Abstract
African-American (AA) women have earlier menarche on average than women of European ancestry (EA), and earlier menarche is a risk factor for obesity and type 2 diabetes among other chronic diseases. Identification of common genetic variants associated with age at menarche has a potential value in pointing to the genetic pathways underlying chronic disease risk, yet comprehensive genome-wide studies of age at menarche are lacking for AA women. In this study, we tested the genome-wide association of self-reported age at menarche with common single-nucleotide polymorphisms (SNPs) in a total of 18 089 AA women in 15 studies using an additive genetic linear regression model, adjusting for year of birth and population stratification, followed by inverse-variance weighted meta-analysis (Stage 1). Top meta-analysis results were then tested in an independent sample of 2850 women (Stage 2). First, while no SNP passed the pre-specified P < 5 × 10−8 threshold for significance in Stage 1, suggestive associations were found for variants near FLRT2 and PIK3R1, and conditional analysis identified two independent SNPs (rs339978 and rs980000) in or near RORA, strengthening the support for this suggestive locus identified in EA women. Secondly, an investigation of SNPs in 42 previously identified menarche loci in EA women demonstrated that 25 (60%) of them contained variants significantly associated with menarche in AA women. The findings provide the first evidence of cross-ethnic generalization of menarche loci identified to date, and suggest a number of novel biological links to menarche timing in AA women.
INTRODUCTION
The timing of the age at first menses (menarche) is one of the primary features shaping female reproductive history and is associated with a number of current and later life health outcomes (1). Earlier age at menarche is associated with increased risk for breast cancer (2–4), reduced stature and increased risk of obesity (5–7) and type 2 diabetes (8), whereas late menarche may be associated with an increased risk of Alzheimer's disease (9) and stroke (10), as well as lower fertility (11,12). Identification of genetic variants influencing variation in the age at menarche may thus shed light on mechanisms involved in a number of chronic diseases in women.
Age at menarche is under relatively strong genetic control, with heritability estimated at ∼50% (13–16). Candidate gene association studies point to the involvement of a number of genetic pathways, most notably those involved in steroid hormone signaling and transport [e.g. estrogen receptor (ER) genes and sex hormone binding globulin gene] and estrogen biosynthesis and metabolism (such as CYP17, CYP19, CYP1A1, and CYP1B1) (17), although many of these associations have not been reliably replicated. In 2009, four large genome-wide association studies (GWASs) in women of European ancestry (EA) were published that together identified two novel genetic loci associated with age at menarche, LIN28B and the intergenic region 9q13.2 (18–21). Some of these variants are probably involved in general growth rate, as LIN28B variants are associated with pubertal timing, height and body mass index (BMI) growth in children (20,22,23) as well as with body size and pubertal traits in animal models (24). Variants near the 9q13.2 SNP are also associated with height in GWAS (25). More recently, a GWAS in over 85 000 EA women identified a further 30 genome-wide significant loci, and 10 suggestive loci, yielding a total of 42 loci associated with menarche timing (26). A number of these had been previously identified as obesity loci, highlighting genetic pleiotropy between female adiposity and timing of menarche, an observation that supports the long-recognized link between these traits from epidemiologic studies (7,27).
There is ethnic variation in the timing of menarche, with African-American (AA) girls currently experiencing menarche ∼4–6 months earlier, on average, than EA girls in the USA (6,28–32). In addition, compared with non-Hispanic White women, non-Hispanic Black women in the USA tend to have twice the prevalence of chronic diseases known to be related to early age at menarche, including childhood obesity (33), the Metabolic Syndrome (34) and diabetes (35), as well as higher prevalence and earlier onset of hypertension (36). Despite the significant heritability of age at menarche and the persistent ethnic variation in both age at menarche and its associated diseases, only one recently published study has sought to identify menarche-related genetic variants in AA women (37).
Here, we present a meta-analysis of GWAS from 15 studies including over 18 000 women to test the association of common genetic variants with age at menarche in AA women. We also conducted a targeted investigation of variants within a ±250 kb region around the 42 SNPs recently reported in EA women, in order to test whether these loci contain variants associated with menarche in AA women and to potentially identify stronger markers of the associations. The study demonstrates (i) suggestive evidence for association of age at menarche in AA with a number of variants in loci involved in growth and insulin signaling, (ii) multiple independent SNP associations in or near RORA, previously identified as a possible menarche locus in EA women and (iii) cross-ethnic generalization of the majority of menarche loci identified to date in EA women.
RESULTS
A total of 18 089 AA women with self-reported age at menarche were included in the Stage 1 meta-analysis. Participants were drawn from seven population-based cohort studies and eight breast-cancer case–control studies, in which association analyses were conducted in cases and controls separately. All Stage 1 studies used agnostic, genome-wide SNP genotyping arrays that were not enriched for SNPs in any particular molecular pathways or candidate regions (see Supplementary Material, Table S1, and Methods). A total of 2850 AA women in the Black Women's Health Study (BWHS) were genotyped de novo as the Stage 2 replication sample. Descriptive characteristics of each study are presented in Table 1 and in Supplementary Material, Text S1. Mean age at menarche in the studies was 12.6 years (range 8–21 years). Not all studies reported the year of birth; for those that did, the year of birth at the individual level ranged from 1908 to 1978, and studies that were born later, on average (e.g. CARDIA) had lower mean age at menarche than studies that were born earlier (e.g. ARIC), which is consistent with the downward secular trend in age at menarche during the 20th century (38).
Table 1.
Description of participating cohorts
| Consortium name/cohort name |
Cohort acronym | Age at menarche |
Birth year |
||||
|---|---|---|---|---|---|---|---|
| n | Mean (SD) | Range | Mean | Range | |||
| AABC (African American Breast Cancer Cohorts) | The Women's Contraceptive and Reproductive Experience Study | CARE (cases) | 357 | 12.4 (1.7) | 8–18 | NA | NA |
| CARE (controls) | 215 | 12.3 (1.74) | 9–18 | NA | NA | ||
| The Carolina Breast Cancer Study | CBCS (cases) | 634 | 12.6 (1.8) | 8–21 | NA | NA | |
| CBCS (controls) | 586 | 12.6 (1.75) | 8–18 | NA | NA | ||
| The Multiethnic Cohort | MEC (cases) | 532 | 12.9 (1.66) | 10–17 | NA | NA | |
| MEC (controls) | 972 | 13.2 (1.6) | 10–17 | NA | NA | ||
| The Nashville Breast Health Study | NBHS (cases) | 304 | 12.6 (1.99) | 8–21 | NA | NA | |
| NBHS (controls) | 182 | 12.4 (1.9) | 8–21 | NA | NA | ||
| Northern California Breast Cancer Family Registry/San Francisco Breast Cancer Study | NC-BCFR/SFBCS (cases) | 575 | 12.6 (1.8) | 8–20 | NA | NA | |
| NC-BCFR/SFBCS (controls) | 269 | 12.6 (1.8) | 8–20 | NA | NA | ||
| The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial | PLCO (Cases) | 56 | 13.3 (1.3) | 9–16 | NA | NA | |
| PLCO (controls) | 116 | 13.1 (1.76) | 9–16 | NA | NA | ||
| The Women's Circle of Health Study | WCHS (cases) | 260 | 12.7 (1.9) | 9–19 | NA | NA | |
| WCHS (controls) | 238 | 12.7 (1.7) | 9–18 | NA | NA | ||
| The Wake Forest University Breast Cancer Study | WFBC (cases) | 112 | 12.5 (1.6) | 8–16 | NA | NA | |
| WFBC (controls) | 116 | 12.7 (1.7) | 9–18 | NA | NA | ||
| CARe (Candidate Gene Association Resource) | Atherosclerosis Risk in Communities | ARIC | 1,690 | 12.87 (1.66) | 9–17 | 1934 | 1921–1945 |
| Coronary Artery Disease in Young Adults | CARDIA | 630 | 12.48 (1.47) | 9–17 | 1959 | 1957–1969 | |
| Cleveland Family Study | CFS | 169 | 12.22 (1.37) | 10–16 | 1964 | 1908–1997 | |
| Jackson Heart Study | JHS | 1,228 | 12.77 (1.70) | 9–17 | 1952 | 1910–1982 | |
| Women's Health Initiative | WHI | 8,086 | 12.6 (1.64) | 9–17 | 1935 | 1913–1948 | |
| Healthy Aging in Neighborhoods of Diversity across the Life Span | HANDLS | 617 | 12.6 (1.80) | 9–17 | 1962 | 1946–1980 | |
| Bogalusa Heart Study | BHS | 145 | 12.5 (1.37) | 9–17 | 1966 | 1959–1978 | |
| TOTAL Stage 1 | 18,089 | ||||||
| Black Women's Health Study (Stage 2 Replication Cohort) | BWHS | 2,850 | 12.4 (1.6) | 9–17 | 1947 | 1925–1974 | |
An overview of the flow of experiments/analyses performed in this study and a summary of their results is provided in Figure 1. The following paragraphs provide details on the results from these two primary experiments: (i) a meta-analysis of GWAS of age at menarche in AA women and (ii) a targeted interrogation of 42 loci previously reported to be associated with age at menarche in EA women.
Figure 1.
Overview of analysis and results. Flow of analyses and brief summary of results from each analysis are presented for the genome-wide meta-analysis of age at menarche in AA women and for the targeted interrogation of 42 menarche loci previously reported in EA women.
Meta-analysis of GWASs of age at menarche in AA women
All Stage 1 studies performed regression analyses to test the linear association of each SNP genotype with age at menarche using an additive genetic model. Covariates included study center (if appropriate), year of birth (or age at study enrollment if birth year not available) to account for known secular trends and the first 10 principal components scores from EIGENSTRAT to adjust for population stratification. Further details of the genotyping, quality control (QC) and analysis methods used for each study are provided in Supplementary Material, Table S1. A quantile-quantile plot of the meta-analysis P-values shows that the test statistics follow the null expectations, with no excess of small P-values beyond that expected by chance (Supplementary Material, Fig. S1; lambda = 1.03). No SNP passed the pre-specified P < 5 × 10−8 threshold for genome-wide significance in Stage 1 (Supplementary Material, Fig. S2).
Table 2 displays SNPs with the lowest P-values from the Stage 1 meta-analysis, using the threshold of P < 1 × 10−5, and other criteria that are described in the Materials and Methods section. Regional association plots for all 20 of these top regions are provided in Supplementary Material, Figure S3. There was little evidence of heterogeneity of SNP effects by study as indicated by P for heterogeneity generally >0.05 and never <0.02. There was no indication of systematic deviation of results in breast cancer cases compared with controls or population-based cohort samples (data not shown).
Table 2.
