit complete loss of Aldh1a1 expression, and ectopic expression of Adh1a3 showed deregulated RA signaling in early eye development (38). Finally, several putative TCF/LEF-binding sites were identified in the Aldh1a1 and Aldh1a3 promoter regions, indicating the possibility of direct Wnt-mediated regulation of RA synthesis during early eye development (38). Alternatively, RA has also been implicated in forebrain patterning and can act by modulating bone morphogenetic protein (BMP) signaling during the presomitic stages of development and Fibroblast Growth Factor 8 (FGF8) and Sonic Hedgehog (SHH) signaling during later development (39).
CONCLUSION
We report three children with A/M due to loss-of-function mutations in ALDH1A3. Our findings support the recent implication of this gene in anophthalmia and severe microphthalmia in humans. Interestingly, an ocular phenotype with the absence of the ventral retina associated with RA inhibition in D. rerio could be partially rescued by provision of vitamin A. Whether a similar therapeutic approach in human would ameliorate A/M remains to be seen.
MATERIALS AND METHODS
Exome sequencing
Following informed consent, high-throughput sequencing was performed in all affected individuals. In the Egyptian family, whole-exome sequencing was performed on the two affected individuals after DNA enrichment using an Agilent SureSelect Human Exon Capture V4 (51 Mbp) kit according to the manufacturer's protocol (Agilent Technologies, Inc., Santa Clara, CA, USA). DNA sequencing was performed on a HiSeq2000 machine (Illumina, Santa Clara, CA, USA), the data were uploaded on DNAnexus, mapped to the hg19 reference genome assembly and the results were viewed with a dedicated browser. Initial analysis was done examining all reported and unreported single-nucleotide polymorphisms (SNPs) to identify common homozygous regions in the affected brothers that were not present in their parents. Three regions sized between 2 and 3 Mb and two regions with a size >3 Mb were identified (Supplementary Table S2). We then searched these regions for homozygous, non-synonymous or frameshift mutations in the coding regions of genes.
Whole-genome sequencing
Genotyping was performed in the sporadic case using Axiom™ Genome-Wide (Human) Array Plates (Affymetrix, Santa Clara, CA, USA) that contain approximately 700 000 SNPs was performed to detect regions of homozygosity/autozygosity. The results were analyzed using the Genotyping Console software (Affymetrix). An amount of 7.5 μg of DNA was submitted for whole-genome sequencing (Complete Genomics, Inc., Mountain View, CA, USA). The methodology for library preparation, mapping of reads to the NCBI reference genome (Build 37, RefSeq accession numbers CM000663–CM000686), local de novo assembly and protocols for variant-calling have previously been published (40,41).
Antisense MO injections in D. rerio
The Committee on Animal Research of the University of California, San Francisco, approved all experiments. Wild-type TL and AB D. rerio were bred and raised at 28.5°C on a 14 h light, 10 h dark cycle as previously described (42). Antisense MOs (Gene Tools LLC, Philomath, OR, USA) were designed to block translation of the D. rerio ortholog of Aldh1a3 (ATG-MO; TATAGTCCCGTTCTGTGCCATAGC) and to disrupt splicing (Splice-MO; TAGTCAAGTTCCTCTCACTTTATCC) (43). The Splice-MO targeted the exon 2, intron 2 boundary to disrupt the exon 2 splice donor site, resulting in a cryptic donor splice site and deletion of the last 20 amino acids of exon 2. A standard mismatch MO was used as a negative control. Embryos were microinjected at the 1–4 cell stage with ATG-MO, Splice-MO or CM at 1.5 to 6 ng/embryo as previously described (43,44). We assessed survival and toxicity at 24 and 48 h.p.f. to verify the correct dose of MOs (data not shown). Embryos were de-chorionated and examined at 24–72 h.p.f. for external phenotypes, including eye defects (microphthalmia, delayed closure of the choroid fissure and coloboma), cardiac edema, pericardial effusion and tail kinking. MO-injected larvae were imaged using a Zeiss Dissection Scope (Leica 1200; Thornwood, NY, USA) and camera, and eye size was measured with the ImageJ (http://rsb.info.nih.gov/ij/) or Leica Application Suite (http://www.leica-microsystems.com/) software. Eye size for injected larvae was represented as mean ±1 standard deviation and statistical significance was taken as P < 0.05.
To verify Aldh1a3 knock-down after the injection of the Splice-MO, RNA was extracted from dechorionated embryos at 30 h.p.f. using TRIzol® (Invitrogen/Life Technologies, Grand Island, NY, USA) and cDNA was prepared (Superscript® III reverse transcriptase; Invitrogen/Life Technologies). Reverse transcription polymerase chain reaction (PCR) was performed using a standard 30 cycle PCR with a forward primer (ATGGCACAGAACGGGACTAT) from exon 1 of D. rerio Aldh1a3 and a reverse primer (ACACCGAGCCTCTCTGACC) from exon 3 of the same gene (data not shown).
