Abstract
Translation of the small G protein RhoA in neurons is regulated by the eukaryotic translation initiation factor eIF4E. Here we show that this translation factor also regulates RhoA expression and activity in breast cancer cells. The introduction of eIF4E into breast tumor cells increased RhoA protein levels, while expression of an eIF4E siRNA reduced RhoA expression. Previous studies indicate that the axon repulsion factor Semaphorin3A (Sema3A) stimulates the eIF4E-dependent translation of RhoA in neurons, and breast tumor cells support autocrine Sema3A signaling. Accordingly, we next examined if autocrine Sema3A signaling drives eIF4E-dependent RhoA translation in breast cancer cells. The incubation of breast tumor cells with recombinant Sema3A rapidly increased eIF4E activity, RhoA protein levels, and RhoA activity. This Sema3A activity was blocked in tumor cells expressing an shRNA-specific for the Sema3A receptor, Neuropilin-1 (NP-1), as well as in cells incubated with an eIF4E inhibitor. Importantly, RhoA protein levels were reduced in Sema3A shRNA-expressing compared to control shRNA-expressing breast tumor cells, demonstrating that autocrine Sema3A increases RhoA expression in breast cancer. Considering that Sema3A suppresses axon extension by stimulating RhoA translation, we next examined if the Sema3A/RhoA axis impacts breast tumor cell migration. The incubation of control breast tumor cells, but not RhoA shRNA-expressing cells, with rSema3A significantly reduced their migration. Collectively, these studies indicate that Sema3A impedes breast tumor cell migration in part by stimulating RhoA. These findings identify common signaling pathways that regulate the navigation of neurons and breast cancer cells, thus suggesting novel targets for suppressing breast tumor cell migration.
Keywords: eIF4E, Neuropilin-1, RhoA, Semaphorin3A, migration, breast tumor
Eukaryotic mRNAs have a 5′ cap structure (m7GpppX) that promotes ribosome binding and is required for efficient translation. Translation of a subset of these mRNAs (weak mRNAs) is inefficient because these mRNAs possess a long, GC-rich 5′ untranslated region (UTR) with significant secondary structure. The translation of these mRNAs is dependent on increased concentrations of eIF4E, a translation initiation factor that is either over-expressed or hyper-activated in tumor cells compared to normal epithelial cells[1]. Notably, tumor growth is dependent on the cap-dependent translation of a number of these weak mRNAs, including c-myc[2]. Importantly, eIF4E is an oncogene, and the overexpression of this translation factor in fibroblasts promotes tumor growth in nude mice[3].
RhoA GTPases convert between an inactive GDP-bound and an active GTP-bound state, and their activation state dramatically influences tumor cell function. RhoA promotes cellular transformation, and stimulates tumor growth in nude mice[4]. Intriguingly, while some studies indicate a critical role for RhoA in tumor cell migration[5], other studies indicate that RhoA suppresses tumor cell migration[6–9].
The axon repulsion factor Semaphorin3A (Sema3A) and its receptor Neuropilin-1 (NP-1) activate the eIF4E translation factor in neurons, resulting in rapid, localized RhoA translation [10, 11]. This Sema3A/eIF4E/RhoA signaling axis promotes growth cone collapse, resulting in the termination of axon extension[11]. Curiously, Sema3A and NP-1 are also expressed in both breast tumor cell lines and primary breast cancers, and function as endogenous suppressors of tumor cell migration[12, 13]. However, to date, the importance of Sema3A/NP-1 signaling in the regulation of eIF4E activity and RhoA expression/activity in tumor cells has not been examined. In the current work, we investigate the hypothesis that common signaling pathways control RhoA expression in neurons and breast cancer cells. Specifically, we examine eIF4E regulation of RhoA expression and activity in breast cancer, and determine the impact of autocrine Sema3A signaling on this pathway.
Materials and Methods
Cell Culture and Reagents
MDA-MB-231 cells were obtained from American Type Culture Collection (ATCC, Rockville, MD). SUM159 cells were kindly provided by Dr. Stephen Ethier (University of Michigan Medical School, Ann Arbor, MI). MDA-MB-231 cells were cultured in low glucose DMEM supplemented with 10% fetal bovine serum, penicillin (100 units/mL), and streptomycin (100ug/mL) (Invitrogen, Carlsbad, CA, USA). SUM159 cells were cultured in Ham’s/F12 supplemented with 5% fetal bovine serum and antibiotics. Recombinant human Semaphorin-3A/Fc, and recombinant human IgG1 Fc were obtained from R&D Systems, Inc. (Minneapolis, MN). Bovine collagen type I was purchased from BD Biosciences (Bedford, MA). Donkey anti-rabbit (Fab)2 and anti-mouse (Fab)2 antibodies conjugated with horseradish peroxidase were from Jackson ImmunoResearch Labs (West Grove, PA). Rabbit anti-eIF4E, anti-eIF4E-BP1, and anti-phospho-(Ser65) eIF4E-BP1 were obtained from Cell Signaling Technology (Danvers, MA). The sources of other antibodies are as follows: mouse anti-RhoA (sc-418, Santa Cruz Biotechnology, Inc. Santa Cruz, CA), rabbit anti-RhoB (#2098; Roche, Indianapolis, IN), rabbit anti-RhoC (#3430; Roche, Indianapolis, IN), rabbit anti-phospho-(Ser209) eIF4E (Invitrogen, Camarillo, CA), rat anti-NP-1 (MAB566, R&D Systems), mouse anti-β-actin antibody (Sigma; Milwaukee, WI), rabbit anti-human Sema3A antibody (ECM Biosciences; Versailles, KY). Western Lightning Chemiluminescence Reagent was from PerkinElmer (Boston, MA). SignalSilence eIF4E siRNA kit was obtained from Cell Signaling Technology (Danvers, MA). 2-Flag-pCDNA3-eIF4E and control vector were gifts from Dr. Lazris-Karatzas (McGill University, Montreal, Canada). Rapamycin was from Calbiochem (San Diego, CA). pcDNA3-EGFP and pcDNA3-EGFP-RhoA-wild-type were obtained from Addgene (Cambridge, MA).