Meta-analysis of age at menarche in AA women: Stage 1 and 2 resultsa
| SNP | Chr. | Nearest gene | Position (Build 36) | Allelesb | EAFc | Stage 1 (maximum n = 18 089) |
Stage 2 (maximum n = 2850) |
Stage 1 and Stage 2 (maximum n = 20 939) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect β (SE)d | P | Phet | Effect β (SE) | P | Effect β (SE) | P | ||||||
| rs4557202e | 3 | B3GALNT1 | 162303218 | C/G | 0.43 | −4.90 (0.96) | 3.51E − 07 | 0.238 | −0.50 (2.24) | 0.822 | −4.21 (0.88) | 1.86E − 06 |
| rs11216435 | 11 | DSCAML1 | 116894142 | T/C | 0.32 | 5.11 (1.02) | 6.33E − 07 | 0.966 | 0.30 (2.49) | 0.904 | 4.41 (0.95) | 3.31E − 06 |
| rs339978 | 15 | RORA | 58724694 | T/C | 0.20 | 5.90 (1.21) | 9.95E − 07 | 0.193 | 2.00 (2.85) | 0.484 | 5.31 (1.11) | 1.76E − 06 |
| rs1476150 | 2 | NAP5 | 133592865 | C/G | 0.65 | 5.69 (1.18) | 1.23E − 06 | 0.683 | −1.36 (2.35) | 0.056 | 4.28 (1.05) | 4.51E − 05 |
| rs7754121 | 6 | HDGFL1 | 23281592 | A/G | 0.10 | 7.69 (1.59) | 1.36E − 06 | 0.434 | −0.28 (3.79) | 0.942 | 6.49 (1.47) | 9.61E − 06 |
| rs320320 | 1 | AKT3 | 241901809 | A/G | 0.48 | 4.55 (0.95) | 1.84E − 06 | 0.117 | 1.03 (2.29) | 0.652 | 4.03 (0.88) | 4.68E − 06 |
| rs12907866 | 15 | CYP19A1 | 49332746 | A/G | 0.84 | 6.15 (1.31) | 2.53E − 06 | 0.587 | 5.63 (3.05) | 0.065 | 6.06 (1.20) | 4.36E − 07 |
| rs6468994 | 8 | ZFPM2 | 106365896 | T/C | 0.64 | 4.67 (1.00) | 2.81E − 06 | 0.309 | −2.69 (2.34) | 0.251 | 3.54 (0.92) | 1.14E − 04 |
| rs11071033 | 15 | UNC13C | 52167492 | T/C | 0.71 | 4.80 (1.03) | 3.54E − 06 | 0.749 | 2.67 (2.50) | 0.286 | 4.49 (0.96) | 2.70E − 06 |
| rs7807441 | 7 | FLJ13195 | 66826223 | T/C | 0.55 | −4.32 (0.94) | 4.14E − 06 | 0.618 | −2.28 (2.25) | 0.311 | −4.02 (0.87) | 3.48E − 06 |
| rs17669535 | 8 | DLGAP2 | 1231631 | C/G | 0.97 | 14.50 (3.15) | 4.20E − 06 | 0.966 | 14.49 (7.61) | 0.057 | 14.50 (2.91) | 6.37E − 07 |
| rs6947406 | 7 | C7orf10 | 41013150 | A/G | 0.87 | −6.43 (1.40) | 4.78E − 06 | 0.963 | −0.16 (3.31) | 0.961 | −5.48 (1.29) | 2.34E − 05 |
| rs980000 | 15 | RORA | 58688255 | T/C | 0.26 | 4.88 (1.07) | 4.86E − 06 | 0.048 | 3.55 (2.57) | 0.168 | 4.69 (0.99) | 2.03E − 06 |
| rs8014131 | 14 | FLRT2 | 85033609 | A/C | 0.42 | −4.48 (0.98) | 5.24E − 06 | 0.157 | −5.29 (2.29) | 0.021 | −4.61 (0.90) | 3.44E − 07 |
| rs7819115 | 8 | DLGAP2 | 1549163 | A/C | 0.36 | −4.52 (0.99) | 5.52E − 06 | 0.471 | −1.19 (2.36) | 0.614 | −4.01 (0.92) | 1.18E − 05 |
| rs7873730 | 9 | ZNF483 | 113343500 | A/T | 0.88 | −7.43 (1.65) | 6.35E − 06 | 0.026 | −4.84 (2.92) | 0.104 | −6.82 (1.44) | 2.17E − 06 |
| rs10441737 | 9 | ZNF483 | 113341406 | T/C | 0.58 | −4.37 (0.97) | 6.55E − 06 | 0.084 | −0.11 (2.24) | 0.961 | −3.70 (0.89) | 3.22E − 05 |
| rs10940138 | 5 | PIK3R1 | 67230225 | T/C | 0.19 | 5.43 (1.21) | 6.78E − 06 | 0.526 | 6.77 (2.87) | 0.018 | 5.64 (1.11) | 4.09E − 07 |
| rs7911165 | 10 | EBF3 | 131516640 | T/C | 0.54 | 4.72 (1.05) | 7.09E − 06 | 0.985 | 1.80 (2.21) | 0.415 | 4.18 (0.95) | 1.05E − 05 |
| rs2796200 | 1 | ZRANB2 | 71431476 | A/G | 0.66 | 4.46 (0.99) | 7.10E − 06 | 0.745 | −2.81 (2.35) | 0.232 | 3.35 (0.92) | 2.45E − 04 |
aTop independent (pairwise r2 < 0.3) SNPs, all with n > 10 000 out of 18 089, MAF > 0.03, and P < 10−5 in meta-analysis.
bEffect/non-effect allele.
cEffect allele frequency.
dEffect β is in weeks.
eStage 2 was conducted using proxy snp rs7651087 (effect allele = C/non-effect allele = T, EAF = 0.4353, r2 = 1.0, 1000 Genomes Project—YRI).
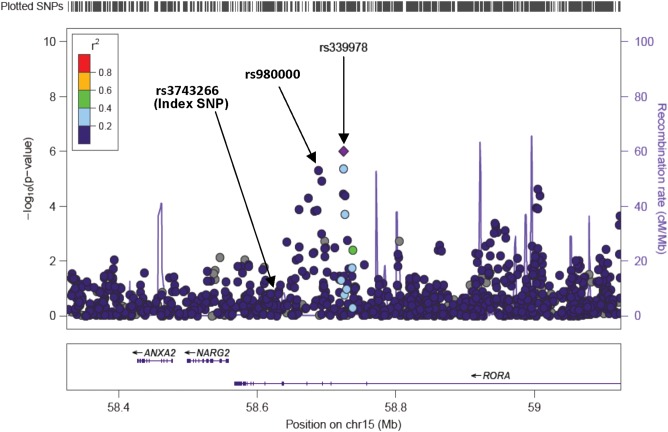
The most statistically significant association was with SNP rs4557202, near B3GALNT3 (P = 3.51 × 10−7), a gene involved in lipid synthesis and metabolic pathways. An intronic enhancer SNP on chromosome 11q23 (rs11216435) near DSCAML1 (Down-syndrome cell-adhesion molecule-like 1) and an SNP on chromosome 15q22 (rs339978) near RORA (the nuclear hormone receptor, RAR-like orphan receptor-alpha) were associated at P < 1 × 10−6. A second SNP (also an intronic enhancer) near RORA (rs980000) was associated at P = 4.86 × 10−6. These latter findings provide evidence of cross-ethnic validation for RORA, which had suggestive (but not confirmed) association with age at menarche in the EA ReproGen analysis (26). The two intronic variants we identified at RORA are common in AA women [minor allele frequency (MAF) of 0.20 for rs339978 and 0.26 for rs980000] but not in EA women (MAF of 0.02 and 0.05 in 1KGP EUR). These SNPs are in low linkage disequilibrium (LD) with one another in HapMap 1000 Genomes Project (1KGP) African samples (AFR in 1KGP, r2 = 0) and are only modestly correlated in EA populations (CEU in 1KGP, r2 = 0.34). Neither of these SNPs were in LD in either population with the index signal (rs3743266) reported previously at RORA in women of EA (r2 = 0).
We also replicated the ZNF483 locus previously reported in EA women (26) in AA women. The two most significant associations in Stage 1 were intronic enhancer variants rs7873730 and rs10441737 near ZNF483 on chromosome 9q31. SNP rs10441737 is a near-perfect proxy for the known menarche variant rs10980926 at this locus in EA women (EUR in 1KGP, r2 = 0.98), with LD also observed in African samples (AFR in 1KGP, r2 = 0.44). The SNP rs7873730 was imputed in all studies (MACH r2 was between 0.72 and 0.86 across studies), was less common in each population than the index SNP (MAF of 0.06 in EUR and 0.10 in AFR) and was only weakly correlated with either of the two SNPs in both EUR and AFR (r2 of 0.08–0.18).
Conditional analysis
To further explore the evidence of multiple independent signals within the RORA and ZNF483 loci, we performed conditional analyses in which both of our top SNPs and the index SNP reported for EA were included as covariates in each of the two independent regression models (Table 3; Fig. 2). For the RORA locus, we found evidence of two independent signals; both rs980000 (P = 6.8 × 10−5) and rs339978 (P = 2.5 × 10−3) remained significant when considered in the same model with the EA index SNP. This finding suggests that there may be multiple functional variants for menarche at this locus in AA women. For ZNF483, we found evidence of a single signal, as only one of the three SNPs (rs10441737) remained nominally significant (P = 0.025) when rs7873730 and rs10980926 (the EA index SNP) were included in the regression model.
Table 3.
Conditional analysis of multiple SNPs near ZNF483 and RORA
| Chr., gene | SNP | Position (Build 36) | Coded allele, frequency in AFR | Marginal betaa, P | Conditional betaa, P |
|---|---|---|---|---|---|
| 9, ZNF483 | rs10980926 (index) | 113333455 | A, 0.61 | 2.34, 0.019 | −0.12, 0.93 |
| 9, ZNF483 | rs7873730 | 113343500 | A, 0.88 | −7.43, 6.4 × 10−6 | −0.56, 0.77 |
| 9, ZNF483 | rs10441737 | 113341406 | T, 0.58 | −4.37, 6.6 × 10−6 | −3.22, 0.025 |
| 15, RORA | rs3743266 (index) | 58568805 | C, 0.33 | −0.69, 0.49 | −0.016, 0.43 |
| 15, RORA | rs339978 | 58724694 | T, 0.20 | 5.90, 1.0 × 10−6 | 3.68, 2.54 × 10−3 |
| 15, RORA | rs980000 | 58688255 | T, 0.26 | 4.88, 4.9 × 10−6 | 4.31, 6.78 × 10−5 |
aBeta values are for effect of SNP on menarche age, in weeks. Conditional analyses were conducted in the largest cohort/studies (WHI, CARe and AABC) using individual-level genotype data in two linear regression models (one for each locus), each of which included the three SNPs, birth year (or enrollment age), study center (if applicable) and the top 10 PCs as covariates. The results were then meta-analyzed using METAL. Bonferroni-corrected P values were used to identify independent signals in the conditional analyses, with P < 0.05 as the criterion for independence.
Figure 2.
Conditional analysis identifies two independent signals for age at menarche near RORA, both of which are different from the previously identified index SNP in EA women. SNPs are plotted using Locus Zoom by position on the chromosome against association with age at menarche (−log10 P). Estimated recombination rates are plotted in blue to reflect local LD structure using the 1KGP AFR reference panel, and the SNPs surrounding the top SNP from the Stage 1 meta-analysis (rs339978, purple diamond) are color coded to reflect their LD with this SNP. Both the first (represented by rs339978, P = 1 × 10−6) and the second signal (represented by rs980000, P = 2 × 10−6) were independent of the index SNP (rs3743266) previously reported for EA women (see Table 3).
Stage 2 analysis in the black women's health study
We examined the 20 top SNPs from the Stage 1 meta-analysis in AA women from the BWHS. For 16 of the 20 SNPs that underwent testing in Stage 2, the direction of effect in the replication sample was consistent with Stage 1 (Table 2). Two SNPs also replicated at a nominal P value (<0.05): rs8014131 near FLRT2 (BWHS P = 0.021; combined P = 3.4 × 10−7) and rs10940138 near PIK3R1 (BWHS P = 0.018; combined P = 4.1 × 10−7), which is involved in the metabolic functions of insulin. Suggestive evidence for replication (consistent direction and nominally significant P value) was also noted for rs12907866 near the aromatase gene CYP19A1 (BWHS P = 0.065; combined P = 4.4 × 10−7) centrally involved in estrogen synthesis, and rs17669535 in DLGAP2 (BWHS P = 0.057; combined P = 6.4 × 10−7). No SNPs passed a Bonferroni-corrected P-value threshold for significance in Stage 2.