For MO rescue, full-length, sequence-verified human ALDH1A3 cDNA clones and mutant clones containing p.Lys190* or p.Lys389* were obtained (Blue Heron Bio, Bothell, WA, USA). Capped, sense mRNA was synthesized in vitro (mMESSAGE mMACHINE® T7 Ultra Kit, Ambion/Life Technologies) and 75–100 pg mRNA was injected into each embryo together with translational MO at a dose of 1.5–2 ng. Larvae were dechorionated and examined at 24 h.p.f. by light microscopy. Eye surface area was calculated and compared between uninjected larvae, larvae injected with ATG-MO alone, larvae injected with ATG-MO and wild-type human mRNA for ALDH1A3, larvae injected with ATG-MO and mutant human mRNA for ALDH1A3/p.Lys190* and larvae injected with ATG-MO and mutant human mRNA for ALDH1A3/p.Lys389*.
For live imaging, larvae were treated with 0.003% 1-phenyl-2-thiourea from 22 h.p.f. and embedded in a 0.5 × 20 mm2 imaging chamber (CoverWell, Grace Bio-labs, Bend, OR, USA) in 1.2% low-melting-point agarose dissolved in E3 medium (45) containing 0.8% norepinephrine and 0.016% tricaine. POU4F3-GFP-expressing larvae were analyzed for retinal structure and optic nerve projection. Immunofluorescence staining was performed after fixation of anesthetized larvae in 4% paraformaldehyde. Larvae were rehydrated in phosphate-buffered saline (PBS) and treated with 0.1% collagenase in PBS to enhance permeability. Incubation times for 5 d.p.f. was 2 h. We used antibody to GFP (anti-GFP raised in chick; Sigma-Aldrich, St Louis, MO, USA) at a concentration of 1:2000. Secondary antibody AlexaFluor 488 (Invitrogen, Carlsbad, CA, USA) was used at a concentration of 1 in 500. Confocal image stacks were acquired using a Zeiss Pascal laser scanning microscope equipped with ×10 and ×20 (air, numerical aperture 0.5) and ×40 (water immersion, numerical aperture 0.8) objectives. To determine axon morphology and arborization, we collected a series of optical planes (z-stack) and collapsed them into a single image (maximum intensity projection) or rendered the volume in three dimensions to provide views of the image stack from different angles. The step size for each z-stack was chosen upon calculation of the theoretical z-resolution of the objective used (typically 0.5–1 mm). The ImageJ plugins StackReg with TurboReg were run with a macro to correct for translational movement in the image acquisition of the stacks.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by The Eunice Kennedy Shriver National Institute of Child Health, National Institutes of Health (K08 HD053476–01A1 to A.S.), the National Eye Institute, National Institutes of Health (R21 EY019999-01 to A.S.) and the Fondation pour la Recherche sur les Maladies Héréditaires to D.F.S. Work in H.B.'s laboratory was supported by a March of Dimes grant.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the families for their participation and to Nadav Ahituv for the use of his fish facilities.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kawaguchi R., Yu J., Honda J., Hu J., Whitelegge J., Ping P., Wiita P., Bok D., Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 2.Duester G. Keeping an eye on retinoic acid signaling during eye development. Chem. Biol. Interact. 2009;178:178–181. doi: 10.1016/j.cbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhinn M., Dollé P. Retinoic acid signalling during development. Development. 2012;139:843–845. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S., Duester G. SnapShot: retinoic acid signaling. Cell. 2011;147:1422. doi: 10.1016/j.cell.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghyselinck N.B., Dupé V., Dierich A., Messaddeq N., Garnier J.M., Rochette-Egly C., Chambon P., Mark M. Role of the retinoic acid receptor beta (RARbeta) during mouse development. Int. J. Dev. Biol. 1997;41:425–447. [PubMed] [Google Scholar]
- 6.Hornby S.J., Ward S.J., Gilbert C.E. Eye birth defects in humans may be caused by a recessively-inherited genetic predisposition to the effects of maternal vitamin A deficiency during pregnancy. Med. Sci. Monit. 2003;9:HY23–HY26. [PubMed] [Google Scholar]