Immunoblotting
Cells (80–90% confluence) were lysed in cell lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% Nonidet P-40, pH 7.8) containing proteinase inhibitor cocktail (Roche) on ice for 30 min. The lysates were clarified at 4°C for 30 min. at 14,000 rpm/min. Protein concentrations were determined using Bio-Rad Protein Assay at OD595nm (Bio-Rad Laboratories, Inc, Hercules, CA). Equivalent amounts of protein for each sample were subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose (Pierce, Rochford, IL). After blocking with 5% nonfat dry milk/PBS, the membranes were incubated with a primary antibody for 2 hours, washed with TTBS (100mM Tris, pH 7.5, NaCl 0.9%, Tween 0.1%) three times, reacted with a secondary antibody for 45 min, and washed with TTBS three times. Protein bands were visualized by ECL (PerkinElmerLife Sciences, Boston, MA). Image intensity was adjusted using Adobe Photoshop, and the relative intensity of protein bands was quantified using Image J software (NIH).
Measuring RhoA activity
RhoA activity was assessed using Rhotekin RBD beads (Cytoskeleton, Denver, CO), which specifically bind to activated (GTP-bound) RhoA. Cell lysate (200 μg) was incubated with 50 μL of Rhotekin-RBD beads on rocker for 1 hour. GTP-Rho bound Rhotekin-RBD beads were pelleted for 1 min by centrifugation at 5,000g at 4°C and washed twice with Cell Lysis Buffer. For the final wash, the GTP-Rho bound Rhotekin-RBD beads were resuspended with 20 μL sample loading buffer and boiled for 2 min. Samples were subjected to 15% SDS-PAGE and immunoblotted with a mouse anti-RhoA antibody (Santa Cruz) or β-actin antibody, followed by HRP-conjugated secondary antibody (Jackson Immunoresearch labs, West Grove, PA). Cell lysates (50 μg) were also immunoblotted with the RhoA and β-actin antibodies. Protein bands were detected by enhanced chemiluminescence.
Migration Assay
Cell migration assays were performed in transwell plates (Corning Inc., Acton, MA) as previously described [12]. In brief, cells were grown in serum-free medium overnight. These cells were transferred to the top chambers either in serum-free medium or in medium containing rSema3A. Culture medium containing 10% FBS was added to the bottom chambers as a chemoattractant source. After 4 hours, the top (non-migrated) cells were removed with cotton swabs, and the migrated cells in the lower membrane were fixed and stained with 0.2% crystal violet. Cell migration assays were carried out in triplicate. The number of migrated cells from five fields of a well were counted, and data were presented as the mean number of cells (in 5 fields) from three wells (+/− standard deviation). Statistical analysis was performed by a Student’s t-test, and a p<0.05 was considered significant.
Small hairpin RNAs (shRNA)
The complementary oligonucleotides to generate the shRNA against human Sema3A, human RhoA, and human NP-1 were designed using BLOCK-iT™ RNAi Designer from Invitrogen (Carlsbad, CA). Importantly, these shRNAs specifically target regions of the Sema3A, RhoA, and NP-1 isoforms that are divergent from their related family isoforms. The oligo sequences were as follows:
Sema3A shRNA1=GGGAAGAACAATGTGCCAAGG
Sema3A shRNA2=GCTAGAATAGGTCAGATATGC
RhoA shRNA=GGCAGATATCGAGGTGGATGG
NP-1 shRNA=GCCTTGAATGCACTTATATTG
The double stranded (ds) oligos were subcloned into pLKOpuro1 (Yang et al., 2004). pLKOpuro1-shLUC was used as a control. pLKOpuro1-shLUC and pLKOpuro1 plasmids were a generous gift from Drs. Yang and Weinberg (Whitehead Institute for Biomedical Research, Cambridge, Massachusetts). pLKOpuro1-shLUC and pLKOpuro1-shSema were co-transfected with pCMV-VSVG and pHR’8.2ΔR (Dr. Weinberg) into 293T cells. After 48 hours, media were collected and centrifuged for 10 min. at 2000 rpm. The supernatants were transferred into targeted cell lines, and these infected cell lines were selected with 1 μg/ml puromycin for one week.