Examination of the Stage 1 findings in EA women
We also evaluated potential associations between the 20 top SNPs from our Stage 1 meta-analysis in AA women and age at menarche in women of EA using data from the ReproGen consortium (Supplementary Material, Table S2). AA associations were directionally consistent with EA associations for 14 of the 20 variants, 3 of which were associated with age at menarche (Bonferoni-corrected P < 0.05). We found suggestive evidence of association near AKT3. The SNP rs320320 was associated in ReproGen at P = 1.06 × 10−3, and the combined Stage 1 AA + ReproGen EA meta-analysis yielded P = 1.18 × 10−7). AKT3 is one of the AKT kinases, which encodes RAC-gamma serine/threonine-protein kinase, and regulates cell signaling in response to insulin and growth factors. In addition to AKT3, both of the SNPs near ZNF483 (rs7873730 and rs10441737) also replicated in EA women, which was expected given that ZNF483 was originally identified as a menarche locus in the ReproGen cohorts (described above) and because these SNPs were in high LD with the index SNP in this locus.
Interrogation of previously published menarche-associated Loci
The second goal of our study was to systematically interrogate the set of 42 menarche loci discovered in EA populations in AA women to confirm cross-ethnic replication of these loci and to identify possible stronger markers of the signals. First we examined the 42 EA index SNPs in these loci and found 26 had the same direction of effect on age at menarche in AA women, although only four of these were also nominally associated with age at menarche in our AA samples (P < 0.05). Other than ZNF483 (discussed above), these included SNPs near CCDC85A, C6orf173, and RXRG (Supplementary Material, Table S3). None of these passed the Bonferroni-adjusted P-value threshold for significance (P < 0.0011).
We next investigated SNPs in a 250 kb region surrounding the 42 index SNPs that were associated with age at menarche in EA (see Materials and Methods) (Table 4), using a Bonferroni correction for the number of effective SNPs queried in each region. Generally, the SNP with the lowest P-value in African populations was in low LD with the index EA SNP in AA women (r2 < 0.2) but was often in high LD with the index SNP in European populations (r2 > 0.8), reflecting the same signal but better localizing it than was possible in the EA studies. Supplementary Material, Figure S4 shows regional association plots of age at menarche in AA women with SNPs within each 42 of the interrogated loci. The main finding from this analysis was that we found cross-population validation for a large proportion of the 42 loci (25 of 42, or 60%). The strongest evidence of association was found for SNPs in RORA and ZNF483 (as discussed above in relation to the top Stage 1 results), but there was also some evidence for cross-population locus replication of seven obesity-associated loci (26,39), including FTO (rs12149832, P = 1.6 × 10−3), SEC16B (rs543874; P = 1.47 × 10−2), STK33/TRIM66 (rs12575252; P = 3.27 × 10−2) and RXRG (rs3767342; P = 4.49 × 10−2) where SNPs were in high LD with the index SNP in EUR (r2 > 0.8), as well as BSX (P = 4.39 × 10−3), TMEM18 (rs2685252; P = 2.06 × 10−2) and LRP1B (rs7607295; P = 4.74 × 10−2), which were not in high LD with the index SNP in EUR (r2 < 0.1). In addition, as in EA women, age at menarche was associated with genetic variants near LIN28B, PLCL1, NR4A2, MKL2 (corrected P value <5 × 10−3 for all), and with variants in INHBA (P = 1.3 × 10−2). Inhibin A is secreted by the granulosa cells of the ovarian follicles in the ovaries to provide negative feedback on follicle-stimulating hormone and is a strong candidate gene for pubertal timing.
Table 4.
Twenty-five of 42 menarche loci identified in European Americans generalize to AA womena
| Nearest genes | Chr | Index SNP identified in EA womenb | Best SNP in the region in AA womenc | LD (index SNP—AA best SNP), YRI Reference panel [R2] | LD (index SNP—AA best SNP), CEU reference panel [R2] | Best SNP coded allele (frequency) | Best SNP β (weeks) | Adjusted P-valued |
|---|---|---|---|---|---|---|---|---|
| RXRG | 1 | rs466639 | rs3767342 | 0.839 | 0.877 | T (0.85) | 4.26 | 0.045 |
| SEC16B | 1 | rs633715 | rs543874 | 0.144 | 0.917 | A (0.75) | 3.77 | 0.015 |
| LRP1B | 2 | rs12472911 | rs7607295 | 0.206 | NA | T (0.90) | −8.83 | 0.047 |
| PLCL1 | 2 | rs12617311 | rs7557664 | 0.037 | 0.11 | A (0.48) | −3.62 | 0.003 |
| NR4A2 | 2 | rs17188434 | rs1113060 | NA | 0.002 | T (0.36) | −4.25 | 0.0004 |
| CCDC85A | 2 | rs17268785 | rs17047854 | 0.318 | 0.943 | A (0.34) | 3.41 | 0.010 |
| TMEM18 | 2 | rs2947411 | rs2685252 | 0.004 | 0.031 | T (0.75) | 3.41 | 0.021 |
| SFRS10 | 3 | rs2002675 | rs4686718 | 0.002 | 0.073 | T (0.35) | −3.23 | 0.05 |
| EEFSEC | 3 | rs2687729 | rs9819578 | 0.543 | 0.075 | T (0.23) | 5.47 | 0.024 |
| ECE2 | 3 | rs3914188 | rs6770142 | 0.044 | 0 | A (0.30) | 2.87 | 0.093 |
| 3q13.32 | 3 | rs6438424 | rs16827902 | 0.012 | NA | T (0.94) | 12.58 | 0.036 |
| TMEM108 | 3 | rs6439371 | rs7613434 | 0.129 | 0.005 | A (0.13) | −4.65 | 0.029 |
| RBM6;RBM5 | 3 | rs6762477 | rs12629572 | 0.003 | 0.071 | T (0.76) | 3.68 | 0.075 |
| CCDC71 | 3 | rs7617480 | rs1464567 | 0.113 | 0.243 | C (0.81) | −2.22 | 0.49 |
| VGLL3 | 3 | rs7642134 | rs2879790 | 0.017 | 0.046 | A (0.21) | −8.18 | 0.012 |
| PHF15 | 5 | rs13187289 | rs12655967 | 0.023 | 0.017 | A (0.53) | −7.71 | 0.029 |
| JMJD1B | 5 | rs757647 | rs11750854 | 0.013 | 0.082 | A (0.67) | −4.27 | 0.46 |
| C6orf173 | 6 | rs1361108 | rs9401888 | 0.404 | 0.806 | A (0.78) | 3.62 | 0.018 |
| PRDM13 | 6 | rs4840086 | rs7740247 | 0.000 | 0.009 | C (0.02) | −15.27 | 0.085 |
| LIN28B | 6 | rs7759938 | rs9386427 | 0.229 | 0.075 | T (0.29) | 4.10 | 0.0025 |
| INHBA | 7 | rs1079866 | rs17171859 | 0.097 | NA | C (0.95) | 11.89 | 0.010 |
| PXMP3 | 8 | rs7821178 | rs6473010 | 0.042 | 0.006 | A (0.99) | −47.64 | 0.097 |
| ZNF483 | 9 | rs10980926 | rs7873730 | 0.093 | 0.118 | A (0.88) | −7.43 | 0.0021 |
| TMEM38B | 9 | rs2090409 | rs7041138 | 0.002 | 0.129 | T (0.51) | 3.31 | 0.015 |
| NARS2 | 11 | rs10899489 | rs1006441 | 0.105 | 0.841 | C (0.15) | 3.90 | 0.10 |
| PHF21A | 11 | rs16938437 | rs11600515 | 0.014 | 0.007 | C (0.04) | 8.61 | 0.01 |
| STK33 | 11 | rs4929923 | rs12575252 | 0.439 | 0.959 | C (0.51) | 3.10 | 0.033 |
| BSX | 11 | rs6589964 | rs17126930 | 0.128) | 0 | T (0.91) | −6.42 | 0.0044 |
| ARNTL | 11 | rs900145 | rs7925241 | 0.025 | NA | A (0.87) | −6.22 | 0.094 |
| C13orf16 | 13 | rs9555810 | rs1163630 | 0.024 | 0.028 | C (0.66) | −3.11 | 0.16 |
| BEGAIN | 14 | rs6575793 | rs941930 | 0.003 | 0.015 | A (0.16) | 4.73 | 0.011 |
| RORA | 15 | rs3743266 | rs339978 | 0.012 | 0.008 | T (0.20) | 5.90 | 0.00004 |
| IQCH | 15 | rs7359257 | rs7174933 | 0.00 | NA | A (0.02) | −12.76 | 0.11 |
| NFAT5 | 16 | rs1364063 | rs8054051 | 0.015 | NA | A (0.97) | −14.49 | 0.74 |
| MKL2 | 16 | rs1659127 | rs39826 | 0.035 | 0.019 | A (0.26) | −3.84 | 0.0082 |
| FTO | 16 | rs9939609 | rs12149832 | 0.057 | 0.934 | A (0.12) | −5.54 | 0.0016 |
| CA10 | 17 | rs9635759 | rs12452390 | 0.001 | 0.001 | T (0.86) | 4.98 | 0.011 |
| FUSSEL18 | 18 | rs1398217 | rs1036349 | 0.026 | 0.133 | T (0.08) | −4.22 | 0.32 |
| SLC14A2 | 18 | rs2243803 | rs9973059 | 0.004 | 0.001 | C (0.84) | 3.88 | 0.092 |
| CRTC1 | 19 | rs10423674 | rs875396 | 0.003 | 0.067 | A (0.27) | −2.55 | 0.54 |
| PIN1 | 19 | rs1862471 | rs10425175 | 0.032 | 0 | T (0.27) | 3.30 | 0.079 |
| PCSK2 | 20 | rs852069 | rs4814606 | 0.006 | 0.011 | A (0.13) | 4.44 | 0.034 |
aLocus generalization defined as Bonferonni-corrected P value for best SNP in the region <0.05.
cAll SNPs in a 250 kb region in either direction of the index SNP were interrogated for association with age at menarche and the SNP with the lowest Bonferonni-correct P value was considered the best.
dBonferonni-corrected P value (0.05/n), based upon number of n independent tests (SNPs) within each region; corrected P values < 0.05 are shown in bold.