- 7.See A.W., Clagett-Dame M. The temporal requirement for vitamin A in the developing eye: mechanism of action in optic fissure closure and new roles for the vitamin in regulating cell proliferation and adhesion in the embryonic retina. Dev. Biol. 2009;325:94–105. doi: 10.1016/j.ydbio.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Casey J., Kawaguchi R., Morrissey M., Sun H., McGettigan P., Nielsen J.E., Conroy J., Regan R., Kenny E., Cormican P., et al. First implication of STRA6 mutations in isolated anophthalmia, microphthalmia, and coloboma: a new dimension to the STRA6 phenotype. Hum. Mutat. 2011;32:1417–1426. doi: 10.1002/humu.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chassaing N., Golzio C., Odent S., Lequeux L., Vigouroux A., Martinovic-Bouriel J., Tiziano F.D., Masini L., Piro F., Maragliano G., et al. Phenotypic spectrum of STRA6 mutations: from Matthew-Wood syndrome to non-lethal anophthalmia. Hum. Mutat. 2009;30:E673–E681. doi: 10.1002/humu.21023. [DOI] [PubMed] [Google Scholar]
- 10.Pasutto F., Sticht H., Hammersen G., Gillessen-Kaesbach G., Fitzpatrick D.R., Nürnberg G., Brasch F., Schirmer-Zimmermann H., Tolmie J.L., Chitayat D., et al. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am. J. Hum. Genet. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golzio C., Martinovic-Bouriel J., Thomas S., Mougou-Zrelli S., Grattagliano-Bessieres B., Bonniere M., Delahaye S., Munnich A., Encha-Razavi F., Lyonnet S., et al. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am. J. Hum. Genet. 2007;80:1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West B., Bove K.E., Slavotinek A.M. Two novel STRA6 mutations in a patient with anophthalmia and diaphragmatic eventration. Am. J. Med. Genet. A. 2009;149A:539–542. doi: 10.1002/ajmg.a.32682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segel R., Levy-Lahad E., Pasutto F., Picard E., Rauch A., Alterescu G., Schimmel M.S. Pulmonary hypoplasia-diaphragmatic hernia-anophthalmia-cardiac defect (PDAC) syndrome due to STRA6 mutations—what are the minimal criteria? Am. J. Med. Genet. A. 2009;149A:2457–2463. doi: 10.1002/ajmg.a.33038. [DOI] [PubMed] [Google Scholar]
- 14.Fantes J., Ragge N.K., Lynch S.A., McGill N.I., Collin J.R., Howard-Peebles P.N., Hayward C., Vivian A.J., Williamson K., van Heyningen V., et al. Mutations in SOX2 cause anophthalmia. Nat. Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 15.Schneider A., Bardakjian T., Reis L.M., Tyler R.C., Semina E.V. Novel SOX2 mutations and genotype phenotype correlation in anophthalmia and microphthalmia. Am. J. Med. Genet. A. 2009;149A:2706–2715. doi: 10.1002/ajmg.a.33098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verma A.S., Fitzpatrick D.R. Anophthalmia and microphthalmia. Orphanet. J. Rare Dis. 2007;2:47. doi: 10.1186/1750-1172-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardakjian T., Weiss A., Schneider A.S. Anophthalmia/microphthalmia overview. In: Pagon R.A., Bird T.C., Dolan C.R., Stephens K., editors. GeneReviews. Seattle, WA: University of Washington; [Internet] 1993–2004. [Google Scholar]
- 18.Gonzalez-Rodriguez J., Pelcastre E.L., Tovilla-Canales J.L., Garcia-Ortiz J.E., Amato-Almanza M., Villanueva-Mendoza C., Espinosa-Mattar Z., Zenteno J.C. Mutational screening of CHX10, GDF6, OTX2, RAX and SOX2 genes in 50 unrelated microphthalmia-anophthalmia-coloboma (MAC) spectrum cases. Br. J. Ophthalmol. 2010;94:1100–1104. doi: 10.1136/bjo.2009.173500. [DOI] [PubMed] [Google Scholar]
- 19.Slavotinek A.M. Eye development genes and known syndromes. Mol. Genet. Metab. 2011;104:448–456. doi: 10.1016/j.ymgme.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr A., Dubé M.P., Zenteno J.C., Jiang H., Asselin G., Evans S.C., Caqueret A., Lakosha H., Letourneau L., Marcadier J., et al. Mutations in a novel serine protease PRSS56 in families with nanophthalmos. Mol. Vis. 2011;17:1850–1861. [PMC free article] [PubMed] [Google Scholar]
- 21.Balikova I., de Ravel T., Ayuso C., Thienpont B., Casteels I., Villaverde C., Devriendt K., Fryns J.P., Vermeesch J.R. High frequency of submicroscopic chromosomal deletions in patients with idiopathic congenital eye malformations. Am. J. Ophthalmol. 2011;151:1087–1094. doi: 10.1016/j.ajo.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Raca G., Jackson C.A., Kucinskas L., Warman B., Shieh J.T., Schneider A., Bardakjian T.M., Schimmenti L.A. Array comparative genomic hybridization analysis in patients with anophthalmia, microphthalmia, and coloboma. Genet. Med. 2011;13:437–442. doi: 10.1097/GIM.0b013e318204cfd2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y. A fast computation of pairwise sequence alignment scores between a protein and a set of single-locus variants of another protein. Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine (BCB'12); New York, NY, USA: ACM; 2012. pp. 414–417. [Google Scholar]