Results
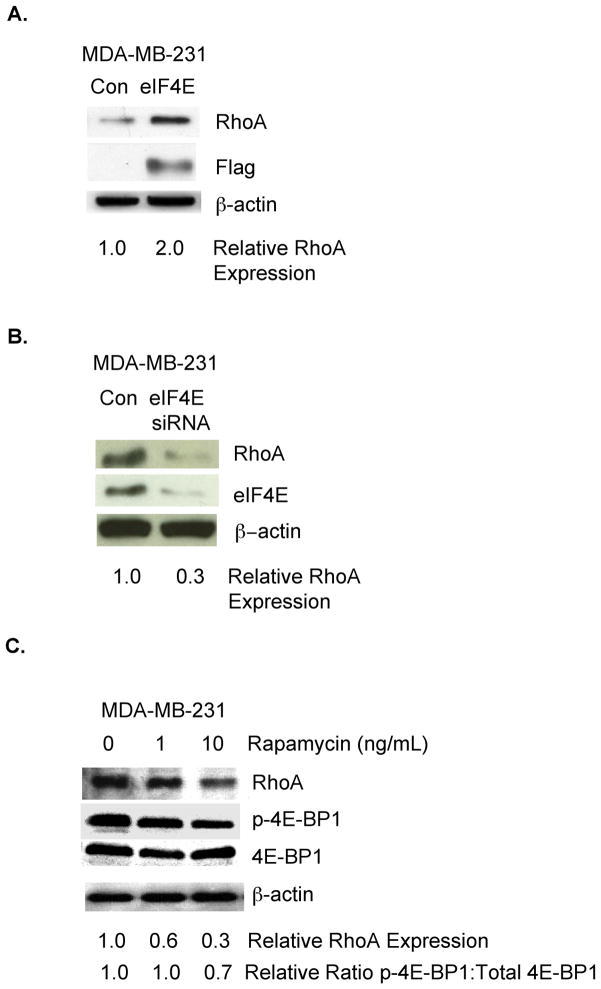
Eukaryotic initiation factor 4E (eIF4E) is an important determinant of RhoA expression in neurons[11] and eIF4E is expressed in breast cancer[1, 14]. Accordingly, we investigated the impact of eIF4E on RhoA expression in a human breast cancer cell line lacking Estrogen Receptor, Progesterone Receptor, and HER2 amplification (MDA-MB-231). As shown in Fig. 1A, the introduction of exogenous eIF4E to MDA-MB-231 breast tumor cells increased RhoA protein levels. To determine if endogenous eIF4E in breast tumor cells controls RhoA protein levels, MDA-MB-231 cells were transfected transiently with an eIF4E siRNA or control siRNA. As shown in Fig. 1B, reducing eIF4E levels in these cells resulted in a 3.3-fold decrease in RhoA protein levels.
Figure 1. The translation initiation factor eIF4E regulates RhoA protein levels in breast tumor cells.
A. MDA-MB-231 breast tumor cells were transfected transiently with a flag-tagged eIF4E-expressing or control vector. Equivalent amounts of protein from these cells were subjected to SDS-PAGE, and immunoblotted with antibodies specific for RhoA, the flag tag, or β–actin, followed by the appropriate species secondary antibody. B. MDA-MB-231 cells were transfected transiently with an eIF4E-specific or control siRNA. Equivalent amounts of protein from these cells were subjected to SDS-PAGE and immunoblotted with antibodies specific for RhoA, eIF4E, or β–actin, followed by the appropriate species secondary antibody. C. MDA-MB-231 cells were incubated in medium containing Rapamycin at the indicated concentration for 4 hours. Equivalent amounts of protein from these cells were subjected to SDS-PAGE and immunoblotted with antibodies specific for RhoA, phospho(Ser65)-eIF4E binding protein (p-4E-BP1), representing the inactive form of the the eIF4E inhibitor eIF4E-BP1, eIF4E-BP1, or β–actin, followed by the appropriate species secondary antibody. For A-C, relative RhoA expression, as well as the ratio of phospho-eIF4E-BP1 to total cellular eIF4E-BP1, was quantified by densitometry using ImageJ software (NIH). Similar results were observed in at least 3 independent trials.
mTOR is a serine/threonine kinase that phosphorylates the eIF4E inhibitor eIF4E binding protein 1 (eIF4E-BP1), resulting in: 1) eIF4E-BP1 dissociation from eIF4E and 2) eIF4E activation[15]. To provide further evidence that the eIF4E pathway regulates RhoA expression in breast cancer, we examined the effect of incubating breast tumor cells with Rapamycin, an established mTOR inhibitor[15], on RhoA protein levels. As shown in Fig. 1C, the incubation of MDA-MB-231 cells with Rapamycin reduced RhoA protein levels three-fold. Confirming that Rapamycin influenced the eIF4E pathway in these breast tumor cells, we observed reduced ratios of phosphorylated eIF4E-BP1 to total cellular eIF4E-BP1 in Rapamycin-treated compared to control MDA-MB-231 (Fig. 1C). Collectively, the studies in Fig. 1 demonstrate that eIF4E is a determinant of RhoA protein levels in MDA-MB-231 breast cancer cells.