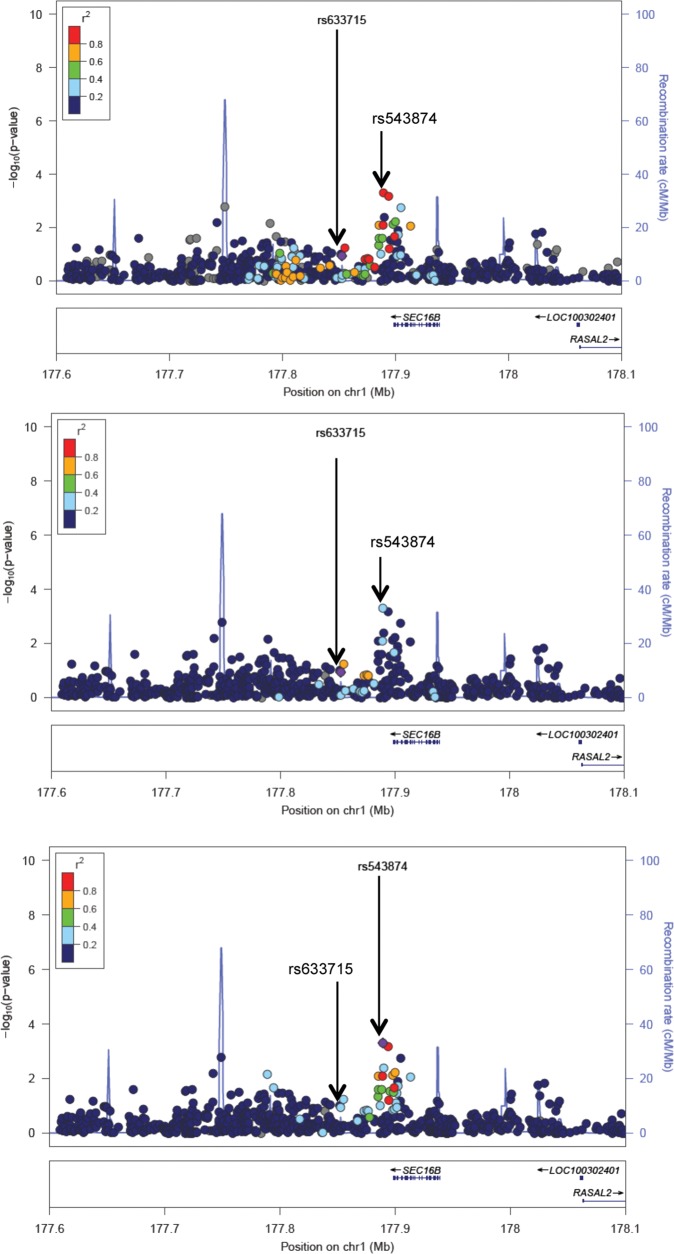
Next, we sought to identify stronger markers of the index signals in AAs through additional fine-mapping (see details of the approach in Materials and Methods). We found SNPs in 8 of the 42 EA regions that were more strongly associated with age at menarche in AA women when compared with the index signal in EA women (i.e. had a P-value for association with menarche <0.004, and a P value at least 1 degree of magnitude lower than the P for the corresponding index association in EA women), and also were in moderate to strong LD with the EA index SNP with r2 > 0.4 with the index SNP in EA (i.e. represented the same genetic ‘signal’) (Table 5). These included SEC16B, CCDC85A, EEFSEC, LIN28B, BSX, NARS2, STK33, and FTO. For instance, at the obesity-related locus SEC16B, we detected a variant (rs543874) approximately 50 kb upstream of the index signal previously identified in EAs (rs633715) that was more strongly associated with menarche in AA women (P = 4.9 × 10−4) than was the index signal (P = 0.12). SNP rs543874 is more strongly correlated with rs633715 in EA populations (r2 = 0.91 in 1KGP EUR) than in AA populations (r2 = 0.18 in 1KGP AFR), which suggests rs543874 may be a better marker of the putatively functional variant in AAs. These relationships are illustrated for the index SNP and the stronger marker in SEC16B in a regional association plot (Fig. 3). Similarly, in the widely replicated obesity locus FTO, rs12149832 may be a stronger marker of the functional variant due to its stronger evidence of association in AA women (P = 2.0 × 10−4) when compared with the index SNP identified in EA women (rs9939609, P = 0.83), and because it again represents the same signal as the index SNP in EUR populations (r2 = 0.88 in 1KGP EUR) but not in AFR populations (r2 = 0.05 in 1KGP AFR). Of interest, five of the eight stronger marker SNPs in this analysis were found in/near loci previously implicated in adiposity via GWA (SEC16B, FTO) (39) were associated with BMI in the GIANT consortium (STK33/TRIM66, NARS2/GAB2) (26), or are involved in neuronal feeding-control circuits (BSX) (40).
Table 5.
Fine-mapping of 42 putative menarche loci localized in EA women: identification of stronger markers of association in AA womena
| Chr., Nearest Gene | Index SNP identified in EA women | Coded allele, frequency in AA | Index SNP beta (weeks), P in AA | Stronger marker | Coded allele, frequency in AA | Stronger marker beta (weeks), P in AA | r2 EURb | r2 AFR b |
|---|---|---|---|---|---|---|---|---|
| 1, SEC16B | rs633715 | C, 0.10 | −2.5, 0.12 | rs543874 | A/G, 0.75 | 3.8, 4.9 × 10−4 | 0.91 | 0.18 |
| 2, CCDC85A | rs17268785 | A, 0.74 | −3.1, 0.017 | rs17047854 | A/G, 0.34 | 3.4, 5.8 × 10−4 | 0.99 | 0.52 |
| 3, EEFSEC | rs2687729 | A, 0.66 | −1.8, 0.075 | rs2075402 | T/C, 0.27 | −3.2, 2.2 × 10−3 | 0.56 | 0.14 |
| 6, LIN28B | rs7759938 | C, 0.53 | 2.5, 0.15 | rs314266 | T/C, 0.33 | −3.8, 2.9 × 10−4 | 0.65 | 0.53 |
| 11, BSX | rs6589964 | A, 0.38 | 0.3, 0.72 | rs1461499 | A/C, 0.63 | 3.5, 3.8 × 10−4 | 0.41 | 0.06 |
| 11, NARS2 | rs10899489 | A, 0.31 | 0.7, 0.48 | rs1006441 | C/G, 0.15 | 3.9, 3.8 × 10−3 | na | na |
| 11, STK33 | rs4929923 | C, 0.55 | −1.5, 0.11 | rs12575252 | C/G, 0.51 | 3.1, 9.9 × 10−4 | 0.92 | 0.55 |
| 16, FTO | rs9939609 | A, 0.47 | −0.2, 0.83 | rs12149832 | A/G, 0.12 | −5.5, 2.0 × 10−4 | 0.88 | 0.05 |
aSNPs selected were those within ±250 kb of index signal, with r2 > 0.4 with index SNP in EUR, P value for marker association <0.004 and at least 1 degree of magnitude lower than p for index SNP.
bLD (r2) between Index SNP and Stronger Marker SNP is based on 1000 Genome Project.
Figure 3.
Fine-mapping of the SEC16B locus in AA women reveals a stronger genetic marker for age at menarche than the index SNP identified in European American women. (A) SNP associations with age at menarche near SEC16B in AAs, using LD from the 1KGP EUR reference panel, in relation to the index SNP rs633715 identified in EA women (purple diamond). The SNP with the lowest P value in the AA meta-analysis (rs543874) is 0.5 Mb upstream from the index SNP and has significantly lower P value (4.9 × 10−4 versus 0.12). The two SNPs are in strong LD in EUR populations (red color indicates r2 > 0.8), showing that it would be considered the same signal in EA women. (B) SNP associations with age at menarche near SEC16B in AAs, using LD from the 1KGP AFR reference panel, in relation to the index SNP rs633715 identified in EA women (purple diamond). The SNP with the lowest P value in the AA meta-analysis (rs543874) was in relatively weak LD (light blue color indicates r2 between 0.2 and 0.4) with the index SNP in AFR, suggesting they lie on different haplotypes. (C) SNP associations with age at menarche near SEC16B in AAs, using LD from the 1KGP AFR reference panel, in relation to the putatively stronger marker SNP rs543874 (purple diamond). Strong LD in AFR populations between rs543874 and a cluster of SNPs surrounding it, but low LD with SNPs near the index SNP rs633715 is seen. This suggests rs543874 may better localize the causal variant giving rise to the association of menarche age with SNPs near SEC16B previously reported in EA women. Note: SNPs are plotted using Locus Zoom by position on the chromosome against association with age at menarche (−log10 P). Estimated recombination rates are plotted in blue to reflect local LD structure, and the SNPs surrounding the index SNP (or most significant SNP) in each case are color coded to reflect their LD with this SNP, marked as a purple diamond.
eQTL analysis of lead AA SNPs
For the 42 EA loci, there were 29 SNPs with at least one UCSC, Vega or RefSeq transcript showing expression in Yoruban African lymphoblastoid cell lines (LCLs). A total of 109 cis-regulatory SNPs were compared for the analysis of the index SNPs, while a total of 111 cis-regulatory SNPs were compared for the AA top SNP analysis. While 11.4% of the queried index SNPs positively overlapped with cis-regulatory SNPs observed in YRI LCLs, twice as many (23.4%) of the top AA SNPs from the regional analysis had positive overlap with cis-regulatory SNPs [nominal P value for χ2 = 0.026; P (Permutation) <0.05]. This provides some evidence for the greater functional relevance of the SNPs identified in AA women when compared with the EA index SNPs. When limiting the comparison to SNPs that were significantly stronger markers of the index signal in the fine-mapping experiment, their overlap with cis-regulatory SNPs was not found to be significantly greater than for the index SNPs.
Discussion
Identification of genetic variation controlling the development of chronic disease risk factors in childhood, such as early menarche, is important because it may point to effective targets for environmental and behavioral interventions in early life, before disease processes are fully entrenched. AA women now experience significantly earlier sexual development (28) and carry a much higher burden of obesity and diabetes than EA women (33,35); therefore, the search for genetic determinants of menarche timing may be of particular value in this population. Nonetheless, virtually all GWAS to date have been conducted in individuals of EA (41), and this is true of menarche GWAS as well; a recent systematic review on the genetics of menarche (42) found only one existing study that provided any estimates of measured genotype effects on age at menarche for AA women. Subsequently, there has been one study published using a targeted genome-wide approach [i.e. using the Metabochip (43) in ∼4000 AA women (37)]. In that study, no SNP association passed correction for multiple testing, and relatively poor coverage by the Metabochip of the previously reported menarche loci in EA women meant that cross-ethnic replication and generalization study was hampered (37).
Meta-analysis of GWASs of age at menarche
The present study remains the largest and most comprehensive genetic examination of menarche timing in AA women to date, including all known available data (from 15 observational cohort and case–control studies) in AA women having both age at menarche information and genome-wide genotype data. Nonetheless, it included far fewer samples than are now available for EA women (≥100 000 in ReproGen). Effect sizes of the variants found via GWA in EA women (26) were fairly small (e.g. accounting for between 1 and 6 weeks variation in age at menarche per copy of the risk allele); therefore, our lack of genome-wide significant associations could stem from a lack of statistical power. Our power calculations show (Supplementary Material, Fig. S5) that for SNPs with MAF > 0.2, we had ≥80% power to detect a relatively small effect size [e.g. 6.5 weeks (0.12 years) earlier age at menarche per copy of the risk allele]. This is an effect size at the upper end of the range observed previously in EA populations [e.g. in LIN28B, a ∼6.9-week reduction in age at menarche per allele copy has been seen (18,20,21,26)]. As many of the SNPs identified in EAs were novel, it is also possible that the reported effect sizes in EA women were overestimated (i.e. the phenomenon of the ‘winner's curse’), which would indicate that while our study was adequately powered to test variants having effects in the range of previously reported SNPs, it was in reality substantially underpowered.