- 25.Le H.G., Dowling J.E., Cameron D.J. Early retinoic acid deprivation in developing zebrafish results in microphthalmia. Vis. Neurosci. 2012;29:219–228. doi: 10.1017/S0952523812000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupé V., Matt N., Garnier J.M., Chambon P., Mark M., Ghyselinck N.B. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl Acad. Sci. USA. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molotkov A., Molotkova N., Duester G. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development. 2006;133:1901–1910. doi: 10.1242/dev.02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao T., Roeser T., Staub W., Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005;132:2955–2967. doi: 10.1242/dev.01861. [DOI] [PubMed] [Google Scholar]
- 29.Fares-Taie L., Gerber S., Chassaing N., Clayton-Smith J., Hanein S., Silva E., Serey M., Serre V., Gérard X., Baumann C., et al. ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am. J. Hum. Genet. 2013;92:265–270. doi: 10.1016/j.ajhg.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang D., Zhang M., Bao J., Zhang L., Xu X., Gao X., Zhao Q. Expressions of Raldh3 and Raldh4 during zebrafish early development. Gene Expr. Patterns. 2008;8:248–253. doi: 10.1016/j.gep.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Isken A., Golczak M., Oberhauser V., Hunzelmann S., Driever W., Imanishi Y., Palczewski K., von Lintig J. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7:258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biehlmaier O., Lampert J.M., von Lintig J., Kohler K. Photoreceptor morphology is severely affected in the beta, beta-carotene-15,15′-oxygenase (bcox) zebrafish morphant. Eur. J. Neurosci. 2005;21:59–68. doi: 10.1111/j.1460-9568.2004.03830.x. [DOI] [PubMed] [Google Scholar]
- 33.Marsh-Armstrong N., McCaffery P., Gilbert W., Dowling J.E., Dräger U.C. Retinoic acid is necessary for development of the ventral retina in zebrafish. Proc. Natl Acad. Sci. USA. 1994;91:7286–7290. doi: 10.1073/pnas.91.15.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulik K.K., Dehart D.B., Rogers J.M., Chernoff N. Teratogenicity of low doses of all-trans retinoic acid in presomite mouse embryos. Teratology. 1995;51:398–403. doi: 10.1002/tera.1420510605. [DOI] [PubMed] [Google Scholar]
- 35.Lohnes D., Mark M., Mendelsohn C., Dollé P., Dierich A., Gorry P., Gansmuller A., Chambon P. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- 36.Matt N., Dupé V., Garnier J.M., Dennefeld C., Chambon P., Mark M., Ghyselinck N.B. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- 37.Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou C.J., Molotkov A., Song L., Li Y., Pleasure D.E., Pleasure S.J., Wang Y.Z. Ocular coloboma and dorsoventral neuroretinal patterning defects in Lrp6 mutant eyes. Dev. Dyn. 2008;237:3681–3689. doi: 10.1002/dvdy.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halilagic A., Ribes V., Ghyselinck N.B., Zile M.H., Dollé P., Studer M. Retinoids control anterior and dorsal properties in the developing forebrain. Dev. Biol. 2007;303:362–367. doi: 10.1016/j.ydbio.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 40.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 41.Roach J.C., Glusman G., Smit A.F., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M., et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao T., Baier H. Lamina-specific axonal projections in the zebrafish tectum require the type IV collagen Dragnet. Nat. Neurosci. 2007;10:1529–1537. doi: 10.1038/nn2002. [DOI] [PubMed] [Google Scholar]
- 43.Bill B.R., Petzold A.M., Clark K.J., Schimmenti L.A., Ekker S.C. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao R., Nevin L., Agarwal P., Riemer J., Bai X., Delaney A., Akana M., JimenezLopez N., Bardakjian T., Schneider A., et al. A male with unilateral microphthalmia reveals a role for TMX3 in eye development. PLoS One. 2010;5:e10565. doi: 10.1371/journal.pone.0010565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muto A., Orger M.B., Wehman A.M., Smear M.C., Kay J.N., Page-McCaw P.S., Gahtan E., Xiao T., Nevin L.M., Gosse N.J., et al. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005;1:e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.