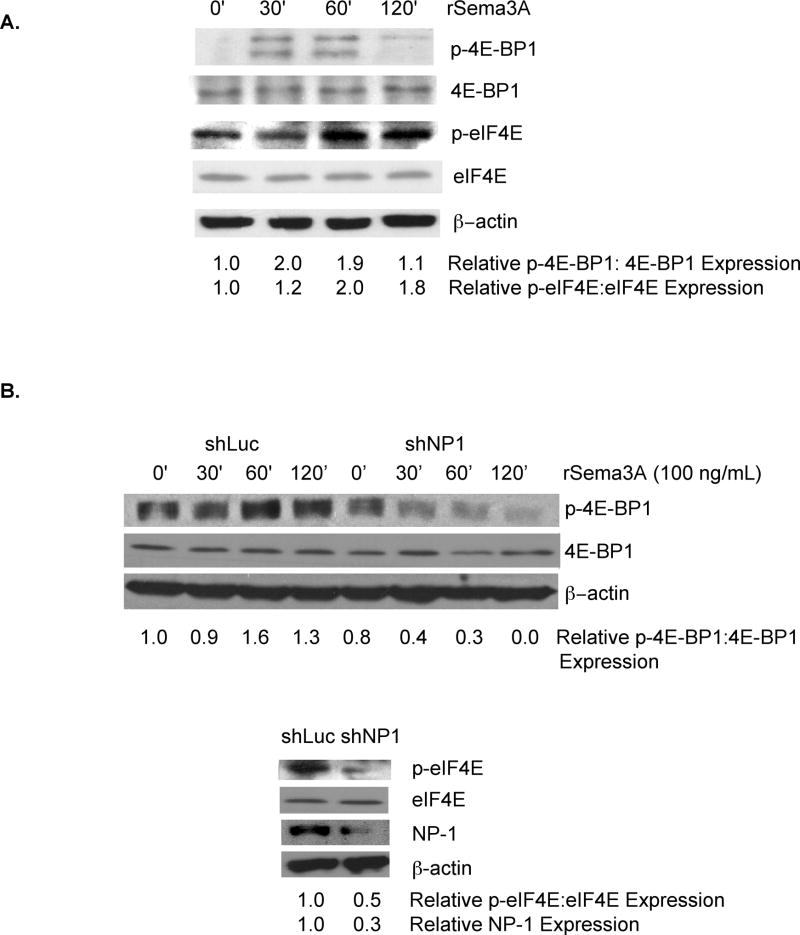
In neurons, the axon repulsion factor Sema3A stimulates the rapid translation of cap-dependent mRNAs by activating eIF4E[11]. Based on our previous demonstration that Sema3A and NP-1 are expressed in breast tumor cell lines and in primary breast cancers[12], we next investigated the impact of Sema3A on eIF4E activity in breast tumor cells. MDA-MB-231 cells with incubated with recombinant Sema3A (rSema3A) for various times, and the level of phoshorylated eIF4E-BP-1, representing the inactivated form of this eIF4E inhibitor[16–18], was measured. As shown in Fig. 2A, phospho-eIF4E-BP1 levels, but not total cellular eIF4E-BP1 levels, were increased two-fold in MDA-MB-231 cells 30 and 60 min following their incubation with rSema3A. This stimulation was transient, as evidenced by reduced levels of phospho-eIF4E-BP-1 in MDA-MB-231 cells incubated with rSema3A for 120 min. As further evidence that Sema3A stimulates the eIF4E pathway in breast cancer cells, we measured the level of phoshorylated eIF4E, representing the activated form of this translation factor[19, 20], in rSema3A-stimulated and control MDA-MB-231 cells. As shown in Fig. 2A, the level of phosphoryated eIF4E was increased approximately two-fold in MDA-MB-231 cells following their incubation with rSema3A for 60 or 120 minutes. In contrast, total cellular levels of eIF4E in MDA-MB-231 cells were unaffected by rSema3A treatment. Notably, rSema3A-induced increases in phospho-eIF4E levels were observed later than rSema3A-stimulated increases in phospho-eIF4E-BP1 levels. These dynamics suggest that a gap exists between the time that Sema3A induces the phosphorylation and inactivation of the eIF4E inhibitor eIF4E-BP-1 and the time that Sema3A increases the level of phosphorylated eIF4E, representing the activated from of this translation factor.
Figure 2. Sema3A stimulates eIF4E activity in breast cancer cells in a manner dependent on the Sema3A receptor, Neuropilin-1.
A. MDA-MB-231 cells (80% confluent) were incubated in serum-free medium overnight prior to their incubation for the indicated times in medium containing rSema3A (100 ng/mL). Equivalent amounts of protein from these cells were subjected to SDS-PAGE and immunoblotted with antibodies specific for phospho(Ser65)-eIF4E binding protein 1 (p-4E-BP1, the inactive form of this eIF4E inhibitor), 4E-BP1, phospho(Ser209)-eIF4E (p-eIF4E, representing the activated form of eIF4E), eIF4E, or β–actin, followed by the appropriate secondary antibody. B. MDA-MB-231 cells were infected with lentiviruses expressing shLuciferase (shLuc) or shNP-1. Top panel: shLuc-expressing and shNP-1-expressing cells (80% confluent) were serum-starved overnight, then incubated +/− rSema3A (100 ng/mL) for the indicated times. Equivalent amounts of protein extracted from these cells were subjected to SDS-PAGE and immunoblotted with phospho(Ser65)-eIF4E-BP-1 (p-4E-BP1), 4E-BP1, or β–actin antibodies, followed by the appropriate species secondary antibody. Bottom panel: Equivalent amounts of total cellular protein extracted from these shRNA-transduced cells were subjected to SDS-PAGE and immunoblotted with antibodies specific for phospho(Ser209)-eIF4E (p-eIF4E), eIF4E, NP-1, or β–actin, followed by the appropriate species secondary antibody. For A-C, relative p-4E-BP1, p-eIF4E, 4EBP1, eIF4E, and NP-1 levels were quantified by densitometry. Similar results were observed in two independent trials.