The primary outcome of our GWA experiment was to provide the first cross-ethnic validation of RORA, strengthening the evidence for its role in menarche. Genetic variants near RORA were previously reported to influence age at menarche in EA women, but at P-values below genome-wide significant thresholds (26). RORA encodes one of the ROR nuclear receptors that regulate the transcription of numerous other genes and is expressed in human endometrium (44). Recently, RORA expression has been found to regulate aromatase (CYP19A1), which converts testosterone to estrogen (45). A SNP in CYP19A1 was among our top Stage 1 results and was marginally associated in Stage 2 (P = 0.065). Variants in genes in the CYP19 gene family have been found to be associated with age at menarche (46,47) as well as other reproductive traits in women (17,18,48,49). Furthermore, a conditional analysis identified two independent signals in RORA in AA women, both of which were independent of the previously reported index SNP identified in EA women. Localization of multiple independent variants that are statistically associated with disease traits is an important first step toward identifying causal variants. Our results suggest two independent signals in/near RORA, refining this putative menarche locus.
In addition, results of the GWA meta-analysis highlight biological pathways that may be important in AA women. A number of the most strongly associated variants from this meta-analysis implicate growth factor and insulin signaling in menarche timing. SNPs in Stage 1 that were also associated with menarche age in Stage 2 included rs10940138 near PIK3R1 (phosphatidylinositol 3-kinase receptor 1, alias p13k in mice), which is part of the PI3K/AKT/mTOR inflammatory pathway. Enhanced activity of this pathway is strongly implicated in ER-positive breast cancer, ovarian cancer and endometrial cancer (50), is involved in over 30 insulin-signaling networks (51) and contains variants associated with body fatness and leptin levels (52). The second SNP from Stage 1 that was associated with menarche in the Stage 2 sample (rs8014131) is located near FLRT2, encoding the fibronectin leucine-rich transmembrane protein 2. FLRT2 acts as a cell-adhesion or signaling molecule and interacts with numerous growth factors including FGFR1, GnRH and GnRHR to control diverse developmental processes. Lastly, a suggestive association was noted with SNP rs320320 near AKT3 in Stage 1 and was also associated with menarche in our sample of EA women in the ReproGen study (P = 1 × 10−3, combined P-value = 1 × 10−7). AKT3 (also known as protein kinase B) is a member of the serine/threonine-protein kinase family, and functions to regulate extracellular signals including platelet-derived growth factor, insulin and insulin-like growth factor 1 (53,54).
Interrogation of menarche loci reported in EA women
While there are numerous pitfalls in the use of diverse populations for GWA at present, including poorer genomic coverage with existing SNP panels and lower imputation quality (55) and complex admixture patterns across regions of the genome (56), diverse populations are very important in building on the findings in individuals of EA (41,55,57). Owing to wide population variation in allele frequencies, sampling of diverse populations is critical, and African ancestry populations in particular should theoretically yield greater resolution on the location of causal variants influencing a trait, given their lower average LD (56). We undertook two investigations to leverage these properties of AA populations to expand the information on menarche variants already identified in EA women. First, we hypothesized that we would find locus replication (association of SNPs in the same region), but not necessarily SNP replication in AA women. Therefore, we examined SNPs in a 250 kb region of the previously reported menarche loci in EAs. Secondly, we hypothesized that by taking advantage of the lower LD structure in AAs, we could gain insight into the fine structure of these loci and localize potentially causal variants.
As recently shown for lipid traits, significant inter-population differences exist in the contributions of individual SNPs within a given locus as well as their magnitude of effect on a given trait (57,58). Similarly, in the present analysis, none of the 42 index SNPs identified in EA women was associated with age at menarche in AA women after Bonferroni correction for multiple testing. In contrast, 60% of the 42 loci contained SNPs (within ±250 kb of the index SNP) were associated with menarche after region-based Bonferroni correction, showing significant overlap in the genes involved in menarcheal timing across race/ethnicity. This finding is important, first, because it strengthens the evidence for these particular loci being involved in menarcheal timing generally. We found, further, that the SNP with the lowest P-value in AA women in each region generally represented the same signal as in EA women (was in high LD with the index SNP in EA populations), although it was in low LD with the index EA SNP in AA populations. These findings point to the value of examining African Ancestry populations to better localize associations identified in EA populations. We also showed modest evidence in our eQTL analysis that particular SNPs in the 42 loci that were associated with menarche in AA women were more likely to influence local gene expression than were the index SNPs in EA women. A limitation of eQTL analysis is that expression is tissue and cell-type specific, and the cell-type (LCL) used here, while from Yoruban (African) ancestry samples, may not be as informative for investigation of gene variants regulating reproductive timing as hypothalamic or ovarian tissues would be, if available.
In a second analysis to identify specific SNPs in the 42 loci that may better capture the association of the EA signal in AAs, we targeted only SNPs in LD (r2 > 0.4) with the index signal in EAs. Through this fine-mapping work, we identified SNPs in at least eight regions that better captured the association with age at menarche in AAs than the SNPs identified in EAs, most of which were in obesity-related loci such as FTO and SEC16B. In the case of SEC16B, the LD structure of AA women was particularly helpful in localizing the signal to a smaller region. The results suggest a close link between female adiposity and the timing of pubertal development in AA women as was found for EA women (26) and again showcase the value of examining AA populations to narrow the subset of potentially functional alleles in loci identified via GWA in EA populations. In future, this work may be enhanced through trans-ethnic meta-analysis (59), which takes into account the expected similarity in allelic effects in more closely related populations while allowing for heterogeneity between more diverse ethnic groups. This approach has already been shown to both increase power to detect association and improve localization of causal variants by combining diverse population data in a single meta-analysis (60,61).
Interpretation
The lack of strong genome-wide significant associations, in combination with significant overlap in loci associated with age at menarche in both AA and EA women, should not be interpreted to mean that ethnic differences in menarche timing are driven solely by environmental factors. First, African ancestry populations have greater haplotype diversity than European and Asian populations, which yields lower sensitivity for GWA because lower genome-wide LD makes the identification of loci less sensitive given the same degree of genomic coverage on a given genotyping array. Better imputation strategies are needed using population-specific sequencing data to detect low-frequency variants and provide better coverage of genomic regions for African ancestry populations (62,63). However, greater haplotypic diversity also allows greater refinement of loci of interest (64), and may have contributed to our greater success in refining multiple RORA signals than in the European cohort studies of menarche (26). Secondly, there are other classes of genetic variants (e.g. less common alleles, copy number variants, other structural variants) that were not assayed here that may be shown to play a role in age at menarche. Thirdly, gene-by-environment interactions may have masked the effect of genetic variants; the presence of such interactions on a genome-wide basis requires much larger sample size than is available at this time for AA women. Therefore, expansion to additional cohorts to increase the sample size to reach the sizes now available for EA individuals (>100 000) will also be necessary before a full assessment of genetic and environmental contributors to ethnic variation in menarche timing can be made.
Limitations
In addition to the issue of sample size and the focus on common variants discussed above, there are a number of limitations of the study. The participating studies used SNP arrays that were designed to capture common variants in populations of EA and thus, a substantial fraction of common variation in AA populations is likely to have been missed or imprecisely tagged following imputation to a reference source such as HapMap (64). The trait under investigation was self-reported; except for subjects in the Bogalusa Health Study, the subjects included in this meta-analysis were adults when detailed reproductive history data were collected. However, recalled age at menarche is highly correlated with observed age at menarche (65), even 30 years later (66). We did not detect significant heterogeneity across cohorts in our genetic meta-analysis, but heterogeneity in data collection methods may have nonetheless contributed to lower precision of our estimates.
Finally, the timing of menarche is sensitive to early nutritional status (including body fatness), economic disadvantage and, more recently, exposure to endocrine-disrupting chemicals (67), but data on these factors prior to or at the time of menarche were not available for our study cohorts. Environmental heterogeneity between AA and EA women may explain the lack of replication of some loci, as we did not have adequate data to adjust for such factors in our analysis. The rate of decline in age at menarche over the 20th century was more rapid in AA than EA women (68), highlighting the potential effects of the changing nutritional environment for our study population. This is important because environmental variation between and within populations may mask genetic effects when those differences are not accounted for (69,70). Birth year is a potentially useful proxy for numerous nutritional (e.g. protein intake) and non-nutritional (e.g. endocrine disruptors) exposures that have changed over time and may influence developmental timing. It is possible that one reason for the lack of genome-wide significant findings in the present analysis is that birth year heterogeneity (and the environmental variation it may index) could have masked genetic associations and contributed to our lack of genome-wide significant SNP discovery. In this regard, we recently showed a menarche genetic risk score-by-birth year interaction effect on childhood BMI, in which the aggregate effect of 42 menarche-related SNPs was greater in those born recently when compared with those born earlier in the 20th century in the same cohort (71). However, in the present study, we had insufficient statistical power to conduct an SNP × birth year interaction analysis at the genome-wide level, and it was furthermore unlikely that this would have significantly altered our meta-analysis results, as we found little evidence for effect heterogeneity across cohorts that varied widely in mean birth year. Nonetheless, this is a limitation of the present analysis, and an interesting avenue for future investigation.
Conclusions
In summary, we confirmed that many menarche loci identified in EA women generalize to AA women, and for some of these loci, examination of AA samples allowed resolution of multiple signals, better localization of their respective signals and stronger associations with menarche than the originally reported SNPs. We present findings from the largest genome-wide association meta-analysis of age at menarche in AA women to date and, although no single SNP reached genome-wide significance, we identified a number of suggestive associations that may help define novel biological pathways involved in this important early life risk factor.
MATERIALS AND METHODS
Subjects
A total of 18 089 AA women with self-reported age at menarche collected at the baseline visit for each study were included in the Stage 1 meta-analysis. Participants were drawn from seven population-based cohort studies including the Women's Health Initiative (WHI, n = 8086), four cohorts within the Candidate Gene Association Resource (CARe): Atherosclerosis Risk in Communities (ARIC; n = 1690), Coronary Artery Risk Development in young Adults (CARDIA; n = 630), Cleveland Family Study (CFS; n = 169) and Jackson Heart Study (JHS; n = 1228); the Bogalusa Heart Study (BHS; n = 145), the Healthy Aging in Neighborhoods of Diversity across the Life Span study (HANDLS; n = 617) and eight breast cancer case–control studies in the African American Breast Cancer Consortium (AABC) (72)(73), including the Carolina Breast Cancer Study (CBCS; n = 634 cases/586 controls), the Los Angeles component of the Women's Contraceptive and Reproductive Experience Study (CARE; n = 357 cases/215 controls), the Multiethnic Cohort (MEC; n = 532 cases/972 controls), the Nashville Breast Health Study (NBHS; n = 304 cases/182 controls), the Northern California Breast Cancer Family Registry/San Francisco Breast Cancer Study (NC-BCFR/SFBCS; n = 575 cases/269 controls), the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO; n = 56 cases/116 controls), the Wake Forest University Breast Cancer study (WFBC; n = 112 cases/116 controls), and the Women's Circle of Health Study (WCHS; n = 260 cases/238 controls). A further 2850 AA women in the Black Women's Health Study (BWHS) were included in the Stage 2. Detailed descriptions of all studies are provided in Supplementary Material, Text S1 and Table 1.
Phenotypes
Age at menarche was reported to the whole year, and ranged from 8 to 21 years of age, except in the case of the MEC, in which age at menarche was reported within 2-year age groups, the mid-point of which was used in the analysis. Self-reported age at menarche in adult women has been shown to be a valid proxy for prospectively collected age at menarche (66,74,75). Age at menarche is a normally distributed trait and therefore was not transformed prior to analysis.