Neuropilin-1 is the exclusive Sema3A receptor in neurons that supports Sema3A signaling[21, 22]. To examine the importance of NP-1 for Sema3A stimulation of the eIF4E pathway in breast cancer cells, we measured rSema3A-induced eIF4E activation in MDA-MB-231 cells expressing a NP-1 shRNA, which reduced NP-1 expression three-fold (Fig. 2B, bottom panel). As shown in Fig. 2B (top panel), the incubation of control MDA-MB-231 cells, but not NP-1 shRNA-expressing MDA-MB-231 cells, with rSema3A for 60 min. significantly increased phospho-eIF4E-BP1 levels. Furthermore, we observed a 2-fold decrease in the level of phosphorylated eIF4E, representing the activated form of eIF4E[19, 20], in rSema3A-stimulated shNP-1-expressing MDA-MB-231 cells compared to rSema3A-treated control cells (Fig. 2B- bottom panel). Notably, rSema3A did not influence total cellular 4EBP1 or eIF4E levels in these cells. These studies indicate that, similar to its activities in neurons, Sema3A stimulates eIF4E activity in breast tumor cells in a NP-1-dependent manner.
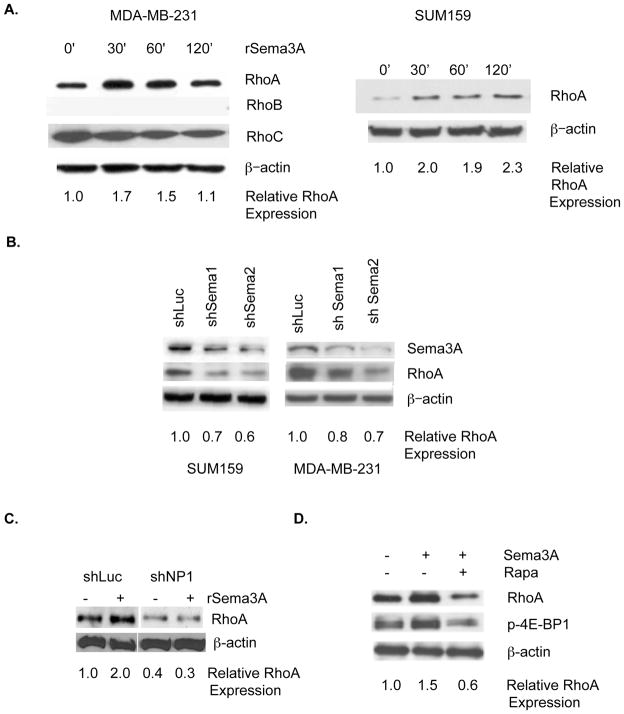
Based on our demonstration that the eIF4E pathway is a determinant of RhoA protein levels in breast cancer cells (Fig. 1), we next investigated the effect of Sema3A on RhoA protein levels in breast cancer. The incubation of either MDA-MB-231 or SUM159 breast tumor cells (both lacking Estrogen Receptor, Progesterone Receptor, and HER2 amplification) with rSema3A increased RhoA protein levels, as assessed by immunoblotting (Fig. 3A). However, rSema3A did not alter RhoA mRNA levels in these cells, as determined by real time PCR (data not shown). The effect of rSema3A on RhoA expression in breast tumor cells was specific because rSema3A did not increase RhoB or RhoC protein levels in MDA-MB-231 cells (Fig. 3A). As evidence that autocrine Sema3A regulates RhoA protein levels in breast cancer cells, we observed that the introduction of either of two Sema3A-targeting shRNAs to either of two breast cancer cell lines (MDA-MB-231 and SUM159) reduced Sema3A and RhoA protein levels (Fig. 3B). We next sought to determine the importance of NP-1 in Sema3A stimulation of RhoA expression. As shown in Fig. 3C, the incubation of control shRNA-expressing MDA-MB-231 cells, but not of NP-1 shRNA-expressing MDA-MB-231 cells with rSema3A increased RhoA protein expression two-fold. To assess the importance of the eIF4E pathway for rSema3A regulation of RhoA protein levels in breast tumor cells, MDA-MB-231 cells were pre-incubated in the presence or absence of the mTOR inhibitor Rapamycin prior to their incubation with rSema3A. As shown in Fig. 3D, the incubation of control MDA-MB-231 cells with rSema3A increased phosphorylated eIF4E-BP1 and RhoA levels, confirming our findings in Fig. 2. However, rSema3A did not increase phosho-eIF4E-BP1 or RhoA levels in Rapamycin-pretreated MDA-MB-231 cells, indicating that rSema3A stimulation of RhoA protein expression is dependent on the eIF4E pathway.
Figure 3. Sema3A signaling in breast tumor cells increases RhoA protein levels in a manner dependent on NP-1 and the eIF4E pathway.
A. MDA-MB-231 and SUM159 breast tumor cells were serum-starved as described in Fig. 2, and stimulated with rSema3A (100 ng/mL) for the indicated times. Equivalent amounts of protein from these cells were subjected to SDS-PAGE and immunoblotted with RhoA, RhoB, RhoC, or β–actin antibodies, followed by the appropriate species secondary antibody. B. MDA-MB-231 breast tumor cells were stably transduced with lentiviruses expressing a control shRNA (shLuc) or either of two Sema3A-specific shRNAs (shSema1 and shSema2). Equivalent amounts of protein extracted from these cells were subjected to SDS-PAGE and probed with Sema3A, RhoA, or β–actin-specific antibodies followed by the appropriate species secondary antibodies. C. shLuc and shNP-1-expressing MDA-MB-231 cells (80% confluent) were serum starved overnight prior to their incubation in medium +/− rSema3A for 60 min., as described for A. Equivalent amounts of protein from these cells were subjected to SDS-PAGE and immunoblotted as described for A. D. MDA-MB-231 cells were incubated +/− rSema3A for 60 min. Equivalent amounts of protein from these cells were subjected to SDS-PAGE and immunoblotted with antibodies specific for RhoA, phospho(Ser65) eIF4E-BP1 (p-4E-BP1), or β-actin, followed by the appropriate species secondary antibody.
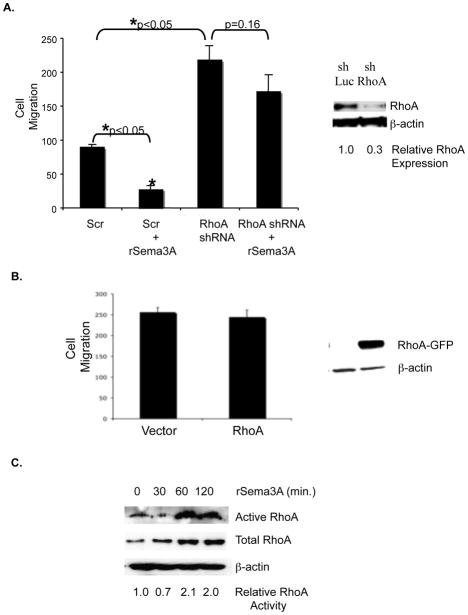
By stimulating RhoA translation and activity in neurons, Sema3A induces growth cone collapse, thus impeding axon extension[11]. Accordingly, we next investigated the impact of Sema3A/RhoA signaling on breast tumor cell migration. The incubation of MDA-MB-231 cells with rSema3A significantly reduced their chemotaxis in a transwell assay (Fig. 4A). The transduction of MDA-MB-231 cells with a RhoA-specific shRNA, which reduced RhoA expression levels three-fold, significantly increased their chemotactic potential (Fig. 4A). Importantly, the incubation of these RhoA shRNA-expressing MDA-MB-231 cells with rSema3A did not significantly reduce their migratory potential (Fig. 4A). These results indicate that RhoA is one determinant of Sema3A migration-suppressing activity.
Figure 4. Sema3A stimulates RhoA activity and suppresses breast tumor cell migration in a RhoA-dependent manner.
A. Right panel: Equivalent amounts of protein from control shRNA-expressing (shLuc) and RhoA shRNA-expressing (shRhoA) MDA-MB-231 cells were subjected to SDS-PAGE and immunoblotted with RhoA or β–actin specific antibodies followed by the appropriate secondary antibody. Left panel: shLuciferase (shLuc)- and shRhoA-expressing MDA-MB-231 cells (80% confluent) were serum starved overnight prior to being incubated +/− rSema3A (100 ng/mL) for 40 min. The ability of these cells to migrate toward 5% FBS culture medium was assessed in a 4 hour assay. Data are presented as the mean number (+/− SD) of migrated cells from three wells (five fields/well). *p<0.05 in a Student’s t test. Similar data were obtained in three independent trials. B. MDA-MB-231 cells were transfected with pcDNA3-EGFP or pcDNA3-EGFP-RhoA-wild-type using Fugene HD (Roche, Indianapolis, IN) After selection in neomycin for 1 week, the migratory potential of these transfectants was assessed in transwells, as described for A. C. MDA-MB-231 cells were incubated +/− rSema3A for the indicated times, and RhoA activity was measured using Rhotekin RBD beads, which bind specifically to activated (GTP-bound) RhoA. These immunoprecipitates were subjected to SDS-PAGE, and immunoblotted with a RhoA antibody, followed by the appropriate species secondary antibody. As a loading control, lysates from these samples were also immunoblotted with a RhoA or β–actin antibody. Similar data were obtained in three independent trials.
If Sema3A suppresses migration in part due to its stimulation of RhoA translation, then the over-expression of RhoA in breast tumor cells should suppress migration. To test this possibility, we transfected MDA-MB-231 cells with a wild-type RhoA or control expression vector, and assessed the relative migratory potential of these transfectants in transwells. As shown in Fig. 4B, over-expression of wild-type RhoA in MDA-MB-231 cells did not impact their migration. Furthermore, expression of wild-type RhoA in these cells did not increase RhoA activity (data not shown). This finding suggests that the ability of Sema3A to suppress breast tumor cell migration cannot be attributed solely to its ability to drive RhoA translation. Accordingly, we next investigated if Sema3A, in addition to driving RhoA translation, also increases RhoA activity in breast tumor cells. As shown in Fig. 4C, the incubation of MDA-MB-231 cells with rSema3A for 60 or 120 minutes increased the level of activated (GTP-bound) RhoA two-fold. Collectively, these studies indicate that Sema3A represses breast tumor cell migration in part by stimulating RhoA expression/activity.