Genotyping and QCs
The Affymetrix Genome-Wide Human SNP array 6.0 (for ARIC, CARDIA, CFS, JHS, and WHI), the Illumina Human1M-Duo BeadChip array (for HANDLS, WFBC, WCHS, NBHS, PLCO, MEC, CBCS, CARE, NC-BCFR, SFBCS) or the Illumina 610K/Illumina CVD SNP array (BHS) is used according to the manufacturer's protocol for genome-wide genotyping. De novo genotyping was conducted at the Broad Institute for the replication samples using a custom-designed Sequenom chip and Taqman assays for SNPs that could not be multiplexed. Several QC filters were applied to the genome-wide genotype data: DNA concordance checks; sample and SNP genotyping success rate [>95%, MAF > 1%, minor allele count > 3]; sample heterozygosity rate, identity-by-descent analysis to identify population outliers, problematic samples and cryptic relatedness. A detailed description of the QC checks applied to the genotypes in each study and consortium is provided in Supplementary Material, Table S1.
SNP imputation
To increase coverage and facilitate comparison with other datasets, imputed genotype data were obtained using MACH (76,77), using all SNPs that passed the QC steps described above, and employing a 1:1 mixture of HapMap phase II CEU and YRI data as the reference panel for imputation.
Ancestry estimation
In all cohorts, SNPs on the GWA arrays were subjected to principal components analysis using EIGENSTRAT (78) to infer genetic ancestry. The top 10 principal components were included in the study-specific genetic association models as covariates to correct for population stratification.
Association analysis
Within each cohort (and within the breast cancer cases and controls separately in AABC), we tested associations between the imputed and genotyped SNPs with age at menarche using an additive genetic model. Linear regression analysis in PLINK (version 1.07) (79) or ProAbel (80) was used for cohorts of unrelated individuals (ARIC, CARDIA, JHS, WHI, BHS, HANDLS and all AABC studies) and linear mixed-effect models in R were used to model family structure for cohorts including related individuals (CFS). Covariates included the woman's year of birth (or age at diagnosis or recruitment for AABC and WHI), if available, to account for the known secular trends in age at menarche, and the first ten principal components from EIGENSTRAT to account for global population stratification.
Meta-analysis
Cohort-specific association results were combined using an inverse variance-weighted meta-analysis approach as implemented in METAL (81). A genome-wide significance threshold was set at P ≤ 5 × 10−8.
Stage 2 analysis
AA women in the Black Women's Health Study (BWHS) were included in the replication stage (description provided in Supplementary Material, Text S1). Given the lack of genome-wide significant findings, we used the following serial inclusion criteria to select top SNPs from the meta-analysis for presentation (Table 2) and for replication testing: SNPs with P < 1 × 10−5 (43 SNPs), SNPs tested in >10 000 women in Stage 1 (yielding 35 SNPs), SNPs with an MAF >0.03 (yielding 34 SNPs) and including only SNPs in relatively low LD with other top SNPs within a 500 kb region (r2 < 0.3) (yielding 20 SNPs). Genotyping of these 20 SNPs was carried out at the Broad Institute Center for Genotyping and Analysis using the Sequenom MassArray iPLEX technology. An average reproducibility of 99.6% was obtained among the blinded duplicates. The call rate was 98.6% or higher for each SNP. A total of 2850 samples were included in the final analyses. The top 30 ancestral informative markers (AIMs) from the Phase 3 admixture panel (82) were genotyped to estimate and control for population stratification due to European admixture; these 30 AIMs are highly correlated with estimates from the whole admixture panel and thus provide efficient and valid adjustment for stratification (83). An additive linear model for associations of age at menarche with genotype was used, adjusting for year of birth and percent EA as continuous variables. All regression models were run using the SAS statistical software version 9.1.3 (SAS Institute, Inc., Cary, NC, USA). One SNP could not be accommodated on the panel (rs4557202) and therefore a proxy SNP in high LD with it (r2 = 0.967 in 1KG-AFR) was chosen (rs7651087).
Replication of SNP associations in EA women
The 20 SNPs genotyped in the AA replication sample were also examined for association with age at menarche in the 32-study Stage 1 meta-analysis results of EA women (26) in over 87 000 women. A Bonferroni-corrected significance threshold of P < 0.05/20 was applied.
Analysis of secondary signals at known loci
Multiple signals within a single locus in the top GWA results were evaluated through conditional analyses in the largest cohort/studies (WHI, CARe and AABC) using individual-level genotype data. Linear regression analyses were conducted that included all such SNPs, birth year, study center (if applicable) and the top 10 PCs as covariates. The results were then meta-analyzed using METAL and the resulting beta coefficients and P-values were compared to assess whether there were multiple independent risk variants within the regions. We applied a Bonferroni-corrected P-value, correcting for the number of comparisons to determine the significance of independent signals.
Interrogation of 42 menarche loci identified in GWAS of EA women
Our second aim was to interrogate the 42 loci previously reported to be associated with age at menarche in EA women in our AA sample (26); 32 of which were genome-wide significant and 10 demonstrated suggestive associations in the previous study. First, we developed a set of criteria to validate the EA index SNPs and interrogate regions around each of these 42 loci. For each index SNP in EA, we looked-up the respective association result with age at menarche in AA. To accommodate the difference of LD structure and possible allelic heterogeneity across different ethnicities, we then interrogated the 250 kb flanking region around each lead SNP for locus replication to determine whether there exist other SNPs in the locus with stronger associations in AA with the outcome. We used the following criteria to identify the top AA SNP: (i) the SNP with the smallest association P-value within the region; (ii) MAF > 0.01; (iii) location of the AA lead SNP within the same recombination block of the lead EA SNP, where the recombination block was defined as a 20% recombination rate. The statistical significance of each identified SNP was evaluated using a region-specific Bonferroni correction for the multiple comparisons. We determined the number of independent SNPs based on the variance inflation factor, which was calculated recursively within a sliding window with size 50 SNPs and pairwise r2 value of 0.2 using PLINK. If a SNP was identified with Bonferroni-adjusted P-value <0.05 within a locus, then this served as evidence of locus replication in AAs.
Secondly, we interrogated all common genotyped and imputed SNPs (MAF > 0.01) within the ±250 kb flanking region of the index SNPs to identify, specifically (a) variants that capture the association in the region in AA women significantly better than the index SNP and (b) variants that may represent secondary signals. We have previously estimated (84) a threshold of significance for (a) as P < 0.004, which is a correction based on the number of tag SNPs in the HapMap YRI population needed to capture (r2 ≥ 0.8) all SNPs that are correlated with the index signal in the HapMap CEU (r2 ≥ 0.2). In an attempt to eliminate minor fluctuations in P-values for correlated SNPs, we took a more conservative approach than for the conditional analyses and further required the P-value to decrease by more than one order of magnitude compared with the association of the EA index signal in AAs. We also required an r2 > 0.4 between the index marker and the more associated marker in AAs and we assessed phase to ensure that the more associated marker is on the same haplotype as the GWAS-reported risk allele in the HapMap CEU population.
For all of the remaining markers that were weakly correlated (r2 < 0.20) with the index signal (in Europeans), and thus may define secondary signals, we applied a more stringent α level for defining statistical significance. Here we set the threshold as 5.6 × 10−6, which is a correction for the number of tag SNPs needed to capture all common alleles (MAF > 0.05, with r2 > 0.8) in the YRI HapMap population. Both (a) and (b) were estimated empirically based on ∼30 regions of 500 kb in size in a previous study of the prostate cancer risk loci (84).
Expression database analysis of menarche SNPs
We queried existing human lymphocyte gene expression databases to determine whether the top SNPs that we identified in each of the 42 loci (Table 4) were more likely to be associated with the expression of nearby genes than the originally identified SNPs in these regions from studies in women of EA. To do so, we applied a sensitive technique for mapping cis-regulatory SNPs (85) in 56 unrelated Yoruban African LCLs (YRI LCLs) used by the HapMap consortium (86).
Statistical power
The Stage 1 meta-analysis of GWA results had ≥80% power to detect relatively small effect sizes (e.g. 0.12 years, or 6.5 weeks earlier menarche per copy of the risk allele) for SNPs with MAF > 0.2 at P < 5 × 10−8 (Supplementary Material, Fig. S5).
The Stage 2 replication sample in 2850 women in the BWHS provided >80% power to detect an SNP having an effect of 7 weeks earlier menarche per copy of the risk allele for alleles with MAF > 0.25 (which included 13 of the 20 queried variants) at P < 0.05 corrected for 20 comparisons (Supplementary Material, Fig. S6).
Ethics statement
All participants gave informed written consent for the use of their genomic material in studies of cardiovascular disease, cancer and aging risk factors, and the project was approved by the institutional review boards at all participating institutions.
SUPPLEMENTARY MATERIAL
FUNDING
Nine parent studies contributed parent study data, ancillary study data and DNA samples through the Massachusetts Institute of Technology-Broad Institute (N01-HC-65226) to create the Candidate Gene Association Resource (CARe) genotype/phenotype database for wide dissemination to the biomedical research community. Of these, four parent studies (ARIC, CARDIA, CFS and JHS) participated in this study of age at menarche. Analysis support came through National Institutes of Health (HHSN268200900055C and 5215810-550000234). Additional support for this menarche project came from R21AG032598. Information on the CARe parent studies follows here. The Atherosclerosis Risk in Communities Study (ARIC) is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), (R01HL087641), (R01HL59367 and R01HL086694); National Human Genome Research Institute (U01HG004402); and National Institutes of Health contract (HHSN268200625226C). Infrastructure was partly supported by a component of the National Institutes of Health and NIH Roadmap for Medical Research Grant (UL1RR025005). Coronary Artery Risk in Young Adults (CARDIA): Work on this manuscript was supported (or partially supported) by contracts from the National Heart, Lung and Blood Institute (NHLBI): University of Alabama at Birmingham, Coordinating Center (N01-HC-95095); University of Alabama at Birmingham, Field Center (N01-HC-48047); University of Minnesota, Field Center (N01-HC-48048); Northwestern University, Field Center (N01-HC-48049); Kaiser Foundation Research Institute (N01-HC-48050); Harbor-UCLA Research and Education Institute (N01-HC-05187), University of California, Irvine (N01-HC-45134, N01-HC-95100); Wake Forest University (Year 20 Exam) (N01-HC-45205); New England Medical Center (Year 20 Exam) (N01-HC-45204). M.W.'s effort is supported by the National Heart, Lung and Blood Institute (K23-HL-87114). Cleveland Family Study (CFS): The Cleveland Family Study was supported by the National Heart, Lung and Blood Institute (RO1-HL46380, M01-RR-00080). Jackson Heart Study (JHS): The Jackson Heart Study is supported by the National Heart, Lung, and Blood Institute and the National Center on Minority Health and Health Disparities through National Institutes of Health contracts (N01-HC-95170, N01-HC-95171 and N01-HC-95172).
AABC Studies (CARE, CBCS, MEC, NBHS, NC-BCFR/SFBCS, PLCO, WCHS, WFBC): This work was supported by a Department of Defense Breast Cancer Research Program Era of Hope Scholar Award to C.A.H. and the Norris Foundation. Each of the participating studies was supported by the following grants: CARE - National Institute for Child Health and Development grant (NO1-HD-3-3175), CBCS – National Institutes of Health Specialized Program of Research Excellence in Breast Cancer (P50-CA58223) and Center for Environmental Health and Susceptibility, National Institute of Environmental Health Sciences, National Institutes of Health, grant (P30-ES10126); MEC – National Institutes of Health grants (R01-CA63464 and R37-CA54281); NHBS – National Institutes of Health grant (R01-CA100374); NC-BCFR - National Institutes of Health grant (U01-CA69417). SFBCS - National Institutes of Health grant (R01-CA77305) and United States Army Medical Research Program grant (DAMD17-96-6071). The Breast Cancer Family Registry (BCFR) was supported by the National Cancer Institute, National Institutes of Health (RFA CA-95-011) and through cooperative agreements with members of the Breast Cancer Family Registry and Principal Investigators; PLCO - Intramural Research Program, National Cancer Institute, National Institutes of Health; WCHS - U.S. Army Medical Research and Material Command (USAMRMC) grant (DAMD-17-01-0-0334), the National Institutes of Health grant (R01-CA100598) and the Breast Cancer Research Foundation; and WFBC—National Institutes of Health grant (R01-CA73629).
Bogalusa Heart Study (BHS): E.N.S., N.J.S. and S.S.M. are supported in part by National Institutes of Health (grant 1U54RR025204-01). W.C., S.R.S. and G.S.B. are supported from National Institute of Environmental Health Science (ES-021724); from the National Institute of Child Health and Human Development (HD-061437 and HD-062783) and from the National Institute on Aging (AG-16592). E.N.S., S.S.M. and N.J.S. are supported in part by National Institute of Health/National Center for Research Resources Grant (UL1RR025774).
Black Women's Health Study (BWHS): BWHS. research was supported from the National Cancer Institute, Division of Cancer Control and Population Science (R01 CA058420 and R01 CA098663); and by a grant from the Susan G. Komen for the Cure Foundation.
Health Across the Lifespan (HANDLS): This research was supported by the Intramural Research Program of the National Institute of Health, National Institute on Aging and the National Center on Minority Health and Health Disparities (Z01-AG000513) and human subjects protocol (# 2009-149).
Women's Health Initiative (WHI): The WHI program is supported by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services (HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C and HHSN271201100004C). Funding for WHI SHARe genotyping was provided by the National Heart, Lung, and Blood Institute Contract (N02-HL-64278).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mrs. Laurie Zurbey at the University of Minnesota School of Public Health for her patient and highly competent editorial assistance in the preparation of this manuscript. We acknowledge all the subjects for their participation.
AABC Studies: The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the BCFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR.
ARIC: The authors thank the staff and participants of the ARIC study for their important contributions.
CARDIA: NHLBI had input into the overall design and conduct of the CARDIA study.
HANDLS: Data analyses for the HANDLS study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
WHI: This manuscript was prepared in collaboration with investigators of the WHI, and has been reviewed and/or approved by the Women's Health Initiative (WHI). WHI investigators are listed at https://cleo/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hartge P. Genetics of reproductive lifespan. Nat. Genet. 2009;41:637–638. doi: 10.1038/ng0609-637. [DOI] [PubMed] [Google Scholar]
- 2.Peeters P.H., Verbeek A.L., Krol A., Matthyssen M.M., De Waard F. Age at menarche and breast cancer risk in nulliparous women. Breast Cancer Res. Treat. 1994;33:55–61. doi: 10.1007/BF00666071. [DOI] [PubMed] [Google Scholar]
- 3.Kotsopoulos J., Lubinski J., Lynch H.T., Neuhausen S.L., Ghadirian P., Isaacs C., Weber B., Kim-Sing C., Foulkes W.D., Gershoni-Baruch R., et al. Age at menarche and the risk of breast cancer in BRCA1 and BRCA2 mutation carriers. Cancer Causes Control. 2005;16:667–74. doi: 10.1007/s10552-005-1724-1. [DOI] [PubMed] [Google Scholar]
- 4.Rockhill B., Moorman P.G., Newman B. Age at menarche, time to regular cycling, and breast cancer (North Carolina, United States) Cancer Causes Control. 1998;9:447–453. doi: 10.1023/a:1008832004211. [DOI] [PubMed] [Google Scholar]
- 5.Biro F.M., McMahon R.P., Striegel-Moore R., Crawford P.B., Obarzanek E., Morrison J.A., Barton B.a., Falkner F. Impact of timing of pubertal maturation on growth in black and white female adolescents: The National Heart, Lung, and Blood Institute Growth and Health Study. J. Pediatr. 2001;138:636–643. doi: 10.1067/mpd.2001.114476. [DOI] [PubMed] [Google Scholar]
- 6.Freedman D.S., Khan L.K., Serdula M.K., Dietz W.H., Srinivasan S.R., Berenson G.S. Relation of age at menarche to race, time period, and anthropometric dimensions: the Bogalusa Heart Study. Pediatrics. 2002;110:e43. doi: 10.1542/peds.110.4.e43. [DOI] [PubMed] [Google Scholar]
- 7.Freedman D.S., Khan L.K., Serdula M.K., Dietz W.H., Srinivasan S.R., Berenson G.S. The relation of menarcheal age to obesity in childhood and adulthood: the Bogalusa heart study. BMC Pediatr. 2003;3 doi: 10.1186/1471-2431-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakshman R., Forouhi N., Luben R., Bingham S., Khaw K., Wareham N., Ong K.K. Association between age at menarche and risk of diabetes in adults: results from the EPIC-Norfolk cohort study. Diabetologia. 2008;51:781–786. doi: 10.1007/s00125-008-0948-5. [DOI] [PubMed] [Google Scholar]
- 9.Rees M. The age of menarche. ORGYN. 1995;4:2–4. [PubMed] [Google Scholar]
- 10.Cui R., Iso H., Toyoshima H., Date C., Yamanoto A., Kikuchi S., Kondo T., Watanabe Y., Koizumi A., Inaba Y., et al. Relationships of age at menopause and reproductive year with mortality from cardiovascular disease in Japanese Postmenopausal women: The JACC Study. J. Epidemiol. 2006;16:177–184. doi: 10.2188/jea.16.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Presser H.B. Age at menarche, socio-sexual behavior, and fertility. Soc. Biol. 1978;25:94–101. doi: 10.1080/19485565.1978.9988327. [DOI] [PubMed] [Google Scholar]
- 12.Komura H., Miyake A., Chen C.F., Tanizawa O., Yoshikawa H. Relationship of age at menarche and subsequent fertility. Eur. J. Obstet. Gynecol. Reprod. Biol. 1992;44:201–203. doi: 10.1016/0028-2243(92)90099-k. [DOI] [PubMed] [Google Scholar]
- 13.Anderson C.A., Duffy D.L., Martin N.G., Visscher P.M. Estimation of variance components for age at menarche in twin families. Behav. Genet. 2007;37:668–677. doi: 10.1007/s10519-007-9163-2. [DOI] [PubMed] [Google Scholar]
- 14.Van den Berg S.M., Boomsma D.I. The familial clustering of age at menarche in extended twin families. Behav. Genet. 2007;37:661–667. doi: 10.1007/s10519-007-9161-4. [DOI] [PubMed] [Google Scholar]
- 15.Towne B., Czerwinski S.A., Demerath E.W., Blangero J., Roche A.F., Siervogel R.M. Heritability of age at menarche in girls from the Fels Longitudinal Study. Am. J. Phys. Anthropol. 2005;128:210–219. doi: 10.1002/ajpa.20106. [DOI] [PubMed] [Google Scholar]
- 16.Kaprio J., Rimpela A., Winter T., Viken R.J., Rimpela M., Rose R.J. Comom genetic influences on BMI and age at menarche. Hum. Biol. 1995;67:739–753. [PubMed] [Google Scholar]
- 17.He C., Kraft P., Buring J.E., Chen C., Hankison S.E., Pare G., Chanock S., Ridker P.M., Hunter D.J. A large-scale candidate-gene association study of age at menarche and age at natural menopause. Hum. Genet. 2010;128:515–527. doi: 10.1007/s00439-010-0878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He C., Kraft P., Chen C., Buring J.E., Hankinson S.E., Chanock S.J., Ridker P.M., David J., Chasman D.I. Genome-wide association studies identify novel loci associated with age at menarche and age at natural menopause. Nat. Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry J.R.B., Stolk L., Franceschini N., Lunetta K.L., Zhai G., McArdle P.F., Smith A.V., Aspelund T., Bandinelli S., Boerwinkle E., et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat. Genet. 2009;41:648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong K.K., Elks C.E., Li S., Zhao J.H., Luan J., Andersen B., Bingham S.A., Brage S., Smith G.D., Ekelund U., et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat. Genet. 2009;41:729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulem P., Gudbjartsson D.F., Rafnar T., Holm H., Olafsdottir E.J., Olafsdottir G.H., Jonsson T., Alexandersen P., Feenstra B., Boyd H.A., et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat. Genet. 2009;41:734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 22.Widén E., Ripatti S., Cousminer D.L., Surakka I., Lappalainen T., Järvelin M.R., Eriksson J.G., Raitakari O., Salomaa V., Sovio U., et al. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am. J. Hum. Genet. 2010;86:773–782. doi: 10.1016/j.ajhg.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong K.K., Elks C.E., Wills A.K., Wong A., Wareham N.J., Loos R.J.F., Kuh D., Hardy R. Associations between the pubertal timing-related variant in LIN28B and BMI vary across the life course. J. Clin. Endocrinol. Metab. 2011;96:E125–E129. doi: 10.1210/jc.2010-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu H., Shah S., Shyh-chang N., Shinoda G., Einhorn W.S., Viswanathan S.R., Takeuchi A., Grasemann C., Rinn J.L., Lopez M.F., et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 26.Elks C.E., Perry J.R.B., Sulem P., Chasman D.I., Franceschini N., He C., Lunetta K.L., Visser J.A., Byrne E.M., Cousminer D.L., et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplowitz P.B. Link between body fat and the timing of puberty. Pediatrics. 2008;121(Suppl):S208–S217. doi: 10.1542/peds.2007-1813F. [DOI] [PubMed] [Google Scholar]
- 28.Chumlea W.C., Schubert C.M., Roche A.F., Kulin H.E., Lee P.A., Himes J.H., Sun S.S. Age at menarche and racial comparisons in US girls. Pediatrics. 2003;111:110–113. doi: 10.1542/peds.111.1.110. [DOI] [PubMed] [Google Scholar]
- 29.Kimm S.Y.S., Barton B.A., Obarzanek E., McMahon R.P., Sabry Z.I., Waclawiw M.A., Schreiber G.B., Morrison J.A., Similo S., Daniels S.R. Racial divergence in adiposity during adolescence: the NHLBI growth and health study. Pediatrics. 2001;107:e34. doi: 10.1542/peds.107.3.e34. [DOI] [PubMed] [Google Scholar]
- 30.Anderson S.E., Dallal G.E., Must A. Relative weight and race influence average age at menarche: results from two nationally representative surveys of US girls studied 25 years apart. Pediatrics. 2003;111:844–850. doi: 10.1542/peds.111.4.844. [DOI] [PubMed] [Google Scholar]
- 31.Herman-Giddens M.E., Slora E.J., Wasserman R.C., Bourdony C.J., Bhapkar M.V., Koch G.G., Hasemeier C.M. Secondary sexual characteristics and menses in young girls seen in office practice: a study from the Pediatric Research in Office Settings Network. Pediatrics. 1997;99:505–512. doi: 10.1542/peds.99.4.505. [DOI] [PubMed] [Google Scholar]
- 32.Wu T., Mendola P., Buck G.M. Ethnic differences in the presence of secondary sex characteristics and menarche among US girls: the Third National Health and Nutrition Examination Survey, 1988–1994. Pediatrics. 2002;110:752–757. doi: 10.1542/peds.110.4.752. [DOI] [PubMed] [Google Scholar]
- 33.Ogden C.L., Carroll M.D., Kit B.K., Flegal K.M. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999–2010. J. Am. Med. Assoc. 2012;307:483–490. doi: 10.1001/jama.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ervin R.B. Prevalence of metabolic syndrome among adults 20 years of age and over, by sex, age, race and ethnicity, and body mass index: United States, 2003–2006. Natl. Health Stat. Reports. 2009;5:1–7. [PubMed] [Google Scholar]