Discussion
eIF4E is an important determinant of RhoA expression in neurons[11]. In the current study, we demonstrate that RhoA expression and activity are also regulated by eIF4E in breast cancer. Intriguingly, RhoA protein levels, but not mRNA levels, are increased in breast tumors relative to normal breast epithelium[23]. Our studies provide an explanation for this finding. Specifically, we show that eIF4E, the activity of which is elevated in breast tumors compared to normal breast epithelium[14], drives RhoA protein expression in tumor cells. Weak mRNAs (i.e. those dependent on increased concentrations of eIF4E for efficient translation) are characterized by long 5′ untranslated regions with high GC content, resulting in extensive secondary structure that impedes translation efficiency[24]. Notably, the RhoA 5′ untranslated region is not long (48 base pairs), but is characterized by high GC content[25].
Our finding that eIF4E regulates RhoA in breast cancer is important because both eIF4E [14] and RhoA[23] are over-expressed in breast cancer relative to normal breast epithelium, and both eIF4E[3] and RhoA[4] possess oncogenic activity. However, to date, no study has linked these proteins in a common pathway in cancer cells. Our findings link eIF4E and RhoA in a common signaling axis, and suggest that RhoA may be a target of eIF4E activity that is relevant to its oncogenic activity, a topic for future study.
Confirming the findings of previous investigators [6–9], our studies indicate that RhoA suppresses breast cancer cell motility. It is important to note that our introduction of a RhoA shRNA into MDA-MB-231 breast tumor cells reduced, but did not abolish RhoA expression (Fig. 4B). Thus, our findings are also concordant with previous work indicating that a baseline level of RhoA is essential for breast tumor cell migration[5].
At face value, it is surprising that RhoA can both drive cellular transformation[4] and suppress cell motility considering that a migratory phenotype is essential for tumor metastasis. To reconcile these opposing RhoA functions, we postulate that, although RhoA activity is required for tumor growth, this small G protein is downregulated during tumor metastatic progression, thus promoting tumor cell migratory and invasive behavior. Supporting this model, a number of pathways have been identified in invasive cancers that downregulate RhoA expression/activity. For example, Smurf1 is a ubiquitin ligase that is recruited to cellular protrusions, driving localized RhoA degradation[26]. In addition, a RhoA-targeting micro-RNA has been identified in invasive breast cancer cells, and this micro-RNA is: 1) critical for breast tumor cell migration and invasion, and 2) upregulated in invasive breast cancers compared to non-invasive cancers [27].
It is well established that eIF4E is over-expressed in metastatic breast tumors, and contributes to tumor growth by inducing the expression of proteins that support tumor cell survival[28]. Considering this literature, our finding that eIF4E drives the expression of a protein that suppresses breast tumor cell migration is somewhat surprising. We postulate that eIF4E can stimulate the expression of proteins that drive tumor growth as well as proteins that suppress tumor cell migration. Of note, other genes have been described that promote cell survival, but impede cell migratory/invasive behavior, such as E-cadherin[29, 30]. We also predict that during their metastatic progression, eIF4E over-expressing tumors must acquire the ability to support pathways that downregulate RhoA expression/activity (e.g. Smurf1, as discussed above). Further studies are needed to investigate these dual activities for eIF4E during breast cancer progression.
Our studies are important in defining an autocrine pathway in breast tumor cells that stimulates eIF4E-dependent RhoA translation and activation. This autocrine pathway is comprised of the axon repulsion factor Sema3A and its receptor NP-1. Considering that NP-1 lacks consensus signaling sequences, further studies are needed to identify the NP-1 co-factors that support Sema3A stimulation of RhoA. Importantly, members of the plexin family, which serve as NP-1 co-receptors in axon repulsion[31, 32], can activate RhoA by stimulating a Rho guanine nucleotide exchange factor[33–35]. We are currently investigating the importance of plexin family members for Sema3A stimulation of eIF4E activity, RhoA translation, and RhoA activity in breast tumor cells.
Our results indicate that Sema3A suppression of breast tumor cell chemotaxis is mediated in part by its stimulation of RhoA expression/activity. Of note, we have previously identified the alpha2 beta1 integrin to be a target of Sema3A that suppresses breast tumor cell chemotaxis[13]. Collectively, these studies suggest that multiple targets of the Sema3A signaling pathway in breast tumor cells repress their chemotactic migration. Interestingly, the RhoA GDP exchange factor Dbl stimulates alpha2 integrin subunit expression in fibroblasts[36]. Accordingly, we are currently investigating if the ability of Sema3A to increase alpha2 beta1 integrin expression in breast cancer cells is dependent on its stimulation of eIF4E-dependent RhoA translation.
Our finding that Sema3A inhibits breast tumor cell migration in part by stimulating RhoA expression/activity has important clinical implications. First, we predict that this autocrine pathway, by suppressing tumor cell migration, may reduce tumor metastatic potential. This prediction rests upon the knowledge that metastatic tumors are comprised of motile and invasive breast cancer cells. Future studies are needed to address the impact of reducing autocrine Sema3A/NP-1 signaling in breast tumor cells on their metastasis in xenograft models. Such studies will establish if neuronal molecules such as Sema3A and NP-1 represent logical therapeutic targets for breast cancer.
Acknowledgments
This work was supported by NIH grant CA93855 (REB)
Abbreviations
- Sema3A
Semaphorin3A
- NP-1
Neuropilin-1
- eIF4E
Eukaryotic Initiation Factor 4E
- eIF4E-BP1
eukaryotic initiation factor 4E Binding Protein-1
References
- 1.Li BD, Liu L, Dawson M, De Benedetti A. Overexpression of eukaryotic initiation factor 4E (eIF4E) in breast carcinoma. Cancer. 1997;79(12):2385–2390. [PubMed] [Google Scholar]
- 2.De Benedetti A, Harris AL. eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol. 1999;31(1):59–72. doi: 10.1016/s1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- 3.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345(6275):544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 4.del Peso L, Hernandez-Alcoceba R, Embade N, Carnero A, Esteve P, Paje C, Lacal JC. Rho proteins induce metastatic properties in vivo. Oncogene. 1997;15(25):3047–3057. doi: 10.1038/sj.onc.1201499. [DOI] [PubMed] [Google Scholar]
- 5.O’Connor KL, Nguyen BK, Mercurio AM. RhoA function in lamellae formation and migration is regulated by the alpha6beta4 integrin and cAMP metabolism. J Cell Biol. 2000;148(2):253–258. doi: 10.1083/jcb.148.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur WT, Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell. 2001;12(9):2711–2720. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur WT, Petch LA, Burridge K. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr Biol. 2000;10(12):719–722. doi: 10.1016/s0960-9822(00)00537-6. [DOI] [PubMed] [Google Scholar]
- 8.Bellovin DI, Simpson KJ, Danilov T, Maynard E, Rimm DL, Oettgen P, Mercurio AM. Reciprocal regulation of RhoA and RhoC characterizes the EMT and identifies RhoC as a prognostic marker of colon carcinoma. Oncogene. 2006;25(52):6959–6967. doi: 10.1038/sj.onc.1209682. [DOI] [PubMed] [Google Scholar]
- 9.Simpson KJ, Dugan AS, Mercurio AM. Functional analysis of the contribution of RhoA and RhoC GTPases to invasive breast carcinoma. Cancer Res. 2004;64(23):8694–8701. doi: 10.1158/0008-5472.CAN-04-2247. [DOI] [PubMed] [Google Scholar]
- 10.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32(6):1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 11.Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436(7053):1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachelder RE, Lipscomb EA, Lin X, Wendt MA, Chadborn NH, Eickholt BJ, Mercurio AM. Competing autocrine pathways involving alternative neuropilin-1 ligands regulate chemotaxis of carcinoma cells. Cancer Res. 2003;63(17):5230–5233. [PubMed] [Google Scholar]
- 13.Pan H, Wanami LS, Dissanayake TR, Bachelder RE. Autocrine semaphorin3A stimulates alpha2 beta1 integrin expression/function in breast tumor cells. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerekatte V, Smiley K, Hu B, Smith A, Gelder F, De Benedetti A. The proto-oncogene/translation factor eIF4E: a survey of its expression in breast carcinomas. Int J Cancer. 1995;64(1):27–31. doi: 10.1002/ijc.2910640107. [DOI] [PubMed] [Google Scholar]
- 15.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. Embo J. 1996;15(3):658–664. [PMC free article] [PubMed] [Google Scholar]
- 16.Fadden P, Haystead TA, Lawrence JC., Jr Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J Biol Chem. 1997;272(15):10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]
- 17.Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, Lawrence JC., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266(5185):653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 18.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 19.Minich WB, Balasta ML, Goss DJ, Rhoads RE. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci U S A. 1994;91(16):7668–7672. doi: 10.1073/pnas.91.16.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whalen SG, Gingras AC, Amankwa L, Mader S, Branton PE, Aebersold R, Sonenberg N. Phosphorylation of eIF-4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. J Biol Chem. 1996;271(20):11831–11837. doi: 10.1074/jbc.271.20.11831. [DOI] [PubMed] [Google Scholar]
- 21.He Z, Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell. 1997;90(4):739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- 22.Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell. 1997;90(4):753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 23.Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87(6):635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graff JR, Zimmer SG. Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin Exp Metastasis. 2003;20(3):265–273. doi: 10.1023/a:1022943419011. [DOI] [PubMed] [Google Scholar]
- 25.Kaarbo M, Crane DI, Murrell WG. RhoA is highly up-regulated in the process of early heart development of the chick and important for normal embryogenesis. Dev Dyn. 2003;227(1):35–47. doi: 10.1002/dvdy.10283. [DOI] [PubMed] [Google Scholar]
- 26.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302(5651):1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 27.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28(22):6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23(18):3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 29.Kantak SS, Kramer RH. E-cadherin regulates anchorage-independent growth and survival in oral squamous cell carcinoma cells. J Biol Chem. 1998;273(27):16953–16961. doi: 10.1074/jbc.273.27.16953. [DOI] [PubMed] [Google Scholar]
- 30.Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66(1):107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi T, Fournier A, Nakamura F, Wang LH, Murakami Y, Kalb RG, Fujisawa H, Strittmatter SM. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell. 1999;99(1):59–69. doi: 10.1016/s0092-8674(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 32.Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99(1):71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 33.Swiercz JM, Kuner R, Behrens J, Offermanns S. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 2002;35(1):51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 34.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc Natl Acad Sci U S A. 2002;99(19):12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J Biol Chem. 2002;277(45):43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- 36.Defilippi P, Olivo C, Venturino M, Dolce L, Silengo L, Tarone G. Actin cytoskeleton organization in response to integrin-mediated adhesion. Microsc Res Tech. 1999;47(1):67–78. doi: 10.1002/(SICI)1097-0029(19991001)47:1<67::AID-JEMT7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]