- 35.Cowie C., Rust K., Ford E., Eberhardt M., Byrd-Holt D., Li C., Willaims D., Gregg E., Bainbridge K., Saydah S., et al. Full accounting of diabetes and pre-diabetes in the U.S. population in 1988–1994 and 2005–2006. Diabetes Care. 2009;32:287–294. doi: 10.2337/dc08-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roger V.L., Go A.S., Lloyd-Jones D.M., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., et al. Heart disease and stroke statistics—2012 update: a report from the American Heart Association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer K.L., Malinowski J., Carty C.L., Franceschini N., Fernández-Rhodes L., Young A., Cheng I., Ritchie M.D., Haiman C.A., Wilkens L., et al. Genetic variation and reproductive timing: African American women from the Population Architecture Using Genomics and Epidemiology (PAGE) study. PloS One. 2013;8:e55258. doi: 10.1371/journal.pone.0055258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Euling S.Y., Herman-Giddens M.E., Lee P.A., Selevan S.G., Juul A., Sørensen T.I.A., Dunkel L., Himes J.H., Teilmann G., Swan S.H. Examination of US puberty-timing data from 1940 to 1994 for secular trends: panel findings. Pediatrics. 2008;121(Suppl):S172–S191. doi: 10.1542/peds.2007-1813D. [DOI] [PubMed] [Google Scholar]
- 39.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J., Mägi R., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakkou M., Wiedmer P., Anlag K., Hamm A., Seuntjens E., Ettwiller L., Tscho M.H., Treier M. A role for brain-specific homeobox factor Bsx in the control of hyperphagia and locomotory behavior. Cell Matab. 2007;5:450–463. doi: 10.1016/j.cmet.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Bustamante C.D., Burchard E.G., De la Vega F.M. Genomics for the world. Nature. 2011;475:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dvornyk V., Waqar-ul-Haq Genetics of age at menarche: a systematic review. Hum. Reprod. Update. 2012;18:198–210. doi: 10.1093/humupd/dmr050. [DOI] [PubMed] [Google Scholar]
- 43.Voight B.F., Kang H.M., Ding J., Palmer C.D., Sidore C., Chines P.S., Burtt N.P., Fuchsberger C., Li Y., Erdmann J., et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zenri F., Hiroi H., Momoeda M., Tsutsumi R., Hosokawa Y., Koizumi M., Nakae H., Osuga Y., Yano T., Taketani Y. Expression of retinoic acid-related orphan receptor alpha and its responsive genes in human endometrium regulated by cholesterol sulfate. J. Steroid Biochem. Mol. Biol. 2012;128:21–28. doi: 10.1016/j.jsbmb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Sarachana T., Xu M., Wu R.C., Hu V.W. Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PloS One. 2011;6:e17116. doi: 10.1371/journal.pone.0017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Y., Xiong D.H., Yang T.L., Guo Y.F., Recker R.R., Deng H.W. Plymorphisms ofestrogen-biosynthesis genes CYP17 and CYP19 may influence age at menarche: a genetic association study in Caucasian females. Hum. Mol. Genet. 2006;15:2401–2408. doi: 10.1093/hmg/ddl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xita N., Chatzikyriakidou A., Stavrou I., Zois C., Georgiou I., Tsatsoulis A. The (TTTA)n polymorphism of aromatase (CYP19) gene is associated with age at menarche. Hum. Reprod. (Oxford, England) 2010;25:3129–3133. doi: 10.1093/humrep/deq276. [DOI] [PubMed] [Google Scholar]
- 48.Dunning A.M., Dowsett M., Healey C.S., Tee L., Luben R.N., Folkerd E., Novik K.L., Kelemen L., Ogata S., Pharoah P.D.P., et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J. Natl. Cancer Inst. 2004;96:936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 49.Haiman C.A., Dossus L., Setiawan V.W., Stram D.O., Dunning A.M., Thomas G., Thun M.J., Albanes D., Altshuler D., Ardanaz E., et al. Genetic variation at the CYP19A1 locus predicts circulating estrogen levels but not breast cancer risk in postmenopausal women. Cancer Res. 2007;67:1893–1897. doi: 10.1158/0008-5472.CAN-06-4123. [DOI] [PubMed] [Google Scholar]
- 50.Urick M.E., Rudd M.L., Godwin A.K., Sgroi D., Merino M., Bell D.W. PIK3R1 (p85) Is somatically mutated at high frequency in primary endometrial cancer. Cancer Res. 2011;71:4061–4067. doi: 10.1158/0008-5472.CAN-11-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasche A., Al-Hasani H., Herwig R. Meta-analysis approach identifies candidate genes and associated molecular networks for type-2 diabetes mellitus. BMC Genomics. 2008;9:310. doi: 10.1186/1471-2164-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jamshidi Y., Snieder H., Wang X., Pavitt M.J., Spector T.D., Carter N.D., O'Dell S.D. Phosphatidylinositol 3-kinase p85α regulatory subunit gene PIK3R1 haplotype is associated with body fat and serum leptin in a female twin population. Diabetologia. 2006;49:2659–2667. doi: 10.1007/s00125-006-0388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakatani K., Sakaue H., Thompson D.A., Weigel R.J., Roth R.A. Identification of a Human Akt3 (Protein Kinase B y) which contains the regulatory serine phosphorylation site. Biochem. Biophys. Res. Commun. 1999;910:906–910. doi: 10.1006/bbrc.1999.0559. [DOI] [PubMed] [Google Scholar]
- 54.Wickenden J.A., Watson C.J. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts. Breast Cancer Res. 2010;12 doi: 10.1186/bcr2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenberg N.A., Huang L., Jewett E.M., Szpiech Z.A., Jankovic I., Boehnke M. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bryc K., Auton A., Nelson M.R., Oksenberg J.R., Hauser S.L., Williams S., Froment A., Bodo J.M., Wambebe C., Tishkoff S.A., et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl. Acad. Sci. U. S. A. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musunuru K., Romaine S.P.R., Lettre G., Wilson J.G., Volcik K.A., Tsai M.Y., Taylor H.A., Schreiner P.J., Rotter J.I., Rich S.S., et al. Multi-ethnic analysis of lipid-associated loci: the NHLBI CARe Project. PloS One. 2012;7:e36473. doi: 10.1371/journal.pone.0036473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adeyemo A., Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13:72–79. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franceschini N., Van Rooij F.J.A., Prins B.P., Feitosa M.F., Karakas M., Eckfeldt J.H., Folsom A.R., Kopp J., Vaez A., Andrews J.S., et al. Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am. J Hum. Genet. 2012;91:744–753. doi: 10.1016/j.ajhg.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dastani Z., Hivert M.F., Timpson N., Perry J.R.B., Yuan X., Scott R.A., Henneman P., Heid I.M., Kizer J.R., Lyytikäinen L.P., et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45 891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auer P.L., Johnsen J.M., Johnson A.D., Logsdon B.A., Lange L.A., Nalls M.A., Zhang G., Franceschini N., Fox K., Lange E.M., et al. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet. 2012;91:794–808. doi: 10.1016/j.ajhg.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K., et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teo Y.Y., Small K.S., Kwiatkowski D.P. Methodological challenges of genome-wide association analysis in Africa. Nat. Rev. Genet. 2010;11:149–160. doi: 10.1038/nrg2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koprowski C., Coates R.J., Bernstein L. Ability of young women to recall past body size and age at menarche. Obesity. 2001;9:478–485. doi: 10.1038/oby.2001.62. [DOI] [PubMed] [Google Scholar]
- 66.Must A., Phillips S.M., Naumova E.N., Blum M., Harris S., Dawson-Hughes B., Rand W.M. Recall of early menstrual history and menarcheal body size: after 30 years, how well do women remember? Am. J. Epidemiol. 2002;155:672–679. doi: 10.1093/aje/155.7.672. [DOI] [PubMed] [Google Scholar]
- 67.Euling S.Y., Selevan S.G., Pescovitz O.H., Skakkebaek N.E. Role of environmental factors in the timing of puberty. Pediatrics. 2008;121(Suppl):S167–S171. doi: 10.1542/peds.2007-1813C. [DOI] [PubMed] [Google Scholar]
- 68.Mcdowell M.A., Brody D.J., Hughes J.P. Has age at menarche changed? Results from the National Health and Nutrition Examination Survey (NHANES) 1999–2004. J. Adolesc. Health. 2007;40:227–231. doi: 10.1016/j.jadohealth.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 69.Hetherington M.M., Cecil J.E. Gene–environment interactions in obesity. Forum Nutr. 2010;63:195–203. doi: 10.1159/000264407. [DOI] [PubMed] [Google Scholar]
- 70.Kang S.J., Chiang C.W., Palmer C.D., Tayo B.O., Lettre G., Butler J.L., Hackett R., Adeyemo A.A., Guiducci C., Berzins I., et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum. Mol. Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson W., Choh A.C., Curren J., Czerwinski S.A., Bellis C., Dyer T.D., Blangero J., Towne B., Demerath E.W. Genetic risk for earlier menarche also influences peri-pubertal body mass index. Am. J. Phys. Anthropol. 2013;150:10–20. doi: 10.1002/ajpa.22121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen F., Chen G.K., Millikan R.C., John E.M., Ambrosone C.B., Bernstein L., Zheng W., Hu J.J., Ziegler R.G., Deming S.L., et al. Fine-mapping of breast cancer susceptibility loci characterizes genetic risk in African Americans. Hum. Mol. Genet. 2011;20:4491–4503. doi: 10.1093/hmg/ddr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haiman C.A., Chen G.K., Vachon C.M., Canzian F., Dunning A., Millikan R.C., Wang X., Ademuyiwa F., Ahmed S., Ambrosone C.B., et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat. Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Damon A., Bajema C. Age at menarche: accuracy of recall after thirty-nine years. Hum. Biol. 1974;46:381–384. [PubMed] [Google Scholar]
- 75.Cooper R., Blell M., Hardy R., Black S., Pollard T.M., Wadsworth M.E.J., Pearce M.S., Kuh D. Validity of age at menarche self-reported in adulthood. J. Epidemiol. Community Health. 2006;60:993–997. doi: 10.1136/jech.2005.043182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype to estimate haplotypes and unobserved genetypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y., Willer C.J., Sanna S., Abecasis G.R. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 79.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., De Bakker P.I.W., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aulchenko Y.S., Struchalin M.V., Van Duijn C.M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reich D., Patterson N., De Jager P.L., McDonald G.J., Waliszewska A., Tandon A., Lincoln R.R., DeLoa C., Fruhan S.A., Cabre P., et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat. Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 83.Ruiz-Narváez E.A., Rosenberg L., Wise L.A., Reich D., Palmer J.R. Validation of a small set of ancestral informative markers for control of population admixture in African Americans. Am. J. Epidemiol. 2011;173:587–592. doi: 10.1093/aje/kwq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haiman C.A., Chen G.K., Blot W.J., Strom S.S., Berndt S.I., Kittles R.A., Rybicki B.A., Isaacs W.B., Ingles S.A., Stanford J.L., et al. Characterizing genetic risk at known prostate cancer susceptibility loci in African Americans. PLoS Genet. 2011;7:e1001387. doi: 10.1371/journal.pgen.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ge B., Pokholok D.K., Kwan T., Grundberg E., Morcos L., Verlaan D.J., Le J., Koka V., Lam K.C.L., Gagné V., et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 86.The International Hap-Map Consortium. Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.